Abstract
We are conducting a genome scan at an average resolution of 10 centimorgans (cM) for type 2 diabetes susceptibility genes in 716 affected sib pairs from 477 Finnish families. To date, our best evidence for linkage is on chromosome 20 with potentially separable peaks located on both the long and short arms. The unweighted multipoint maximum logarithm of odds score (MLS) was 3.08 on 20p (location, x̂ = 19.5 cM) under an additive model, whereas the weighted MLS was 2.06 on 20q (x̂ = 57 cM, recurrence risk, λ̂s = 1.25, P = 0.009). Weighted logarithm of odds scores of 2.00 (x̂ = 69.5 cM, P = 0.010) and 1.92 (x̂ = 18.5 cM, P = 0.013) were also observed. Ordered subset analyses based on sibships with extreme mean values of diabetes-related quantitative traits yielded sets of families who contributed disproportionately to the peaks. Two-hour glucose levels in offspring of diabetic individuals gave a MLS of 2.12 (P = 0.0018) at 9.5 cM. Evidence from this and other studies suggests at least two diabetes-susceptibility genes on chromosome 20. We have also screened the gene for maturity-onset diabetes of the young 1, hepatic nuclear factor 4-a (HNF-4α) in 64 affected sibships with evidence for high chromosomal sharing at its location on chromosome 20q. We found no evidence that sequence changes in this gene accounted for the linkage results we observed.
Type 2 diabetes is a common multifactorial heterogeneous disease with both genetic and environmental determinants and an uncertain mode of inheritance (1). At least three groups have recently completed genome scans for type 2 diabetes and many are nearing completion. Hanis et al. (2) reported genome-wide significance on chromosome 2q37 on a combined data set of 440 Mexican-American affected sib pairs (ASPs). In a sample from Botnia, Western Finland, a small number of selected pedigrees with the lowest quartile for mean 30-min insulin levels after oral glucose tolerance tests showed significant evidence for linkage to type 2 diabetes on chromosome 12q (3). More recently, evidence for linkage was obtained on chromosome 11q for both diabetes and body mass index (BMI) in 264 Pima Indian families (4).
In contrast, maturity-onset diabetes of the young (MODY) is a rare monogenic form of type 2 diabetes that has an autosomal dominant mode of inheritance. At least five different genes, located on chromosomes 20, 7, 12, 13, and 17, independently cause MODY within single pedigrees (5–9). MODY genes may also play a minor role in the common form of type 2 diabetes (10).
Several groups have reported modest evidence for linkage on chromosome 20 for type 2 diabetes (11–13). All of these linkage peaks cover broad regions and appear to include the MODY1 gene (hepatic nuclear factor-4a or HNF-4α). Here, we report our results from chromosome 20 as part of an ongoing genome scan in a large Finnish sample of affected sibships and extended families (14). Together with results from previous studies, our findings support the evidence for more than one diabetes-predisposing gene on chromosome 20. We also show that variants in the HNF-4α gene for MODY1 do not explain the suggestive logarithm of odds (lod) scores detected on chromosome 20q.
MATERIALS AND METHODS
Study Design.
The Finland–U.S. Investigation of Non-Insulin-Dependent Diabetes Mellitus Genetics (FUSION) Study is an international collaborative effort with the goal of mapping and cloning the genes predisposing to type 2 diabetes and intermediate quantitative traits in Finnish subjects (14). Briefly, index cases were ascertained with age of onset 35–60 years and with at least one type 2 diabetic sibling. Fasting glucose, fasting insulin, C-peptide, and glutamic acid decarboxylase antibody were measured in all affected individuals. An oral glucose tolerance test conforming to World Health Organization standards (15) was performed on all unaffected subjects and on untreated or diet-treated diabetic subjects.
In a subset of families, unaffected spouses and offspring of an index case or affected sibling were also invited to undergo a tolbutamide-modified frequently sampled intravenous glucose tolerance test. Minimal model analysis (16, 17) was used to derive estimates for glucose effectiveness (SG), insulin sensitivity (SI), acute insulin response to glucose (AIRG) (18), and the disposition index (DI = SI × AIRG) (17). Lacking direct assessments of these metabolic parameters in our affected subjects, we derived surrogate empirical indices to allow us to analyze these traits. We assessed insulin sensitivity using [1/(fasting glucose × fasting insulin)] (19) and refer to this as SI (estimated). We assessed insulin secretion using two different indices: IRI = fasting insulin/fasting glucose and IRC = fasting C-peptide/fasting glucose. These empirical indices were also computed and analyzed for unaffected subjects.
Before statistical analysis, we excluded 134 family members in which an affected sibling or first-degree relative met criteria for probable type 1 diabetes (14). We used the program relpair (20) to estimate identity by descent (IBD) sharing of marker genotypes to identify likely misspecified relationships. Using this method we identified and excluded four monozygous cotwins, 14 probable half-sibs, and four suspected duplicate samples. In the quantitative trait locus (QTL) analysis, individuals who took medication relevant to the analysis variable of interest were also excluded but on a case-by-case basis.
Marker Typing on Chromosome 20.
Thirty-eight loci were selected from among dinucleotide and tetranucleotide repeat markers located on chromosome 20. Primers were designed from GenBank database sequences. All reverse primers were “G-ended” on the 5′ end to promote nontemplated addition of adenine by Taq DNA polymerase (21). Detailed genotyping methodology appears elsewhere (22) and primer sequences are available on our website (http://genome.nhgri.nih.gov/FUSION).
Statistical Analysis.
(i) ASP analysis. We estimated marker order and intermarker distances from 210 extended FUSION families and the Centre d’Etude des Polymorphismes Humain (CEPH) reference pedigrees using crimap and multimap (23, 24). We used gene counting to estimate allele frequencies for each marker using the complete FUSION family data set and 231 unrelated elderly Finnish normoglycemic controls. These estimates were used in subsequent linkage analyses. As our primary linkage analysis we used a mode-of-inheritance free ASP method introduced by Risch (25–27), as implemented in the computer program siblink (28, 29). We carried out maximum likelihood estimation of the IBD sharing vector for an additive genetic model and under the “possible triangle constraints” (30). Analyses were performed both by (a) treating all ASPs as independent and (b) by correcting for the dependence among ASPs from the same sibship by weighting the contribution of each ASP by 2/s, where s = number of affected sibs in a family (31).
The analyzed dataset consisted of 385 nuclear families with two affected sibs, 83 with three, six with four, and one each with five, six, or seven. From our 477 families we constructed a total of 716 ASPs that, with weighting, were approximately equivalent to 584 (s−1) sib pairs (31). Because the distribution of the MLS is unknown for this problem, we estimated empirical P values using computer simulation of 10,000 replicates of 584 independent ASPs.
In an attempt to reduce the impact of genetic heterogeneity, we explored the ASP-specific lod scores subsetting on diabetes-related quantitative traits that may reflect different underlying susceptibility genes influencing type 2 diabetes in these families. The ASP families were ranked by the mean value of diabetes-related quantitative traits in the affected sibs and the cumulative sum of the lod score grid was evaluated consecutively after each family was added in order. This analysis was performed twice, once starting with the lowest-ranked family to the highest ranked and once from the highest-ranked family to the lowest ranked. The maximum of the cumulative MLS was recorded each time. We performed this ordered subset analysis using fasting glucose, fasting C-peptide, fasting insulin, SI (est.) IRI, IRC, age of diagnosis, BMI, and number of sibs. We generated the empirical distribution to estimate the P values under a permutation test framework (32).
(ii) QTL Linkage Analysis.
We used a mode-of-inheritance free method for QTL analysis using multipoint variance components (33, 34). We modeled affected and unaffected subjects separately because of the large differences in trait variance observed between these groups (14). In addition, we computed analyses on unaffected offspring alone to avoid difficulties because of intergenerational differences in variance. We transformed all data to approximate univariate normality before analysis and modeled mean levels of each quantitative trait value as a linear function of age and gender. Because obesity is a known risk factor for type 2 diabetes (35) and genes for obesity may overlap with those for type 2 diabetes, we tested models with and without BMI as a predictor. We implemented the variance components approach using a version of the pedigree analysis program fisher (36) that incorporates estimation of IBD probabilities based on the multipoint algorithm used in siblink (29). For our strongest multipoint linkage results, we computed empirical P values based on 10,000 simulation replicates each.
Single-Stranded Conformational Polymorphism (SSCP) and Sequence Analysis of HNF-4α.
One individual from each of 64 type 2 diabetic sibships with a high degree of IBD sharing in the approximate location of the HNF-4α gene was chosen from the sample for the initial SSCP screening. The estimated IBD proportion for the selected sibships ranged from 0.64 to 0.99. Sixteen individuals who were 70 years of age with no history of type 2 diabetes and with two recent negative oral glucose tolerance test results were selected as controls. A total of 192 controls with similar characteristics were typed in cases where there was a rare allele or an initial association with 16 controls.
Twenty-four primer sets were designed to screen the entire coding region (12 exons) for sequence variation of the HNF-4α gene as well as 1 kb of the near promoter region and the 3′ untranslated region. The sequences used for primer design were from Yamagata et al. (GenBank accession no. U72960), and several of the coding region primer sets were taken from published material (5). The primer sequences are available on our website. The amplification conditions and the direct DNA sequencing protocol have been described previously (37).
RESULTS
In our genome scan for type 2 diabetes (now over 90% complete), only two regions, to date, had weighted multipoint lod scores of 2 or higher. Both were located on chromosome 20. Results from the rest of the genome scan will be reported elsewhere on completion.
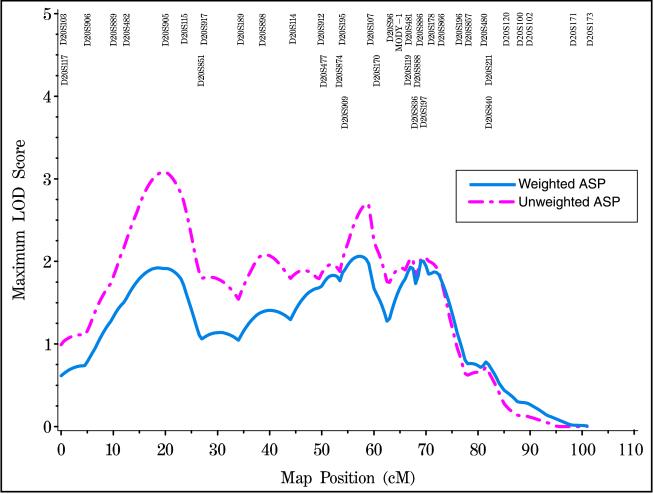
We estimated the chromosome 20 genetic map length to be 100.9 centimorgans (cM). Our map includes 38 markers at an average density of 2.6 cM. Single locus MLSs and heterozygosities are presented for each marker in Table 1. Using multipoint analysis under an additive model, we observed weighted multipoint lod score peaks of 1.92 (P = 0.013) at 18.5 cM, 2.06 (P = 0.009) at 57 cM, and 2.00 (P = 0.01) at 69.5 cM. The corresponding unweighted multipoint MLSs were 3.08 at 19.5 cM, 2.74 at 58.5 cM, and 2.05 at 69.5 cM (Fig. 1).
Table 1.
Unweighted and weighted single-point MLS for 716 affected sib pairs
| Locus | Market het | Map, cM | Unweighted MLS | Weighted MLS |
|---|---|---|---|---|
| D20S103 | 0.73 | 0.0 | 0.057 | 0.064 |
| D20S117 | 0.87 | 0.2 | 1.186 | 0.430 |
| D20S906 | 0.79 | 4.6 | 0.525 | 0.344 |
| D20S889 | 0.84 | 9.6 | 1.324 | 1.112 |
| D20S482 | 0.69 | 12.1 | 0.780 | 0.323 |
| D20S905 | 0.64 | 19.6 | 1.836 | 1.046 |
| D20S115 | 0.65 | 23.2 | 0.541 | 0.419 |
| D20S851 | 0.75 | 26.4 | 0.532 | 0.236 |
| D20S917 | 0.88 | 27.0 | 0.847 | 0.620 |
| D20S189 | 0.68 | 34.0 | 0.055 | 0.051 |
| D20S898 | 0.75 | 38.2 | 1.051 | 0.652 |
| D20S114 | 0.82 | 44.0 | 0.844 | 0.510 |
| D20S912 | 0.83 | 49.4 | 1.187 | 1.414 |
| D20S477 | 0.73 | 49.9 | 0.799 | 0.372 |
| D20S874 | 0.81 | 52.9 | 0.506 | 0.605 |
| D20S195 | 0.84 | 53.5 | 0.527 | 0.556 |
| D20S909 | 0.70 | 54.0 | 0.317 | 0.452 |
| D20S107 | 0.83 | 58.9 | 3.968 | 2.851 |
| D20S170 | 0.81 | 60.1 | 1.343 | 1.166 |
| D20S96 | 0.82 | 62.7 | 1.278 | 0.478 |
| D20S119 | 0.80 | 66.1 | 0.104 | 0.395 |
| D20S481 | 0.83 | 66.2 | 0.928 | 1.106 |
| D20S836 | 0.82 | 67.4 | 2.811 | 2.568 |
| D20S888 | 0.88 | 67.9 | 1.156 | 0.916 |
| D20S886 | 0.84 | 68.4 | 0.439 | 0.652 |
| D20S197 | 0.76 | 69.1 | 2.117 | 1.787 |
| D20S178 | 0.80 | 70.6 | 1.528 | 1.116 |
| D20S866 | 0.85 | 72.5 | 2.900 | 2.264 |
| D20S196 | 0.79 | 75.9 | 0.511 | 0.877 |
| D20S857 | 0.84 | 77.7 | 0.170 | 0.382 |
| D20S480 | 0.77 | 80.7 | 0.996 | 0.760 |
| D20S211 | 0.64 | 81.6 | 2.022 | 1.613 |
| D20S840 | 0.82 | 81.6 | 1.622 | 1.430 |
| D20S120 | 0.85 | 84.9 | 0.119 | 0.113 |
| D20S100 | 0.74 | 87.6 | 0.000 | 0.000 |
| D20S102 | 0.47 | 89.5 | 0.165 | 0.347 |
| D20S171 | 0.81 | 97.8 | 0.000 | 0.000 |
| D20S173 | 0.73 | 100.9 | 0.000 | 0.000 |
het = heterozygosity
Figure 1.
ASP linkage analysis of 716 ASPs from 477 Finnish families. Unweighted (magenta) and weighted (blue) analyses are shown.
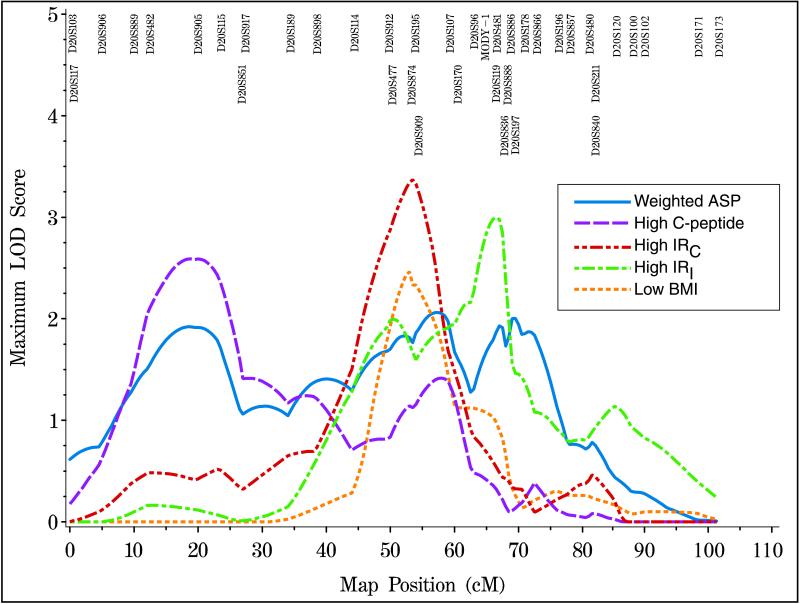
We also performed ordered subset analysis on nine different quantitative traits. We describe results only in cases where small numbers of families contributed disproportionately to the high lod scores generated from the whole dataset. The 15 sibships (3% of the total number of families) with the lowest mean BMI (19.81–23.62 kg/m2) produced a weighted multipoint MLS of 2.56 at 53 cM. Similarly, the 19 sibships (4%) with the highest IRC (3.5–6.3 × 10−7) gave a weighted multipoint MLS of 3.46 at 53.5 cM (Fig. 2). Thus, these different subsets of the data give MLSs in the neighborhood of our second peak at 57 cM when using all families. The 70 sibships (15%) with the highest IRI (17.13–42.90 × 10−9) yielded a lod score of 3.06 at 66 cM, close to our third peak at 69.5 cM for the overall analysis (Fig. 2). Finally, 89 sibships (19%) with the highest mean fasting C-peptide (2.24–4.46 nmol/l) gave a lod score of 2.93 at 21 cM, in close proximity to our first peak using the whole dataset (Fig. 2). In no cases were the empirical P values for the ordered subset analyses significant at the 0.05 level (data not shown), but we report the analyses because of their potential biological interest.
Figure 2.
Ordered subset analyses of families who give weighted MLSs near the three linkage peaks using the whole dataset. The analyses show (i) 89 families with the highest mean fasting C-peptide (purple), (ii) 19 families with the highest mean IRC (red), (iii) 70 families with the highest mean IRI (green) and (iv) 15 families with the lowest mean BMI (orange). The solid line (blue) depicts the weighted analyses using 716 ASPs.
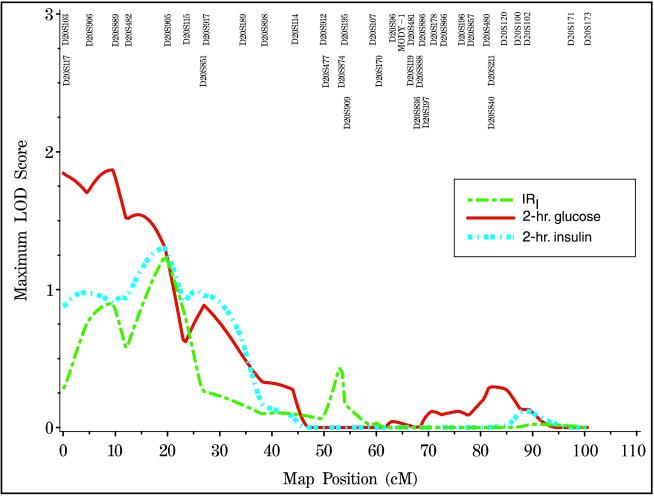
We performed variance components analyses of quantitative traits on a total of 194 unaffected spouse and 520 unaffected offspring of diabetic subjects from 210 extended families, after adjusting for age and gender. A multipoint MLS of 1.06 at 19 cM was observed for 2-hr insulin and this increased to 1.30 at 19.5 cM in the offspring-only analysis (Fig. 3). A MLS of 1.12 was generated at 19.5 cM for IRI by using all unaffecteds, and the MLS increased to 1.24 at the same location in the offspring-only analysis. Finally, a MLS of 1.59 was observed at 9.5 cM for 2-hr glucose while a MLS of 1.87 was observed at the same location in the offspring-only analysis. When an additional adjustment was made for BMI, we observed a MLS of 1.78 (empirical P value = 0.0008) at 9.5 cM for 2-hr glucose. When only offspring were analyzed, the MLS for 2-hr glucose increased to 2.12 (empirical P value = 0.0018). The other quantitative measures did not yield higher lod scores after the additional adjustment with BMI. No other quantitative measures in affecteds or unaffecteds (including results from the frequently sampled intravenous glucose tolerance test) gave multipoint lod scores over 1 in these analyses.
Figure 3.
Variance components QTL linkage analysis, adjusted for age and gender, in unaffected offspring. IRI (green), 2-hr glucose (red) and 2-hr insulin (light blue) are shown.
We investigated the possibility that the lod score peak at 69.5 cM could arise from variants in the MODY1 (HNF-4α) gene, which was mapped by us to the D20S96-D20S119 (62.7–66.1 cM) interval using radiation hybrid mapping (data not shown). Analysis of 12 HNF-4α exons and the promoter region in at least 16 controls and 64 individuals from affected sibships, who showed high sharing at MODY1, revealed 14 base substitutions (three altering amino acids) and three deletions (Table 2). Twelve of these changes have not been described previously.
Table 2.
DNA variants and polymorphisms in HNF-4α gene in the FUSION study
| Location | Nucleotide* | Change | Amino acid change‡ | Diabetic chromosome | Control chromosome |
|---|---|---|---|---|---|
| Promoter | −926 | del3† | 8/126 | 14/288 | |
| −920 | del3† | 3/126 | 2/288 | ||
| −943 | C→T† | 1/126 | 0/288 | ||
| −625 | A→C† | 58/126 | 13/32 | ||
| Exon 1a | 38 | del7† | 5′ UTR | 2/128 | 9/300 |
| 125 | G→A† | 5′ UTR or V8I§ | 1/128 | 0/300 | |
| Exon 1c | 57 | G→A | V49M | 12/116 | 3/28 |
| Intron 1b | +64 | C→A† | 0/124 | 1/32 | |
| −38 | T→C‡ | 41/120 | 143/310 | ||
| −5 | C→T | 20/120 | 65/310 | ||
| −4 | G→A† | 1/120 | 0/310 | ||
| Exon 2 | 86 | C→T | A58A¶ (A109A‖) | 8/120 | 20/310 |
| Intron 2 | −8 | C→G† | 1/128 | 0/288 | |
| Exon 4 | 31 | C→T | T130I¶ (T181I)‖) | 5/116 | 2/32 |
| Intron 4 | +6 | G→A† | 1/116 | 0/372 | |
| 3′ Untranslated region | +299** | G→A† | 7/126 | 1/32 | |
| +536** | G→T† | 9/126 | 2/30 |
Genomic sequence based on Yamagata et al. (5). Relative position in introns is with respect to splice donor (+) or acceptor (−) site. Promoter region numbering is with respect to transcription start site. Exon numbering is with respect to cDNA sequence.
New variants not previously reported by others.
P = 0.03
Upstream of an alternate translation start site. Affects coding region only in the Drews et al. submission (41).
Is with respect to a cDNA sequence excluding exon 1b and 1c.
Is with respect to a cDNA sequence including exon 1b and 1c.
Is relative to stop codon in mRNA.
Five substitutions were unique to single diabetic families and were not found in at least 144 controls that were successfully typed (Table 2). The specific rare variant was associated with affection status in each of these families, whereas the wild-type allele was always present in unaffected spouses. Of these rare variants found in single diabetic families, three occurred near splice junctions, but not in the conserved donor or acceptor sites. The intron 1b C(+64)A substitution was also relatively rare, found in one control but in no diabetic individuals.
Six new and more common changes were identified in both affecteds and control individuals (Table 2). None of the common variants were significantly associated with diabetes. The intron 1b substitution, T(-38)C, was significantly less frequent in diabetic individuals compared with controls (P = 0.03; Table 2). Two nucleotide substitutions that resulted in amino acid changes were found in both affecteds and in controls (V49M and T130I). These two variants have been reported previously (5, 38, 39). The third nucleotide substitution, which was unique to one diabetic family, occurred in exon 1a (40) and could lead to a valine-to-isoleucine change (V8I). However, since there is still controversy as to the possible start sites in HNF-4α gene, it is unclear whether this change is at the 5′ untranslated region or in the coding region (40, 41).
When the five families with unique variants in HNF-4α were removed, the MLS on chromosome 20q dropped to 1.74 at 69.5 cM, under the weighted additive model.
DISCUSSION
We analyzed a large group of Finnish type 2 diabetic families and found evidence for linkage to chromosome 20. Three linkage peaks were seen after analyses of diabetes and diabetes-related traits. These linkages were at approximately 0–25 cM, 50–60 cM, and 63–72 cM respectively from the marker D20S103. Although the second and third peaks could be explained by a single susceptibility locus, evidence for linkage on both arms on chromosome 20 argues for the presence of more than one susceptibility locus. As far as we know, we are the first group to show evidence for linkage to the proximal p arm of chromosome 20 in type 2 diabetes. Most of our evidence comes from families with affected sibships greater than two. Ordered subset analyses of our data revealed that a small number of families, with high or low values of important diabetes-related traits, give rise to large lod scores near the three peaks. These analyses provide additional evidence for more than one susceptibility locus on this chromosome. Caution is required in interpreting these results, however, since none of the subset analyses resulted in P values approaching traditional levels of statistical significance.
At least three other groups have reported similar results on 20q (11–13). Ji et al. (11) examined 29 extended Caucasian families comprising 498 individuals, 159 of whom had type 2 diabetes. Although there was weak evidence for linkage in the total data set, sibs with an age of diagnosis over 47 years (14 families with 54 sib pairs) yielded a multipoint nonparametric linkage score of 3.30 (equivalent to a lod score of 2.36; P = 0.009), very near D20S197 (69.1 cM on our map). Bowden et al. (12) analyzed 53 ASPs from 21 Caucasian families, each with at least one member with diabetic nephropathy. They found a MLS of 1.48 in the D20S197/D20S178 interval (69.1–70.6 cM on our map). Zouali and colleagues (13) studied 148 type 2 diabetes pedigrees comprising 301 ASPs. Their total sample gave a weighted MLS of 1.81 (P = 0.003) near the RPN II locus (54–58.9 cM on our map). In 42 early-onset families (55 sib pairs with age of diagnosis <45 years), the weighted MLS increased to 2.34 (P = 0.0009) but maximized near the phosphoenolpyruvate carboxykinase (PCK) locus (87.6–100.9 cM on our map). Finally, in a nondiabetic obese sample of 258 sib pairs from 152 pedigrees, there was some evidence for linkage over a broad area on most of the q arm for percent body fat (P < 0.004), BMI (P < 0.008), and fasting insulin (P < 0.0005) (42). Two of the above studies found evidence for a locus very near our third lod score peak (11, 12), whereas results from Zouali et al. (13) may suggest the existence of two loci, one near our second peak and one close to the PCK locus where we observe no evidence for linkage. In contrast to two of the above studies (11, 13), we also find no evidence for linkage when we performed ordered subset analysis based on age of diagnosis.
HNF-4α, the gene for MODY1, is a member of the nuclear receptor superfamily, a family of transcription factors that play an important role in cellular regulation linking extracellular signals and transcription responses. Five newly identified rare variants were present in one diabetic family each and were not found in at least 70 controls. Three of these were near splice junctions, but it is not expected that these variants will disrupt the splicing machinery because of the distance from the conserved splice junction sequences. Unfortunately, reverse transcription–PCR on RNA from peripheral blood has, thus far, failed to amplify reliably HNF-4α mRNA, so it has not been possible to determine whether these variants have an effect on normal splicing. We conclude, from the effect of removing these families, that rare diabetes-specific variants in the HNF-4α gene cannot explain the high lod scores in 20q in our data. The observed drop in lod score, when the five families are removed, is consistent with the selection of families for scanning on the basis of high IBD sharing in this region. Furthermore, these families were typical of the 64 families in the evidence they provided for linkage. One variant in intron 1b, T(-38)C, was significantly more common in controls compared with affecteds in our dataset but Malecki et al. (39) found the same variant to be less common in controls, although their result was not significant (39).
We are aware of two other relatively important candidate genes on chromosome 20, agouti signaling protein (ASP, 53.5–58.9 cM on our map) and HNF-3β (approximate location, 50 cM). Mutations of the murine agouti gene are associated with obesity-related type 2 diabetes through possible changes in calcium flux in skeletal muscle (43); HNF-3β has been found to regulate positively the expression of the MODY genes, HNF-4α and HNF-1α, and their downstream targets (44).
In summary, we have found evidence for linkage for diabetes and diabetes-related traits on chromosome 20p and 20q, confirming and strengthening earlier reports. Ordered subset analysis has given corroborative evidence for more than one susceptibility gene for type 2 diabetes and related phenotypes. We have also found five rare variants in the HNF-4α gene, but these are unlikely to account for the linkage results on chromosome 20q. Further analysis of a large replication sample and extended pedigree analysis of families studied in this sample may allow confirmation of linkage. Concurrent linkage disequilibrium analysis, which will include geographical stratification based on birthplaces, may also enhance mapping resolution for eventual positional cloning.
Acknowledgments
The FUSION project is made possible by intramural funds from the National Human Genome Research Institute (NHGRI) (project number OH95-C-N030) and by R01 HG00376. R.M.W. was supported previously by training grant T32 HG00040 and an individual postdoctoral award F32 DK09525 and is currently supported by a Career Development Award from the American Diabetes Association. C.D.L. is supported by training grant T32 HG0040. We thank Edna Ross, Lisa Taylor, Elza Demirchyan, Peggy White, Ed Trager, and the clinical and nursing staff from Finland for their strenuous efforts. We also thank the Finnish families for their cooperation in this study.
ABBREVIATIONS
- cM
centimorgan
- ASP
affected sib pair
- lod
logarithm of odds
- MLS
maximum lod score
- MODY
maturity-onset diabetes of the young
- BMI
body mass index
- IBD
identity by descent
- QTL
quantitative trait locus
References
- 1.Bennett P H, Bogardus C, Tuomilehto J, Zimmett P. In: Epidemiology and Natural History of NIDDM: Nonobese and Obese. Alberti K G M M, DeFronzo R A, Keen H, Zimmett P, editors. New York: Wiley; 1992. pp. 147–176. [Google Scholar]
- 2.Hanis C, Boerwinkle E, Chakraborty R, Ellsworth D, Concannon P, Stirling B, Morrison V A, Wapelhorst B, Spielman R, Gogolin-Ewens K J, et al. Nat Genet. 1996;13:161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- 3.Mahtani M M, Widen E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, et al. Nat Genet. 1996;14:90–94. doi: 10.1038/ng0996-90. [DOI] [PubMed] [Google Scholar]
- 4.Hanson R L, Ehm M G, Pettitt D J, Prochazka M, Thompson D B, Timberlake D, Faroud T, Kobes S, Baier L, Burns D K, et al. Am J Hum Genet. 1998;63:1130–1138. doi: 10.1086/302061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamagata K, Furuta H, Oda N, Kaisaski P, Menzel S, Cox N J, Fajans S S, Signorini S, Stoffel M, Bell G I. Nature (London) 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 6.Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell G I, Zouali H, Lesage S, Velho G, Iris F, Passa P, et al. Nature (London) 1992;356:721–722. doi: 10.1038/356721a0. [DOI] [PubMed] [Google Scholar]
- 7.Yamagata K, Oda N, Kaisaki P J, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox R D, Lathrop G M, Boriraj V V, et al. Nature (London) 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 8.Stoffers D A, Ferrer J, Clarke W L, Habener J F. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 9.Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn B N, Lindner T, Yamagata K, Ogata M, Tomonaga O, et al. Nat Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 10.Shuldiner A R, Silver K D. In: Candidate Genes for Non-Insulin-Dependent Diabetes Mellitus. LeRoith D, Taylor S, Olefsky J M, editors. Philadelphia: Lippincott; 1996. pp. 565–574. [Google Scholar]
- 11.Ji L, Malecki M, Warram J H, Yang Y, Rich S S, Krolewski A S. Diabetes. 1997;46:876–881. doi: 10.2337/diab.46.5.876. [DOI] [PubMed] [Google Scholar]
- 12.Bowden D W, Sale M, Howard T D, Qadri A, Spray B J, Rothschild C B, Akots G, Rich S S, Freedman B I. Diabetes. 1997;46:882–886. doi: 10.2337/diab.46.5.882. [DOI] [PubMed] [Google Scholar]
- 13.Zouali H, Hani E H, Phillipi A, Vionnet N, Beckmann J S, Demenais F, Froguel P. Hum Mol Genet. 1997;6:1401–1408. doi: 10.1093/hmg/6.9.1401. [DOI] [PubMed] [Google Scholar]
- 14.Valle T, Tuomilehto J, Bergman R N, Ghosh S, Hauser E R, Eriksson J, Nylund S J, Kohtamaki K, Toivanen L, Vidgren G, et al. Diabetes Care. 1998;21:949–958. doi: 10.2337/diacare.21.6.949. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization Study Group on Diabetes Mellitus. World Health Organization: WHO Expert Committee on Diabetes Mellitus. Vol. 646. Geneva: WHO; 1980. [PubMed] [Google Scholar]
- 16.Bergman R N, Phillips L S, Cobelli C. J Clin Invest. 1981;1981:1456–1467. doi: 10.1172/JCI110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergman R N, Prager R, Volund A, Olefsky J M. J Clin Invest. 1987;79:790–800. doi: 10.1172/JCI112886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward W K, Bolgiano D C, McKnight B, Halter J B, Porte D., Jr J Clin Invest. 1984;74:1318–1328. doi: 10.1172/JCI111542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sluiter W, Erkelens D, Terpstra P, Weitsma W, Doorenbos H. Diabetes. 1976;25:245–249. doi: 10.2337/diab.25.4.245. [DOI] [PubMed] [Google Scholar]
- 20.Boehnke M, Cox N. Am J Hum Genet. 1997;61:423–429. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnuson V, Ally D, Nylund S, Karanjawala Z, Rayman J, Knapp J, Lowe A, Ghosh S, Collins F. BioTechniques. 1996;21:2–10. doi: 10.2144/96214rr03. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh S, Karanjawala Z, Hauser E R, Ally D, Knapp J I, Rayman J B, Musick A, Tannenbaum J, Te C, Shapiro S, et al. Genome Res. 1997;7:165–178. doi: 10.1101/gr.7.2.165. [DOI] [PubMed] [Google Scholar]
- 23.Lander E S, Green P. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matise T C, Perlin M, Chakravarti A. Nat Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- 25.Risch N. Am J HumGenet. 1990;46:222–228. [Google Scholar]
- 26.Risch N. Am J Hum Genet. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- 27.Risch N. Am J Hum Genet. 1990;46:242–253. [PMC free article] [PubMed] [Google Scholar]
- 28.Hauser E R, Boehnke M, Guo S-W, Risch N. Genet Epidemiol. 1996;13:117–137. doi: 10.1002/(SICI)1098-2272(1996)13:2<117::AID-GEPI1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Hauser E R, Boehnke M. Biometrics. 1998;54:1238–1246. [PubMed] [Google Scholar]
- 30.Holmans P. Am J Hum Genet. 1993;52:362–374. [PMC free article] [PubMed] [Google Scholar]
- 31.Suarez B K, Hodge S E. Clin Genet. 1979;15:126–136. doi: 10.1111/j.1399-0004.1979.tb01751.x. [DOI] [PubMed] [Google Scholar]
- 32.Hauser E R, Watanabe R M, Duren W L, Boehnke M. Am J Hum Genet Suppl. 1998;63:A45. [Google Scholar]
- 33.Amos C I. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- 34.Lange K, Westlake J, Spence M A. Ann Hum Genet. 1976;39:485–491. doi: 10.1111/j.1469-1809.1976.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 35.Knowler W C, Pettitt D J, Saad M F, Bennett P H. Diabetes Metab Rev. 1990;6:1–27. doi: 10.1002/dmr.5610060101. [DOI] [PubMed] [Google Scholar]
- 36.Lange K, Weeks D, Boehnke M. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 37.Castilla L H, Couch F J, Erdos M R, Hoskins K F, Calzone K, Garber J E, Boyd J, Lubin M B, Deshano M L, Brody L C, et al. Nat Genet. 1994;8:387–391. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- 38.Moller A M, Urhammer S A, Dalgaard L T, Reneland R, Berglund L, Hansen T, Clausen J O, Lithell H, Pedersen O. Diabetologia. 1997;40:980–983. doi: 10.1007/s001250050778. [DOI] [PubMed] [Google Scholar]
- 39.Malecki M T, Antonellis A, Casey P, Ji L, Wantman M, Warram J H, Krolewski A S. Diabetes. 1998;47:970–972. doi: 10.2337/diabetes.47.6.970. [DOI] [PubMed] [Google Scholar]
- 40.Furuta H, Iwasaki N, Oda N, Hinokio Y, Horikawa Y, Yamagata K, Yano N, Sugahiro J, Ogata M, Ohgawara H, et al. Diabetes. 1997;46:1652–1657. doi: 10.2337/diacare.46.10.1652. [DOI] [PubMed] [Google Scholar]
- 41.Drewes T, Senkel S, Holewa B, Ryffel G U. Mol Cell Biol. 1996;16:925–931. doi: 10.1128/mcb.16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lembertas A V, Perusse L, Chagnon Y C, Fisier J S, Warden C H, Purcell-Huynh D A, Dionne F T, Gagnon J, Nadeau A, Lusis A J, et al. J Clin Invest. 1997;100:1240–1247. doi: 10.1172/JCI119637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zemel M B, Kim J H, Woychik R P, Michaud E J, Kadwell S H, Patel I R, Wilkison W O. Proc Natl Acad Sci USA. 1995;92:4733–4737. doi: 10.1073/pnas.92.11.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan S A, Angeles Navas M, Dufort D, Rossant J, Stoffel M. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]