Abstract
Aberrant transcriptional repression through chromatin remodeling and histone deacetylation has been postulated as the driving force for tumorigenesis. FBI-1 (formerly called Pokemon) is a member of the POK family of transcriptional repressors. Recently, FBI-1 was characterized as a critical oncogenic factor that specifically represses transcription of the tumor suppressor gene ARF, potentially leading indirectly to p53 inactivation. Our investigations on transcriptional repression of the p53 pathway revealed that FBI-1 represses transcription of ARF, Hdm2 (human analogue of mouse double minute oncogene), and p21CIP1 (hereafter indicated as p21) but not of p53. FBI-1 showed a more potent repressive effect on p21 than on p53. Our data suggested that FBI-1 is a master controller of the ARF-Hdm2-p53-p21 pathway, ultimately impinging on cell cycle arrest factor p21, by inhibiting upstream regulators at the transcriptional and protein levels. FBI-1 acted as a competitive transcriptional repressor of p53 and Sp1 and was shown to bind the proximal Sp1–3 GC-box and the distal p53-responsive elements of p21. Repression involved direct binding competition of FBI-1 with Sp1 and p53. FBI-1 also interacted with corepressors, such as mSin3A, NCoR, and SMRT, thereby deacetylating Ac-H3 and Ac-H4 histones at the promoter. FBI-1 caused cellular transformation, promoted cell cycle proliferation, and significantly increased the number of cells in S phase. FBI-1 is aberrantly overexpressed in many human solid tumors, particularly in adenocarcinomas and squamous carcinomas. The role of FBI-1 as a master controller of the p53 pathway therefore makes it an attractive therapeutic target.
The BTB/POZ2 domain, originally identified in Drosophila melanogaster bric-à-brac, tramtrack, and broad complex transcription regulators, and in pox virus zinc finger proteins (1, 2), is an evolutionarily conserved protein-protein interaction domain. About 1,000 distinct BTB/POZ entries exist in sequence data bases (3, 4). Among 194 human BTB/POZ domain regulatory proteins, about 40 proteins are POK proteins. POK proteins consist of an N-terminal POZ domain and a C-terminal Krüppel-type (C2H2) zinc finger domain. The C-terminal zinc fingers recognize and bind specific DNA sequences, and the POZ domain mediates homo- or heterodimerization and interacts with other proteins, such as corepressors, histone deacetylase, and other transcription factors, to regulate transcription (2–4).
BTB/POZ domain regulatory proteins have various cellular regulatory functions. In particular, some of the POK proteins with a BTB/POZ domain and Krüppel-like zinc finger are major determinants in apoptosis (5), development (6, 7), transcription (8–14), and oncogenesis (8, 11, 15, 16). The oncogenic members of the POK family include promyelocytic leukemia zinc finger (PLZF) (11, 16), BCL-6 (B cell lymphoma-6) (17), and HIC-1 (hypermethylated in cancer) (18). Another interesting, newly identified POK transcription factor with proto-oncogenic activity is FBI-1 (mouse LRF), encoded by the ZBTB7A gene (also named Pokemon/rat OCZF), which acts as a transcriptional repressor or activator depending on the promoter context (8, 19–24).
We have been investigating the biological functions of human FBI-1 (19–22). FBI-1 was originally purified as a transcription factor that binds to the bipartite inducer of short transcripts element of human immunodeficiency virus, type 1, long terminal repeat and also to the proximal promoter of the human ADH5/FDH gene (21, 23). FBI-1 stimulates the Tat activity of human immunodeficiency virus, type 1, long terminal repeat and represses human ADH5/FDH gene expression by interacting with Sp1 zinc fingers (21, 24).
The mouse counterpart of FBI-1, LRF, coimmunoprecipitates and colocalizes with BCL-6 and is involved in chondro-genesis and adipogenesis (25–28). The rat homologue of FBI-1, OCZF, is a transcriptional repressor and is involved in osteoclastogenesis (29). FBI-1 enhances NF-κB-mediated transcription through an interaction between the POZ domain of FBI-1 and the RHD of NF-κB (22). It has also been shown that FBI-1 acts as a transcription regulator in adipocyte differentiation and adipogenesis (27, 28). Maeda et al. (8) colleagues identified a central role for LRF in oncogenic transformation, in which it cooperates with several proto-oncogenes and decreases expression of the tumor suppressor protein p19 (ARF). They also demonstrated that LRF is a key for instructing early lymphoid progenitors in mice to develop into B lineage cells by repressing T cell-instructive signals produced by the cell-fate signal protein, Notch (30). Even more recently, we have shown that FBI-1 represses transcription of the tumor suppressor Rb gene, and we also demonstrated that FBI-1 blocked differentiation of mouse C2C12 myoblast cells into myotubes by repressing transcription of the Rb gene (19). Also, we have demonstrated that expression of FASN (fatty-acid synthase), which is important in palmitate synthesis and cell proliferation in cancer cells, is potently activated by FBI-1 in the presence of SREBP-1 (20).
FBI-1 is overexpressed in some human cancers, adipose tissues isolated from genetically obese mice, and diet-induced obese mice, and prostate LNCaP cancer cells treated with androgen (thus growing fast) (8, 20). FBI-1 can stimulate cell proliferation by repressing expression of tumor suppressors Rb and ARF, and also by increasing the supply of cell membrane lipid component palmitate by activating FASN gene expression (8, 19, 20). FBI-1 overexpression in NIH3T3 cells results in more lipid accumulation during differentiation. FBI-1 may be important for adipocyte differentiation, where it may activate expression of adipogenic genes, including FASN (20, 27, 28). FBI-1 appears to be critically involved in cell proliferation, preadipocyte differentiation, and adipogenesis.
p21 inhibits the activities of cyclin-cdk2 complexes and is a major regulator of mammalian cell cycle arrest (31–33). p21 is primarily regulated at the transcriptional level (Ref. 34 and references therein). Whereas induction of p21 leads predominantly to cell cycle arrest, repression of p21 gene expression may have a variety of outcomes, including cell proliferation, depending on the cell context (Refs. 32, 34 and references therein). p21 gene is a transcriptional target of p53, which acts on its distal regulatory elements (31) and plays a crucial role in mediating G1, G2, or S phase growth arrest upon exposure to DNA-damaging agents (32, 33, 35–37). In addition to p53, a variety of other factors, including Sp1/Sp3, Smads, AP2, STAT, BRCA1, E2F-1/E2F-3, and C/EBPα and -β, activate the transcription of p21 gene (34).
Other major regulators that affect p21 gene expression are the Sp1 family transcription factors, which bind to the proximal promoter (38, 39). The Sp1–3 GC-box bound by Sp1 has been shown to be particularly important; mutation of the site nearly eliminates transcription, and it also disrupts the synergistic transcriptional activation by Sp1 and p53 (38). Sp1 can interact with proteins of the basal transcriptional machinery, as well as transcription factors, coactivators and corepressors, including E2F1, FBI-1, GATA, NF-κB, p53, Rb, SREBP-1, YY1, p300, HDAC, BCL-6 interacting corepressor, NCoR, and SMRT. These interactions and direct binding competition among Sp1 family and Krüppel-like transcription factors are important in the transcriptional regulation of genes with a GC-box in their promoters (38–43).
Expression of proto-oncogenic FBI-1 is increased in multiple cancers (8, 44). FBI-1 was recently shown to repress the tumor suppressor gene ARF, which in turn lowers expression of the tumor suppressor gene p53. FBI-1 overexpression causes oncogenesis in the thymus, liver, and spleen (8). Because FBI-1 is also overexpressed in solid tumors, such as cancers of the colon and bladder, where the normal functions of ARF and p53 are lost (8, 44), it is likely that FBI-1 has additional target genes through which it exerts its oncogenic activity.
In this study, we investigated whether the ARF-Hdm2-p53-p21 pathway (referred to as the p53 pathway) (45), an important element in cell cycle control and oncogenesis, is controlled by FBI-1; in particular, we focused on the cyclin-dependent kinase inhibitor p21 gene. Furthermore, we investigated the mechanism and physiological consequence of FBI-1 action. Our data suggest that FBI-1 is a master regulator of the p53 pathway and plays a critical role in regulating important biological processes controlled by p21 and other members of the p53 pathway, such as oncogenic cellular transformation, cell growth, and proliferation.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents—pGL2-p21-Luc, pGL2-p21-Luc WT –131 bp, and pGL2-p21-Luc WT –101 bp were kindly provided by Dr. Yoshihiro Sowa (Kyoto Perpetual University of Medicine, Japan). Constructions of pcDNA3-FBI-1 and pcDNA3-FBI-1ΔPOZ are reported elsewhere (21). Human p53 cDNA was cloned into pcDNA3.1 (Invitrogen) to prepare pcDNA3.1-p53. Sp1ZFDBD (amino acids 622–778) and FBI-1ZFDBD (amino acids 382–490) were prepared by cloning PCR-amplified cDNA fragments into pGEX4T1. To prepare GST-POZFBI-1, a cDNA fragment encoding the POZ domain of FBI-1 was cloned into pGEX4T3. The lentiviruses M1.4-FBI-1-FLAG, -GFP, or -LacZ were obtained from Vectorcorea (Korea). The sequences of all the constructs used in this study were verified.
Various pGL2 promoter-Luc and expression vectors for p53 and FBI-1 were prepared by standard cloning procedures. Various purified recombinant proteins were prepared from Escherichia coli BL21(DE3) transformed with pGEX4T1 protein expression vectors (Amersham Biosciences).
Primary and secondary antibodies were obtained from Upstate (Charlottesville, VA), Chemicon (Temecula, CA), Calbiochem, Santa Cruz Biotechnology (Santa Cruz, CA), Dako, Vector Laboratories, and Abcam (Cambridge, UK). Lipofectamine reagents (Invitrogen) were used for transfection. Buffer recipes, PCR amplification procedures, siRNA sequences, and primer sequences are available upon request. Unless otherwise noted, all other chemical reagents were purchased from Sigma.
Cell Culture—Human osteosarcoma Saos-2 cell lines, lacking endogenous p53, were purchased from ATCC (Manassas, VA). Saos-2 cells were cultured in McCoy's 5A medium containing 15% fetal bovine serum. All other cell lines were cultured in Dulbecco's modified Eagles medium containing 10% fetal bovine serum (Invitrogen). HeLa cells cultured with Polybrene (8 μg/ml) were incubated (37 °C, 5% CO2) for 6–8 h with 350 μl of 4 × 107 transducing units/ml LentiM1.4-FLAG-FBI-1 (experimental) or LentiM1.4eGFP and LentiM1.4LacZ (control). NIH/3T3 cells were infected with His- and Myc-tagged LentiM1.4-FBI-1 (experimental) or LentiM1.4 (control). Puromycin was used to select stable cells after 2–3 days.
Doxycycline (1 μg/ml)-inducible FBI-1-overexpressing cells were prepared by transfecting mammalian Flp-In T-REx host HEK293T cells (Invitrogen) with pOG44:pcDNA5/FRT/TO-FBI-1 plasmid DNA (9:1). Stable FBI-1-overexpressing cells were selected with hygromycin (300 μg/ml) and blasticidin (15 μg/ml).
Transcriptional Analysis—pGL2-Hdm2-Luc, pGL2-ARF-Luc, and pGL2-p53-Luc were prepared by cloning the promoter fragments into pGL2-Luc Basic (Promega, WI). WT or mutant p21-Luc reporter plasmids, pcDNA3-FBI-1, pcDNA3.1-p53, and pCMV-LacZ were transiently transfected into HeLa cells. Luciferase activity, measured at 36 h, was normalized by cotransfected β-galactosidase activity or protein concentration.
Chromatin Immunoprecipitation (ChIP)—A ChIP assay (Upstate) was used to investigate whether FBI-1 and FRE interact in vivo. Subconfluent HeLa cells and Saos-2 cells were transfected for 48 h with 1 μg of pGL2-p21-Luc and 3 μg of either pcDNA3 or pcDNA3-FBI-1-FLAG. HeLa cells and Drosophila SL2 cells were fixed with 1% formaldehyde, washed, lysed with SDS lysis buffer, and sonicated into DNA fragments of 500–1000 bp. Rest of ChIP assays was performed as described elsewhere (19, 20).
Oligonucleotide primers were designed to amplify the proximal promoter or distal p53-binding region of p21. To examine whether ectopic FBI-1 altered the acetylation of the histone H3 and H4 tails of nucleosomes at the proximal p21 promoter element, we used antibodies specific to Ac-H3 and Ac-H4 and followed the same procedures described above.
To investigate the binding competition between Sp1 and FBI-1, SL2 cells were cotransfected with the pGL2-p21-Luc–2.4-kb reporter plasmid, pPac-Sp1 expression vector and pPac-FBI-1 expression vector (1–4-fold excess). After 36 h, the SL2 cells were fixed, immunoprecipitated, and analyzed as described above.
Site-directed Mutagenesis—Mutations were introduced into the p21 proximal promoter sequence using the QuikChange site-directed mutagenesis kit (Stratagene). Amplified mixtures, incubated (37 °C, 1 h) with DpnI, were used to transform competent E. coli. Constructs were confirmed with an ABI automatic DNA sequencer (Ramsey, MN).
siRNA Experiments—siRNAs for glyceraldehyde-3-phosphate dehydrogenase and the three siRNAs of FBI-1 were designed and purchased from Ambion (Austin, TX). Twenty nanograms of each of three FBI-1 siRNAs were transfected into 6 × 106 HeLa cells. Total RNA was prepared from control and experimental HeLa cells expressing FBI-1, GFP, and LacZ, using TRIzol (Invitrogen). cDNAs were synthesized using reverse transcription with 5 μg of total RNA, random hexamer (10 pmol), and 200 units of Superscript reverse transcriptase II (Invitrogen).
Electromobility Shift Assays (EMSA)—EMSAs were performed as described (21). Binding reactions were performed at room temperature for 30 min in binding buffer (20 μl) with recombinant FBI-1ZFDBD or Sp1ZFDBD (0.1 μg) and labeled probe (10,000 cpm). Where indicated, antibodies against GST-Tag, FBI-1, or Sp1 were also added. To investigate competitive binding between Sp1 and FBI-1, the probe was incubated with recombinant Sp1ZFDBD (75 ng) and increasing amounts of FBI-1 ZFDBD (75–675 ng).
Western Blot Analysis—Cells were lysed in RIPA buffer, and extracts (40 μg) were separated on a 10% SDS-PAGE. Proteins were transferred to an Immun-Blot® polyvinylidene difluoride membrane (Bio-Rad) and blocked for 1 h with 5% skim milk. Blotted membranes were incubated with various primary antibodies diluted 1:2000, followed by horseradish peroxidase-conjugated mouse or goat IgG. Protein bands were visualized with ECL solution (PerkinElmer Life Sciences).
Immunoprecipitation—Doxycycline-induced HEK293Trex-FBI-1-FLAG cells were washed, centrifuged, and resuspended in lysis buffer containing protease inhibitors. Cell lysate was pre-cleared. The supernatant was incubated overnight at 4 °C with M2 anti-FLAG antibody, followed by protein A-Sepharose Fast Flow beads. Beads were collected, washed, and resuspended in an equal volume of 5× SDS loading buffer. Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by Western blot.
FACS Analysis—Washed and fixed HeLa cells were stained in the dark with 50 μg/ml propidium iodide in 100 μg/ml ribonuclease A for 30 min at 37 °C. DNA content, cell cycle profiles, and forward scatter were analyzed using a FACSCalibur (BD Biosciences), with emission detection at 488 nm (excitation) and 575 nm (peak emission). Data were analyzed using ModFit LT 2.0 (Verity Software House, Inc., ME) and WindMDI 2.8 (Joseph Trotter, Scripps Research Institute, CA).
Colony Foci Formation Assay—HeLa cells (1 × 105 cells/well) were transfected with pcDNA3 or pcDNA3-FBI-1 (0.5 μg/μl). Transfected cells were cultured for 2 weeks, and colonies resistant to G418 (800 μg/ml) selection were stained with crystal violet (0.5% in 20% ethanol).
MTT Assay—Various cell lines were plated to ∼30–80% confluence, transfected for various times at 37 °C with FBI-1 siRNA (500 pmol), and incubated for 3 h at 37 °C with 20 μl/well MTT (2 mg/ml). Precipitates were dissolved with 1 ml of dimethyl sulfoxide. Cellular proliferation was determined from the conversion of MTT to formazan using a SpectraMAX 250 (Molecular Device Co., Sunnyvale, CA) at 570 nm.
BrdUrd Incorporation—Various cell lines were plated at 40% confluency and transfected for 48 h with 1.68 μg of siRNA, pcDNA3-FBI-1-FLAG, or pcDNA3. Cells were incubated for 4 h in BrdUrd (20 μm)-containing Dulbecco's modified Eagle's medium, washed, and fixed. Cells were then permeabilized, incubated for 2 h with an anti-BrdUrd monoclonal antibody, washed, and incubated for 1 h with an AlexaFluor 488 goat anti-mouse IgG secondary antibody. Nuclei were stained with DAPI for 10 min. HeLa cells were analyzed using a Radiance 2100 laser scanning system (Bio-Rad).
RESULTS
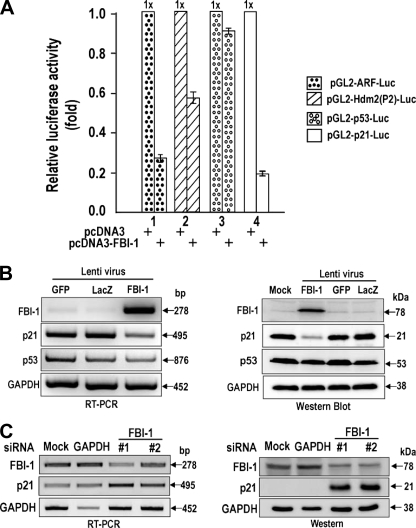
FBI-1 Is a Potent Transcription Repressor of the p53 Pathway—Because FBI-1 causes multiple cancers and is a GC-box-binding transcription factor, we suspected that FBI-1 might regulate one or more genes of the p53 pathway (45). First, we analyzed whether four genes (ARF, Hdm2, p53, and p21) of the p53 pathway were regulated by FBI-1 in HeLa cells cotransfected with promoter-luciferase gene fusion reporter plasmids (supplemental Fig. 1) and an FBI-1 expression vector. FBI-1 potently repressed ARF, as reported previously (8), and also repressed Hdm2 and p21 promoters; however, it did not repress p53. Additionally, overexpression or knockdown of FBI-1 revealed that FBI-1 is a transcriptional repressor of p21 in HeLa cells and also in HCT116 cells (Fig. 1, A–C; supplemental Fig. 2, A and B).
FIGURE 1.
FBI-1 represses transcription of the ARF, Hdm2, and p21 genes of the p53 pathway. A, transcriptional analysis of ARF, Hdm2, p53, and p21 promoters fused with luciferase genes in HeLa cells. B, reverse transcription-PCR and Western blot analysis of stable HeLa cells overexpressing FBI-1, GFP, and LacZ. Overexpression was established by infection with recombinant lentiviruses. GAPDH, control. C, reverse transcription-PCR and Western blot analysis. Knockdown of FBI-1 mRNA by siRNAs #1 and #2 increases p21 gene expression. Mock, negative control. GAPDH, positive control siRNA against GAPDH.
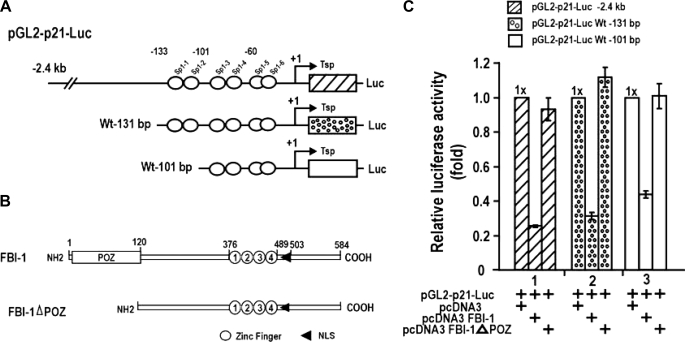
FBI-1 Represses the Transcription of Cell Cycle Arrest p21 Gene, and the POZ Domain Is Important for This Repression—The transcriptional repressions of ARF and Hdm2 are mutually compensatory in the regulation of p53 stability and/or activity. Because FBI-1 represses transcription of three genes of the p53 pathway, and its transcriptional regulatory effects ultimately converge on the regulation of p21 gene expression, we selected and examined the transcriptional regulation of p21 by FBI-1 in detail. We examined which region of the p21 promoter and which domain of FBI-1 are important in transcriptional repression in HeLa cells. FBI-1 repressed transcription of three different p21 promoter constructs (>70–60%), and repression was particularly potent with a –2.4-kb promoter. Deletion of the FBI-1 POZ domain eliminated transcriptional repression, suggesting that the POZ domain is important in transcriptional repression (Fig. 2C). FBI-1 repressed transcription by acting on the proximal promoter, which has six Sp1-binding GC-boxes. FBI-1 may also repress transcription by other mechanisms that involve distal upstream regulatory elements, because FBI-1 repressed transcription even more potently on constructs with longer promoter elements. In human colon cancer HCT116 p53+/+ and p53–/– cells, FBI-1 repressed transcription of p53 pathway genes even more potently and nearly eliminated transcription of p21 (supplemental Fig. 2, A and B).
FIGURE 2.
FBI-1 represses transcription of p21 gene by acting on the GC-rich proximal promoter. A and B, structures of the three pGL2-p21-Luc constructs and the two FBI expression constructs used in transcription assays, shown at right. Open circles, Sp1 binding GC-box; open box, POZ domain; numbered open circles, zinc fingers; arrowhead, nuclear localization sequence. C, transient transcription assays of p21 promoters in HeLa cells.
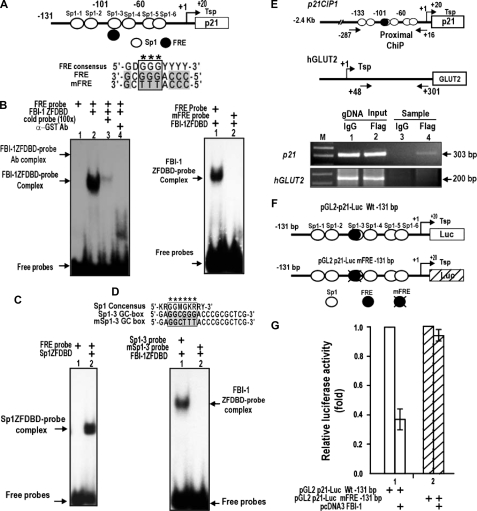
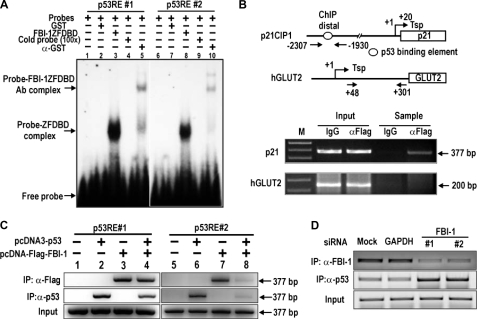
Identification of a Functional GC-box Regulatory Element in the p21 Promoter That Is Critical in Transcriptional Repression by FBI-1—Because FBI-1 is a GC-box-binding transcription factor (8), we reasoned that FBI-1-binding sites may be present on the p21 promoter, and may even overlap with Sp1-binding GC-boxes. We found seven potential FBI-1-binding sites (FREs), with varying degrees of binding affinity, located proximal to or overlapping with six Sp1-binding sites (data not shown). EMSA and transcriptional assays revealed one functionally significant FRE (5′-GCGGGACCC-3′), which overlapped an Sp1–3 GC-box (Fig. 3A). EMSA of Sp1–3 GC-box (or FRE) showed that FBI-1 binds the FRE, and mutation of the core GGG → TTT (mFRE) abolished FBI-1 binding (Fig. 3B). EMSA also showed that Sp1ZFDBD bound to the FRE probe (Fig. 3C). Additional EMSA showed that FBI-1ZFDBD bound to the Sp1–3 GC-box but was unable to bind to the mSp1–3 GC-box (Fig. 3D). Furthermore, ChIP assays of endogenous p21 in HeLa cells showed that FBI-1 bound to the proximal promoter but not to the control GLUT2 promoter (Fig. 3E). We tested whether the FRE overlapping the GC-box-3 is functionally significant in transcriptional repression by FBI-1. In HeLa cells, FBI-1 repressed pGL2-p21-Luc WT –131 bp but could not repress transcription on the mutated (GGG → TTT) promoter, suggesting that FBI-1 binds to the FRE and represses transcription in vivo (Fig. 3, F and G).
FIGURE 3.
FBI-1 binds to the proximal FRE or Sp1 GC-box 3 of the p21 promoter, and the element is functional in transcriptional repression by FBI-1. A, structure of pGL2-p21-Luc WT (–131 bp). Open circles, GC boxes; filled circles, FRE. Sp1–3 GC-box and FRE are overlapping. B, EMSA of Sp1–3 GC-box (FRE). FBI-1 binds the FRE, and mutation of the core GGG → TTT (mFRE) abolishes FBI-1 binding. C, EMSA showing that Sp1ZFDBD binds to FRE probe. D, EMSA. FBI-1ZFDBD bound the Sp1–3 GC-box but was unable to bind the mSp1–3 GC-box. Sequence alignment of the Sp1 consensus sequence, Sp1–3 GC box, and the mutated Sp1–3 GC box is shown above. In mSp1–3 GC-box, the core GGG was mutated into TTT. ZFDBD, zinc finger DNA binding domain. E, ChIP assays in HeLa cells and structures of two endogenous human gene promoters, p21 and hGLUT2. Arrows, locations of PCR primers used in ChIP assays. Open circle, GC boxes; filled circle, FRE; gDNA, genomic DNA. F and G, structures of pGL2-p21-Luc WT (–131 bp) and mutant FRE. Open circles, GC boxes; filled circles, FRE. Sp1–3 GC-box and FRE are overlapping. Transcriptional assays of pGL2-p21-Luc WT FRE or pGL2-p21-Luc mFRE in HeLa cells and identification of functional FBI-1-binding element (FRE) important in transcriptional repression are shown.
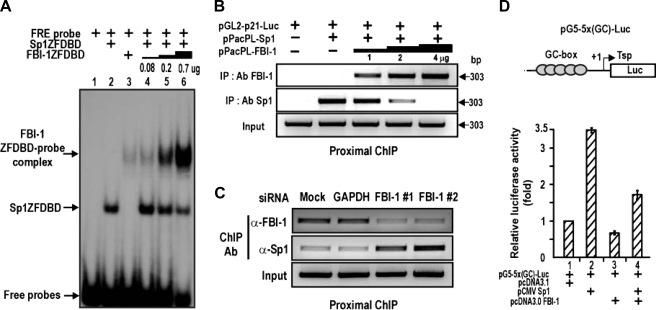
FBI-1 Competes with Sp1 to Repress Transcription–The above data suggested that FBI-1 and Sp1 might compete with each other to bind to the GC-rich FRE/GC-box 3 element. Indeed, EMSA showed that binding of Sp1ZFDBD to the FRE/Sp1–3 GC-box was competed by excess FBI-1ZFDBD in vitro (Fig. 4A). ChIP assays of binding competition between Sp1 and FBI-1 on the proximal promoter of p21 in Drosophila SL2 cells also showed similar binding competition (Fig. 4B). Knockdown of FBI-1 mRNA expression in human HEK293A cells resulted in decreased FBI-1 binding and increased Sp1 binding to the proximal promoter region of endogenous p21 gene (Fig. 4C). Binding competition between these two proteins may be important in the transcriptional repression of p21. In HeLa cells, ectopic Sp1 increased transcription of the reporter pG5–5x(GC)-Luc, and FBI-1 repressed transcriptional activation by Sp1, again suggesting a binding competition not only on the FRE/Sp1–3 box of p21 gene but also on the canonical Sp1-binding GC-box (Fig. 3G and Fig. 4D). Overall, our data indicate that FBI-1 is a GC-box-binding transcription factor that can compete with Sp1 and repress transcription on the FRE/Sp1–3 box that is critical for p21 transcription.
FIGURE 4.
FBI-1 is a GC-box-binding protein that competes with Sp1 for binding to the FRE or Sp1 GC-box 3 of the p21 promoter to repress transcription. A, EMSA of Sp1–3 GC-box (or FRE) and binding competition between Sp1ZFDBD and excess FBI-1ZFDBD. ZFDBD, zinc finger DNA binding domain. B, ChIP assays and binding competition between FBI-1 and Sp1 on the p21 promoter (–2.4 kb) in Drosophila SL2 cells. SL2 cells were transfected with pGL2-p21-Luc, Sp1 expression vector, and excess FBI-1 expression vector. Cells were fixed with formaldehyde and analyzed for promoter binding by ChIP. FBI-1 decreases Sp1 binding. C, ChIP assays and binding competition between FBI-1 and Sp1 on the endogenous p21 promoter in human HEK293A cells. Knockdown of endogenous FBI-1 increased Sp1 binding. D, transcriptional assays. Transcriptional activation of pG5–5x(GC box)-Luc by Sp1 is repressed by FBI-1 in HeLa cells. The GC-box is derived from the well defined Sp1 binding consensus sequence.
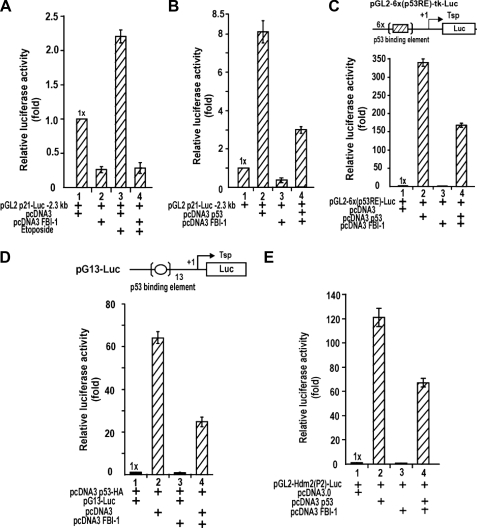
FBI-1 Can Repress Transcription Activation of p53-responsive Genes by Ectopic p53 or by p53 Induced by Etoposide Treatment—FBI-1 yielded more potent transcriptional repression of the p21 promoter containing the longer regulatory sequence with p53-binding elements (Fig. 2C). FBI-1 inhibited the regulatory element of p21 that is critical for transcriptional activation by Sp1 and communication between proximal Sp1 and distal p53 (Fig. 3G). p53 can interact with Sp1 bound on the Sp1–3 GC-box and can potently activate p21 transcription (38). p53 expression is inducible by DNA damage, such as that induced by etoposide treatment. Therefore, we investigated whether FBI-1 could block the transcriptional activation of p21 by etoposide-activated p53 or by ectopic p53 (in HeLa or Saos-2 cells lacking p53). Transcriptional activation by p53 was repressed by FBI-1 (Fig. 5, A and B; supplemental Fig. 2, C and D; and supplemental Fig. 3, A and B). Furthermore, transcription of the p53-responsive genes pGL2–6x(p53RE)-Luc, pG13-Luc, and pGL2-Hdm2(P2)-Luc, which contain the p53-binding elements of p21, canonical p53-binding element, and human Hdm2, respectively, were also repressed by FBI-1 in HeLa, HCT116 p53+/+, and HCT116 p53–/– cells (Fig. 5, C–E; supplemental Fig. 2E; and Fig. 3C).
FIGURE 5.
FBI-1 can repress transcription activation of p53-responsive genes by ectopic p53 or by p53 induced by etoposide treatment. A, induction of p53 by etoposide and transcriptional repression of p21 by FBI-1 in HeLa cells. B, transcriptional activation of p21 by ectopic p53 is inhibited by FBI-1 in Saos-2 p53–/– cells. C, transcriptional activation of pGL2–6x(p53RE)-Luc by ectopic p53 can be repressed by FBI-1 in Saos-2 cells. The p53RE is from the distal p53-binding element of p21. D and E, transcriptional activation of pG13-Luc and pGL2-Hdm(P2)-Luc by ectopic p53 can be repressed by FBI-1 in Saos-2 cells. pG13 contains 13 copies of the p53-binding element. The human Hdm2 gene promoter contains one p53-responsive element.
FBI-1 Binds to the p53-binding Elements of p21 and Competes with p53 to Repress Transcription—The above data, particularly Fig. 5, C and D, suggest that FBI-1 may bind directly to p53-binding elements to repress transcription activation by p53. EMSA and ChIP assays performed on the endogenous p21 gene in HeLa and Saos-2 cells revealed that FBI-1 binds directly to the two p53-binding elements in vitro and in vivo, respectively (Fig. 6, A and B), and that p53 and FBI-1 compete for binding to the p53 regulatory elements (Fig. 6C and supplemental Fig. 3D). The ChIP assays also showed that FBI-1 decreased p53 binding, and vice versa, on the p53-binding elements (Fig. 6C). Alternatively, knockdown of FBI-1 increased p53 binding (Fig. 6D).
FIGURE 6.
FBI-1 competes with p53 for binding to the distal p53-binding elements of p21 to block transcriptional activation by p53. A, EMSA showing that FBI-1 binds two distal p53-binding elements. ZFDBD, recombinant zinc finger DNA binding domain. B, ChIP assay of FBI-1 binding on the endogenous distal p53-binding elements of p21 in HeLa cells transfected with pcDNA3-FLAG-FBI-1. M, DNA size marker. C, ChIP assays and binding competition between FBI-1 and p53 on the distal p53-binding elements of endogenous p21 in Saos-2 cells. Saos-2 cells were transfected with a p53 expression vector, with or without an FBI-1 expression vector. Coexpression of p53 and FBI-1 decreases binding of the other. D, ChIP assays showing that knockdown of endogenous FBI-1 in HEK293A cells increases p53 binding. IP, immunoprecipitation.
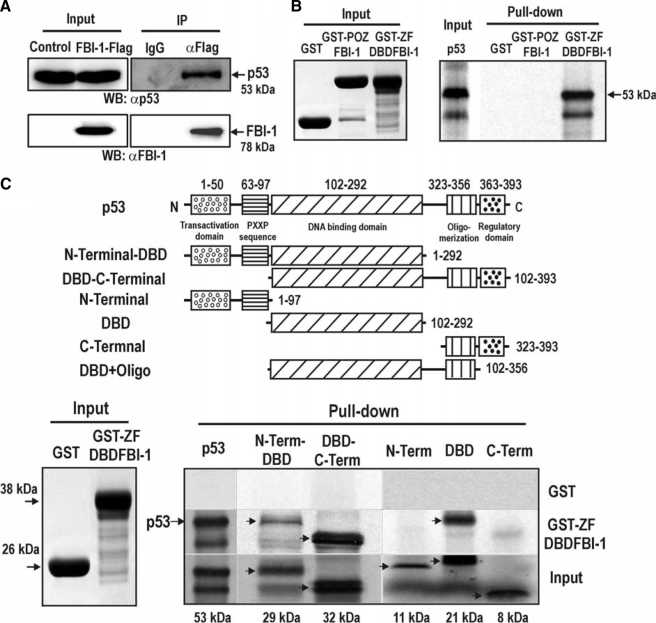
FBI-1 and p53 Interact Directly via Their DNA Binding Domains—FBI-1 and p53 may interact and mutually affect DNA binding activities. Coimmunoprecipitation and Western blot analyses of protein extracts prepared from stable HEK293Trex-FLAG-FBI-1 cells showed that p53 and FBI-1 interact with each other (Fig. 7A). Furthermore, GST fusion protein pulldown assays indicated that the molecular interaction between FBI-1 and p53 was direct and involved the ZFDBD domain of FBI-1 and the p53 polypeptide, each of which contains a DNA binding domain (Fig. 7, B and C). Interaction between FBI-1 and p53 may affect their DNA binding activities, as revealed by ChIP assays with the two p53-binding elements (Fig. 6C).
FIGURE 7.
Zinc finger DNA binding domain of FBI-1 interacts with p53 via the p53 DNA binding domain (DBD). A, coimmunoprecipitation of FBI-1 and p53. Lysates prepared from control HEK293Trex cells and stable 293Trex-FLAG-FBI-1 cells were immunoprecipitated (IP) using anti-FLAG M2 antibody beads and analyzed by Western blotting (WB) with anti-p53 and anti-FBI-1 antibodies. B and C, GST fusion protein pulldown assays. Recombinant GST, GST-POZFBI-1, or GST-ZFDBDFBI-1 were incubated with [35S]methionine-labeled p53 or its deleted polypeptides, pulled down, and resolved by 15% SDS-PAGE. The gel was then exposed to x-ray film.
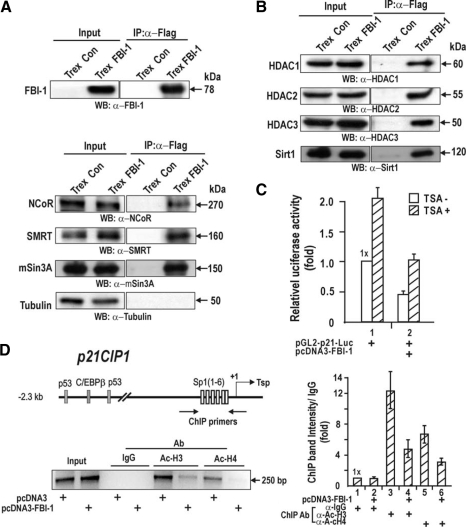
FBI-1 Interacts with Corepressor-HDAC Complexes to Deacetylate Proximal Promoter Histones and to Repress Transcription of Cell Cycle Arrest p21 Gene—Although FBI-1 can repress transcription by competing with Sp1 and p53 for binding to critical transcriptional regulatory elements, the mechanism by which FBI-1 represses transcription remains unclear. Our transcription data suggest that the POZ domain is important in transcriptional repression (Fig. 2, B and C). Transcriptional repressors such as PLZF and BCL-6 repress transcription by interacting with corepressors, such as BCL-6 interacting corepressor, NCoR, mSin3A, and SMRT, via their POZ domains. Coimmunoprecipitation of nuclear extracts prepared from HEK293Trex-FLAG-FBI-1 cells showed that FBI-1 interacted with corepressors such as NCoR, SMRT, and mSin3A (Fig. 8A). Furthermore, class 1–3 HDACs were also present in the FBI-1-corepressor complexes. Treatment of HeLa cells cotransfected with pGL2-p21-Luc and pcDNA3-FBI-1 with the HDAC inhibitor TSA increased transcription 2-fold (Fig. 8, B and C). TSA treatment in the presence of FBI-1 did not fully de-repress transcription to the TSA-treated control level. Overall, our data suggest that transcriptional repression by FBI-1 involves a corepressor-HDAC complex and a binding competition between the activator Sp1 (or p53) and the repressor FBI-1. Using a ChIP assay on HeLa cells, we investigated whether the FRE/Sp1–3 GC-box-bound FBI-1-corepressor-HDAC complex and the distal p53-binding elements could deacetylate Ac-H3 and Ac-H4 histones bound to the proximal promoter of endogenous p21. FBI-1 significantly decreased the acetylation of histones 3 and 4, suggesting that histone deacetylation at the proximal promoter is responsible for transcriptional repression by FBI-1 (Fig. 8D).
FIGURE 8.
FBI-1 interacts with corepressors and deacetylates histones Ac-H3 and Ac-H4 bound at the p21 proximal promoter. A, coimmunoprecipitation of FBI-1 and corepressors NCoR, SMRT, and mSin3A. Nuclear extract prepared from HEK293Trex control and stable HEK293Trex-FLAG-FBI-1 cells were coimmunoprecipitated with anti-FLAG M2 antibody. Precipitates were analyzed by Western blot (WB) using antibodies against FBI-1 and corepressors. Tubulin, control (Con). IP, immunoprecipitated. B, coimmunoprecipitation of FBI-1 and class 1–3 HDACs. Assays were carried out as in A. C, transcriptional repression by FBI-1 involves HDACs. HeLa cells were transfected with pGL2-p21-Luc, with or without an FBI-1 expression vector. Transfected cells treated with TSA for 30 h before harvest were analyzed for luciferase activity. D, ChIP assays for histone modification at the proximal promoter of endogenous p21 using antibodies against Ac-H3 and Ac-H4. HeLa cells were transfected with an FBI-1-FLAG expression plasmid and immunoprecipitated with the antibodies indicated. A histogram of the ChIP assays is shown at the right.
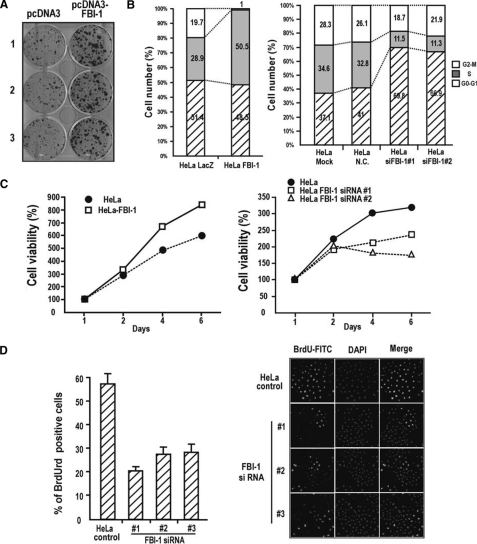
FBI-1 Causes Cellular Transformation and Promotes Cell Proliferation—p21, a major regulator of cell cycle arrest, is potently repressed by FBI-1 at the transcriptional level. Therefore, FBI-1 may cause cellular transformation and promote cell cycle progression. HeLa cells transfected with FBI-1 expression vectors formed substantial numbers of large transformation foci, suggesting that FBI-1 caused cellular transformation (Fig. 9A). FACS analysis showed that FBI-1 overexpression stimulated cell cycle progression and increased the number of HeLa cells in S phase (28.9% in control HeLa-LacZ versus 50.5% in HeLa-FBI-1). Furthermore, the number of cells in G2-M phase decreased from 19.7 to 1%. Knockdown of endogenous FBI-1 mRNA resulted in a decreased number of cells in S phase and a concomitant increase in the number of cells in G0-G1 phase (Fig. 9B). MTT assays also showed that although FBI-1 overexpression significantly increased cell viability or growth, FBI-1 knockdown by RNA interference decreased cell viability or growth (Fig. 9C). Although the percentage of cells incorporating BrdUrd (as an indicator of cell proliferation) significantly increased in NIH/3T3 cells and HEK292Trex cells stably expressing FBI-1 (supplemental Fig. 4, B–F), knockdown of FBI-1 mRNA significantly decreased the number of HeLa cells incorporating BrdUrd (Fig. 9D). In stable human colon cancer HCT116 p53+/+ and p53–/– cells overexpressing FBI-1, FBI-1 decreased p21 expression, stimulated cell cycle progression, and increased the number of cells in S phase, regardless of the presence of p53. Knockdown of FBI-1 mRNA had the opposite effect. These effects were more dramatic following etoposide treatment, when most cells were in S phase and only a few cells were in G0-G1 phase following FBI-1 overexpression (supplemental Fig. 6). Interestingly, FBI-1 expression was decreased by genotoxic stress, suggesting the possibility of protein degradation, as was recently reported for BCL-6 (46) (supplemental Fig. 5, A and B).
FIGURE 9.
FBI-1 causes cellular transformation and promotes cell cycle progression and proliferation. A, foci formation assays. HeLa cells transfected with the FBI-1 expression vector were cultured in G418 medium and stained with 0.1% crystal violet. B, FACS analysis of cell cycle progression. HeLa cells stably overexpressing FBI-1 and LacZ were established by lentivirus infection, or HeLa cells transfected with an siRNA against FBI-1 mRNA were stained with propidium iodide, and cell proliferation was measured. Mock, transfection without siRNA; N.C., scrambled siRNA negative control. C, MTT assay of HeLa and stable HeLa-FBI-1 cells grown for 2, 4, or 6 days. Alternatively, MTT assays were performed in HeLa cells treated with either negative control siRNA or siRNA specific to FBI-1. All assays were performed in triplicate. Error bars are included, but are too tight to see. D, knockdown of FBI-1, and BrdUrd incorporation. HeLa cells were transfected with control siRNA or three different FBI-1 siRNAs. After transfection, cells were grown in media containing BrdUrd and analyzed for BrdUrd incorporation as under “Experimental Procedures.” FITC, fluorescein isothiocyanate; DAPI, 4′,6-diamidino-2-phenylindole.
We analyzed FBI-1 expression in human solid cancer tissues by immunochemical staining of tissue microarrays. Most of the tumors with positive FBI-1 staining were adenocarcinomas or squamous carcinomas of the stomach, pancreas, lung, prostate, etc. In particular, of the 45 colon cancer tissue microarray cores tested, 19 displayed strong FBI-1 expression in the nucleus of cancer tissue regions (data not shown).
DISCUSSION
We have been investigating the novel biological functions of human POK protein FBI-1 (19–22), which is overexpressed in human cancers and prostate LNCaP cancer cells treated with androgen (thus growing fast) (8, 20). Recently, increasing amounts of information obtained by us and others suggested that FBI-1 may be critically involved in cell growth, cell proliferation, oncogenesis, and cancer (8, 19, 20).
Maeda et al. (8) identified a central role for mouse FBI-1/LRP in oncogenic transformation, in which expression of the tumor suppressor protein, ARF, is decreased. ARF is a negative regulator of ubiquitin-protein isopeptide ligase Mdm2, which participates in the degradation of p53 by ubiquitin-dependent degradation by the proteasome. The repression of ARF expression can stimulate cell proliferation by decreasing the stability or activity of p53 via Mdm2.
p53 and NF-κB pathways are intertwined in cellular physiology, and p53 and NF-κB repress the activities of each other (Ref. 47 and references therein). NF-κB represses p53 stability by directly up-regulating Mdm2. Interestingly, whereas FBI-1 down-regulates p53 expression via ARF-Mdm2, it also enhances activity of NF-κB. The NF-κB pathway is recognized to be the central mediator of immune response in mammals. NF-κB proteins are frequently hyperactive in cancer and inflammatory diseases. NF-κB proteins activate their target genes, which regulate diverse physiological processes ranging from cell proliferation and invasion to cell death. There is a strong correlation between constitutively active NF-κB and chronic inflammation, which in turn is a determinant in the development of many cancers (47). Interestingly, FBI-1 was shown to increase the transcription of NF-κB-responsive genes by dramatically increasing nuclear stay of NF-κB (23), which may be important in prolonged inflammatory reactions and perhaps in cell proliferation and the development of cancer.
The Rb protein is a tumor suppressor that plays a pivotal role in the negative control of the cell cycle and tumor progression (48). We recently demonstrated that FBI-1 could repress tumor suppressor Rb gene expression, which can reverse the cell cycle arrest effect of Rb (19). Furthermore, we also showed that FBI-1 plays a critical role in the synthesis of fatty acid palmitate in the presence of SREBP-1 or -2, master controller of fatty acid synthesis, thereby providing more cellular lipid membrane components needed for rapid cell proliferation in cancer cells (20).
All the above data suggest that FBI-1 may regulate a set of genes important in cell growth and proliferation, which may be important in the development of cancer and cell proliferation of cancer cells. We investigated whether FBI-1 could regulate the p53 pathway, a key player in cell cycle regulation. We showed that FBI-1 directly or indirectly represses expression of ARF, Hdm2, and p21 genes of the p53 pathway (Fig. 10A). Because the repression of ARF and Hdm2 genes by FBI-1 is mutually compensatory in terms of p53 expression, and FBI-1 potently stimulates cell cycle progression, we reasoned that the principal target of FBI-1 might be the cell cycle arrest gene p21. In addition to blocking or inhibiting expression of ARF and Hdm2, and thereby eventually affecting p21 expression via regulating p53 expression, FBI-1 directly inhibits the critical elements (i.e. proximal FRE/Sp1–3 GC-box and p53-responsive elements) for transcriptional repression of p21, each of which are important in basal and inducible expression of p21.
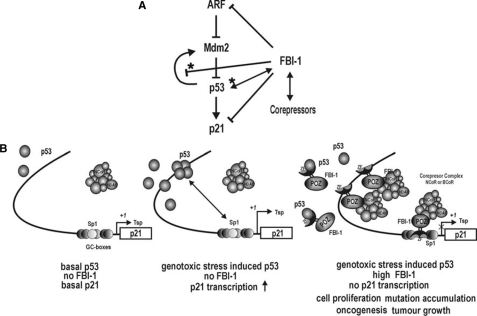
FIGURE 10.
FBI-1 is a master controller of the ARF-Hdm2-p53-p21 pathway. A, p53 pathway and regulation by FBI-1. →, transcriptional activation; solid line with ⊥, transcriptional repression; solid line with ⊥ ★, transcriptional repression by binding competition with p53; ↔, protein-protein interaction; ↔ with ★, inhibition of binding by protein-protein interaction. B, model of transcriptional repression of p21 in three different physiological states. Proto-oncogene FBI-1 attacks the three critical elements (FRE or Sp1–3 GC-box, and two p5- responsive elements) that are important in basal constitutive and inducible expression. FBI-1 recruits corepressor-HDAC complexes and deacetylates histones Ac-H3 and Ac-H4 at the proximal promoter to repress transcription. FBI-1 also interacts with p53 to inhibit p53 binding, and vice versa. Tsp (+1), transcription start site; ZF, zinc finger DNA binding domain.
In particular, our in-depth molecular investigation of p21 transcription revealed novel features of the proto-oncogene FBI-1. First, FBI-1 is a GC-box-binding transcription factor, which can compete with Sp1 for binding to the proximal FRE/Sp1–3 GC-box. This competitive binding by FBI-1 inhibits basal transcriptional activation by Sp1 and abrogates synergistic communication between Sp1 and p53 bound at the two distal p53 binding elements. Second, FBI-1 is a p53-responsive element-binding protein. It inhibits p53 through binding competitively with p53 at p53-responsive elements or by interacting directly with p53.
We propose a model for the transcriptional repression of p21 by FBI-1 (Fig. 10). Under normal cellular conditions where p53 levels are low and no FBI-1 is present, p21 is expressed at a low level and cells proliferate normally. When cells are challenged with genotoxic stress, the tumor suppressor p53 is markedly induced and activates transcription of p21 by interacting with Sp1 bound at the FRE/Sp1–3 GC-box. The induced p21 arrests cell cycle progression and allows cells to repair DNA damage. When cells are under genotoxic stress (and thus have high levels of p53) and FBI-1 expression is high (as is the case in some cancer tissues or highly proliferative tissues), FBI-1 represses transcription by directly binding to both the proximal FRE/Sp1–3 GC-box and the distal p53-binding elements. Although p53 expression is high, it must compete with FBI-1 to bind to the distal p53-binding elements. Molecular interactions between p53 and FBI-1 can also inhibit p53 binding. Because the proximal FRE/Sp1-GC box is occupied by FBI-1, p53 cannot activate transcription. The FBI-1-corepressor-HDAC complexes deacetylate histones H3 and H4 at the proximal promoter, thereby repressing transcription of the p21 gene promoter. Cells proliferate without cell cycle arrest, mutations accumulate, and cells are likely to undergo oncogenic transformations.
MIZ-1 (Myc-interacting zinc finger protein 1), another interesting POK family protein that binds to the proximal promoter of p21 gene, is a transcription activator of the p21 gene (49). c-Myc and BCL-6 were previously shown to repress transcription of the p21 gene after being recruited to the p21 promoter by molecular interaction with MIZ-1 (49, 50). Interestingly, FBI-1 transcription repressor binds to the element intercalated between the two juxtaposed MIZ-1-binding elements (49). Although largely uncharacterized, this complex interaction among MIZ-1, FBI-1, Sp1 and proximal promoter elements may also contribute to the transcriptional repression of cell cycle arrest gene p21.
The molecular mechanism revealed here is quite different from those of other POK or POK-related proteins such as BCL-6, PLZF, and PLZF-RARα. BCL-6 is expected to function by directly binding to specific DNA sequences and suppressing the transcription of target genes, as reported for DNA damage sensor ATR gene (ATM- and Rad3-related) (51), PDCD2 gene (programmed cell death-2) (52), and p53 gene promoter (53), by EMSA and ChIP. However, with the exception of the few cases mentioned above, the molecular action mechanism of true in vivo BCL-6 target-binding sites remains largely elusive. BCL-6 controls the transcription of genes lacking a BCL-6-binding site by an alternative mechanism. BCL-6 interacted with the transcription activator MIZ-1 and, via MIZ-1, bound the proximal promoter and suppressed transcription of the cell cycle arrest gene p21 (50). Through this repression mechanism, BCL-6 may facilitate the proliferative expansion of germinal centers during the normal immune response and, when deregulated, the pathological expansion of B cell lymphomas.
Although potential binding sites for PLZF were identified by CAST assays (54), and PLZF was shown to bind to the promoters and repress transcription of c-Myc and HoxD genes (55, 56), true in vivo regulatory binding sites and the molecular action mechanism of repression by PLZF remain largely unknown, as for BCL-6. Although a transcriptional regulatory mechanism for PLZF-RARα was proposed that involves potential binding to retinoic acid-response element on the genome and participation of corepressors and HDACs, the true target of PLZF-RARα also remains uncharacterized (11, 16). Our unpublished data3 and ongoing investigation indicate that PLZF and PLZF-RARα are also potent regulators of cell growth and proliferation, controlling the p53 pathway in a manner similar to that of FBI-1, as characterized in this report. A recent report by Savage et al. (57) reported that tissues from PLZF-transgenic mice show significant increases in the ethynyl deoxyuridine assay for proliferation, and a microarray experiment of PLZF knock-out mice showed elevated expression of p21 and p53 mRNA, suggesting a cell proliferative effect of PLZF by expression modulation of the p53 pathway (Gene Expression Omnibus, www.ncbi.nlm.nih.gov). The molecular mechanism elucidated in this study for FBI-1 may also underlie the cell proliferation of hematopoietic cells, or acute promyelocytic leukemic cells, caused by abnormal expression of BCL-6, PLZF, and PLZF-RARα.
Overall, the molecular features of FBI-1 revealed in this investigation may explain how FBI-1 acts as a master control gene of cellular transformation and cell proliferation by potently blocking the p53 pathway. Our data should also provide insight into the regulatory role of POK family transcription regulators in cell growth and proliferation, and potentially in oncogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Bert Vogelstein for HCT116 p53+/+ and p53–/– cells.
This work was supported by National Research Laboratory Research Grant ROA-2003-000-10318-0 (to M. W. H.) and Medical Research Center Grant R13-2002-054-05002-0 (to M. W. H.) from the Korea Science and Engineering Foundation of the Korea Ministry of Science and Technology and in part by National R & D Program for Cancer Control Affairs Grant 00001561 (to M. W. H.) from the Korean Ministry for Health, Welfare, and Family.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
Footnotes
The abbreviations used are: BTB/POZ, bric-à-brac, tramtrack, broad complex/poxvirus and zinc finger; ARF, alternative reading frame; ChIP, chromatin immunoprecipitation; EMSA, electromobility shift assay; FACS, fluorescence-activated cell sorter; FBI-1, factor that binds to the inducer of short transcripts of human immunodeficiency virus-1; FBS, fetal bovine serum; FRE, FBI-1 response element; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; NCoR, nuclear receptor corepressor; PLZF, promyelocytic leukemia zinc finger protein; SMRT, silencing mediator for retinoid and thyroid receptors; Sp1, specificity protein 1; WT, wild type; ZFDBD, zinc finger DNA binding domain; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; GFP, green fluorescent protein; siRNA, small interfering RNA; Rb, retinoblastoma; TSA, trichostatin A; HDAC, histone deacetylase.
W.-I. Choi and M.-W. Hur, unpublished data.
References
- 1.Koonin, E. V., Senkevich, T. G., and Chernos, V. I. (1992) Trends Biochem. Sci. 17 213–214 [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and Koonin, E. V. (1999) J. Mol. Biol. 285 1353–1361 [DOI] [PubMed] [Google Scholar]
- 3.Bardwell, V. J., and Treisman, R. (1994) Genes Dev. 8 1664–1677 [DOI] [PubMed] [Google Scholar]
- 4.Albagli, O., Dhordain, P., Deweindt, C., Lecocq, G., and Leprince, D. (1995) Cell Growth & Differ. 6 1193–1198 [PubMed] [Google Scholar]
- 5.Yamochi, T., Kaneita, Y., Akiyama, T., Mori, S., and Moriyama, M. (1999) Oncogene 18 487–494 [DOI] [PubMed] [Google Scholar]
- 6.Farkas, G., Gausz, J., Galloni, M., Reuter, G., Gyurkovics, H., and Karch, F. (1994) Nature 371 806–808 [DOI] [PubMed] [Google Scholar]
- 7.Barna, M., Hawe, N., Niswander, L., and Pandolfi, P. P. (2000) Nat. Genet. 25 166–172 [DOI] [PubMed] [Google Scholar]
- 8.Maeda, T., Hobbs, R. M., Merghoub, T., Guernah, I., Zelent, A., Cordon-Cardo, C., Teruya-Feldstein, J., and Pandolfi, P. P. (2005) Nature 433 278–285 [DOI] [PubMed] [Google Scholar]
- 9.Deltour, S., Guerardel, C., and Leprince, D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhordain, P., Albagli, O., Lin, R. J., Ansieau, S., Quief, S., Leutz, A., Kerckaert, J. P., Evans, R. M., and Leprince, D. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, R. J., Nagy, L., Inoue, S., Shao, W., Miller, W. H., and Evans, R. M. (1998) Nature 391 811–814 [DOI] [PubMed] [Google Scholar]
- 12.Dong, S., Zhu, J., Reid, A., Strutt, P., Guidez, F., Zhong, H. J., Wang, Z. Y., Licht, J., Waxman, S., Chomienne, C., Chen, Z., Zelent, A., and Chen, S. J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 3624–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C. C., Ye, B. H., Chaganti, R. S., and Dalla-Favera, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 6947–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huynh, K. D., Fischle, W., Verdin, E., and Bardwell, V. J. (2000) Genes Dev. 14 1810–1823 [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Z., Brand, N. J., Chen, A., Chen, S. J., Tong, J. H., Wang, Z. Y., Waxman, S., and Zelent, A. (1993) EMBO J. 12 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grignani, F., De Matteis, S., Nervi, C., Tomassoni, L., Gelmetti, V., Cioce, M., Fanelli, M., Ruthardt, M., Ferrara, F. F., Zamir, I., Seiser, C., Grignani, F., Lazar, M. A., Minucci, S., and Pelicci, P. G. (1998) Nature 391 815–818 [DOI] [PubMed] [Google Scholar]
- 17.Kerckaert, J. P., Deweindt, C., Tilly, H., Quief, S., Lecocq, G., and Bastard, C. (1993) Nat. Genet. 5 66–70 [DOI] [PubMed] [Google Scholar]
- 18.Chen, W., Cooper, T. K., Zahnow, C. A., Overholtzer, M., Zhao, Z., Ladanyi, M., Karp, J. E., Gokgoz, N., Wunder, J. S., Andrulis, I. L., Levine, A. J., Mankowski, J. L., and Baylin, S. B. (2004) Cancer Cell 6 387–398 [DOI] [PubMed] [Google Scholar]
- 19.Jeon, B. N., Yoo, J. Y., Choi, W. I., Lee, C. E., Yoon, H. G., and Hur, M. (2008) J. Biol. Chem. 283 33199–33210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi, W. I., Jeon, B. N., Park, H., Yoo, J. Y., Kim, Y. S., Koh, D. I., Kim, M. H., Kim, Y. R., Lee, C. E., Kim, K. S., Osborne, T. F., and Hur, M. W. (2008) J. Biol. Chem. 283 29341–29354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, D. K., Suh, D., Edenberg, H. J., and Hur, M. W. (2002) J. Biol. Chem. 277 26761–26768 [DOI] [PubMed] [Google Scholar]
- 22.Lee, D. K., Kang, J. E., Park, H. J., Kim, M. H., Yim, T. H., Kim, J. M., Heo, M. K., Kim, K. Y., Kwon, H. J., and Hur, M. W. (2005) J. Biol. Chem. 280 27783–27791 [DOI] [PubMed] [Google Scholar]
- 23.Pessler, F., Pendergrast, P. S., and Hernandez, N. (1997) Mol. Cell. Biol. 17 3786–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pendergrast, P. S., Wang, C., Hernandez, N., and Huang, S. (2002) Mol. Biol. Cell 13 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies, J. M., Hawe, N., Kabarowski, J., Huang, Q. H., Zhu, J., Brand, N. J., Leprince, D., Dhordain, P., Cook, M., Morriss-Kay, G., and Zelent, A. (1999) Oncogene 18 365–375 [DOI] [PubMed] [Google Scholar]
- 26.Liu, C. J., Prazak, L., Fajardo, M., Yu, S., Tyagi, N., and Cesare, P. E. D. (2004) J. Biol. Chem. 279 47081–47091 [DOI] [PubMed] [Google Scholar]
- 27.Laudes, M., Christodoulides, C., Sewter, C., Rochford, J. J., Considine, R. V., Sethi, J. K., Vidal-Puig, A., and O'Rahilly, S. (2004) J. Biol. Chem. 279 11711–11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laudes, M., Bilkovski, R., Oberhauser, F., Droste, A., Gomolka, M., Leeser, U., Udelhoven, M., and Krone, W. (2008) J. Mol. Med. 86 597–608 [DOI] [PubMed] [Google Scholar]
- 29.Kukita, A., Kukita, T., Ouchida, M., Maeda, H., Yatsuki, H., and Kohashi, O. (1999) Blood 94 1987–1997 [PubMed] [Google Scholar]
- 30.Maeda, T., Merghoub, T., Hobbs, R. M., Dong, L., Maeda, M., Zakrzewski, J., van den Brink, M. R., Zelent, A., Shigematsu, H., Akashi, K., Teruya-Feldstein, J., Cattoretti, G., and Pandolfi, P. P. (2007) Science 316 860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.el-Deiry, W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W., and Vogelstein, B. (1993) Cell 75 817–825 [DOI] [PubMed] [Google Scholar]
- 32.el-Deiry, W. S., Harper, J. W., O'Connor, P. M., Velculescu, V. E., Canman, C. E., Jackman, J., Pietenpol, J. A., Burrell, M., Hill, D. E., Wang, Y., Wiman, K. G., Mercer, W. E., Kastan, M. B., Kohn, K. W., Elledge, S. J., Kinzler K. W., and Vogelstein, B. (1994) Cancer Res. 54 1169–1174 [PubMed] [Google Scholar]
- 33.Brugarolas, J., Moberg, K., Boyd, S. D., Taya, Y., Jacks, T., and Lees, J. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gartel, A. L., and Radhakrishnan, S. K. (2005) Cancer Res. 65 3980–3985 [DOI] [PubMed] [Google Scholar]
- 35.Niculescu, A. B., III, Chen, X., Smeets, M., Hengst, L., Prives, C., and Reed, S. I. (1998) Mol. Cell. Biol. 18 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogryzko, V. V., Wong, P., and Howard, B. H. (1997) Mol. Cell. Biol. 17 4877–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radhakrishnan, S. K., Feliciano, C. S., Najmabadi, F., Haegebarth, A., Kandel, E. S., Tyner, A. L., and Gartel, A. L. (2004) Oncogene 23 4173–4176 [DOI] [PubMed] [Google Scholar]
- 38.Koutsodontis, G., Tentes, I., Papakosta, P., Moustakas, A., and Kardassis, D. (2001) J. Biol. Chem. 276 29116–29125 [DOI] [PubMed] [Google Scholar]
- 39.Kardassis, D., Papakosta, P., Pardali, K., and Moustakas, A. (1999) J. Biol. Chem. 274 29572–29581 [DOI] [PubMed] [Google Scholar]
- 40.Lee, J. A., Suh, D. C., Kang, J. E., Kim, M. H., Park, H., Lee, M. N., Kim, J. M., Jeon, B. N., Roh, H. E., Yu, M. Y., Choi, K. Y., Kim, K. Y., and Hur, M. W. (2005) J. Biol. Chem. 280 28061–28071 [DOI] [PubMed] [Google Scholar]
- 41.Kwon, H. S., Kim, M. S., Edenberg, H. J., and Hur, M. W. (1999) J. Biol. Chem. 274 20–28 [DOI] [PubMed] [Google Scholar]
- 42.Kaczynski, J., Cook, T., and Urrutia, R. (2004) Genome Biol. 4 206.1–206.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lomberk, G., and Urrutia, R. (2005) Biochem. J. 392 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeda, T., Hobbs, R. M., and Pandolfi, P. P. (2005) Cancer Res. 65 8575–8578 [DOI] [PubMed] [Google Scholar]
- 45.Toledo, F., and Wahl, G. M. (2006) Nat. Rev. Cancer 6 909–923 [DOI] [PubMed] [Google Scholar]
- 46.Phan, R. T., Saito, M., Kitagawa, Y., Means, A. R., and Dalla-Favera, R. (2007) Nat. Immunol. 8 1132–1139 [DOI] [PubMed] [Google Scholar]
- 47.Dey, A., Tergaonkar, V., and Lane, D. P. (2008) Nat. Rev. Drug Discov. 7 1031–1040 [DOI] [PubMed] [Google Scholar]
- 48.Giacinti, C., and Giordano, A. (2006) Oncogene 25 5220–5227 [DOI] [PubMed] [Google Scholar]
- 49.Seoane, J., Le, H. V., and Massagué, J. (2002) Nature 419 729–734 [DOI] [PubMed] [Google Scholar]
- 50.Phan, R. T., Saito, M., Basso, K., Niu, H., and Dalla-Favera, R. (2005) Nat. Immunol. 6 1054–1060 [DOI] [PubMed] [Google Scholar]
- 51.Ranuncolo, S. M., Polo, J. M., Dierov, J., Singer, M., Kuo, T., Greally, J., Green, R., Carroll, M., and Melnick, A. (2007) Nat. Immunol. 8 705–714 [DOI] [PubMed] [Google Scholar]
- 52.Baron, B. W., Zeleznik-Le, N., Baron, M. J., Theisler, C., Huo, D., Krasowski, M. D., Thirman, M. J., Baron, R. M., and Baron, J. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7449–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phan, R. T., and Dalla-Favera, R. (2004) Nature 432 635–639 [DOI] [PubMed] [Google Scholar]
- 54.Li, J. Y., English, M. A., Ball, H. J., Yeyati, P. L., Waxman, S., and Licht, J. D. (1997) J. Biol. Chem. 272 22447–22455 [DOI] [PubMed] [Google Scholar]
- 55.McConnell, M. J., Chevallier, N., Berkofsky-Fessler, W., Giltnane, J. M., Malani, R. B., Staudt, L. M., and Licht, J. D. (2003) Mol. Cell. Biol. 23 9375–9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barna, M., Merghoub, T., Costoya, J. A., Ruggero, D., Branford, M., Bergia, A., Samori, B., and Pandolfi, P. P. (2002) Dev. Cell 3 499–510 [DOI] [PubMed] [Google Scholar]
- 57.Savage, A. K., Constantinides, M. G., Han, J., Picard, D., Martin, E., Li, B., Lantz, O., and Bendelac, A. (2008) Immunity 29 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.