Abstract
Mycobacterium tuberculosis possesses an unusual cell wall that is replete with virulence-enhancing lipids. One cell wall molecule unique to pathogenic M. tuberculosis is polyacyltrehalose (PAT), a pentaacylated, trehalose-based glycolipid. Little is known about the biosynthesis of PAT, although its biosynthetic gene cluster has been identified and found to resemble that of the better studied M. tuberculosis cell wall component sulfolipid-1. In this study, we sought to elucidate the function of papA3, a gene from the PAT locus encoding a putative acyltransferase. To determine whether PapA3 participates in PAT assembly, we expressed the protein heterologously and evaluated its acyltransferase activity in vitro. The purified enzyme catalyzed the sequential esterification of trehalose with two palmitoyl groups, generating a diacylated product similar to the 2,3-diacyltrehalose glycolipids of M. tuberculosis. Notably, PapA3 was selective for trehalose; no activity was observed with other structurally related disaccharides. Disruption of the papA3 gene from M. tuberculosis resulted in the loss of PAT from bacterial lipid extracts. Complementation of the mutant strain restored PAT production, demonstrating that PapA3 is essential for the biosynthesis of this glycolipid in vivo. Furthermore, we determined that the PAT biosynthetic machinery has no cross-talk with that for sulfolipid-1 despite their related structures.
Mycobacterium tuberculosis, the bacterium that causes tuberculosis in humans, has a complex cell wall that contains a number of unique glycolipids intimately linked to mycobacterial pathogenesis (1, 2). The biosynthesis of many of these virulence factors, including the trehalose mycolates, phenolic glycolipids, and sulfolipid-1 (SL-1),3 is largely understood (3–5). In contrast, relatively little is known about the biosynthesis of other prominent M. tuberculosis glycolipids, such as di-, tri-, and polyacyltrehaloses. These acyltrehaloses are located in the outer surface of the cell wall and contain di- and tri-methyl branched fatty acids that are only found in pathogenic species of mycobacteria (6, 7). Previous studies suggest a role for these glycolipids in anchoring the bacterial capsule, which impedes phagocytosis by host cells (6).
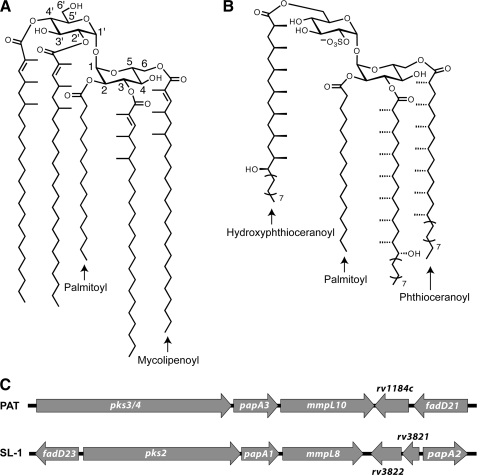
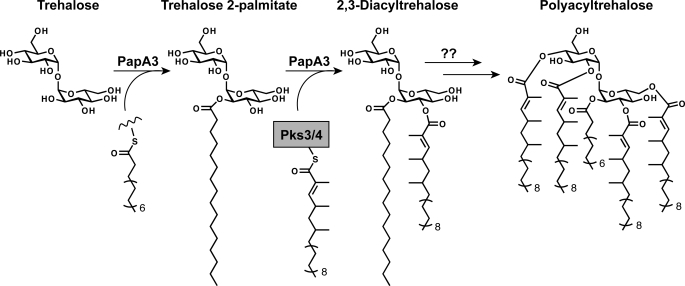
The major polyacyltrehalose (PAT) of M. tuberculosis, also referred to as pentaacyl or polyphthienoyl trehalose, consists of five acyl chains, four mycolipenic (phthienoic) acids and one fully saturated fatty acid, linked to trehalose (Fig. 1A) (8). The mycolipenic acid side chains of PAT are products of the polyketide synthase gene pks3/4 (7). Disruption of pks3/4 (also referred to as msl3 (7)) abolishes PAT biosynthesis and causes cell aggregation. At present, the remaining proteins required for PAT assembly have not been characterized.
FIGURE 1.
PAT and SL-1 share related structures and biosynthetic gene clusters. A, structure of PAT. B, structure of SL-1. C, genomic arrangement of the PAT and SL-1 biosynthetic gene clusters.
Interestingly, the PAT biosynthetic gene cluster strongly resembles that of SL-1, which is a structurally similar trehalose-based glycolipid unique to pathogenic mycobacteria (Fig. 1B) (9). Both gene clusters contain polyketide synthase (pks), acyltransferase (pap), and lipid transport (mmpL) genes in a similar genomic arrangement (Fig. 1C). The SL-1 locus encodes two acyltransferase genes, papA1 and papA2, which are required for SL-1 biosynthesis (5, 10). These proteins belong to the mycobacterium-specific polyketide-associated protein (Pap) family of acyltransferases, which share a conserved HX3DX14Y motif that is required for activity (11). The PapA2 enzyme catalyzes the esterification of the 2′-position of trehalose 2-sulfate with a saturated fatty acid. PapA1 mediates the subsequent esterification of this intermediate with a hydroxyphthioceranoyl group produced by Pks2 (5). Interestingly, the PAT locus contains a gene, Rv1182, that is homologous to both papA1 and papA2 (55 and 53% amino acid identity, respectively). This gene is annotated as papA3 in the genome and was previously shown to encode a protein bearing the signature Pap motif (11).
Here we demonstrate that papA3 encodes an acyltransferase essential for the biosynthesis of PAT. Deletion of the papA3 gene resulted in loss of the glycolipid from M. tuberculosis lipid extracts, as determined by high resolution mass spectrometry. Moreover, the purified enzyme was shown to selectively and sequentially acylate trehalose in vitro, generating a diacylated product similar to the 2,3-diacyltrehaloses of M. tuberculosis. Together, these data confirm that PapA3 plays a crucial role in PAT biosynthesis and highlight its potential involvement in the biosynthesis of related M. tuberculosis acyltrehaloses.
EXPERIMENTAL PROCEDURES
Reagents and Chemicals—Pfu DNA polymerase was from Stratagene (La Jolla, CA). Oligonucleotides were from Elim Biopharmaceuticals, Inc. (Hayward, CA). Restriction enzymes were from New England Biolabs (Ipswich, MA). Qiagen (Valencia, CA) kits were used for plasmid DNA purification and the extraction of DNA from agarose gels. T4 DNA ligase and BL21(DE3) chemically competent cells were purchased from Invitrogen. DNA sequencing was performed by Elim Biopharmaceuticals, Inc. 14C-Palmitoyl coenzyme A (14C-PCoA), 14C-palmitic acid, 14C-butyryl coenzyme A, 14C-crotonoyl coenzyme A, and 14C-docosanoyl coenzyme A were purchased from ARC Radiolabeled Chemicals (St. Louis, MO; 50–55 mCi/mmol). Unlabeled palmitoyl coenzyme A (PCoA) was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Trehalose 2-palmitate (T2P) and trehalose 2-sulfate were synthesized as described previously (12–14). The synthesis and structural characterization of trehalose 3-palmitate (T3P) are reported in the supplemental material. Glass-backed silica gel 60 HPTLC plates were purchased from EMD Chemicals (Gibbstown, NJ). All other chemicals were purchased from Sigma or Fluka (St. Louis, MO) and used without further purification.
Electrospray Ionization Fourier Transform Ion Cyclotron Mass Spectrometry (ESI-FT-ICR MS)—Mass spectra were obtained on an Apex II FT-ICR mass spectrometer equipped with a 7-tesla actively shielded superconducting magnet (Bruker Daltonics, Billerica, MA). Ions were introduced into the ion source via direct injection at a rate of 1 μl/min. Ions were generated with an Apollo pneumatically assisted electrospray ionization source (Analytica, Branford, CT) operating in the negative ion mode and were accumulated in an RF-only external hexapole for 0.5–2 s before being transferred to the ion cyclotron resonance cell for mass analysis. Mass spectra consist of 512,000 data points and are an average of between 28 and 128 scans. The spectra were acquired using XMASS version 6.0.0 or 7.0.8 (Bruker Daltonics). All spectra were internally calibrated with at least four known compounds.
ESI Linear Ion Trap MS—Additional mass spectra were obtained on an LTQ ion trap mass spectrometer equipped with an ESI source (ThermoFinnigan) operating in either the negative or positive ion mode. Ions were introduced into the ion source via direct injection at a rate of 5 μl/min. For linear ion trap tandem mass spectrometry (MSn) experiments, the precursor ions were isolated with an isolation width of 1–3 Da; the ions were activated with a 13–20% normalized collision energy for 100 ms, and the qz value was maintained at 0.250. Spectra are an average of 100 scans, acquired using Xcalibur, version 1.4 (ThermoFinnigan).
Preparation of Protein Expression Vector—The papA3 gene (Rv1182, encoding residues 2–472) was amplified from M. tuberculosis H37Rv genomic DNA with the primers 5′-TACTGTAGTCGAATTCTTGCGGGTTGGACCGTTGAC-3′ (EcoRI) and 5′-GATTACAGGTCTGCAGTCAGGCAACATTCTGCTGCT-3′ (PstI). Rv1182 was ligated into a modified pMAL-C2X vector (New England Biolabs) encoding an N-terminal His7 tag and TEV cleavage site. The TEV cleavage site was introduced by site-directed mutagenesis using the QuikChange PCR mutagenesis kit (Stratagene), and the His7 tag was annealed to the 5′ end of the maltose-binding protein (MBP)-coding region. DNA sequencing was performed to confirm the successful construction of the protein-encoding plasmid.
Protein Expression and Purification—E. coli BL21(DE3) cells were transformed with the protein expression plasmid. Transformants were used to inoculate 1-liter cultures of LB medium containing 100 mg/liter ampicillin and 2 g/liter glucose. The cultures were incubated at 37 °C for ∼2.5 h with shaking until an A600 of 0.6–0.8 was attained. Protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 100 μm. After 18–20 h at 18 °C, cells from each liter of culture were harvested and suspended in 30 ml of lysis buffer (20 mm Tris, pH 7.4, 200 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm TCEP) supplemented with 5 μg/ml lysozyme and 5 μg/ml DNase. Cells were lysed using a high pressure homogenizer (Avestin EmulsiFlex-C5). The lysate was cleared by centrifugation and applied to amylose resin (New England Biolabs) equilibrated with lysis buffer. After washing the resin with additional lysis buffer, MBP-PapA3 was eluted in lysis buffer containing 10 mm maltose. The eluted protein was diluted ∼1:15 in low salt buffer (50 mm Tris, pH 7.4, 1 mm DTT, 1 mm TCEP, 10% glycerol) and loaded onto a MonoQ HR 5/5 column (GE Healthcare) equilibrated with the same buffer. The protein was purified using a gradient of 1.5–100% high salt buffer (50 mm Tris, pH 7.4, 1 m NaCl, 1 mm DTT, 1 mm TCEP, 10% glycerol) over 40 min at a constant temperature of 4 °C. Fractions (0.5 ml each) were analyzed by SDS-PAGE, and those containing pure protein were pooled and analyzed by the Bradford protein assay to determine protein concentration. MBP was cleaved from PapA3 using AcTEV protease (Invitrogen) and removed using nickel-nitrilotriacetic acid resin (Qiagen), which bound the His-tagged MBP and protease while leaving the desired protein in solution. PapA3 was subsequently concentrated and stored at –80 °C.
To confirm the identity of the protein, the purified sample was desalted on a microbore reversed-phase column (Bruker Agilent) and characterized using a Bruker Hewlett-Packard ESI-ion trap mass spectrometer. Tryptic digestion and mass fingerprinting using an LTQ mass spectrometer provided further confirmation of the identity of the protein. Protein concentration was determined by UV absorption at 280 nm using a calculated extinction coefficient of 57,340 m–1 cm–1.
Biochemical Characterization of PapA3—A previously described TLC-based assay (5) was used to characterize the acyltransferase activity of PapA3. Briefly, PapA3 (2 μm) was incubated with 20 μm 14C-PCoA and 1 mm of the desired sugar substrate in reaction buffer (100 mm ammonium bicarbonate, pH 7.2) for 2 h at room temperature. Alternative 14C-labeled acyl donors were screened at a concentration of 20 μm. The reactions were quenched by the addition of an equal volume of ethanol and subsequently analyzed by TLC (35:65 methanol: chloroform) and phosphorimaging. Stocks of each sugar substrate were prepared in water with the exception of T2P and T3P, which were dissolved in dimethyl sulfoxide (DMSO). The amount of DMSO in any given reaction did not exceed 5% of the reaction volume.
MS Characterization of Reaction Products—PapA3 (1 μm) was incubated with 20 μm unlabeled PCoA and 1 mm trehalose or T2P in reaction buffer for 6 h at room temperature. Control reactions without enzyme were also prepared. Samples were lyophilized and stored at –20 °C prior to analysis by ESI-FT-ICR and linear ion trap MS.
Construction of M. tuberculosis Mutants—M. tuberculosis cells (Erdman strain) were cultured in 7H9 medium supplemented with 10% OADC, 0.5% glycerol, and 0.05% Tween 80 or on 7H10 solid agar medium supplemented with 10% OADC and 0.5% glycerol. Hygromycin (50 μg/ml) or kanamycin (25 μg/ml) was used when necessary. The ΔpapA3 mutant strain was created by homologous recombination as described previously (15). Briefly, specialized transduction phage phMWS120 was incubated with concentrated wild-type Erdman M. tuberculosis cells for 4 h at 39 °C. Cells were then plated on 7H10 plates containing hygromycin. Colonies were picked and screened for the deletion by PCR. The resulting deletion replaced 1136 bp of papA3 (amino acids 45–423) with a hygromycin resistance cassette. The ΔpapA3::papA3 complementation strain was created by cloning the papA3 gene from M. tuberculosis (Erdman strain) into the mycobacterial expression vector pMV261 (16) under the control of the groEL promoter, resulting in the complementation plasmid pMWS149. This plasmid was electroporated into the ΔpapA3 strain, and transformants were selected on kanamycin-containing plates. The Δstf0 mutant was created by homologous recombination using transduction phage phMWS102 as described above. The resulting deletion replaced 600 bp of stf0 (amino acids 22–222) with a hygromycin resistance cassette.
Preparation of M. tuberculosis Lipid Extracts—M. tuberculosis cultures were synchronized and grown in 7H9 media to A600 = 0.6. Cultures (50 ml) were transitioned to Tween 80-free 7H9 media and grown at 37 °C for 1 day. Surface lipids were extracted with hexane (1 ml) as described previously (17). The remaining cell pellets were extracted with 4 ml of chloroform: methanol (1:1).
Sample Preparation for Mass Spectrometry—PapA3 reaction product samples were resuspended in 1 ml of 100% methanol (MeOH) for ESI-FT-ICR MS analysis. For linear ion trap MS analysis of PapA3 reaction product samples, 500 μl of the resuspended volume were concentrated to dryness and resuspended in 3 ml of chloroform (CHCl3):isopropyl alcohol (IPA) (2:1). M. tuberculosis cell surface extracts were concentrated to dryness under nitrogen and resuspended in 3 ml of CHCl3:IPA (2:1).
Lipids in each sample were separated using a modification of the method of Kaluzny et al. (18). Samples were passed over a solid-phase extraction column (Sep-Pak Vac, NH2 resin, Waters) that had been pre-charged with 3 ml of 0.1 m ammonium acetate in MeOH. The column was eluted using 3 ml of each of the following solvents: 1) CHCl3:IPA (2:1); 2) diethyl ether:acetic acid (98:2); 3) 100% MeOH; and 4) 0.1 m ammonium acetate in MeOH. Column fractions from the PapA3 reaction product samples were concentrated to dryness and resuspended in 1 ml of CHCl3:MeOH (2:1). Acyltrehaloses eluted in solvent 3. For linear ion trap MSn experiments, lithium acetate was added to the PapA3 reaction products and the T2P and T3P standards to a final concentration of 1 mm. Column fractions from M. tuberculosis cell surface extracts were concentrated to dryness under nitrogen and resuspended in either 200 μl CHCl3:MeOH (2:1) (fractions from solvents 1 and 2) or 400 μl of CHCl3:MeOH (2:1) (fractions from solvents 3 and 4) for mass spectrometry analysis. PAT eluted in solvent 1, and SL-1 eluted in solvent 4.
RESULTS
Genomic Analysis of the PAT Biosynthetic Locus—The pap gene family encodes polyketide synthase-associated acyltransferases that are involved in the synthesis of some of the complex lipids produced by M. tuberculosis (5, 11). In the M. tuberculosis genome, papA3 is clustered with the polyketide synthase-encoding gene pks3/4. In some strains, including the sequenced H37Rv strain, there is an intervening stop codon in pks3/4 that results in two separate open reading frames (termed pks3 and pks4) (7). Strains containing this mutation do not synthesize PAT (19), indicating that an intact pks3/4 gene is essential for the biosynthesis of this glycolipid. Within the same gene cluster resides mmpL10, which encodes a putative lipid transporter. MmpL10 belongs to the same protein family as MmpL8, which is required for SL-1 biosynthesis (17, 19, 20). The genomic organization of pks3/4, papA3, and mmpL10 parallels that of pks2, papA1, and mmpL8 in the SL-1 biosynthetic gene cluster (Fig. 1C). We previously demonstrated that papA1 is the acyltransferase responsible for coupling the polyketide product of Pks2 to a trehalose-based acceptor (5). By analogy, we hypothesized that PapA3 is essential for PAT biosynthesis, catalyzing trehalose acylation.
PapA3 Is an Acyltransferase That Esterifies Trehalose and T2P—The papA3 gene from the H37Rv M. tuberculosis strain was expressed in Escherichia coli BL21(DE3) as an N-terminal MBP fusion protein. SDS-PAGE analysis revealed an apparent molecular mass of 95 kDa for the purified protein. Following TEV cleavage and subsequent removal of the protease and MBP by Ni2+-affinity chromatography (supplemental Fig. S1), the identity of the protein was confirmed by mass spectrometry. The measured mass (51,809 ± 3 Da) was consistent with the predicted molecular mass of the protein (51,777.5 Da) oxidized at a single methionine residue. In addition, tryptic digestion and mass fingerprinting of the purified protein generated ∼61% sequence coverage, providing further confirmation of the identity of the protein (data not shown).
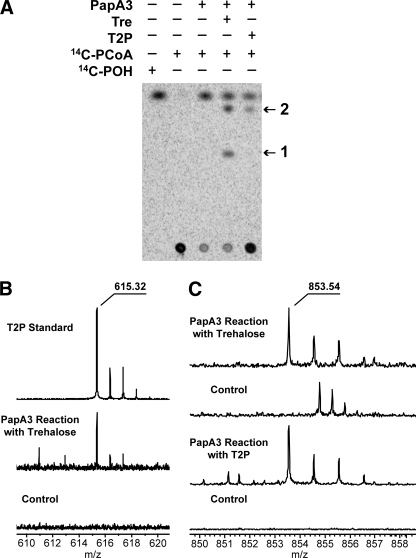
Incubation of PapA3 with 14C-PCoA and trehalose resulted in the formation of two unique products, as determined by silica gel TLC and phosphorimaging (Fig. 2A). Only the less polar product was formed by the reaction of PapA3 with 14C-PCoA and synthetic T2P (12, 13). PapA3 showed no activity against several other saccharides, including T3P, trehalose 2-sulfate, α,β-trehalose, glucose, and maltose, suggesting that the enzyme is selective for trehalose and T2P (Table 1). Notably, PapA3 hydrolyzes 14C-PCoA to 14C-palmitic acid in the absence of another substrate (Fig. 2A), similar to the other characterized Pap enzymes. Further kinetic analysis of the PapA3 reaction containing trehalose or T2P was precluded by the complexity of the product mixture.
FIGURE 2.
PapA3 is an acyltransferase that sequentially palmitoylates trehalose in vitro. A, PapA3 was incubated with 14C-PCoA and either trehalose (Tre) or T2P. The reactions were analyzed by TLC and phosphorimaging. Two new products (1 and 2) were observed in the reaction with trehalose, but only product 2 was observed in the reaction with T2P. B, ESI-FT-ICR MS analysis of product 1 from the PapA3 reaction with trehalose. A product ion with m/z 615.32, corresponding to the m/z of a chloride adduct of synthetic T2P, was observed in the PapA3 reaction. In contrast, the control reaction lacking PapA3 showed no product at m/z 615. C, ESI-FT-ICR MS analysis of product 2 from the PapA3 reaction with Tre and T2P. An ion with m/z 853.54 was observed in both reactions but was not present in control reactions lacking PapA3.
TABLE 1.
Substrate specificity of PapA3
ND means not detectable.
| Substrate | Product formationa |
|---|---|
| Nucleophileb | |
| Trehalose | Yes |
| T2P | Yes |
| T3P | ND |
| Trehalose 2-sulfate | ND |
| α,β-Trehalose | ND |
| Glucose | ND |
| Maltose | ND |
| Acyl-CoAc | |
| Palmitoyl-CoA | Yes |
| Docosanoyl-CoA | Yes |
| Butyryl-CoA | ND |
| Crotonoyl-CoA | ND |
Product formation was assessed by TLC and phosphorimaging.
Reactions were performed with 2 μm enzyme, 20 μm 14C-palmitoyl-CoA, and 1 or 10 mm of each substrate in 100 mm ammonium bicarbonate, pH 7.2, at room temperature for 2 h.
Reactions were performed with 2 μm enzyme, 20 μm 14C-acyl-CoA, and 1 mm of trehalose or T2P in 100 mm ammonium bicarbonate, pH 7.2, at room temperature for 2 h.
Interestingly, no activity was detected with 14C-butyryl coenzyme A or 14C-crotonoyl coenzyme A, which contains a trans-2-ene functionality like the mycolipenoyl groups of PAT (Table 1). However, product formation was observed upon incubation of PapA3 with trehalose and 14C-docosanoyl coenzyme A, which consists of a 22-carbon saturated fatty acid conjugated to coenzyme A (data not shown). This suggests that PapA3 may also accept the 24-carbon backbone of mycolipenic acid. Taken together, these findings indicate that lipid chain length influences the substrate specificity of PapA3.
T2P and Trehalose Dipalmitate Are Products of PapA3—Products from the reaction of PapA3 with PCoA and either trehalose or T2P were characterized by ESI-FT-ICR and linear ion trap MS operating in the negative ion mode. The reaction of PapA3 with trehalose yielded two unique products measured via FT-ICR MS at m/z 615.3153 and m/z 853.5447, corresponding to the exact masses of the chloride adducts of trehalose palmitate and trehalose dipalmitate, respectively (Fig. 2, B and C). The accurate mass spectra were calibrated internally, and both reaction products were measured to sub-ppm accuracy. The product ion at m/z 853.54 was also observed in the reaction of PapA3 with T2P (Fig. 2C). Importantly, these products were not observed in the absence of enzyme. Linear ion trap tandem mass spectrometry (MSn) of the ion at m/z 615.32 yielded dissociation ions consistent with those derived from synthetic T2P (supplemental Fig. S2). MSn of the ion at m/z 853.54 was consistent with 2,3-dipalmitoylation of a single pyranose ring of trehalose (supplemental Fig. S3), which was confirmed in the positive ion mode using lithium-ion coordination (supplemental Fig. S4). Together, these data demonstrate that PapA3 sequentially acylates trehalose to form trehalose dipalmitate.
T3P Is Not Produced by PapA3—To rule out the possibility that the initial acylation product of PapA3 is T3P or a mixture of 2- and 3-palmitoylated species, we synthesized T3P for additional biochemical and structural studies. To determine whether T3P is a viable intermediate in the biosynthesis of trehalose dipalmitate, we incubated T3P with PapA3 and 14C-PCoA. No product formation was detected following TLC and phosphorimaging (supplemental Fig. S5), indicating that T3P is not a substrate for PapA3. Interestingly, the presence of T3P in the reaction mixture appears to diminish the hydrolysis of 14C-PCoA by PapA3, suggesting it may instead inhibit the enzyme.
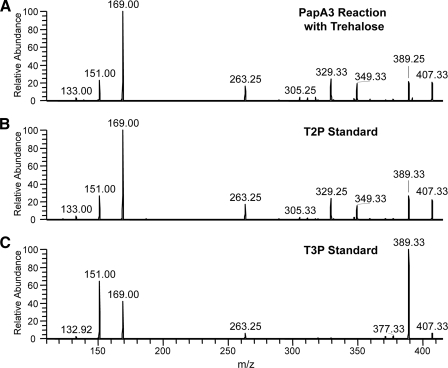
For further confirmation that the monoacyl product of PapA3 is not T3P, we analyzed T3P by MSn in the positive ion mode using lithiumcation coordination (21, 22) and compared its fragmentation to that of T2P and the PapA3 monoacyl reaction product ion (Fig. 3). T2P, T3P, and the monoacyl PapA3 reaction product were observed at m/z 587, corresponding to the lithium adducts of these molecules. Two dissociation ions at m/z 425 and 407 were observed in the MS2 spectra of these ions, corresponding to the cleavage of the glycosidic bond between the pyranose rings of trehalose (data not shown). The MS3 spectrum of the monoacyl PapA3 reaction product dissociation ion at m/z 407 was identical to that derived from synthetic T2P (Fig. 3, A and B) and dramatically distinct from the MS3 spectrum derived from synthetic T3P (Fig. 3C). These data clearly demonstrate that the monoacyl PapA3 reaction product is not T3P. Combined with the finding that T3P is not a substrate for PapA3, these data support the assignment of T2P as the initial acylation product of PapA3.
FIGURE 3.
Linear ion trap MSn of the monoacyl product ion from the reaction of PapA3 with trehalose (A) is consistent with that of synthetic T2P (B) and not that of synthetic T3P (C). MSn analysis was performed in the positive ion mode using lithium-cation coordination. Shown are the MS3 spectra of the dissociation ions obtained from the cleavage of the glycosidic bond between the individual pyranose rings of trehalose.
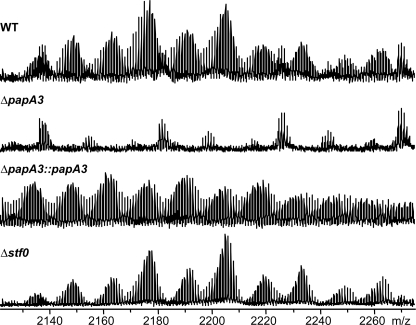
PapA3 Is Required for PAT Biosynthesis in Vivo—To determine whether PapA3 is required for PAT biosynthesis in vivo, a papA3 deletion mutant, ΔpapA3, was generated in the Erdman strain of M. tuberculosis. The H37Rv strain sequenced by Cole et al. (23) contains a stop codon in pks3/4, which truncates the encoded polyketide synthase and abolishes PAT biosynthesis. However, sequencing confirmed that the Erdman strain used in these studies encodes a single open reading frame for pks3/4, yielding a functional polyketide synthase (data not shown). Extraction of the crude lipids from wild-type Erdman cells with organic solvents followed by ESI-FT-ICR MS analysis confirmed the presence of the PAT lipid envelope in this strain of M. tuberculosis (Fig. 4). In contrast, PAT was not observed in extracts from the ΔpapA3 mutant strain. Complementation of the ΔpapA3 mutant strain with a plasmid encoding papA3 restored PAT production, demonstrating that PapA3 is essential for the biosynthesis of this glycolipid in M. tuberculosis.
FIGURE 4.
PAT biosynthesis requires papA3, but not stf0, in vivo. ESI-FT-ICR MS analysis of lipid extracts from wild type, ΔpapA3::papA3, and Δstf0 M. tuberculosis strains revealed the presence of characteristic PAT lipoforms that are absent from ΔpapA3 M. tuberculosis extracts.
PAT Biosynthesis Is Independent of SL-1 Biosynthesis—Given the similarities between the PAT and SL-1 genetic loci, we sought to determine whether the two biosynthetic pathways shared common intermediates. The first committed step in SL-1 biosynthesis is the sulfation of trehalose at the 2-position of one of the glucose moieties by the sulfotransferase Stf0 (24). Thus, we analyzed the M. tuberculosis Δstf0 mutant strain, which lacks SL-1 as well as its upstream biosynthetic precursors, for the presence of PAT. FT-ICR MS analysis of crude lipid extracts clearly showed the presence of PAT in the Δstf0 mutant strain (Fig. 4). Also, SL-1 synthesis was not perturbed in the ΔpapA3 mutant strain. These data indicate that the PAT biosynthetic pathway is independent of the SL-1 pathway and that these pathways share no intermediates other than the cellular pool of trehalose.
DISCUSSION
The data presented here establish that PapA3 is an acyltransferase required for PAT biosynthesis in M. tuberculosis. Recombinant PapA3 selectively acylates trehalose and T2P in a manner consistent with the structure of PAT. Also, deletion of papA3 from M. tuberculosis prevents PAT synthesis in vivo. Furthermore, despite the genetic and structural similarities between PAT and SL-1, the biosynthetic pathways of these metabolites are independent. However, many questions remain about PapA3 and PAT biosynthesis.
In vitro, PapA3 catalyzes the sequential transfer of two palmitoyl groups onto a single glucose residue of trehalose, suggesting that PapA3 installs both the palmitoyl group at the 2-position of PAT and the 3-mycolipenoyl group (Fig. 1A). It is possible that PapA3 associates with other proteins in the PAT biosynthetic pathway, forming a coordinate synthetic complex similar to that described for phthiocerol dimycocerosate biosynthesis (25). Such supramolecular assemblies may influence substrate availability and orientation in the PapA3 active site, thereby conferring this unique activity.
Whether PapA3 is truly a bifunctional acyltransferase remains unclear. PapA3 may only install the palmitoyl group of PAT in vivo, in which case the formation of trehalose dipalmitate by the purified enzyme is an in vitro artifact. Such activity has been observed with the lauroyltransferase of E. coli lipid A biosynthesis, which transfers two lauroyl groups in vitro but only one in vivo (26). Alternatively, the mycolipenoyl groups found at the 3-, 6-, 2′-, and 4′-positions of PAT may all be physiological products of PapA3. This hypothesis is supported by the genetic association between papA3 and the mycolipenate synthase pks3/4 and the promiscuity of PapA3 toward the acylation state of trehalose. Unfortunately, our analysis of the acylation events catalyzed by PapA3 is limited by the lack of a commercially available mycolipenoyl substrate and our limited capability to detect either T2P or trehalose mycolipenates in vivo.
The chemical similarities between PAT and SL-1 suggest that the biosynthesis of these molecules may be comparable. However, the SL-1 gene locus encodes two Pap proteins, PapA2 and PapA1, which sequentially install a palmitoyl group and a methyl-branched hydroxyphthioceranoyl group, respectively. By comparison, the PAT gene locus encodes only PapA3. Previously, we showed that PapA2 does not recognize trehalose and cannot account for T2P synthesis (5). We initially hypothesized that the SL-1 precursor trehalose-2-sulfate-2′-palmitate (termed SL659) may be desulfated by a sulfatase to form T2P, thus negating the need for a committed trehalose palmitoyl-transferase. However, the Δstf0 mutant strain, which lacks SL659, maintained the ability to synthesize PAT. We therefore conclude that SL-1 and PAT biosynthesis are independent.
On the basis of our biochemical and genetic data, we propose the model for PAT biosynthesis shown in Fig. 5. Initially, PapA3 modifies the 2-position of one of the glucose residues of trehalose with a palmitoyl group from an unknown acyl donor, most likely PCoA or an acyl pantotheine-based cofactor. A mycolipenoyl group is then transferred to the 3-position of T2P by PapA3, which may associate directly with Pks3/4 or an unknown acyl carrier protein to initiate this second acylation step. The resulting 2,3-diacyltrehalose may be transported to the cell surface without further modification through an unknown pathway. Alternatively, it may serve as a biosynthetic intermediate that is elaborated either intracellularly or extracellularly with the three remaining mycolipenoyl groups of PAT by means of PapA3 or an unidentified acyltransferase. By analogy to other M. tuberculosis lipid biosynthetic pathways, transport of PAT or its precursor to the cell surface is most likely accomplished by MmpL10.
FIGURE 5.
Proposed PAT biosynthetic pathway. PapA3 first acylates the 2-position of one of the glucose residues of trehalose with a palmitoyl group to form T2P. A mycolipenoyl group, synthesized by Pks3/4, is then transferred to the 3-position of T2P by PapA3 to generate 2,3-diacyltrehalose. 2,3-Diacyltrehalose may either be transported to the cell surface or serve as a biosynthetic intermediate that is further elaborated with mycolipenic acids to give PAT.
Clearly, several questions regarding the biosynthesis of PAT remain unanswered. The synthesis of physiological substrates for in vitro assays and the comparative lipid analysis of a panel of PAT biosynthetic mutants, including ΔmmpL10 and Δpks3/4, may resolve some of these issues. Whereas genes from the PAT biosynthetic gene cluster are up-regulated under various conditions of environmental stress, including phagosomal acidification and nutrient starvation (27, 28), the role of PAT in M. tuberculosis pathogenesis remains a mystery. Notably, a recent study of an M. tuberculosis strain deficient in PAT biosynthesis suggests PAT does not contribute to virulence in mice (29). However, the phenotype of SL-1-deficient M. tuberculosis strains is also indistinguishable from wild type in the murine model of infection (5), suggesting the function of these glycolipids may be host-specific. Thus, a more appropriate model of tuberculosis may be key to elucidating the role of these lipids in M. tuberculosis pathogenesis.
Supplementary Material
Acknowledgments
We thank T. Jothi for preparing the complementation plasmid used in this study. We thank D. King (Howard Hughes Medical Institute Mass Spectrometry Laboratory, University of California, Berkeley), D. Rabuka, and B. Smart for technical contributions. We also thank S. Gilmore and J. Seeliger for technical advice and helpful discussions.
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grant AI51622 (to C. R. B.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S5.
Footnotes
The abbreviations used are: SL-1, sulfolipid-1; PAT, polyacyltrehalose; PCoA, palmitoyl coenzyme A; T2P, trehalose 2-palmitate; T3P, trehalose 3-palmitate; ESI-FT-ICR MS, electrospray ionization Fourier transform ion cyclotron mass spectrometry; MSn, linear ion trap tandem mass spectrometry; MBP, maltose-binding protein; DTT, dithiothreitol; TCEP, tris(2-carboxyethyl)phosphine; DMSO, dimethyl sulfoxide; MeOH, methanol; CHCl3, chloroform; IPA, isopropyl alcohol.
References
- 1.Brennan, P. J., and Nikaido, H. (1995) Annu. Rev. Biochem. 64 29–63 [DOI] [PubMed] [Google Scholar]
- 2.Minnikin, D. E., Kremer, L., Dover, L. G., and Besra, G. S. (2002) Chem. Biol. 9 545–553 [DOI] [PubMed] [Google Scholar]
- 3.Takayama, K., Wang, C., and Besra, G. S. (2005) Clin. Microbiol. Rev. 18 81–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreras, J. A., Stirrett, K. L., Lu, X., Ryu, J. S., Soll, C. E., Tan, D. S., and Quadri, L. E. (2008) Chem. Biol. 15 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar, P., Schelle, M. W., Jain, M., Lin, F. L., Petzold, C. J., Leavell, M. D., Leary, J. A., Cox, J. S., and Bertozzi, C. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rousseau, C., Neyrolles, O., Bordat, Y., Giroux, S., Sirakova, T. D., Prevost, M. C., Kolattukudy, P. E., Gicquel, B., and Jackson, M. (2003) Cell. Microbiol. 5 405–415 [DOI] [PubMed] [Google Scholar]
- 7.Dubey, V. S., Sirakova, T. D., and Kolattukudy, P. E. (2002) Mol. Microbiol. 45 1451–1459 [DOI] [PubMed] [Google Scholar]
- 8.Daffe, M., Lacave, C., Laneelle, M. A., Gillois, M., and Laneelle, G. (1988) Eur. J. Biochem. 172 579–584 [DOI] [PubMed] [Google Scholar]
- 9.Schelle, M. W., and Bertozzi, C. R. (2006) ChemBioChem 7 1516–1524 [DOI] [PubMed] [Google Scholar]
- 10.Bhatt, K., Gurcha, S. S., Bhatt, A., Besra, G. S., and Jacobs, W. R., Jr. (2007) Microbiology 153 513–520 [DOI] [PubMed] [Google Scholar]
- 11.Onwueme, K. C., Ferreras, J. A., Buglino, J., Lima, C. D., and Quadri, L. E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4608–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace, P. A., Minnikin, D. E., and Ridell, M. (1994) J. Chem. Soc. Chem. Commun., 329–330
- 13.Wallace, P. A., and Minnikin, D. E. (1994) Carbohydr. Res. 263 43–59 [DOI] [PubMed] [Google Scholar]
- 14.Langston, S., Bernet, B., and Vasella, A. (1994) Helv. Chim. Acta 77 2341–2353 [Google Scholar]
- 15.Glickman, M. S., Cox, J. S., and Jacobs, W. R., Jr. (2000) Mol. Cell 5 717–727 [DOI] [PubMed] [Google Scholar]
- 16.Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., Bansal, G. P., Young, J. F., Lee, M. H., Hatfull, G. F., Snapper, S. B., Barletta, R. G., Jacobs, W. R., Jr., and Bloom, B. R. (1991) Nature 351 456–460 [DOI] [PubMed] [Google Scholar]
- 17.Converse, S. E., Mougous, J. D., Leavell, M. D., Leary, J. A., Bertozzi, C. R., and Cox, J. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaluzny, M. A., Duncan, L. A., Merritt, M. V., and Epps, D. E. (1985) J. Lipid Res. 26 135–140 [PubMed] [Google Scholar]
- 19.Domenech, P., Reed, M. B., and Barry, C. E., III (2005) Infect. Immun. 73 3492–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domenech, P., Reed, M. B., Dowd, C. S., Manca, C., Kaplan, G., and Barry, C. E., III (2004) J. Biol. Chem. 279 21257–21265 [DOI] [PubMed] [Google Scholar]
- 21.Zhou, Z., Ogden, S., and Leary, J. A. (1990) J. Org. Chem. 55 5444–5446 [Google Scholar]
- 22.Hofmeister, G. E., Zhou, Z., and Leary, J. A. (1991) J. Am. Chem. Soc. 113 5964–5970 [Google Scholar]
- 23.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, Tekaia, F., Badcock, K., Basham, D., Brown, D., Chillingworth, T., Connor, R., Davies, R., Devlin, K., Feltwell, T., Gentles, S., Hamlin, N., Holroyd, S., Hornsby, T., Jagels, K., Krogh, A., McLean, J., Moule, S., Murphy, L., Oliver, K., Osborne, J., Quail, M. A., Rajandream, M. A., Rogers, J., Rutter, S., Seeger, K., Skelton, J., Squares, R., Squares, S., Sulston, J. E., Taylor, K., Whitehead, S., and Barrell, B. G. (1998) Nature 393 537–544 [DOI] [PubMed] [Google Scholar]
- 24.Mougous, J. D., Petzold, C. J., Senaratne, R. H., Lee, D. H., Akey, D. L., Lin, F. L., Munchel, S. E., Pratt, M. R., Riley, L. W., Leary, J. A., Berger, J. M., and Bertozzi, C. R. (2004) Nat. Struct. Mol. Biol. 11 721–729 [DOI] [PubMed] [Google Scholar]
- 25.Jain, M., and Cox, J. S. (2005) PLoS Pathog. 1 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Six, D. A., Carty, S. M., Guan, Z., and Raetz, C. R. (2008) Biochemistry 47 8623–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohde, K. H., Abramovitch, R. B., and Russell, D. G. (2007) Cell Host Microbe 2 352–364 [DOI] [PubMed] [Google Scholar]
- 28.Hampshire, T., Soneji, S., Bacon, J., James, B. W., Hinds, J., Laing, K., Stabler, R. A., Marsh, P. D., and Butcher, P. D. (2004) Tuberculosis 84 228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesne-Seck, M. L., Barilone, N., Boudou, F., Gonzalo Asensio, J., Kolattukudy, P. E., Martin, C., Cole, S. T., Gicquel, B., Gopaul, D. N., and Jackson, M. (2008) J. Bacteriol. 190 1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.