Abstract
Inhibitor of apoptosis (IAP) proteins are widely expressed throughout nature and suppress cell death under a variety of circumstances. X-linked IAP, the prototypical IAP in mammals, inhibits apoptosis largely through direct inhibition of the initiator caspase-9 and the effector caspase-3 and -7. Two additional IAP family members, cellular IAP1 (cIAP1) and cIAP2, were once thought to also inhibit caspases, but more recent studies have suggested otherwise. Here we demonstrate that cIAP1 does not significantly inhibit the proteolytic activities of effector caspases on fluorogenic or endogenous substrates. However, cIAP1 does bind to caspase-3 and -7 and does so, remarkably, at distinct steps prior to or following the removal of their prodomains, respectively. Indeed, cIAP1 bound to an exposed IAP-binding motif, AKPD, on the N terminus of the large subunit of fully mature caspase-7, whereas cIAP1 bound to partially processed caspase-3 in a manner that required its prodomain and cleavage between its large and small subunits but did not involve a classical IAP-binding motif. As a ubiquitin-protein isopeptide ligase, cIAP1 ubiquitinated caspase-3 and -7, concomitant with binding, in a reaction catalyzed by members of the UbcH5 subfamily (ubiquitin carrier protein/ubiquitin-conjugating enzymes), and in the case of caspase-3, differentially by UbcH8. Moreover, wild-type caspase-7 and a chimeric caspase-3 (bearing the AKPD motif) were degraded in vivo in a proteasome-dependent manner. Thus, cIAPs likely suppress apoptosis, at least in part, by facilitating the ubiquitination and turnover of active effector caspases in cells.
Apoptosis is a programmed form of cell death that is generally executed through the activation of caspases,2 cysteine proteases that exhibit an almost absolute preference for cleavage after aspartate residues. Caspases are synthesized as single-chain zymogens, containing a prodomain, as well as large and small subunits that include residues required for substrate recognition and cleavage (1). During death receptor or mitochondria-dependent apoptosis, the long prodomain-containing initiator caspase-8/10 and -9 are recruited via their adapter proteins, Fas-associated death domain and apoptotic protease-activating factor-1 (Apaf-1), to multimeric caspase-activating complexes known as the death-inducing signaling complex and the apoptosome, respectively (1, 2). In the latter case, mitochondrial outer membrane permeabilization (MOMP) is required to mediate the release of cytochrome c from the intermembrane space into the cytosol, where it stimulates dATP/ATP-dependent oligomerization of Apaf-1 into the apoptosome (2). Once recruited, all initiator caspases are concentrated within their respective complexes and are thought to be activated as a result of dimerization, with concomitant autocatalytic cleavage of the activation loops that separate their large and small subunits (1). However, unlike caspase-8 and -10, caspase-9 must remain bound to the apoptosome to exhibit significant catalytic activity, so that in addition to promoting dimerization, the apoptosome may also induce conformational changes in caspase-9 that are necessary for its activation (3–6).
In contrast to initiator caspases, effector caspases, such as caspase-3 and -7, contain short prodomains and exist normally as latent dimers, wherein their activation loops sterically hinder substrate access and hold the substrate binding pocket in an inactive conformation (1). Effector caspases are directly activated by caspase-8, -9, and -10, and following cleavage of caspase-3 between its large and small subunits, the two-chain p20/p12 form becomes a catalytically active heterotetramer and undergoes subsequent autocatalytic processing between its prodomain and large subunits to generate the fully mature p17/p12 form of the enzyme (7). Similarly, procaspase-7 is also activated following cleavage of its activation loop to generate its two-chain p22/p12 form; however, it remains unclear whether removal of its prodomain in cells (to generate its p19/p12 form) is accomplished primarily via autocatalysis, active caspase-3, or perhaps by serine proteases at a non-aspartate residue (8, 9). Caspase-3 and -7 exhibit significant sequence and structural homology, differing primarily in their short prodomains. Despite this fact, caspase-3 processes a wider array of protein substrates during apoptosis and is largely responsible for dismantling the cell (10). Thus, interesting questions remain regarding the physiological roles of caspase-7, whether caspase-7 activity is differentially regulated compared with caspase-3, and what structural features determine (and in some cases limit) its substrate specificity.
Given the devastating consequences of unfettered caspase activation, cells have evolved mechanisms to regulate caspase activity. For example, IAPs, originally identified in baculoviruses, possess one or more baculovirus IAP repeat (BIR) domains, and at least one of the eight family members, XIAP, selectively inhibits the activities of caspase-9, -3, and -7 (1, 11). Mechanistically, the BIR3 domain in XIAP binds to an exposed IBM on the N terminus of the small subunit of processed caspase-9, situated directly above the active site, and limits the access of substrates (12, 13). By contrast, the linker region (located between the BIR1 and BIR2 domains in XIAP) lies across the active sites of caspase-3 and -7 and binds in a reverse orientation to substrates, thereby preventing cleavage of the linker while simultaneously preventing the access of substrates (14, 15). The BIR2 domain then stabilizes the linker-caspase-3 (and linker-caspase-7) interactions further by binding to an exposed IBM on the N terminus of the small subunit in the adjacent caspase dimer (14, 16). Importantly, IAP antagonists, such as Smac/DIABLO and Omi/HtrA2, are normally sequestered to the intermembrane space of mitochondria and are released (along with cytochrome c) into the cytoplasm during apoptosis. As IAP antagonists also possess IBMs, they bind to BIR domains and prevent or relieve the inhibition of caspases by IAPs (1).
Previously, two additional IAP family members, cIAP1 and cIAP2, were also thought to inhibit caspases, but more recent studies suggest that these IAPs bind but do not inhibit caspases (17–19). Nevertheless, various studies have shown that cIAPs can protect cells from apoptosis, are overexpressed or mutated in some cancers, and can promote tumorigenesis (20–25), raising questions as to how these IAPs inhibit cell death or whether they have additional functions (26). XIAP, cIAP1, and cIAP2 possess C-terminal RING zinc finger domains with E3 ubiquitin (Ub) ligase activities capable of catalyzing the ubiquitination and subsequent proteasomal degradation of cellular targets, including themselves (27, 28). Moreover, cIAPs have been shown to ubiquitinate several factors, including TNF receptor-associated factor 2, the serine/threonine kinase NIK, receptor-interacting protein 1, and the IAP antagonist Smac (29–34). However, although there is some evidence to support a direct role for ubiquitination in the regulation of effector caspases by XIAP (35, 36), the role of cIAPs in this process remains unclear, particularly in vivo. We demonstrate herein that cIAP1 binds to caspase-3 and -7 at unique steps in their processing, prior to or following the removal of their prodomains, respectively. Moreover, rather than directly inhibiting these effector caspases, cIAP1 ubiquitinates them and targets them for proteasome-dependent degradation, thereby suppressing apoptosis.
EXPERIMENTAL PROCEDURES
Reagents—Antibodies to caspase-3 and -7 were obtained from Cell Signaling Technology (catalog numbers 9665, 9661, and 9491; Danvers, MA) or BD Biosciences (catalog number 551238; San Jose, CA). Actin antibody was purchased from Oncogene (catalog number CP01; Cambridge, MA), Myc antibody from Cell Signaling Technology (catalog number 2276), and HA antibody from Covance (catalog number MMS-101R; Emeryville, CA). Horseradish peroxidase-conjugated secondary antibodies to rabbit and mouse IgG were obtained from DakoCytomation (catalog number P0448; Carpinteria, CA) and Sigma (catalog number A8924), respectively. Mouse IgM was purchased from Calbiochem (catalog number 401225), and anti-FLAG® M2 monoclonal antibody-agarose affinity gel (catalog number A2220) was obtained from Sigma.
Plasmids—Procaspase-3 and -7 were cloned into BamHI-XhoI sites of pET21b vector (Novagen, Gibbstown, NJ). SGIS procaspase-7, Δ24–27 procaspase-7, SGVD procaspase-7, AKPD procaspase-3, SGPI procaspase-3, and various doubly swapped chimeras were then generated by PCR-SOE and cloned into pET21b using the following primers: C7-US, 5′-GTCGGGATCCGATGGCAGA-3′; C7-DS, 5′-GGTGCTCGAGTTGACTGAAG-3′; C7-SGIS-US, 5′-GTGGATTCTGGAATATCCCGGTCCTCGTTTGTACCGTC-3′; C7-SGIS-DS, 5′-GGACCGGGATATTCCAGAATCCACTGAATCTTCATTTGCTG-3′; C7-Δ24–27-US, 5′-AATGAAGATTCAGTGGATCGGTCCTCGTTTGTACCGTC-3′; C7-Δ24–27-DS, 5′-CGGTACAAACGAGGACCGATCCACTGAATCTTCATTTGCTG-3′; C7-SGVD-US, 5′-AGGCCGACTCGGGGGTCGACAATGACACAGATGC-3′; C7-SGVD-DS, 5′-GCATCTGTGTCATTGTCGACCCCCGAGTCGGCCT-3′; C7-D28A-US, 5′-AATGAAGATTCAGTGGCTGCTAAGCCAGACCG-3′; C7-D28A-DS, 5′-CGGTCTGGCTTAGCAGCCACTGAATCTTCATT-3′; C3-US, 5′-GTCGGGATCCCATGGAGAAC-3′; C3-DS, 5′-GGTGCTCGAGGTGATAAAAATA-3′; C3-AKPD-US, 5′-ATGGACGCTAAGCCAGACCTGGACAACAGTTATAAAATGGAT-3′; C3-AKPD-DS, 5′-GTCCAGGTCTGGCTTAGCGTCCATTGATTCGCTTCCATGT-3′; C3-SGPI-US, 5′-GAGACAGACAGTGGTCCTATTGATGACATGGCG-3′; C3-SGPI-DS, 5′-ACGCCATGTCATCAATAGGACCACTGTCTGTCTC-3′; C3-D23A-US, 5′-GGAAGCGAATCAATGGCCTCTGGAATATCCCTG-3′; C3-D23A-DS, 5′-CAGGGATATTCCAGAGGCCATTGATTCGCTTCC-3′; C3-GGVD-US, 5′-TGGCATTGAGACAGACGGTGGTGTTGATGATGA-3′; and C3-GGVD-DS, 5′-TCATCATCAACACCACCGTCTGTCTCAATGCCA-3′. WT procaspase-3 and AKPD procaspase-3 were then subcloned into BamHI-XhoI of pcDNA3.1-Myc-His6 (Invitrogen). Finally, for all of the caspase-3 constructs, the caspase cleavage site NSVD9↓S was mutated (D9A) to ensure that autocatalytic removal of the prodomain occurred only at Asp-28.
To generate Ub fusion constructs of SGIS-caspase-3-myc, AKPD-caspase-3-myc, AKPD-caspase-7-myc, and SGIS-caspase-7-myc, residues 33–276 of caspase-3 and residues 28–303 of caspase-7 were subcloned into BamHI-XhoI of pDhaUb (kindly provided by Prof. Alexander Varshavsky, California Institute of Technology, Pasadena), containing an N-terminal mouse dihydrofolate reductase (DHFR), HA tag, and Ub. The BamHI site was subsequently replaced with either an AKPD or SGIS by site-directed mutagenesis, so that the final glycine residue in Ub was immediately followed by an alanine or serine residue, respectively. Each of the DHFR-HA-Ub-caspase constructs was then subcloned into the EcoRI/XhoI sites of pcDNA3.1-Myc-His6. The FLAG-Ub construct was generated by cloning Ub into the BamHI/NotI sites of pEBB-FLAG.
XIAP and cIAP1 were cloned into EcoRI-NotI sites of pGEX-4T-1 (Amersham Biosciences). XIAP-BIR1-BIR2 was then generated by introducing a stop codon into XIAP at residue 242 by site-directed mutagenesis. p23 cochaperone in pET45b and truncated PARP (residues 1–373) in pET24b were kindly provided by Prof. Seamus J. Martin (Trinity College, Ireland) and Prof. Hung-wen Liu (University of Texas, Austin), respectively. All of the E2 constructs were kindly provided by Prof. Aaron Ciechanover (Technion-Israel Institute of Technology, Haifa, Israel).
Recombinant Protein Expression—Protein expression was induced in bacterial cultures with 0.1–1 mm isopropyl β-d-thiogalactopyranoside when bacterial densities reached an OD of 0.4–0.6. Cultures were then incubated as follows: 16 h at 27 °C for GST-XIAP-BIR1-BIR2; 16 h at 24 °C for GST-cIAP1; 4 h at 28 °C for caspase-3; 16 h at 37 °C for caspase-7; 16 h at 37 °C for all E2 Ub-conjugating enzymes; and 16 h at 18 °C for PARP-(1–373). GST-tagged proteins were purified by fast protein liquid chromatography using glutathione-Sepharose resin (Novagen), and His6-tagged proteins were purified by fast protein liquid chromatography using Ni2+-charged resin (Qiagen, Valencia, CA). Stock protein concentrations were determined by the Bradford assay or, in the case of active caspases, were determined by active site titration using Z-VAD-fmk and/or DEVD-CHO (37, 38).
Protein Binding Assays—GST-tagged cIAP1 and XIAP-BIR1-BIR2 (4 μm for Coomassie; 400 nm for Western blotting) were bound to glutathione-Sepharose beads (Novagen) for 4 h at 4 °C. The beads were then washed three times with GST-phosphate-buffered saline (PBS) (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4) and incubated with recombinant caspases (4 μm for Coomassie; 12 nm for Western blotting) in 500 μl of GST-PBS for 2 h at 4 °C. Finally, after extensive washing with assay buffer (150 mm NaCl, 2 mm DTT, 25 mm Tris, pH 8.0), bound proteins were eluted by incubating the protein-bound beads in GSH elution buffer (50 mm Tris, pH 8.0, 40 mm GSH reduced) for 20 min at 25 °C. Proteins were then subjected to SDS-PAGE and Coomassie Blue staining.
Caspase Assays—WT and chimeric caspase-3 and -7 (10 nm) were incubated with IAPs (40 nm) in caspase buffer (50 mm PIPES/KOH, 2 mm EDTA, 0.1% (w/v) CHAPS, 5 mm DTT, pH 7.2) for 30 min at 37 °C. The caspase-3/7 substrate, DEVD-AMC (final 20 nm; Biomol International, Plymouth Meeting, PA) was then added, and the release of AMC was monitored (λex/λem = 380 nm/460 nm) using a Victor3 plate reader.
PARP and p23 Cochaperone Cleavage Assays—WT and chimeric caspases-3 and -7 (400 nm) were incubated ±cIAP1 or XIAP-BIR1-BIR2 (800 nm) in buffer (10 mm PIPES/KOH, 100 mm NaCl, 10% (w/v) sucrose, 0.1% (w/v) CHAPS, 10 mm DTT, pH 7.2) for 30 min at 25 °C. Truncated PARP (10 μg; 49 kDa) was then added and incubated at 25 °C for indicated times. Similarly, WT and chimeric caspase-3 and -7 (150 nm) were incubated with p23 cochaperone (10 μg) in buffer at 25 °C for indicated times, and the mixtures were subjected to SDS-PAGE and Coomassie Blue staining.
Ubiquitination Assays—Reactions (20 μl) containing wild-type or methylated Ub (2 μm; Biomol), His-tagged E1 (60 nm; Biomol), and His-tagged human UbcH3, UbcH5a-c, UbcH6, UbcH7, or UbcH8 (200 nm; expressed and purified in-house) were incubated along with Mg2+/ATP (4 mm), caspase proteins (100 nm), and cIAP1 protein (100 nm) in reaction buffer (50 mm Tris, pH 7.6, 10% glycerol, 0.5 mm DTT, and supplemented with protease inhibitor mixture). In some incubations the polycaspase inhibitor Z-VAD-fmk (25 μm) was also included, as indicated. Reactions were incubated at 37 °C for 40 min and then quenched with 2× Laemmli loading dye and subjected to SDS-PAGE and immunoblotting.
Cell Culture and Transfections—MCF-7 human breast adenocarcinoma cells and HEK293 human embryonic kidney cells were grown in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 5% fetal bovine serum (Atlanta Biologicals, Norcross, GA), 5% FetalPlex (Gemini Bio-Products, West Sacramento, CA), 100 IU/ml penicillin, 100 μg/ml streptomycin (Mediatech, Manassas, VA), and 4 mm glutamine (Invitrogen). Cells were maintained at 37 °C in humidified air containing 5% CO2 and passaged every 3 days. Cells were transfected with 1.5 μg/ml plasmid DNA using Lipofectamine™ 2000 (Invitrogen), according to the manufacturer's protocol, and for stable transfections, cells were subsequently selected in 1.5 mg/ml G418. Individual clones were then isolated and tested for the expression of caspase-3 by immunoblotting.
Cell Viability Assays—MCF-7 cells were harvested by trypsinization, washed with PBS, and resuspended in annexin-V binding buffer (10 mm HEPES, pH 7.4, 140 mm NaCl, 2.5 mm CaCl2) containing annexin-V-fluorescein isothiocyanate and propidium iodide (Roche Applied Science). Fluorescein isothiocyanate- and/or propidium iodide-labeled cell populations were counted by flow cytometer (Beckman-Coulter, Fullerton, CA).
Western Blotting—Cells were collected by trypsinization, washed with PBS, and lysed in RIPA buffer (10 mm Na2PO4, 0.3 m NaCl, 0.1% SDS, 1% Nonidet P-40, 1% deoxycholate, 2 mm EDTA, pH 7.2) with protease inhibitors (10 mg/ml leupeptin, 10 mg/ml pepstatin, 10 mg/ml aprotinin, 200 mm phenylmethylsulfonyl fluoride). Lysates were assayed for protein concentration (Bradford assay), mixed with Laemmli loading dye, and boiled for 10 min. Proteins were then resolved by SDS-PAGE, transferred to Hybond C nitrocellulose (Amersham Biosciences), and blocked in Tris-buffered saline containing 5% nonfat dry milk and 0.1% Tween 20. Blots were then exposed to primary and secondary antibodies as indicated, and protein-antibody complexes were visualized by enhanced chemiluminescence (Amersham Biosciences).
Statistics—All experiments were performed at least three times. Each data point represents the mean ± S.E. Multiple group comparisons were performed by one-way analysis of variance, followed by a Student-Neuman-Keuls post hoc analysis (p < 0.05 was considered statistically significant).
RESULTS
cIAP1 Binds to Active Caspase-3 and -7 through Distinct Mechanisms at Steps Prior to or Following Removal of Their Prodomains—Although once reported to inhibit caspase-3 and -7, more recent studies suggest that cIAPs bind to these effector caspases but do not inhibit their activities (17–19). In an effort to further clarify the binding mechanisms of cIAP1 for caspase-3 and -7, we began by examining the protein-protein interactions between fully processed recombinant caspase-7 and full-length GST-cIAP1. As expected, cIAP1 interacted with WT active caspase-7, resulting in the pulldown of the p19 large subunit (LS) and the associated p12 small subunit (SS) (Fig. 1, A and B, lane 2). Surprisingly, however, cIAP1 failed to bind WT p17/p12 caspase-3 (Fig. 1C, lane 1). Consistent with a previous report (18), we found that removal of a putative IBM (Δ24–27, i.e. ΔAKPD) from the large subunit of caspase-7 resulted in an active enzyme that could no longer bind to cIAP1 (Fig. 1, A and B, lane 3).
FIGURE 1.
Binding mechanisms for cIAP1 with caspase-3 and -7. A, schematic of caspase-3 and -7 (Casp-3 and Casp-7) primary sequences. AKPD and SGPI motifs from the LS and small subunits (SS) of caspase-7 were swapped with the corresponding SGIS and SGVD motifs in caspase-3 to generate various chimeric caspase-3/7 proteins. B–E, GST-tagged cIAP1 was immobilized on glutathione-Sepharose beads and incubated with wild-type (WT) and mutant caspase-3 and caspase-7 proteins, as described under “Experimental Procedures.” After extensive washing, bead-bound proteins were eluted in Laemmli loading dye and subjected to SDS-PAGE/Coomassie Blue staining or Western blotting (WB). F, table of binding results for cIAP1 with caspases (+ indicates binding; – indicates no binding).
To determine whether the AKPD motif was sufficient to mediate IAP binding to effector caspases, we generated caspase-3/7 chimeric enzymes, in which the AKPD motif from caspase-7 was replaced with the corresponding motif in caspase-3 and vice versa, resulting in SGIS caspase-7 and AKPD caspase-3 (Fig. 1A). Following expression in bacteria, AKPD caspase-3 and SGIS caspase-7 underwent autocatalytic activation, wherein their prodomains were removed, exposing the AKPD and SGIS motifs, respectively. Remarkably, in subsequent pulldown experiments, we observed that GST-cIAP1 could now bind to AKPD caspase-3 (Fig. 1C, lane 2), whereas SGIS caspase-7 exhibited significantly reduced binding compared with the wild-type enzyme (Fig. 1B, lane 4). Thus, despite their high degree of sequence homology, caspase-3 and -7 differed dramatically in their abilities to bind cIAP1, and the mere presence of the AKPD motif in caspase-7 was required and sufficient to mediate this interaction.
During our studies, Salvesen and co-workers (14, 16) confirmed an earlier report indicating that the N termini of the small subunits of human caspase-3 and -7 (exposed following cleavage of these caspases at Asp-175 and Asp-198, respectively), likewise functioned as weak IBMs and stabilized the interactions of active caspase-3 and -7 with XIAP. Therefore, to determine whether this second site might also participate in binding to cIAP1, we generated caspase-3/7 chimeras in which the SGPI motif in caspase-7 was replaced with the SGVD motif in caspase-3 and vice versa (Fig. 1A). Interestingly, in subsequent pulldown experiments, cIAP1 bound to SGPI caspase-3 with higher affinity than WT caspase-3 but with lower affinity than AKPD caspase-3 (Fig. 1C, lanes 1–3). Conversely, cIAP1 interacted with SGVD caspase-7 with a slightly lower affinity than WT caspase-7 (Fig. 1B, lanes 2 and 5). Similarly, the doubly swapped SGIS/SGVD caspase-7 could no longer bind to cIAP1, whereas AKPD/SGPI caspase-3 now displayed prominent binding (Fig. 1, B, lane 7, and C, lane 4). Thus, the AKPD motif in caspase-7 played the dominant role in its association with cIAP1, whereas the SGPI motif most likely participated in stabilizing the interaction.
To this point, our data indicated that cIAP1 could bind to fully mature WT caspase-7 but not caspase-3. However, we knew that the initial cleavage of procaspase-3 within its activation loop (Asp-175) by the initiator caspase-8 and -9 produced an intermediate p20/p12 caspase-3, which in turn underwent autocatalysis to remove its prodomain and generate the p17/p12 form of the enzyme (7). Given that we had examined only p17/p12 caspase-3 in our previous pulldown assays, we decided to test the ability of cIAP1 to bind p20/p12 caspase-3, compared with p17/p12 caspase-3. Recombinant p20/p12 caspase-3 was generated by expressing a mutant of procaspase-3 (D9A/D28A), which could undergo normal processing at Asp-175 but could not remove its prodomain (Fig. 1A). As expected, cIAP1 failed to bind to the fully mature p17/p12 form of caspase-3 but remarkably did bind to the intermediate p20/p12 form of the enzyme (Fig. 1E, lanes 2 and 3).
The presence of alanine mutations at Asp-9 and Asp-28 in p20/p12 caspase-3 did not confer binding to cIAP1, as identical results were obtained when p20/p12 caspase-3 was generated via Apaf-1·caspase-9 apoptosome-dependent cleavage of catalytically inactive procaspase-3 (supplemental Fig. S1, lanes 2 and 3). Moreover, although processing of procaspase-3 at Asp-175 was required to bind cIAP1 (supplemental Fig. S1, lanes 3 and 4), the putative IBM on the N terminus of the small subunit of p20/p12 caspase-3 did not participate in binding, as mutation of the SGVD motif to GGVD failed to disrupt the interaction of p20/p12 caspase-3 with cIAP1 (Fig. 1E, lanes 3 and 4). Finally, we performed similar pulldown experiments with recombinant p22/p12 caspase-7, generated by expressing procaspase-7 (D23A) in bacteria (Fig. 1A). As before, cIAP1 readily bound to p19/p12 caspase-7 but failed to bind to the intermediate processed p22/p12 form of caspase-7 (Fig. 1D, lanes 2 and 3). Thus, in summary, cIAP1 utilized different binding mechanisms to associate with caspase-3 and -7; the first required the prodomain of caspase-3, and the second involved a classical IBM that was exposed following removal of the prodomain from caspase-7 (Fig. 1F).
Presence of AKPD in Wild-type Caspase-7 and in Chimeric Caspase-3 Facilitates Only Weak Inhibition by cIAP1—Because cIAP1 does not bind directly to the active site of caspase-3 or -7, there has been some controversy as to its ability to inhibit these effector caspases (17–19). Given that the AKPD motif in caspase-7 is positioned near the active site of the adjacent dimer in the heterotetramer (see Fig. 5 for model), we questioned whether cIAP1 might limit the access of caspase-7 to its substrates. Therefore, we initially examined fully mature WT caspase-3 and -7, as well as each of the chimeric enzymes (described above), for their capacity to cleave the fluorescent substrate DEVD-AMC in the presence and absence of cIAP1 (Fig. 2A). We speculated that this small peptide substrate might gain access to the active site of each enzyme irrespective of cIAP1, and as predicted, cIAP1 failed to inhibit the DEVDase activity of caspase-7, caspase-3, or any of the chimeras tested (Fig. 2A).
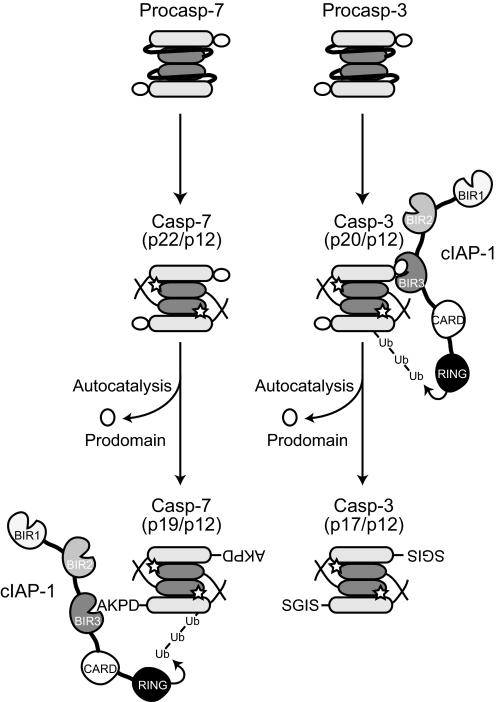
FIGURE 5.
Scheme for regulation of caspase-3 and -7 by cIAP1. Initiator caspases (Casp)-8 and -9 initiate the processing of procaspase-3 and -7 between their large and small subunits, resulting in the formation of p20/p12 caspase-3 and p22/p12 caspase-7, which in turn undergo further processing (via autocatalysis or the action of other proteases) to generate the fully mature p17/p12 caspase-3 and p19/p12 caspase-7, respectively. cIAP1 exhibits remarkable selectivity in that it binds to the intermediate processed p20/p12 caspase-3 (but not p22/p12 caspase-7) and the fully processed p19/p12 caspase-7 (but not p17/p12 caspase-3). Once bound, cIAP1 does not directly inhibit the proteolytic activities caspase-3 and -7, but rather ubiquitinates them and mediates their degradation via the proteasome, thereby suppressing apoptotic cell death.
FIGURE 2.
cIAP1 does not significantly inhibit the proteolytic activities of wild-type or chimeric caspase-3 and -7. A, wild-type and chimeric caspases (Casp)-3 and -7 were incubated in the absence (open circles) or presence (closed circles) of cIAP1, and their proteolytic activities were monitored by measuring the liberation of AMC from the fluorogenic caspase substrate DEVD-AMC. XIAP-BIR1-BIR2 (closed diamonds) was utilized as a positive control. B, expression of GST-tagged BIR1-BIR2 of XIAP and full-length cIAP1. C–I, wild-type and chimeric caspase-3 and -7 were incubated with PARP or p23 cochaperone (Cochap) ± cIAP1 or XIAP-BIR1-BIR2, as described under “Experimental Procedures.” Cleavage of PARP or p23 cochaperone was then visualized by SDS-PAGE/Coomassie Blue staining. Frags, fragments.
Notably, however, in every instance where AKPD was present on the N terminus of the large subunit, the corresponding enzyme exhibited reduced proteolytic activity (Fig. 2A). Indeed, replacement of AKPD in caspase-7 with SGIS resulted in enhanced activity (Fig. 2A, compare the initial velocities of panel a, 17.0 FU/s, with panel b, 29.7 FU/s), whereas replacement of SGIS in caspase-3 with AKPD suppressed activity (Fig. 2A, compare panel e, 33.8 FU/s, with panel f, 13.4 FU/s). By contrast, the presence of SGVD or SGPI on the N termini of the small subunits of caspase-3 and -7 had no effect (Fig. 2A, compare panels c with a and g with e). It is worth noting that previous reports utilizing the same substrate indicate that caspase-3 possesses a kcat/Km value of 1.4 m–1 s–1 compared with 0.063 m–1 s–1 for caspase-7 and that the aldehyde-based inhibitor DEVD-CHO inhibits caspase-3 with an ∼7-fold lower Ki value than caspase-7 (37, 39). Our data suggested that the presence of AKPD in caspase-7 was responsible, at least in part, for its reduced DEVDase activity compared with caspase-3 (Fig. 2A).
Next, to determine whether the presence of AKPD affected the activity of caspase-7 or caspase-3 on a relevant endogenous substrate, or the ability of cIAP1 to inhibit proteolysis, we expressed and purified an N-terminal fragment of human PARP (amino acids 1–373; 49 kDa) containing the caspase-3/7 cleavage site, DEVD214↓G (40, 41). WT caspase-7, SGIS/SGVD caspase-7, WT caspase-3, and AKPD/SGPI caspase-3 were then incubated individually with PARP ± cIAP1 (or the positive control XIAP-BIR1-BIR2; Fig. 2C) and monitored for the processing of PARP over time (note the differing time frames for caspase-3 and -7) (Fig. 2, D–G). Under these conditions, WT caspase-3 exhibited greater proteolytic activity on PARP compared with WT caspase-7; however, removal or addition of AKPD had little effect on the rate of PARP cleavage by SGIS/SGVD caspase-7 or AKPD/SGPI caspase-3 compared with WT caspase-7 or WT caspase-3, respectively (Fig. 2, D and F). Thus, the AKPD motif altered the activity of caspase-7 against the small peptide substrate DEVD-AMC but not against an endogenous substrate containing the same caspase cleavage site.
In the presence of cIAP1, PARP cleavage by AKPD/SGPI caspase-3 was only slightly inhibited compared with WT caspase-3, whereas the inhibition of SGIS/SGVD caspase-7 by cIAP1 was only marginally reduced compared with WT caspase-7 (Fig. 2, E and G). Moreover, cIAP1 had no effect on the activity of p20/p12 caspase-3, the intermediate processed form of the enzyme (Fig. 2H). XIAP-BIR1-BIR2, on the other hand, which occupies the active sites of both caspase-3 and -7, fully inhibited each of the WT and chimeric enzymes (Fig. 2C). Thus, it appears that cIAP1 partially occludes (and weakly inhibits) the access of larger endogenous substrates to the active site of caspase-7 by binding to the AKPD motif located on the N terminus of the large subunit.
Given that PARP is an excellent substrate for both caspase-3 and -7, we still questioned whether the presence of AKPD might alter substrate specificity. We therefore performed cleavage assays with recombinant p23 cochaperone, recently described as a preferred caspase-7 substrate (10). As expected, caspase-7 processed p23 cochaperone more efficiently than did caspase-3; however, the presence of AKPD in WT caspase-7 or AKPD/SGPI caspase-3 had no apparent effect on their proteolytic activities compared with SGIS/SGVD caspase-7 or WT caspase-3, respectively (Fig. 2I). Whether the AKPD motif in caspase-7 restricts or confers activity against other endogenous substrates remains unknown, but at this point it appears unlikely.
cIAP1 Ubiquitinates Distinct Forms of Caspase-3 and -7 and Displays Context-dependent Preferences for E2 Ub-conjugating Enzymes—Despite its ability to inhibit apoptosis, our data suggested that cIAP1 was unlikely to inhibit cell death solely through the weak inhibition of active caspase-3 and -7 (Fig. 2). Nevertheless, cIAP1 is an E3 Ub ligase, and because it bound to caspase-3 and -7, we sought to examine its potential for ubiquitinating these effector caspases. We incubated p19/p12 caspase-7 with cIAP1, in the presence of an E1 Ub-activating enzyme, the E2 Ub-conjugating enzyme UbcH5b, recombinant Ub, and ATP. cIAP1 catalyzed the ubiquitination of the p19 LS of caspase-7 (Fig. 3A, lanes 1 and 2). Interestingly, addition of Z-VAD-fmk (an irreversible inhibitor of caspase-3 and -7) to the incubation enhanced ubiquitination, indicating that cIAP1-dependent ubiquitination of caspase-7 did not involve an interaction between cIAP1 and the active site of caspase-7 and that one or more of the components in the ubiquitination reaction were likely proteolytic substrates for caspase-7 (Fig. 3A, lane 3). When Ub was substituted in the reaction with methyl-Ub, a modified form of Ub that cannot undergo chain extension at Lys-48, eight distinct bands were observed, indicating that the p19 LS of caspase-7 was modified on at least 8 of the available 20 lysine residues (Fig. 3A, lane 6).
FIGURE 3.
Ubiquitination of wild-type and chimeric caspase-3 and -7 by cIAP1. A and B, ubiquitination assays were performed using cIAP1 as the E3 Ub ligase and either processed p19/p12 caspase (Casp)-7, p20/p12 caspase-3, or p17/p12 caspase-3 as the substrate, along with an E1 Ub-activating enzyme, the E2 Ub-conjugating enzyme UbcH5b, Ub, and ATP. Asterisks denote catalytically inactive (C163A) forms of caspase-3. Where indicated, the caspase inhibitor Z-VAD-fmk was added to prevent any potential inactivation of ubiquitinating factors, and methylated-Ub was utilized to determine the number of ubiquitination sites in the large subunits of caspase-3 and -7. Immunoblotting was performed using antibodies to caspase-3 or -7. C and D, UbcH family members were screened in cIAP1-dependent ubiquitination assays of p19/p17 caspase-7 and p20/p12 caspase-3. Immunoblotting was performed using an antibody to caspase-3 or -7 and exposed to film for 1 min. E and F, chimeric caspases-3 and -7 were examined in cIAP1-dependent ubiquitination assays, as described for A and B.
cIAP1 also readily modified the p20 LS of p20/p12 caspase-3 on at least 7 of the available 14 lysine residues (Fig. 3B, compare lanes 2, 4, 6, and 7). For these reactions, p20/p12 caspase-3 was generated by incubating catalytically inactive procaspase-3 (C163A) with active caspase-8. Some of the p20/p12 caspase-3 was also processed between its prodomain and large subunit by caspase-8 to generate smaller amounts of p17/p12 caspase-3 (Fig. 3B, lanes 2, 4, and 5–7). However, it was clear that cIAP1 did not catalyze the ubiquitination of p17/p12 caspase-3, as pure preparations of p17/p12 caspase-3 (generated by expressing WT procaspase-3 in bacteria) were not ubiquitinated even in the presence of Z-VAD-fmk (Fig. 3B, compare lanes 1 and 3; data not shown). Therefore, consistent with its binding profile, cIAP1 ubiquitinated the p20/p12 form of caspase-3 but not the fully processed p17/p12 form of the enzyme.
We also examined several of the E2 Ub-conjugating enzymes for their substrate preferences and found that each member of the UbcH5 subfamily (and to a lesser extent UbcH6 but not UbcH3 or UbcH7) could support the ubiquitination of p17/p12 caspase-7 and p20/p12 caspase-3 (Fig. 3, C and D). In contrast to caspase-7, however, UbcH8 also readily catalyzed the ubiquitination of p20/p12 caspase-3 (Fig. 3D), indicating that in addition to the cleavage status of caspase-3 and -7, the tissue distribution of cIAPs and UbcH family members may well dictate the extent of effector caspase ubiquitination in vivo. Notably, cIAP2 and XIAP also catalyzed the ubiquitination of p19/p12 caspase-7 and p20/p12 caspase-3 (and exhibited similar E2 preferences) but required significantly longer exposure times (>10 min compared with 1 min for cIAP1) to detect ubiquitinated caspases (Fig. 3, C and D, and supplemental Fig. S2, A–D).
Finally, we examined each of the caspase-3/7 chimeric enzymes in our ubiquitination assay using cIAP1 and UbcH5b. A previous report suggested that the RING domain of cIAP2 alone was sufficient to catalyze the ubiquitination of caspase-7 at high concentrations in vitro (42). However, we predicted that cIAP1 would have to bind to caspase-7 through BIR domain-IBM interactions to polyubiquitinate the enzyme. As anticipated, WT caspase-7 and SGVD caspase-7, both of which contained AKPD motifs, underwent robust cIAP1-dependent ubiquitination, whereas SGIS caspase-7 and SGIS/SGVD caspase-7 did not (Fig. 3E). Similarly, catalytically active AKPD caspase-3 and AKPD/SGPI caspase-3 also underwent ubiquitination, whereas WT caspase-3 and SGPI caspase-3, both of which lacked the AKPD motif, failed to do so (Fig. 3F). Therefore, the presence of AKPD was required and sufficient for mediating the cIAP1-dependent interaction and ubiquitination of WT caspase-7 and the AKPD-containing chimeric caspase-3 enzymes.
Procaspase-7 (but Not Procaspase-3) Is Activated following Removal of Its Prodomain and Is Ubiquitinated and Degraded by the Proteasome in an AKPD-dependent Manner—Speculation that IAPs might inhibit apoptosis, at least in part, through ubiquitination and degradation of caspases has existed for some time, with limited evidence to support this supposition. For XIAP, this is understandable, given that it inhibits the activities of caspase-3 and -7 with Ki values in the low nanomolar range (16). Indeed, the direct inhibition of caspases by XIAP, particularly following overexpression, likely masks any antiapoptotic effect that might be mediated by its E3 Ub ligase activity (43). We have demonstrated herein that cIAP1 is a poor inhibitor of caspase-3 and -7, but it does efficiently ubiquitinate them in vitro. Nevertheless, even in the case of cIAPs, proving that ubiquitination of caspases is important in vivo is complicated by the fact that XIAP and cIAPs likely exhibit redundancy (44) and can ubiquitinate multiple cellular targets, any number of which might alter the sensitivity of cells to apoptosis (26). Moreover, mitochondrial release of the IAP antagonists, Smac and Omi, during apoptosis disrupts the interaction of IAPs with caspases (45–47), further complicating efforts to assess the importance of IAP-dependent ubiquitination of caspases in vivo.
Given the aforementioned difficulties, we took another approach to address the importance of ubiquitination and degradation of caspases in cells. We began by generating fusion constructs, in which the prodomains of caspase-3 and -7 were replaced with DHFR, followed by a hemagglutinin tag and monomeric Ub. This DHFR-HA-Ub (hereafter referred to as the “tracer”) was then fused to the large and small subunits of either caspase-3 or caspase-7 (Fig. 4A). Previous studies have shown that Ub fusion proteins are cotranslationally processed by Ub-specific proteases, so that cleavage occurs immediately following the final glycine in Ub (48, 49), which in our case would liberate the tracer and an LS-small subunit single-chain protease containing either the freshly exposed AKPD or SGIS motif. By monitoring the expression level of the tracer, one can also ensure that the initial expression levels of all constructs are comparable.
FIGURE 4.
IBM-dependent degradation of caspase-7 and chimeric AKPD caspase-3 suppresses TRAIL-induced apoptosis. A–D, Ub fusion constructs of AKPD-caspase (Casp)-7, SGIS-caspase-7, AKPD-caspase-3, and SGIS-caspase-3 were transfected into HEK293 cells, and the degradation of caspase-3 or caspase-7 was examined (in the presence or absence of the proteasome inhibitor MG132) by immunoblotting with caspase-3 or caspase-7-specific antibodies to their large subunits and a Myc antibody to their epitope-tagged small subunits (SS). Vec-Ctrl, vector control. In some experiments (C), AKPD-caspase-7 and SGIS-caspase-7 were cotransfected with FLAG-Ub. Total ubiquitinated proteins were then isolated using anti-FLAG M2-agarose beads and immunoblotted with an anti-active caspase-7 antibody. WB, Western blot. *, p < 0.5. E and F, MCF-7 cells were stably transfected with wild-type caspase-3 or AKPD caspase-3 constructs and treated with TRAIL (500 ng/ml) for 1–2 h. The cells were subsequently washed, replaced with fresh medium for 22 h, and examined for cell viability by flow cytometry. Caspase-3 processing was also assessed at indicated time points following TRAIL treatment (500 ng/ml; 2 h). G, wild-type and AKPD caspase-3 processing in bacteria was examined at indicated time points following induction with isopropyl β-d-thiogalactopyranoside (IPTG).
As expected, upon expression of these constructs in HEK293 cells, the tracers were efficiently removed from each of the caspase-3 and caspase-7 proteins, indicating that all constructs were expressed at equivalent levels (Fig. 4, B and D). Interestingly, however, only the AKPD caspase-7 and chimeric SGIS caspase-7 constructs underwent spontaneous cleavage between their large and small subunits to generate the fully mature p19/p12 caspase-7 enzymes (Fig. 4B). The SGIS caspase-3 and chimeric AKPD caspase-3 constructs remained uncleaved as single-chain proteins (ΔproCasp3) (Fig. 4D). Thus, consistent with previous reports, removal of the prodomain from caspase-7 (but not caspase-3) was sufficient to induce its complete processing and activation (8). More importantly, once formed in cells, both the p19 and p12 subunits of AKPD caspase-7 (i.e. WT caspase-7) were degraded in a proteasome-dependent manner, as pretreatment of cells with MG132 prevented the loss of both subunits (Fig. 4B, compare lanes 2 and 5). By contrast, neither subunit of SGIS caspase-7 was degraded (Fig. 4B, compare AKPD caspase-7 in lane 2 with SGIS caspase-7 in lane 3), indicating that the presence of the AKPD motif was essential for the turnover of active caspase-7. Notably, the single-chain version of AKPD caspase-7 (ΔproCasp7), which had yet to undergo processing between its large and small subunits, was not readily degraded in cells, indicating that cleavage between these two subunits was required for efficient binding to and ubiquitination by IAPs. Finally, to confirm that AKPD caspase-7 was selectively ubiquitinated in vivo, we also cotransfected AKPD caspase-7 and SGIS caspase-7 with a FLAG-Ub construct and isolated total ubiquitinated proteins using anti-FLAG M2 affinity gel. As expected, when the precipitates were immunoblotted with an antibody to cleaved caspase-7, only AKPD caspase-7 was readily ubiquitinated (Fig. 4C).
Wild-type Caspase-3 Is More Potent than Chimeric AKPD Caspase-3 during TRAIL-induced Cell Death—Because the DHFR-HA-Ub-caspase-3 fusion proteins did not undergo spontaneous cleavage between their large and small subunits following removal of their tracers, we were unable to determine whether processed caspase-3 underwent ubiquitination and degradation in vivo (Fig. 4D). We therefore generated stable MCF-7 cell lines expressing WT caspase-3 or AKPD caspase-3 (Fig. 4, E and F). MCF-7 cells were chosen because they do not express caspase-3 and thus exhibit resistance to certain proapoptotic stimuli (50). These cells were then incubated with TRAIL for 1–2 h, after which the ligand was removed, and the cells were incubated in fresh medium for an additional 22 h. As predicted, TRAIL-induced cell death was enhanced dramatically in cells expressing WT caspase-3 versus the vector control plasmid (Fig. 4E). However, there was a significant reduction in the level of cell death in cells expressing the chimeric AKPD caspase-3 compared with those expressing WT caspase-3, and correspondingly, levels of the processed p17 large subunit were largely absent from the AKPD caspase-3-transfected cells, even though the levels of procaspase-3 were clearly diminished (Fig. 4, E and F). Thus, similar to WT caspase-7, the presence of AKPD in chimeric caspase-3 mediated its turnover in vivo and consequently suppressed cell death induced by TRAIL.
Attempts to inhibit the degradation of the p17 large subunit in caspase-3 with MG132 were not possible, as the combination of MG132 with TRAIL dramatically increased the overall level of caspase processing and cell death (data not shown). Therefore, to verify that swapping the SGIS motif for AKPD in WT caspase-3 did not merely inhibit the autocatalytic removal of its prodomain, we expressed full-length WT and AKPD caspase-3 in bacteria. As shown in Fig. 4G, WT caspase-3 and AKPD caspase-3 underwent time-dependent activation, and the subsequent autocatalytic conversion from their p20/p12 to their fully mature p17/p12 forms occurred with essentially identical kinetics. Thus, the presence of AKPD in chimeric caspase-3 did not inhibit the autocatalytic removal of its prodomain, consistent with the proposed role for caspase-3 in removing the prodomain of caspase-7 during apoptosis (8).
DISCUSSION
Here we have demonstrated that cIAP1 binds but does not inhibit the proteolytic activities of caspase-3 and -7 (Fig. 2), consistent with a recent report (17). Moreover, we have provided the strongest evidence to date that effector caspases are in fact ubiquitinated and degraded in vivo in an IBM-dependent manner (Figs. 3 and 4). Perhaps our most striking observations, however, were that cIAP1 bound to caspase-3 and -7 at distinct steps prior to or following the removal of their prodomains, respectively (Fig. 1 and Fig. 5). Indeed, cleavage of their activation loops was required for both caspases to bind cIAP1. However, for caspase-7, removal of its prodomain was also essential, as this resulted in the exposure of an AKPD motif on the N terminus of its large subunit, whereas the intermediate processed p20/p12 form of caspase-3 readily bound to cIAP1, but failed to do so following autocatalytic removal of its prodomain (Figs. 1 and 5). Although it remains unclear precisely how cIAP1 binds to p20/p12 caspase-3, it is worth noting that the BIR2 domain in Drosophila IAP1 (DIAP1) binds to the initiator caspase DRONC (homolog of caspase-9) by binding to the flexible linker between the prodomain and large subunit of the unprocessed enzyme (51, 52). Regardless, the unique differences in the binding mechanisms for cIAP1 to caspase-3 and -7 raise a number of intriguing questions. For example, what is the point in having an IAP that binds effector caspases but does not directly inhibit their activities? Moreover, why would cIAP1 bind to two effector caspases that are highly homologous through such unique mechanisms and at distinct steps in their activation?
The answers to these questions and others remain speculative; however, there are some provocative possibilities. For example, it is clear that active caspase-3 plays a number of nonapoptotic roles in everything from embryonic and neural stem cell differentiation, macrophage differentiation, and lens cell differentiation to erythropoiesis, sometimes through cleavage of specific transcription factors (53–59). The activity of caspase-3 in these processes must be sequestered, limited, and controlled to prevent the onslaught of full-blown apoptosis, but it remains unclear how this is achieved. Notably, cIAPs localize in part to the nucleus (60, 61) and thus might perform such a task. But even if this were true, why would cIAP1 bind to only the intermediate p20/p12 caspase-3 and not the fully mature form? We and others have shown that p20/p12 caspase-3 is catalytically active and normally accumulates in apoptotic cells following the initial cleavage of procaspase-3 within its activation loop by initiator caspases. At some point, however, when sufficient p20/p12 caspase-3 is present within the cell, this intermediate form undergoes autocatalytic removal of its prodomain to generate the fully mature p17/p12 caspase-3. This suggests that by specifically targeting the p20/p12 form of caspase-3, cIAP1 might regulate, in a spatiotemporal manner, the amount of p20/p12 caspase-3 present within a given compartment, such as the nucleus. In doing so, cIAP1 could allow p20/p12 caspase-3 to carry out its nonapoptotic roles by regulating either the number and/or the extent to which certain substrates are processed. Under conditions of a strong apoptotic stimulus, however, p20/p12 caspase-3 would accumulate too rapidly to be adequately controlled by cIAP1, thereby resulting in the autocatalytic conversion of p20/p12 caspase-3 to a form of active caspase-3 that is no longer subject to regulation by cIAP1. In instances where cIAPs are overexpressed, such as in many cancers, they might be expected to inhibit not only apoptosis but also the nonapoptotic roles of caspase-3, which as noted above, often involve the activation of differentiation pathways.
With regard to caspase-7, our findings were similarly thought-provoking. In the experiments involving the Ub fusion constructs of caspase-3 and -7, we found that cotranslational removal of the tracer from ΔproCasp7 resulted in apparently spontaneous processing between its large and small subunits, whereas removal of the tracer from ΔproCasp3 failed to undergo processing (Fig. 4, A–C). Importantly, Denault and Salvesen (8) previously reported that removal of the prodomain from caspase-7 was required for its efficient activation by initiator caspases in vivo (but not in vitro), leading them to speculate that procaspase-7 may be sequestered via its prodomain to a cytosolic location inaccessible to initiator caspases. If so, this may explain why cIAP1 fails to bind the p22/p12 form of caspase-7, which in our hands was generated in bacteria by expressing the D23A mutant of caspase-7 (Fig. 1D). Indeed, if removal of the prodomain of caspase-7 in apoptotic cells precedes cleavage of its activation loop by initiator caspases, then this intermediate would rarely (if ever) be formed in cells, and thus, there would be no reason for cIAP1 to bind this form of caspase-7.
In agreement with Tenev et al. (18), we found instead that cIAP1 bound to an AKPD motif, on the N terminus of the large subunit of caspase-7, that was exposed following the removal of its prodomain. More importantly, we observed that AKPD caspase-7, but not the chimeric SGIS caspase-7, underwent ubiquitination and proteasome-dependent degradation in vivo and that placement of this AKPD motif in caspase-3 suppressed its ability to execute the apoptotic program following treatment with the death ligand TRAIL (Fig. 4, A–E). Consistent with our results, Denault and Salvesen (8) found that transfection of a pro-domainless version of caspase-7 was highly toxic to cells, and it is notable that Ala-24 was substituted with a methionine in their construct, resulting in an MKPD motif that almost certainly would have disrupted its interaction with cIAP1 (1, 8). As to why cIAP1 might regulate caspase-7 in the first place, once again there is evidence that caspase-7 participates in nonapoptotic pathways. Caspase-7 has recently been identified as a target of the caspase-1 inflammasome (62), and it proteolytically activates the cytokine, endothelium-monocyte-activating polypeptide II (63).
Finally, as discussed previously, XIAP suppresses apoptosis in large part by directly inhibiting the effector caspase-3 and -7, and this inhibition is relieved upon the release of IAP antagonists, such as Smac/DIABLO and Omi/HtrA2, from mitochondria (1). Unfortunately, many cancers overexpress antiapoptotic Bcl-2 family members, rendering them resistant to MOMP. Therefore, a number of groups have recently developed cell-permeable Smac mimetics to antagonize XIAP, particularly in cancer cells resistant to MOMP. Surprisingly, however, Smac mimetics were found to induce apoptosis by stimulating the autoubiquitination and degradation of cIAPs. Depletion of cIAPs, in turn, initiated a paracrine-mediated TNF-dependent cell death pathway (31–34). Nevertheless, because TNF induces apoptosis via the sequential activation of the initiator caspase-8 and the effector caspase-3 and -7, disruption of IAP-caspase interactions remains a critical step in death receptor-mediated cell death (64). Based upon our data, cIAP1 does not directly inhibit caspase-3 and -7, but does bind and ubiquitinate these caspases and targets them for destruction via the proteasome. Thus, the degradation of cIAPs by Smac mimetics likely promotes cell death by simultaneously initiating a TNF-dependent cell death pathway and suppressing the degradation of active effector caspases.
Supplementary Material
Acknowledgments
We thank Prof. Aaron Ciechanover, Prof. Hungwen Liu, Prof. Seamus J. Martin, and Prof. Alexander Varshavsky for generously providing plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant CA129521 from NCI (to S. B. B.). This work was also supported by American Cancer Society Grant RSG-05-029-01-CCG and PhRMA Foundation (to S. B. B.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: caspase, cysteinyl aspartate-specific protease; BIR, baculovirus inhibitor of apoptosis repeat; cIAP1, cellular inhibitor of apoptosis 1; cIAP2, cellular inhibitor of apoptosis 2; DHFR, dihydrofolate reductase; IBM, inhibitor of apoptosis binding motif; Ub, ubiquitin; Ubc, ubiquitin-conjugating enzyme; PARP, poly(ADP-ribose)polymerase; XIAP, X-linked inhibitor of apoptosis; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; DTT, dithiothreitol; PBS, phosphate-buffered saline; GST, glutathione S-transferase; HA, hemagglutinin; WT, wild type; PIPES, 1,4-piperazinediethanesulfonic acid; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; TNF, tumor necrosis factor; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; MOMP, mitochondrial outer membrane permeabilization; LS, large subunit; AMC, 7-amino-4-methylcoumarin; FU, fluorescence unit.
References
- 1.Fuentes-Prior, P., and Salvesen, G. S. (2004) Biochem. J. 384 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bratton, S. B., MacFarlane, M., Cain, K., and Cohen, G. M. (2000) Exp. Cell Res. 256 27–33 [DOI] [PubMed] [Google Scholar]
- 3.Bratton, S. B., Walker, G., Srinivasula, S., Sun, X.-M., Butterworth, M., Alnemri, E. S., and Cohen, G. M. (2001) EMBO J. 20 998–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez, J., and Lazebnik, Y. (1999) Genes Dev. 13 3179–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pop, C., Timmer, J., Sperandio, S., and Salvesen, G. S. (2006) Mol. Cell 22 269–275 [DOI] [PubMed] [Google Scholar]
- 6.Chao, Y., Shiozaki, E. N., Srinivasula, S. M., Rigotti, D. J., Fairman, R., and Shi, Y. (2005) PLoS Biol. 3 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han, Z., Hendrickson, E. A., Bremner, T. A., and Wyche, J. H. (1997) J. Biol. Chem. 272 13432–13436 [DOI] [PubMed] [Google Scholar]
- 8.Denault, J. B., and Salvesen, G. S. (2003) J. Biol. Chem. 278 34042–34050 [DOI] [PubMed] [Google Scholar]
- 9.Zhou, Q., and Salvesen, G. S. (1997) Biochem. J. 324 361–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh, J. G., Cullen, S. P., Sheridan, C., Luthi, A. U., Gerner, C., and Martin, S. J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 12815–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deveraux, Q. L., and Reed, J. C. (1999) Genes Dev. 13 239–252 [DOI] [PubMed] [Google Scholar]
- 12.Srinivasula, S. M., Hegde, R., Saleh, A., Datta, P., Shiozaki, E., Chai, J., Lee, R. A., Robbins, P. D., Fernandes-Alnemri, T., Shi, Y., and Alnemri, E. S. (2001) Nature 410 112–116 [DOI] [PubMed] [Google Scholar]
- 13.Shiozaki, E. N., Chai, J., Rigotti, D. J., Riedl, S. J., Li, P., Srinivasula, S. M., Alnemri, E. S., Fairman, R., and Shi, Y. (2003) Mol. Cell 11 519–527 [DOI] [PubMed] [Google Scholar]
- 14.Riedl, S. J., Renatus, M., Schwarzenbacher, R., Zhou, Q., Sun, C., Fesik, S. W., Liddington, R. C., and Salvesen, G. S. (2001) Cell 104 791–800 [DOI] [PubMed] [Google Scholar]
- 15.Chai, J., Shiozaki, E., Srinivasula, S. M., Wu, Q., Datta, P., Alnemri, E. S., and Shi, Y. (2001) Cell 104 769–780 [DOI] [PubMed] [Google Scholar]
- 16.Scott, F. L., Denault, J. B., Riedl, S. J., Shin, H., Renatus, M., and Salvesen, G. S. (2005) EMBO J. 24 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckelman, B. P., and Salvesen, G. S. (2006) J. Biol. Chem. 281 3254–3260 [DOI] [PubMed] [Google Scholar]
- 18.Tenev, T., Zachariou, A., Wilson, R., Ditzel, M., and Meier, P. (2005) Nat. Cell Biol. 7 70–77 [DOI] [PubMed] [Google Scholar]
- 19.Roy, N., Deveraux, Q. L., Takahashi, R., Salvesen, G. S., and Reed, J. C. (1997) EMBO J. 16 6914–6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu, Z. L., McKinsey, T. A., Liu, L., Gentry, J. J., Malim, M. H., and Ballard, D. W. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10057–10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, C. Y., Mayo, M. W., Korneluk, R. G., Goeddel, D. V., and Baldwin, A. S., Jr. (1998) Science 281 1680–1683 [DOI] [PubMed] [Google Scholar]
- 22.Zender, L., Spector, M. S., Xue, W., Flemming, P., Cordon-Cardo, C., Silke, J., Fan, S. T., Luk, J. M., Wigler, M., Hannon, G. J., Mu, D., Lucito, R., Powers, S., and Lowe, S. W. (2006) Cell 125 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keats, J. J., Fonseca, R., Chesi, M., Schop, R., Baker, A., Chng, W. J., Van Wier, S., Tiedemann, R., Shi, C. X., Sebag, M., Braggio, E., Henry, T., Zhu, Y. X., Fogle, H., Price-Troska, T., Ahmann, G., Mancini, C., Brents, L. A., Kumar, S., Greipp, P., Dispenzieri, A., Bryant, B., Mulligan, G., Bruhn, L., Barrett, M., Valdez, R., Trent, J., Stewart, A. K., Carpten, J., and Bergsagel, P. L. (2007) Cancer Cell 12 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai, Z., Zhu, W. G., Morrison, C. D., Brena, R. M., Smiraglia, D. J., Raval, A., Wu, Y. Z., Rush, L. J., Ross, P., Molina, J. R., Otterson, G. A., and Plass, C. (2003) Hum. Mol. Genet. 12 791–801 [DOI] [PubMed] [Google Scholar]
- 25.Imoto, I., Tsuda, H., Hirasawa, A., Miura, M., Sakamoto, M., Hirohashi, S., and Inazawa, J. (2002) Cancer Res. 62 4860–4866 [PubMed] [Google Scholar]
- 26.Salvesen, G. S., and Duckett, C. S. (2002) Nat. Rev. Mol. Cell Biol. 3 401–410 [DOI] [PubMed] [Google Scholar]
- 27.Silke, J., Kratina, T., Chu, D., Ekert, P. G., Day, C. L., Pakusch, M., Huang, D. C., and Vaux, D. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373–428 [DOI] [PubMed] [Google Scholar]
- 29.Li, X., Yang, Y., and Ashwell, J. D. (2002) Nature 416 345–347 [DOI] [PubMed] [Google Scholar]
- 30.Hu, S., and Yang, X. (2003) J. Biol. Chem. 278 10055–10060 [DOI] [PubMed] [Google Scholar]
- 31.Wang, L., Du, F., and Wang, X. (2008) Cell 133 693–703 [DOI] [PubMed] [Google Scholar]
- 32.Bertrand, M. J. M., Milutinovic, S., Dickson, K. M., Ho, W. C., Boudreault, A., Durkin, J., Gillard, J. W., Jaquith, J. B., Morris, S. J., and Barker, P. A. (2008) Mol. Cell 30 689–700 [DOI] [PubMed] [Google Scholar]
- 33.Varfolomeev, E., Blankenship, J. W., Wayson, S. M., Fedorova, A. V., Kayagaki, N., Garg, P., Zobel, K., Dynek, J. N., Elliott, L. O., Wallweber, H. J., Flygare, J. A., Fairbrother, W. J., Deshayes, K., Dixit, V. M., and Vucic, D. (2007) Cell 131 669–681 [DOI] [PubMed] [Google Scholar]
- 34.Vince, J. E., Wong, W. W., Khan, N., Feltham, R., Chau, D., Ahmed, A. U., Benetatos, C. A., Chunduru, S. K., Condon, S. M., McKinlay, M., Brink, R., Leverkus, M., Tergaonkar, V., Schneider, P., Callus, B. A., Koentgen, F., Vaux, D. L., and Silke, J. (2007) Cell 131 682–693 [DOI] [PubMed] [Google Scholar]
- 35.Suzuki, Y., Nakabayashi, Y., and Takahashi, R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, L., Smith, L., Wang, Z., and Smith, J. B. (2003) Mol. Pharmacol. 64 334–345 [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Calvo, M., Peterson, E. P., Rasper, D. M., Vaillancourt, J. P., Zamboni, R., Nicholson, D. W., and Thornberry, N. A. (1999) Cell Death Differ. 6 362–369 [DOI] [PubMed] [Google Scholar]
- 38.Stennicke, H. R., and Salvesen, G. S. (1999) Methods (San Diego) 17 313–319 [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Calvo, M., Peterson, E. P., Leiting, B., Ruel, R., Nicholson, D. W., and Thornberry, N. A. (1998) J. Biol. Chem. 273 32608–32613 [DOI] [PubMed] [Google Scholar]
- 40.Lazebnik, Y. A., Kaufmann, S. H., Desnoyers, S., Poirier, G. G., and Earnshaw, W. C. (1994) Nature 371 346–347 [DOI] [PubMed] [Google Scholar]
- 41.Germain, M., Affar, E. B., D'Amours, D., Dixit, V. M., Salvesen, G. S., and Poirier, G. G. (1999) J. Biol. Chem. 274 28379–28384 [DOI] [PubMed] [Google Scholar]
- 42.Huang, H., Joazeiro, C. A., Bonfoco, E., Kamada, S., Leverson, J. D., and Hunter, T. (2000) J. Biol. Chem. 275 26661–26664 [DOI] [PubMed] [Google Scholar]
- 43.Bratton, S. B., Lewis, J., Butterworth, M., Duckett, C. S., and Cohen, G. M. (2002) Cell Death Differ. 9 881–892 [DOI] [PubMed] [Google Scholar]
- 44.Rumble, J. M., Bertrand, M. J., Csomos, R. A., Wright, C. W., Albert, L., Mak, T. W., Barker, P. A., and Duckett, C. S. (2008) Biochem. J. 415 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du, C., Fang, M., Li, Y., Li, L., and Wang, X. (2000) Cell 102 33–42 [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, Y., Imai, Y., Nakayama, H., Takahashi, K., Takio, K., and Takahashi, R. (2001) Mol. Cell 8 613–621 [DOI] [PubMed] [Google Scholar]
- 47.Verhagen, A. M., Ekert, P. G., Pakusch, M., Silke, J., Connolly, L. M., Reid, G. E., Moritz, R. L., Simpson, R. J., and Vaux, D. L. (2000) Cell 102 43–53 [DOI] [PubMed] [Google Scholar]
- 48.Turner, G. C., and Varshavsky, A. (2000) Science 289 2117–2120 [DOI] [PubMed] [Google Scholar]
- 49.Hunter, A. M., Kottachchi, D., Lewis, J., Duckett, C. S., Korneluk, R. G., and Liston, P. (2003) J. Biol. Chem. 278 7494–7499 [DOI] [PubMed] [Google Scholar]
- 50.Janicke, R. U., Sprengart, M. L., Wati, M. R., and Porter, A. G. (1998) J. Biol. Chem. 273 9357–9360 [DOI] [PubMed] [Google Scholar]
- 51.Chai, J., Yan, N., Huh, J. R., Wu, J. W., Li, W., Hay, B. A., and Shi, Y. (2003) Nat. Struct. Biol. 10 892–898 [DOI] [PubMed] [Google Scholar]
- 52.Meier, P., Silke, J., Leevers, S. J., and Evan, G. I. (2000) EMBO J. 19 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujita, J., Crane, A. M., Souza, M. K., Dejosez, M., Kyba, M., Flavell, R. A., Thomson, J. A., and Zwaka, T. P. (2008) Cell Stem Cell 2 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernando, P., Brunette, S., and Megeney, L. A. (2005) FASEB J. 19 1671–1673 [DOI] [PubMed] [Google Scholar]
- 55.Netea, M. G., Lewis, E. C., Azam, T., Joosten, L. A., Jaekal, J., Bae, S. Y., Dinarello, C. A., and Kim, S. H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3515–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zandy, A. J., Lakhani, S., Zheng, T., Flavell, R. A., and Bassnett, S. (2005) J. Biol. Chem. 280 30263–30272 [DOI] [PubMed] [Google Scholar]
- 57.Weber, G. F., and Menko, A. S. (2005) J. Biol. Chem. 280 22135–22145 [DOI] [PubMed] [Google Scholar]
- 58.Ishizaki, Y., Jacobson, M. D., and Raff, M. C. (1998) J. Cell Biol. 140 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krauss, S. W., Lo, A. J., Short, S. A., Koury, M. J., Mohandas, N., and Chasis, J. A. (2005) Blood 106 2200–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samuel, T., Okada, K., Hyer, M., Welsh, K., Zapata, J. M., and Reed, J. C. (2005) Cancer Res. 65 210–218 [PubMed] [Google Scholar]
- 61.Vischioni, B., Giaccone, G., Span, S. W., Kruyt, F. A., and Rodriguez, J. A. (2004) Exp. Cell Res. 298 535–548 [DOI] [PubMed] [Google Scholar]
- 62.Lamkanfi, M., Kanneganti, T. D., Van Damme, P., Vanden Berghe, T., Vanoverberghe, I., Vandekerckhove, J., Vandenabeele, P., Gevaert, K., and Nunez, G. (2008) Mol. Cell. Proteomics 7 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shalak, V., Kaminska, M., Mitnacht-Kraus, R., Vandenabeele, P., Clauss, M., and Mirande, M. (2001) J. Biol. Chem. 276 23769–23776 [DOI] [PubMed] [Google Scholar]
- 64.Bratton, S. B., and Cohen, G. M. (2003) Cell Death Differ. 10 4–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.