Abstract
The activation of LFA-1 (lymphocyte function-associated antigen) is a critical event for T cell co-stimulation. The mechanism of LFA-1 activation involves both affinity and avidity regulation, but the role of each in T cell activation remains unclear. We have identified antibodies that recognize and block different affinity states of the mouse LFA-1 I-domain. Monoclonal antibody 2D7 preferentially binds to the low affinity conformation, and this specific binding is abolished when LFA-1 is locked in the high affinity conformation. In contrast, M17/4 can bind both the locked high and low affinity forms of LFA-1. Although both 2D7 and M17/4 are blocking antibodies, 2D7 is significantly less potent than M17/4 in blocking LFA-1-mediated adhesion; thus, blocking high affinity LFA-1 is critical for preventing LFA-1-mediated adhesion. Using these reagents, we investigated whether LFA-1 affinity regulation affects T cell activation. We found that blocking high affinity LFA-1 prevents interleukin-2 production and T cell proliferation, demonstrated by TCR cross-linking and antigen-specific stimulation. Furthermore, there is a differential requirement of high affinity LFA-1 in the activation of CD4+ and CD8+ T cells. Although CD4+ T cell activation depends on both high and low affinity LFA-1, only high affinity LFA-1 provides co-stimulation for CD8+ T cell activation. Together, our data demonstrated that the I-domain of LFA-1 changes to the high affinity state in primary T cells, and high affinity LFA-1 is critical for facilitating T cell activation. This implicates LFA-1 activation as a novel regulatory mechanism for the modulation of T cell activation and proliferation.
LFA-1 (lymphocyte function-associated antigen), an integrin family member, is important in regulating leukocyte adhesion and T cell activation (1, 2). LFA-1 consists of the αL (CD11a) and β2 (CD18) heterodimer. The ligands for LFA-1, including intercellular adhesion molecule ICAM3-1, ICAM-2, and ICAM-3, are expressed on antigen-presenting cells (APCs), endothelial cells, and lymphocytes (1). Mice that are deficient in LFA-1 have defects in leukocyte adhesion, lymphocyte proliferation, and tumor rejection (3–5). Blocking LFA-1 with antibodies can prevent inflammation, autoimmunity, organ graft rejection, and graft versus host disease in human and murine models (6–10).
LFA-1 is constitutively expressed on the surface of leukocytes in an inactive state. Activation of LFA-1 is mediated by inside-out signals from the cytoplasm (1, 11). Subsequently, activated LFA-1 binds to the ligands and transduces outside-in signals back into the cytoplasm that result in cell adhesion and activation (12, 13). The activation of LFA-1 is a critical event in the formation of the immunological synapse, which is important for T cell activation (2, 14, 15). The active state of LFA-1 is regulated by chemokines and the T cell receptor (TCR) through Rap1 signaling (16). LFA-1 ligation lowers the activation threshold and affects polarization in CD4+ T cells (17). Moreover, productive LFA-1 engagement facilitates efficient activation of cytotoxic T lymphocytes and initiates a distinct signal essential for the effector function (18–20). Thus, LFA-1 activation is essential for the optimal activation of T cells.
The mechanism of LFA-1 activation involves both affinity (conformational changes within the molecule) and avidity (receptor clustering) regulation (21–23). The I-domain of the LFA-1 αL subunit is the primary ligand-binding site and has been proposed to change conformation, leading to an increased affinity for ligands (24–26). The structural basis of the conformational changes in the I-domain of LFA-1 has been extensively characterized (27). Previously, we have demonstrated that the conformation of the LFA-1 I-domain changes from the low affinity to the high affinity state upon activation. By introducing disulfide bonds into the I-domain, LFA-1 can be locked in either the closed or open conformation, which represents the “low affinity” or “high affinity” state, respectively (28, 29). In addition, we identified antibodies that are sensitive to the affinity changes in the I-domain of human LFA-1 and showed that the activation-dependent epitopes are exposed upon activation (30). This study supports the presence of the high affinity conformation upon LFA-1 activation in cell lines. It has been demonstrated recently that therapeutic antagonists, such as statins, inhibit LFA-1 activation and immune responses by locking LFA-1 in the low affinity state (31–34). Furthermore, high affinity LFA-1 has been shown to be important for mediating the adhesion of human T cells (35, 36). Thus, the affinity regulation is a critical step in LFA-1 activation.
LFA-1 is a molecule of great importance in the immune system, and its activation state influences the outcome of T cell activation. Our previous data using the activating LFA-1 I-domain-specific antibody MEM83 indicate that avidity and affinity of the integrin can be coupled during activation (37). However, whether affinity or avidity regulation of LFA-1 contributes to T cell activation remains controversial (23, 38, 39). Despite the recent progress suggesting that conformational changes represent a key step in the activation of LFA-1, there are considerable gaps to be filled. When LFA-1 is activated, the subsequent outside-in signaling contributes to T cell activation via immunological synapse and LFA-1-dependent signaling. It is critical to determine whether high affinity LFA-1 participates in the outside-in signaling and affects the cellular activation of T cells. Nevertheless, the rapid and dynamic process of LFA-1 activation has hampered further understanding of the role of high affinity LFA-1 in primary T cell activation. The affinity of LFA-1 for ICAM-1 increases up to 10,000-fold within seconds and involves multiple reversible steps (23). In addition, the activation of LFA-1 regulates both adhesion and activation of T cells, two separate yet closely associated cellular functions. When LFA-1 is constitutively expressed in the active state in mice, immune responses are broadly impaired rather than hyperactivated, suggesting the complexity of affinity regulation (40). Therefore, it is difficult to dissect the mechanisms by which high affinity LFA-1 regulates stepwise activation of T cells in the whole animal system.
In the present study, we identified antibodies recognizing and blocking different affinity states of mouse LFA-1. These reagents allowed us to determine the role of affinity regulation in T cell activation. We found that blocking high affinity LFA-1 inhibited IL-2 production and proliferation in T cells. Furthermore, there is a differential requirement of high affinity LFA-1 in antigen-specific activation of CD4+ and CD8+ T cells. The activation of CD4+ T cells depends on both high and low affinity LFA-1. For CD8+ T cell activation, only high affinity LFA-1 provides co-stimulation. Thus, affinity regulation of LFA-1 is critical for the activation and proliferation of naive T cells.
EXPERIMENTAL PROCEDURES
Retrovirus Infection—Wild type, high affinity, or low affinity mouse αL was constructed in retrovirus expression vector pMSCVneo (BD Biosciences). Retroviral supernatants were generated by transient transfection of 293T cells. Transduction of primary mouse lymphocytes was performed as previously described (41). Briefly, splenocytes were harvested from CD11a-deficient mice and cultured at 2 × 106 cell/ml in the presence of anti-CD3 mAb (catalog number 145-2C11; BD Biosciences) at 1 μg/ml for 3 days. On day two, the cells were spin-infected with retroviral supernatant in the presence of 2.5 μg/ml Polybrene for 90 min at 2,500 rpm at 37 °C. After infection, the retroviral supernatant was replaced. On day three, the cells were collected and used to assay the expression of αL by flow cytometry.
Homotypic Aggregation Assay—Murine EL-4 cells (1 × 106 cells/well) were stimulated with phorbol 12-myristate 13-acetate (PMA) at a final concentration of 50 ng/ml. The reactions were performed in flat bottom 96-well plates at 37 °C for 2 h. Then aggregation was determined using light microscopy. The degree of aggregation of EL-4 was scored as follows: 0, no cells were clustered; 1, less than 10% of cells were aggregated; 2, clustering of less than 50% of cells; 3, nearly 100% of cells were in small, loose aggregates; 4, nearly 100% of cells were in large clusters (42).
Static Adhesion Assay—Binding of primary mouse lymphocytes to ICAM-1 was examined as described briefly, and purified mouse recombinant ICAM-1/FC (R&D Systems) was coated on flat-bottom 96-well plates overnight at 4 °C. Mouse lymphocytes were pretreated with mAb 2D7 or M17/4 or isotype control in the presence or absence of Mn2+ for 30 min at room temperature and loaded into ICAM-1-coated wells at a concentration of 1 × 106 cells/well. The bound cells were counted under a microscope in representative fields.
Mixed Lymphocyte Culture—CFSE stock (Molecular Probes) was added to the responder cells at a concentration of 0.5 μm. The experiment was performed in 48-well microtiter plates (Costar). CFSE-labeled C57BL/6 responder cells were plated at 1 × 106 cells/ml in a volume of 500 μl/well and cocultured at a ratio of 2:1 with 3400-centigray irradiated C57BL/6 or Balb/C stimulator cells. The plates were then placed in a humidified incubator.
OT-I and OT-II T Cell Activation—Both OT-I and OT-II mice were purchased from Jackson Laboratories. The Ova (ISQAVHAAHAEINEAGR) and SIINFEKL peptides were ordered with 90% purity (SynPep). Each peptide preparation was tested for optimal biological activity before being used for experiments. TCR transgenic CD4+ (OT-II) or CD8+ (OT-I) T cells from lymph nodes of 6–8-week-old mice were positively sorted using CD4- or CD8-microbeads (Miltenyi Biotec). Splenic APCs from C57BL/6 mice were prepared by complement-mediated lysis of Thy1+ T cells. OT-II or OT-I T cells (1 × 106 cells/well) were stimulated with Ova (1, 5, and 20 μg/ml) or SIINFEKL peptide (0.1, 0.01, and 0.001 μg/ml), respectively, in the presence of irradiated splenic APCs (1 × 106/ml) in 96-well plates. Culture supernatants were collected after 24 h to determine IL-2 expression. Proliferation was assayed on day 3 by adding [3H]thymidine to the culture for the last 8 h.
RESULTS
mAbs 2D7 and M17/4 Bind to Different Affinity States of LFA-1 I-domain—mAb M17/4 has been used successfully to inhibit LFA-1-mediated immune responses in various animal disease models (6–9). Therefore, we sought to determine whether the potency of M17/4 is due to its ability to block high affinity LFA-1. According to our previous study, disulfide bonds were used to lock the I-domain of human LFA-1 in the low affinity (closed) or high affinity (open) conformation (29, 30). Because the sequence homology of human and mouse LFA-1 I-domains is 72.8%, we predicted that creating disulfide bonds at the equivalent sites in the mouse sequence would similarly produce either a locked low affinity or high affinity LFA-1 I-domain (supplemental Fig. 1A). We introduced cysteine mutations in mouse αL to form a disulfide bond and generate the locked closed (L289C/K294C) or open (K287C/K294C) I-domain. The subsequent models of the resulting mouse low affinity and high affinity I-domain structures are illustrated in supplemental Fig. 1B.
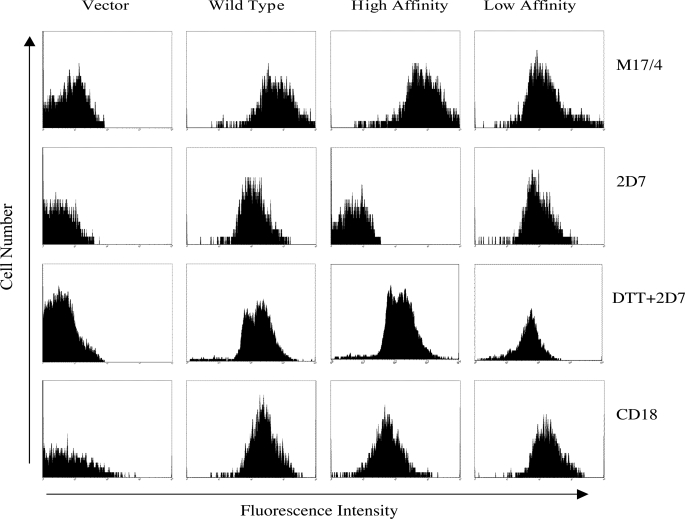
The mutants and wild-type αL were cloned into a retrovirus vector and transduced into lymphocytes from the αL-deficient mouse. The expression level of LFA-1 was measured by both anti-CD11a and anti-CD18 antibodies. As shown in Fig. 1, two rat anti-mouse αL mAbs, M17/4 and 2D7, were used to examine the expression of mouse LFA-1. We found M17/4 can bind to wild type, high affinity, and low affinity αL. Although 2D7 bound to both wild type and low affinity αL, it did not bind to the high affinity conformation. The difference was not attributed to the lack of LFA-1 expression, because both anti-CD18 antibody and green fluorescent protein were used to confirm the protein expression level in the cells transduced with the high affinity αL. In addition, green fluorescent protein was used to confirm the protein expression level in the cells transduced with the high affinity αL (data not shown). We further investigated whether the absence of 2D7 binding is due to the conformational changes within the high affinity I-domain. Disulfide bond reduction in the locked high affinity human LFA-1 was shown to result in the conversion to the low affinity conformation (28, 30). After treating the cells with dithiothreitol, the binding of 2D7 to LFA-1 was restored to a level similar to that of wild type and low affinity LFA-1 (Fig. 1). Therefore, 2D7 preferentially binds to the low affinity conformation, and this specific binding is abolished when LFA-1 is locked in the high affinity conformation. In contrast, M17/4 can bind both the locked high and low affinity forms of LFA-1.
FIGURE 1.
Antibody 2D7 binds to mouse LFA-1 in the locked low affinity conformation. The wild type, high affinity, or low affinity mouse αL was expressed on the surface of CD11a-deficient splenocytes using retrovirus transduction. The level of cell surface expression was determined by flow cytometry using mAbs M/17 and 2D7 with or without dithiothreitol treatment and anti-CD18 (filled histograms). The binding of M17/4 and 2D7 to different forms of αL on the gated GFP+CD3+ cells was determined by flow cytometry.
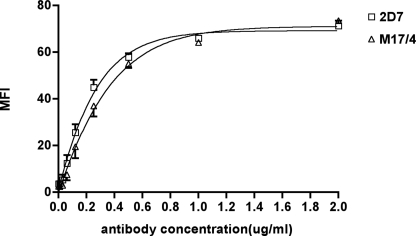
Next, we mapped the epitopes of 2D7 and M17/4. Since neither 2D7 nor M17/4 cross-reacts with human αL, we used the previously described human-mouse chimeric αL mutants to map the epitopes (43). The 11 αL chimeras containing segments of the mouse αL swapped into human αL or vice versa were cotransfected with mouse β2 into 293T cells (supplemental Table 1). 2D7 binds to residues 118–153, which are located at the N terminus of the I-domain. M17/4 binds to residues 249–303, which are at the C terminus of the I-domain. Thus, both 2D7 and M17/4 recognize the I-domain, but they bind to different regions. We compared the binding affinity of 2D7 and M17/4 to the inactive form of LFA-1 on resting T cells. As shown in Fig. 2, the binding curves are similar for 2D7 and M17/4, and the saturating binding dose is ∼1 μg/ml.
FIGURE 2.
M17/4 and 2D7 have the same binding affinity for the inactive LFA-1. Primary mouse lymphocytes were incubated with a serial diluting concentration of M17/4-fluorescein isothiocyanate or 2D7-fluorescein isothiocyanate in the presence of a saturating concentration of CD3-PE. Cells were washed and subjected to flow cytometry. CD3+ cells were gated to obtain the mean fluorescent intensity (MFI) of M17/4 and 2D7. The data points represent the mean of three individual experiments. The binding curves were generated using a linear regression model (GraphPad Prism version 2.0).
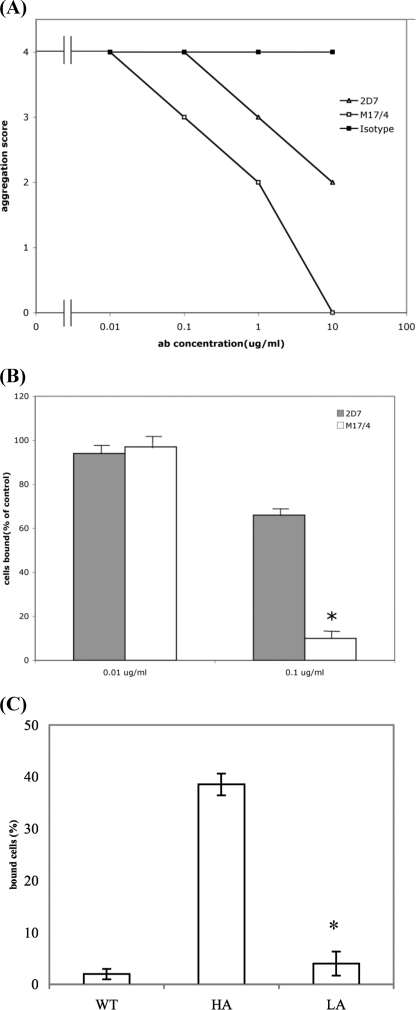
Blocking High Affinity LFA-1 Prevents Lymphocyte Adhesion—The function of I-domain mAbs is complicated by the fact that they can either inhibit or activate the binding of LFA-1 to its ligand ICAM-1 through various mechanisms (37, 44). We further examined the effect of 2D7 and M17/4 on LFA-1-mediated adhesion with aggregation assay and static adhesion assay. The purpose is to address whether blocking high affinity LFA-1 prevents LFA-1-mediated cell adhesion. In the EL-4 aggregation assay, cells were stimulated with PMA at a final concentration of 50 ng/ml in flat-bottom 96-well plates at 37 °C for 2 h in the presence of either 2D7, M17/4, or isotype control. As shown in Fig. 3A, both 2D7 and M17/4 blocked homotypic aggregation of cells stimulated by PMA, but 2D7 was less efficient compared with M17/4 at equivalent concentrations. Similar results were obtained with primary mouse lymphocyte aggregation (data not shown). We then tested whether these mAbs can block the binding of primary mouse lymphocytes to ICAM-1 in the static adhesion assay. Primary mouse lymphocytes were preincubated with 2D7, M17/4, or isotype control and then loaded into 96-well flat bottom plates coated with purified mouse ICAM-1 in the presence of Mn2+. As shown in Fig. 3B, both 2D7 and M17/4 inhibited lymphocyte adhesion to ICAM-1 at a concentration of 0.1 μg/ml. M17/4 almost completely inhibited the adhesion, whereas 2D7 only partially blocked the adhesion with about 70% cells remaining bound to ICAM-1. Although both 2D7 and M17/4 are blocking antibodies, 2D7 is significantly less potent than M17/4 in blocking LFA-1-mediated adhesion. To confirm that high affinity LFA-1 is important for lymphocyte adhesion, we first purified αL-deficient lymphocytes transduced with either wild type, high affinity, or low affinity αL (from Fig. 1) by using a cell sorter to collect the retrovirus-transfected cells with equivalent green fluorescent protein expression level. As shown in Fig. 3C, lymphocytes expressing either wild type (WT) or low affinity (LA) LFA-1 were incapable of adhering to the coated ICAM-1 in the absence of Mn2+, whereas 38% cells with high affinity (HA) LFA-1 remained bound in the static adhesion assay. Thus, the high affinity form of αL is critical for LFA-1-mediated adhesion.
FIGURE 3.
Blocking of LFA-1-mediated adhesion with M17/4 and 2D7. A, homotypic aggregation. Murine EL-4 cells activated by PMA were treated with 2D7, M17/4, or isotype control. Aggregation was scored as described under “Experimental Procedures.” Three independent experiments showed identical results. B and C, static adhesion assay. Primary mouse lymphocytes were preincubated with 2D7, M17/4, or isotype control in the presence of Mn2+ and then loaded into a 96-well flat bottom plate coated with purified mouse ICAM-1 (B). Purified αL-deficient lymphocytes transduced with wild type (WT), high affinity (HA), or low affinity (LA) LFA-1 were loaded into a 96-well flat bottom plate coated with purified mouse ICAM-1 (C). Binding to ICAM-1 was measured by counting cells adhered to the bottom after washes. Results are mean and S.D. of three independent experiments normalized to that of isotype control. The asterisk represents data with p value less than 0.05 in Student's t test.
High Affinity LFA-1 Facilitates IL-2 Production and Proliferation in T Cells—We identified mAbs M17/4 and 2D7, which recognize and block different affinity states of mouse LFA-1. M17/4 binds to the I-domain in both high and low affinity conformations, whereas 2D7 is an activation-sensitive mAb that preferentially binds to the low affinity LFA-1 but not to the high affinity conformation. Utilizing these reagents, we investigated whether affinity regulation of LFA-1 participates in and affects cellular activation of naive T cells.
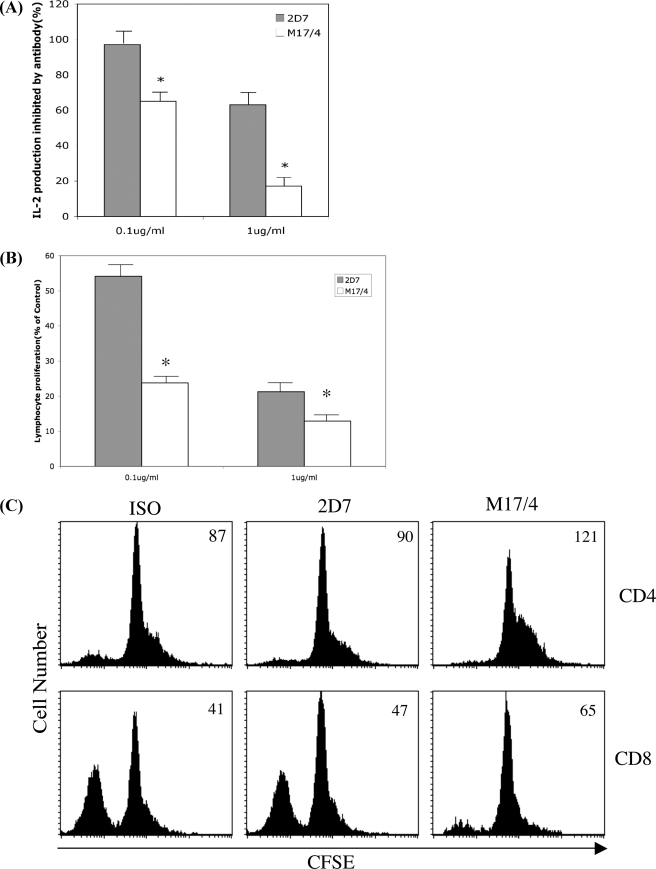
First, we examined the effects of 2D7 and M17/4 on IL-2 production in activated mouse T cells. Primary mouse T cells were activated by plate-coated anti-CD3 antibody. Secreted IL-2 in the supernatant was measured 24 h after activation. As shown in Fig. 4A, M17/4 inhibited IL-2 production at 0.1 μg/ml and almost completely blocked IL-2 production at a concentration of 1 μg/ml, whereas 2D7 only partially reduced IL-2 production to ∼60% of the control at the concentration of 1 μg/ml. Thus, blocking high affinity LFA-1 inhibits IL-2 production.
FIGURE 4.
Inhibition of mouse T cell activation with 2D7 and M17/4. A, IL-2 production in culture supernatant after 24-h activation with coated anti-CD3 antibody in the presence of 2D7, M17/4, or isotype control at the indicated concentration. Asterisk, data with p value less than 0.05 in Student's t test. B, mouse T cell proliferation. Column-purified mouse T cells were activated by coated anti-CD3 antibody in the presence of mAb 2D7 or M17/4 or isotype control at the indicated concentration. Cell proliferation was measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay after 48 h. Results are mean and S.D. of three independent experiments normalized to that of isotype control. Asterisk, data with p value less than 0.05 in Student's t test. C, mixed lymphocyte culture. Responder cells from C57/BL6 spleen were labeled with CFSE and cocultured with irradiated Balb/C stimulator cells for 72 h in the presence 2D7, M17/4, or isotype control at a concentration of 1 μg/ml. The responder cells were stained with CD4-PerCP, CD8-APC, and CD3-PE. The gated CD8+/CD3+ or CD4+/CD3+ cells are displayed in the histogram plots for CFSE. The number at the right top corner of each histogram plot is the MFI of CFSE.
It has been demonstrated that T lymphocyte proliferation is impaired in LFA-1 knock-out mice (3). We therefore investigated whether high affinity LFA-1 contributes to T cell proliferation. As shown in Fig. 4B, both 2D7 and M17/4 can inhibit lymphocyte proliferation after stimulation with coated anti-CD3 antibody, although the potency of 2D7 was significantly less than that of M17/4. We further examined the affinity regulation of LFA-1 on both CD4+ and CD8+ T cell proliferation upon alloactivation in mixed lymphocyte reactions. CFSE-labeled responder cells were plated with irradiated stimulator cells for 72 h, and cell proliferation was measured by CFSE intensity with FACS. As shown in Fig. 4C, M17/4 can inhibit the proliferation of both CD4+ and CD8+ T cells, whereas 2D7 showed only minimal inhibitory effect compared with control. In addition, locking LFA-1 in the low affinity state by treating the cells with lovastatin reduced T cell proliferation, similar to that observed with M17/4 (data not shown). Thus, high affinity LFA-1 is required for T cell proliferation in mixed lymphocyte reactions.
It has been shown that LFA-1 signaling contributes to T cell activation through the Erk1/2 mitogen-activated protein kinase signal pathway in CD4+ T cells (17). Therefore, we examined whether high affinity LFA-1 plays a role in Erk1/2 mitogen-activated protein kinase signaling in both CD4+ and CD8+ T cells. Mouse T lymphocytes were activated, and phosphorylated Erk1/2 was measured by intracellular staining. As shown in supplemental Fig. 3A, 2.28% of CD4+ T cells were positive for phospho-p44/42 after 15 min of stimulation. In comparison, only 1.34% of cells were positive for the phospho-p44/42 in the presence of 2D7, whereas M17/4 reduced the frequency to 0.85%. In contrast to CD4+ T cells, the signaling in the CD8 compartment was robust, with 34.9% cells positive for phospho-p44/42. 2D7 had minimal effect on the Erk1/2 signaling. However, M17/4 reduced the percentage of phosphorylated cells to 18.1%, a 50% reduction compared with isotype control (34.9%). Thus, high affinity LFA-1 contributes to Erk1/2 signal pathway in T cell activation.
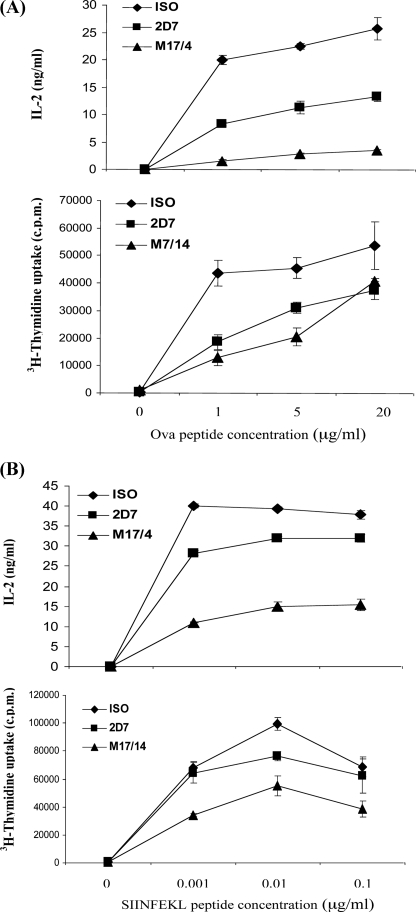
Inhibition of Antigen-specific T Cell Activation with 2D7 and M17/4—LFA-1 co-stimulates lymphocyte activation by participating in the formation of the immunological synapse (2, 14). It has been demonstrated that LFA-1 facilitates T cell activation by promoting adhesion of antigen-specific T cells to APC (46, 47). We and others have shown that high affinity LFA-1 is critical for T cell adhesion. Therefore, we sought to determine whether high affinity LFA-1 is required for the activation of antigen-specific T cells from OT-I and OT-II TCR-transgenic mice.
After CD4+ T cells from OT-II mice were activated by APC loaded with various doses of the Ova peptide, we measured IL-2 production and cell proliferation in the presence of 2D7 or M17/4. As shown in Fig. 5A, M17/4 completely blocked IL-2 production independent of the peptide concentration. Blocking low affinity LFA-1 with 2D7 only reduced IL-2 production to 50% of that in the control. Thus, both high and low affinity LFA-1 were important for IL-2 production in CD4+ T cells. M17/4 prevented CD4+ T cell proliferation more effectively than 2D7 at low antigen stimulation, whereas at high dose peptide stimulation, both M17/4 and 2D7 had similar effects. Next, we examined the role of high affinity LFA-1 in CD8+ T cells. T cells from OT-I mice were stimulated with various concentrations of the SIINFEKL peptide. As shown in Fig. 5B, blocking low affinity LFA-1 with 2D7 had minimal inhibitory effects on both IL-2 production and proliferation of CD8+ T cells. In contrast, there was a 50% reduction of IL-2 production and proliferation observed in the presence of M17/4. Thus, high affinity LFA-1 plays an essential role in CD8+ T cell activation.
FIGURE 5.
Inhibition of antigen-specific T cell activation with 2D7 and M17/4. CD4+ T cells from OT-II mice (A) or CD8+ T cells from OT-I mice (B) were treated with Ova (1, 5, and 20 μg/ml) or SIINFEKL (0.1, 0.01, and 0.001 μg/ml) peptide, respectively, in the presence of APCs. Antibody 2D7 or M17/4 or isotype control at a concentration of 1 μg/ml was added for 4 days. T cells were restimulated with plate-bound anti-CD3. IL-2 was measured 24 h after treatment. Proliferation was assayed at 72 h for OT-II cells and 48 h for OT-I cells after treatment by adding [3H]thymidine to the culture for the last 8 h.
In summary, there is a differential requirement of LFA-1 in CD4+ and CD8+ T cell activation. The IL-2 production of CD4+ T cells depends on both high and low affinity LFA-1. For CD8+ T cell activation, LFA-1 provides co-stimulation for optimal IL-2 production and proliferation, which is delivered only through high affinity LFA-1.
DISCUSSION
In the current study, we have investigated the role of high affinity LFA-1 in T cell activation using antibodies that bind to different affinity states of the LFA-1 I-domain. We demonstrated the functional importance of high affinity LFA-1 in the activation of naive T cells under various conditions. Furthermore, we explored the mechanisms by which high affinity LFA-1 affects T cell signaling. We found that high affinity LFA-1 induces IL-2 production and T cell proliferation. Remarkably, although CD4+ T cell activation relies on both high and low affinity LFA-1, only high affinity LFA-1 provides co-stimulation for CD8+ T cell activation. This implicates a novel regulatory mechanism for the modulation of T cell activation. Together, our data demonstrated that the I-domain of LFA-1 changes to the high affinity state in primary T cells and that high affinity LFA-1 is critical for facilitating T cell activation.
The ICAM-1 ligand binding site of LFA-1 is located on the I-domain. Previous studies have shown that the I-domain changes from a closed to open conformation upon LFA-1 activation (28–30). However, the correlation of the observed conformational states with LFA-1-mediated function in primary T cells is lacking. We characterized two mouse antibodies, 2D7 and M17/4, that recognize different affinity states of the LFA-1 I-domain. Although both 2D7 and M17/4 bind to the inactive LFA-1 with similar affinity, M17/4 binds to the I-domain in both high and low affinity conformations, whereas 2D7 preferentially binds to the low affinity LFA-1 but not to the high affinity conformation. 2D7 binds to the N terminus of the I-domain (residues 118–153), which encompasses most of the metal ion-dependent adhesion site-coordinating residues. Changes in the metal ion-dependent adhesion site that accompany activation may result in an effective conformation-sensitive epitope for LFA-1 (48). Thus, the epitope of 2D7 is conformation-sensitive, and the binding is abolished when LFA-1 is locked in the high affinity conformation. More importantly, 2D7 is less efficient in inhibiting LFA-1-mediated adhesion in comparison with M17/4. This suggests that the 2D7 antibody cannot bind the high affinity LFA-1 and induces a conformational switch back to its low affinity form. Conversely, the M17/4 antibody can recognize both affinity states of the integrin but serves as a more effective inhibition of LFA-1 as it prevents accessibility of the ICAM-1 to the metal ion-dependent adhesion site in LFA-1. Our data not only validated the predicted I-domain conformational switch model but also provided evidence that the high affinity conformation occurs when LFA-1 is activated on the cell surface and is therefore less sensitive to the low affinity-specific antibody 2D7.
In addition to facilitating firm adhesion, LFA-1 plays an important role in T cell activation in the context of the immunological synapse (2, 49). Interestingly, we found that there is a different requirement for high affinity LFA-1 in antigen-specific T cell activation from OT-I and OT-II TCR-transgenic mice. The activation of CD4+ T cells depends on both high and low affinity LFA-1. For CD8+ T cell activation, however, only high affinity LFA-1 is required to provide maximal activation (Fig. 5B). The same trend is also observed in Erk1/2 signaling in anti-CD3 activated T cells (supplemental Fig. 3A), and thus high affinity LFA-1 contributes to the Erk1/2 signal pathway in T cell activation. To investigate whether Erk1/2 signaling directly resulted from TCR ligation, we used pharmacological inhibitors for Lck, ZAP70, and PI3K, molecules downstream of TCR (14, 46). The Lck inhibitor PP1 completely blocked the ERK1/2 signal in both CD4+ and CD8+ T cells, whereas inhibitors for ZAP70 (piceatannol) and phosphatidylinositol 3-kinase (wortmannin) significantly reduced the percentage of phosphorylated cells (supplemental Fig. 3B). Therefore, the LFA-1-dependent Erk1/2 signal pathway is activated by TCR ligation. Our data suggested that TCR stimulation by CD3-ligation activates LFA-1, and subsequently the outside-in signal transduced by high affinity LFA-1 contributes to Erk1/2 phosphorylation in activated T cells.
The function of LFA-1 is to strengthen the central supramolecular activation complex by forming the peripheral supramolecular activation complex, thus supporting and maintaining a mature synapse between T cells and APCs (2, 49). Previous studies provided evidence that the binding of TCR on CD4+ cells with peptide-major histocompatibility complex-II complex is relatively weak and less stringent (50, 51). Our data demonstrated that CD4+ T cell activation is LFA-1-dependant, including both high and low affinity LFA-1. It is possible that the engagement of LFA-1 is indispensable for the formation of a stable immunological synapse for CD4+ T cells. In contrast, the binding of the TCR on CD8+ T cells with peptide-major histocompatibility complex-I is stable, and CD8+ T cell activation requires fewer ligands bound to the TCR compared with CD4+ T cells (13, 52, 54). Even in the presence of mAb M17/4, which blocks both high and low affinity LFA-1 binding, CD8+ T cells can be activated (Fig. 5). Thus, the basal activation of CD8+ T cells does not require LFA-1 engagement, although high affinity LFA-1 optimizes CD8+ T cell activation. It appears that LFA-1 engagement is particularly relevant for facilitating T cell activation in the setting of weak interactions, such as low affinity TCR or low antigen concentration. The differential requirement of LFA-1 in CD4+ and CD8+ T cell activation reflects the inherent nature of their TCR stringencies. It is intriguing to propose that LFA-1 modulates T cell activation in two steps. First, LFA-1 enhances the strength of TCR and antigen interaction to form the central supramolecular activation complex, such as in OT-II CD4+ T cell activation. This is probably delivered through low affinity LFA-1 stimulation. Subsequently, the high affinity LFA-1 provides costimulation for optimal activation in both CD4+ and CD8+ T cells.
In addition to affinity regulation, avidity change or clustering of LFA-1 on the cell surface has been postulated for LFA-1 activation. Studies so far have been unable to distinguish between the functional importance of these two models, although they are not necessarily mutually exclusive. Furthermore, the relative degree to which clustering and avidity is present in the TCR may not necessarily reflect what is observed during LFA-1/ICAM-1-mediated adhesion to the endothelial wall prior to transendothelial migration due to the shear flow environment within the vasculature (55, 56). It is generally agreed that both affinity and avidity are tightly regulated by complex signaling events and cytoskeleton rearrangements, regardless of various working hypotheses (22, 23, 38, 59). A reasonable possibility is that the activation of LFA-1 involves both affinity and avidity regulation together in order to fine-tune the immune responses. Indeed, when microclustering was induced by PMA or CD3 ligation on mouse T cells, we found that M17/4 preferentially bound to the distinct polarized cap regions, whereas 2D7 stained uniformly (supplemental Fig. 2). After stimulation with PMA for 15 or 30 min, there was no distinct clustering detected on T cells following staining with 2D7, the antibody that only binds to low affinity LFA-1. In contrast, 100% of cells displayed the clustering pattern using antibody M17/4, which binds to both high and low affinity LFA-1 (data not shown). Thus, our data suggest that the clustering regions on the surface of activated T cells constitute the high affinity LFA-1. This clustering may well result from ICAM-1-linked heterotetramers due to D4/D4 and D1/D1 dimerization (27, 57–58). In this manner, affinity and avidity are linked, as was observed for the activating mAb MEM83, where cellular activation was induced only by linked array formation with an IgG, and this effect was absent for the Fab (37).
The regulation of LFA-1 activation is critical in inflammatory and immune responses. There has been long standing interest in LFA-1 as a therapeutic target for regulating immunity (60, 61). Although M17/4 has been successful in various animal disease models, the effectiveness of anti-LFA-1 therapy has been limited by the inability to target the activated LFA-1 (53). Efalizumab, a mAb blocking nonselectively the high and low affinity LFA-1, has recently been approved for the treatment of psoriasis (45). Better understanding of the function of high affinity LFA-1 provides a rationale for developing reagents selectively targeting activated LFA-1 (31–33). The second generation anti-LFA-1 therapy may prove clinically advantageous as a result of improved specificity and potency.
In conclusion, our study investigates a fundamental issue of T cell activation and demonstrates for the first time that LFA-1 affinity regulation modulates naive T cell activation. The activation of memory T cells might have different regulatory mechanisms, and the precise role of high affinity LFA-1 in their activation and proliferation remains to be explored. In addition to TCR, chemokines regulate LFA-1 activation as well. It has been demonstrated that high affinity LFA-1 is important for the adhesion mediated by chemokines (35, 36, 39). Further studies will be required to determine whether chemokines and TCR synergistically regulate LFA-1 affinity during T cell activation.
Supplementary Material
Acknowledgments
We thank Dr. Timothy Springer for providing the human-mouse chimeras, Dr. Christie Ballantyne for providing the CD11a-deficient mice, Drs. Krishna Komanduri and Chen Dong for discussion, and Jason Mitchelle for technical assistance. The animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Texas M.D. Anderson Cancer Center.
This work was supported by American Cancer Society Grant RSG-08-183-01-LIB (to Q. M.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
Footnotes
The abbreviations used are: ICAM, intercellular adhesion molecule; APC, antigen-presenting cell; TCR, T cell receptor; mAb, monoclonal antibody; PMA, phorbol 12-myristate 13-acetate; IL, interleukin; CFSE, carboxyfluorescein succinimidyl ester.
References
- 1.Springer, T. A. (1994) Cell 28 301–314 [DOI] [PubMed] [Google Scholar]
- 2.Dustin, M. L. (2003) Ann. N. Y. Acad. Sci. 987 51–59 [DOI] [PubMed] [Google Scholar]
- 3.Ding, Z. M., Babensee, J. E., Simon, S. I., Lu, H., Perrard, J. L., Bullard, D. C., Dai, X. Y., Bromley, S. K., Dustin, M. L., Entman, M. L., Smith, C. W., and Ballantyne, C. M. (1999) J. Immunol. 163 5029–5038 [PubMed] [Google Scholar]
- 4.Schmits, R., Kundig, T. M., Baker, D. M., Shumaker, G., Simard, J. J., Duncan, G., Wakeham, A., Shahinian, A., van der Heiden, A., Bachmann, M. F., Ohashi, P. S., Mak, T. W., and Hickstein, D. D. (1996) J. Exp. Med. 83 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlin-Rufenach, C., Otto, F., Mathies, M., Westermann, J., Owen, M. J., Hamann, A., and Hogg, N. (1999) J. Exp. Med. 189 1467–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harning, R., Pelletier, J., Lubbe, K., Takei, F., and Merluzzi, V. J. (1991) Transplantation 52 842–845 [DOI] [PubMed] [Google Scholar]
- 7.van Kooyk, Y., de Vries-van der Zwan, A., de Waal, L. P., and Figdor, C. G. (1994) Transplant. Proc. 26 401–403 [PubMed] [Google Scholar]
- 8.Blazar, B. R., Taylor, P. A., Panoskaltsis-Mortari, A., Gray, G. S., and Vallera, D. A. (1995) Blood 85 2607–2618 [PubMed] [Google Scholar]
- 9.Kootstra, C. J., Van Der Giezen, D. M., Van Krieken, J. H., De Heer, E., and Bruijn, J. A. (1997) Clin. Exp. Immunol. 108 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebwohl, M., Tyring, S. K., Hamilton, T. K., Toth, D., Glazer, S., Tawfik, N. H., Walicke, P., Dummer, W., Wang, X., Garovoy, M. R., and Pariser, D. (2003) N. Engl. J. Med. 349 2004–2013 [DOI] [PubMed] [Google Scholar]
- 11.Butcher, E. C., and Picker, L. J. (1996) Science 5 60–66 [DOI] [PubMed] [Google Scholar]
- 12.Diamond, M. S., and Springer, T. A. (1994) Curr. Biol. 4 506–517 [DOI] [PubMed] [Google Scholar]
- 13.Brower, R. C., England, R., Takeshita, T., Kozlowski, S., Margulies, D. H., Berzofsky, J. A., and Delisi, C. (1994) Mol. Immunol. 31 1285–1293 [DOI] [PubMed] [Google Scholar]
- 14.Huppa, J. B., and Davis, M. M. (2003) Nat. Rev. Immunol. 3 973–983 [DOI] [PubMed] [Google Scholar]
- 15.Dustin, M. L. (2007) Curr. Opin. Cell Biol. 19 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katagiri, K., Maeda, A., Shimonaka, M., and Kinashi, T. (2003) Nat. Immunol. 4 741–748 [DOI] [PubMed] [Google Scholar]
- 17.Perez, O. D., Mitchell, D., Jager, G. C., South, S., Murriel, C., McBride, J., Herzenberg, L. A., Kinoshita, S., and Nolan, G. P. (2003) Nat. Immunol. 4 1083–1092 [DOI] [PubMed] [Google Scholar]
- 18.Huppa, J. B., Gleimer, M., Sumen, C., and Davis, M. M. (2003) Nat. Immunol. 4 749–755 [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson, S. R., Williams, N. A., and Morgan, D. J. (2005) J. Immunol. 174 3401–3407 [DOI] [PubMed] [Google Scholar]
- 20.Anikeeva, N., Somersalo, K., Sims, T. N., Thomas, V. K., Dustin, M. L., and Sykulev, Y. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 6437–6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes, R. O. (2002) Cell 110 673–687 [DOI] [PubMed] [Google Scholar]
- 22.Carman, C. V., and Springer, T. A. (2003) Curr. Opin. Cell Biol. 15 547–556 [DOI] [PubMed] [Google Scholar]
- 23.Dustin, M. L., Bivona, T. G., and Philips, M. R. (2004) Nat. Immunol. 5 363–372 [DOI] [PubMed] [Google Scholar]
- 24.Shimaoka, M., Takagi, J., and Springer, T. A. (2002) Annu. Rev. Biophys. Biomol. Struct. 31 485–516 [DOI] [PubMed] [Google Scholar]
- 25.Shimaoka, M., Xiao, T., Liu, J. H., Yang, Y., Dong, Y., Jun, C. D., McCormack, A., Zhang, R., Joachimiak, A., Takagi, J., Wang, J. H., and Springer, T. A. (2003) Cell 10 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legge, G. B., Morris, G. M., Sanner, M. F., Takada, Y., Olson, A. J., and Grynszpan, F. (2002) Proteins 48 151–160 [DOI] [PubMed] [Google Scholar]
- 27.Luo, B. H., Carman, C. V., and Springer, T. A. (2007) Annu. Rev. Immunol. 25 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, C., Shimaoka, M., Zang, Q., Takagi, J., and Springer, T. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 27 2393–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimaoka, M., Lu, C., Palframan, R. T., von Andrian, U. H., McCormack, A., Takagi, J., and Springer, T. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 22 6009–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, Q., Shimaoka, M., Lu, C., Jing, H., Carman, C. V., and Springer, T. A. (2002) J. Biol. Chem. 22 10638–10641 [DOI] [PubMed] [Google Scholar]
- 31.Weitz-Schmidt, G., Welzenbach, K., Brinkmann, V., Kamata, T., Kallen, J., Bruns, C., Cottens, S., Takada, Y., and Hommel, U. (2001) Nat. Med. 7 687–692 [DOI] [PubMed] [Google Scholar]
- 32.Gadek, T. R., Burdick, D. J., McDowell, R. S., Stanley, M. S., Marsters, J. C., Jr., Paris, K. J., Oare, D. A., Reynolds, M. E., Ladner, C., Zioncheck, K. A., Lee, W. P., Gribling, P., Dennis, M. S., Skelton, N. J., Tumas, D. B., Clark, K. R., Keating, S. M., Beresini, M. H., Tillev, J. W., Presta, L. G., and Bodarv, S. C. (2002) Science 295 1086–1089 [DOI] [PubMed] [Google Scholar]
- 33.Shimaoka, M., and Springer, T. A. (2003) Nat. Rev. Drug. Discov. 2 703–716 [DOI] [PubMed] [Google Scholar]
- 34.Kallen, J., Welzenbach, K., Ramage, P., Geyl, D., Kriwacki, R., Legge, G., Cottens, S., Weitz-Schmidt, G., and Hommel, U. (1999) J. Mol. Biol. 292 1–9 [DOI] [PubMed] [Google Scholar]
- 35.Kim, M., Carman, C. V., and Springer, T. A. (2003) Science 301 1720–1725 [DOI] [PubMed] [Google Scholar]
- 36.Shamri, R., Grabovsky, V., Gauguet, J. M., Feigelson, S., Manevich, E., Kolanus, W., Robinson, M. K., Staunton, D. E., von Andrian, U. H., and Alon, R. (2005) Nat. Immunol. 6 497–506 [DOI] [PubMed] [Google Scholar]
- 37.Carreño, R., Li, D., Sen, M., Nira, I., Yamakawa, T., Ma, Q., and Legge, G. B. (2008) J. Biol. Chem. 283 10642–10648 [DOI] [PubMed] [Google Scholar]
- 38.Hynes, R. O. (2003) Science 300 755–756 [DOI] [PubMed] [Google Scholar]
- 39.Kim, M., Carman, C. V., Yang, W., Salas, A., and Springer, T. A. (2004) J. Cell Biol. 167 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semmrich, M., Smith, A., Feterowski, C., Beer, S., Engelhardt, B., Busch, D. H., Bartsch, B., Laschinger, M., Hogg, N., Pfeffer, K., and Holzmann, B. (2005) J. Exp. Med. 201 1987–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston, S. C., Dustin, M. L., Hibbs, M. L., and Springer, T. A. (1990) J. Immunol. 145 1181–1187 [PubMed] [Google Scholar]
- 42.Wooten, D. K., Teague, T. K., and McIntyre, B. W. (1999) J. Leukocyte Biol. 65 127–136 [DOI] [PubMed] [Google Scholar]
- 43.Huang, C., and Springer, T. A. (1995) J. Biol. Chem. 270 19008–19016 [DOI] [PubMed] [Google Scholar]
- 44.Lu, C., Shimaoka, M., Salas, A., and Springer, T. A. (2004) J. Immunol. 173 3972–3978 [DOI] [PubMed] [Google Scholar]
- 45.Gordon, K. B., Papp, K. A., Hamilton, T. K., Walicke, P. A., Dummer, W., Li, N., Bresnahan, B. W., Menter, A., and Efalizumab Study Group (2003) J. Am. Med. Assoc. 290 3073–3080 [DOI] [PubMed] [Google Scholar]
- 46.Mueller, K. L., Daniels, M. A., Felthauser, A., Kao, C., Jameson, S. C., and Shimizu, Y. (2004) J. Immunol. 173 2222–2226 [DOI] [PubMed] [Google Scholar]
- 47.Bachmann, M. F., McKall-Faienza, K., Schmits, R., Bouchard, D., Beach, J., Speiser, D. E., Mak, T. W., and Ohashi, P. S. (1997) Immunity 7 549–557 [DOI] [PubMed] [Google Scholar]
- 48.Vorup-Jensen, T., Waldron, T. T., Astrof, N., Shimaoka, M., and Springer, T. A. (2007) Biochim. Biophys. Acta 1774 1148–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogsgaard, M., Huppa, J. B., Purbhoo, M. A., and Davis, M. M. (2003) Semin. Immunol. 15 307–315 [DOI] [PubMed] [Google Scholar]
- 50.Demotz, S., Grey, H. M., and Sette, A. (1990) Science 249 1028–1030 [DOI] [PubMed] [Google Scholar]
- 51.Harding, C. V., and Unanue, E. R. (1990) Nature 346 574–576 [DOI] [PubMed] [Google Scholar]
- 52.Irvine, D. J., Purbhoo, M. A., Krogsgaard, M., and Davis, M. M. (2002) Nature 419 845–949 [DOI] [PubMed] [Google Scholar]
- 53.Matthews, J. B., Ramos, E., and Bluestone, J. A. (2003) Am. J. Transplant. 3 794–803 [DOI] [PubMed] [Google Scholar]
- 54.Purbhoo, M. A., Irvine, D. J., Huppa, J. B., and Davis, M. M. (2004) Nat. Immunol. 5 524–530 [DOI] [PubMed] [Google Scholar]
- 55.Astrof, N. S., Salas, A., Shimaoka, M., Chen, J., and Springer, T. A. (2006) Biochemistry 45 15020–15028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreiber, T. H., Shinder, V., Cain, D. W., Alon, R., and Sackstein, R. (2007) Blood 109 1381–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Y., Jun, C. D., Liu, J. H., Zhang, R., Joachimiak, A., Springer, T. A., and Wang, J. H. (2004) Mol. Cell 14 269–276 [DOI] [PubMed] [Google Scholar]
- 58.Aricescu, A. R., and Jones, E. Y. (2007) Curr. Opin. Cell Biol. 19 543–550 [DOI] [PubMed] [Google Scholar]
- 59.Hogg, N., Henderson, R., Leitinger, B., McDowall, A., Porter, J., and Stanley, P. (2002) Immunol. Rev. 186 164–171 [DOI] [PubMed] [Google Scholar]
- 60.Nicolls, M. R., and Gill, R. G. (2006) Am. J. Transplant. 6 27–36 [DOI] [PubMed] [Google Scholar]
- 61.Scheinfeld, N. (2006) Expert Opin. Drug Saf. 5 197–209 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.