Abstract
Glucocorticoid (GC) is known to affect the reproductive system by suppressing the gonadotropin-releasing hormone (GnRH) gene expression in the hypothalamus. However, the mechanism of this effect is poorly understood. We show here that the GC-induced reduction of GnRH mRNA is due to attenuation of a post-transcriptional process i.e. splicing of intron A. Treatment of dexamethasone (DEX), a synthetic GC, lowered GnRH mRNA transcripts and was accompanied by reduced excision of the first intron (intron A) from the GnRH pre-mRNA both in vitro and in vivo. While seeking to identify the splicing factors involved in GC-inhibited GnRH pre-mRNA splicing, we found that DEX down-regulated neuro-oncological ventral antigen-1 (Nova-1) mRNA and protein and that knockdown of Nova-1 reduced intron A excision from GnRH pre-mRNA. Nova-1 overexpression reversed the DEX-induced reduction of intron A excision. Nova-1 appears to promote intron A excision by binding to the distal region of exon 1 of the GnRH pre-mRNA. Taken together, our findings indicate that the intron A excision by Nova-1 is a target of GC for down-regulation of GnRH gene expression, and more importantly, we characterized Nova-1, a brain-enriched splicing regulator responsible for GnRH pre-mRNA splicing.
Many studies have demonstrated that stress affects neuroendocrine systems. Accumulating evidence points to cross-talk between the hypothalamic-pituitary-adrenal axis and the hypothalamic-pituitary-gonadal axis (1, 2). Physical or mental stress activates the hypothalamic-pituitary-adrenal axis, consisting of corticotropin-releasing hormone, adrenocorticotropic hormone (ACTH),3 and glucocorticoid (GC), and the increased GC secretion from the adrenal gland in turn suppresses activity of the hypothalamic-pituitary-gonadal axis (3, 4). GC exerts its action on the hypothalamus, where it interferes with discharge of gonadotropin-releasing hormone (GnRH) (5). Down-regulation of GnRH production by stress leads to functional disorders of sexual development and reproduction (6, 7). For instance, administration of the synthetic GC homolog, dexamethasone (DEX), decreases GnRH mRNA levels, probably by lowering GnRH promoter activity (8, 9). This inhibitory effect of GC on GnRH gene expression and secretion may be mediated by the GC receptor (GR), which is expressed in GnRH neurons of the hypothalamus and in GnRH-producing GT1 cells (8). Recently, DeFranco and co-workers (10) identified a negative GC response element within the mouse GnRH promoter to which GR binds in association with the DNA-binding transcription factor Oct-1. Thus, it is likely that GC regulates GnRH gene expression at the transcriptional level in both the hypothalamus and GT1 cells (10). However, whether GC-mediated down-regulation of GnRH gene expression is associated with a post-transcriptional process (i.e. attenuation of splicing of the GnRH pre-mRNA) is currently unknown.
Post-transcriptional control of the GnRH pre-mRNA splicing is recognized as an important factor in the cell type- and developmental stage-specific expression of GnRH (11, 12), and a subset of SR and SR-related proteins thus far has been shown to be involved in this process (13, 14). Excision of the first intron (intron A) of GnRH pre-mRNA is known to be a rate-limiting step in the production of mature GnRH RNA. Intron A is a weak intron, and its 5′ splice site score is 4.8 (maximum of 12.6 and mean of 8.1), and the 3′ splice site score is 3.7 (maximum of 14.2 and mean of 7.9) according to the splice site score calculation program (available on the World Wide Web) and since it contains tandem repeats of upstream ATG sequences that interfere with proper translation of the mRNA into the GnRH precursor peptide (11, 15). Thus, it is desirable to establish whether GC suppresses GnRH pre-mRNA splicing in addition to GnRH promoter activity and, if so, which splicing factor(s) is involved in the process.
In this study, we demonstrate that GC-evoked down-regulation of GnRH gene expression can occur at the level of GnRH pre-mRNA splicing. More importantly, we determined a target splicing regulator of GC; DEX appears to inhibit splicing by preventing expression of the neuron-specific RNA splicing factor, neuro-oncological ventral antigen 1 (Nova-1), which stimulates intron A excision by binding to exon 1 of the GnRH pre-mRNA.
EXPERIMENTAL PROCEDURES
Reagents—All cell culture materials were obtained from Invitrogen. DEX and DEX 21-phosphate were purchased from Sigma, and when not specified, other chemicals were also from Sigma.
Cell Culture and Transient Transfection—GT1-1 and NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 4 mm glutamine, 1 mm sodium pyruvate, 100 units/ml penicillin/streptomycin, and 10% fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37 °C. The cells were plated in 6-well plates and grown to 60–80% confluence in 1 day. For transfection experiments, cells were washed twice with Dulbecco's phosphate-buffered saline, and the medium was changed before transfection to serum- and antibiotic-free Dulbecco's modified Eagle's medium. Plasmids for transfection experiments were purified with Qiagen columns according to the manufacturer's instructions and dissolved in TE buffer (1 mm Tris, pH 8.0, 0.1 mm EDTA). The cells were transfected with plasmid DNA using Lipofectamine PLUS reagent (Invitrogen). Excess DNA complexes were washed out with Dulbecco's phosphate-buffered saline the next day, and complete Dulbecco's modified Eagle's medium was added. After 48 h of recovery in normal medium, cells were subjected to the indicated treatments and/or analyses.
Northern Blot Hybridization—Total RNA isolation and Northern blotting were carried out as described previously with minor modifications (15). Briefly, 30 μg of each RNA was resolved on a 1.2% formaldehyde-agarose gel and transferred for 16 h by diffusion blotting to a Nytran filter (pore size 0.45 μm; Schleicher & Schuell). cDNA probe complementary to GnRH mRNA or intron A was generated by a random priming method in the presence of [α-32P]dCTP.
Plasmid Constructions and RT-PCR Analysis—The primers used for competitive RT-PCR were as follows: E1-up, 5′-GGAAGACATCAGTGTCCCAGA-3′; IA-up, 5′-TACCTCTGCAGTTTCTGTGA-3′; E3-dn, 5′-GAAGTGCTGGGGTTCTGC-3′; GFP-dn, 5′-GTCCAGCTCGACCAGGAT-3′. Fragments of the middle part of the splicing regulator gene lacking the RS domain coding sequence were subcloned into the truncated form of EGFP in pEGFP-N1 (BD Biosciences), the pEGFP-N1-ΔGFP vector, or pcDNA3.1 (Invitrogen) in the antisense direction, generating the antisense expression plasmids pEGFP-N1-ΔGFP-AS-CELF6, pEGFP-N1-ΔGFP-AS-HuC, pEGFP-N1-ΔGFP-AS-KSRP, pEGFP-N1-ΔGFP-AS-NAPOR-1, pEGFP-N1-ΔGFP-AS-Nova-1, and pEGFP-N1-ΔGFP-AS-SWAP. The primers used for cloning antisense expression constructs and semiquantitative RT-PCR are shown in Table S1. Mutation of the p1A234-GFP reporter construct was carried out with a QuikChange site-directed mutagenesis kit (Stratagene) and specifically designed primers. Total RNA was prepared from the cells, and to eliminate possible DNA contamination, RNA samples of 3 μg were further treated with 10 units of DNase I (Roche Applied Science) for 30 min at 37 °C. The DNase-treated RNA was heated for 10 min at 75 °C and reverse-transcribed using random hexamers; the resulting cDNA was amplified by PCR, as previously described (14). The primer sequences used as an internal control for semiquantitative RT-PCR were as follows: HPRT-up, 5′-CCTGCTGGATTACATTAAAGCACTG-3′; HPRT-dn, 5′-CTTCGTGGGGTCCTTTCACCAGC-3′. The full-length nova-1 cDNA was subcloned into pCMV-Tag3 (Stratagene) to express it in a Myc-tagged form. The primer sequences used were as follows: Nova-1-cds-up, 5′-AAGCTTATGATGGCGGCAGCTCCC-3′; Nova-1-cds-dn, 5′-CTCGAGTCAACCCACTTTCTGAGG-3′.
Western Blot Analysis—For immunoblotting, nuclear proteins were resolved on 8% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore). The nuclear extracts were prepared as described previously (11). Target proteins were detected with anti-Nova-1 (Upstate Biotechnology, Inc., Lake Placid, NY) and anti-lamin B1 (M-20; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies. The immune complexes were visualized with an ECL detection kit (Pierce).
Preparation of Glutathione S-Transferase (GST)-tagged Nova-1 Protein—GST-tagged Nova-1 cloned in baculovirus transfer vector (BD Biosciences) was used to produce recombinant baculoviruses. The fusion proteins were expressed in Sf21 cells. GST-Nova-1 protein was affinity-purified with glutathione-Sepharose 4 Fast Flow according to the manufacturer's instructions (GE Healthcare).
In Vitro Splicing Assay—The in vitro splicing reactions were performed as described previously (11). Briefly, the gel-purified radiolabeled RNA substrates produced by in vitro transcription were subjected to the splicing reaction in 25 μl of reaction mixture containing 5 mm HEPES, pH 7.9, 20 mm creatine phosphate, 0.4 mm ATP, 0.6% polyvinylalcohol, 3 mm MgCl2, and 20 units of RNase inhibitor, supplemented with 50 μg of cytoplasmic S100 extract, at 30 °C for 2 h. After phenol/chloroform extraction and subsequent ethanol precipitation, the RNAs were separated on a 6% denaturing polyacrylamide gel containing 8 m urea. The gels were dried and autoradiographed at –70 °C for 1 day.
UV Cross-linking Assay—RNA-protein binding reactions were performed in reaction mixtures containing 0.4 mm ATP, 20 mm creatine phosphate, 3 mm MgCl2, 20 units of RNase inhibitor, 5 μg of yeast tRNA, 32P-labeled RNA probe, and purified proteins for 30 min at 30 °C as previously described with minor modifications (13). UV cross-linking was performed on ice, 4.5 cm away from a UV source at 1.2 J (Stratagene). Each sample was then incubated with 200 units of RNase T1 and 200 units of RNase A for 10 min at 37 °C. The resulting RNA-protein complexes were denatured and resolved by SDS-PAGE. The gels were dried and autoradiographed at –70 °C for 1 day.
Electrophoretic Mobility Shift Assay—Electrophoretic mobility shift assays were carried out as previously described (13). RNA-protein binding reactions took place in 20-μl volumes containing 0.4 mm ATP, 20 mm creatine phosphate, 3 mm MgCl2, 20 units of RNase inhibitor, 5 μg of yeast tRNA, 32P-labeled RNA probe, and the indicated GST-Nova-1 recombinant protein. After 30 min at 30 °C, 6 μl of glycerol was added to the reaction mixtures, which were immediately fractionated on 5% nondenaturing polyacrylamide gels. These were dried and exposed to x-ray film (Fuji) at –70 °C for 1 day.
Animals and Drug Administration—Adult male C57BL/6J mice, 6 weeks of age, were purchased from the Institute of Laboratory Animal Resources of Seoul National University and used in all experiments. They were kept in temperature-controlled (22–23 °C) quarters under a 12-h light and dark photo-period (light on at 8:00 a.m.) with standard mouse chow and water available ad libitum. The mice (4 per group) were injected intraperitoneally with 4 mg/kg of DEX 21-phosphate or phosphate-buffered saline at 9:00–10:00 a.m. on 14 days in succession. The mice were sacrificed 12 h after the last injection, and their brains were removed on ice. Hypothalamus and cerebral cortex were dissected and quickly frozen in liquid nitrogen. All animal experiments were approved by the Animal Care and Use Committee of Seoul National University.
Real Time RT-PCR Assay—The primers used for real time RT-PCR were as follows: E1-RT-up, 5′-CAACCTACCAACGGAAGCTC-3′; E2-RT-dn, 5′-GCCTTCCAAACACACAGTCA-3′; Nova-1-RT-up, 5′-ACCACCAAGTCCTCTCCATC; Nova-1-RT-dn, 5′-ACAGTAGCACCTCCCTTCC-3′; GAPDH-RT-up, 5′-TGCACCACCAACTGCTTAGC-3′; GAPDH-RT-dn, 5′-GCAGTGATGGCATGGACTGT-3′. Total RNA was prepared, reverse-transcribed, and amplified as above. Real time PCR was performed with SYBR Green I (Invitrogen) using a 7300 Real-Time PCR system (Applied Biosystems).
RESULTS
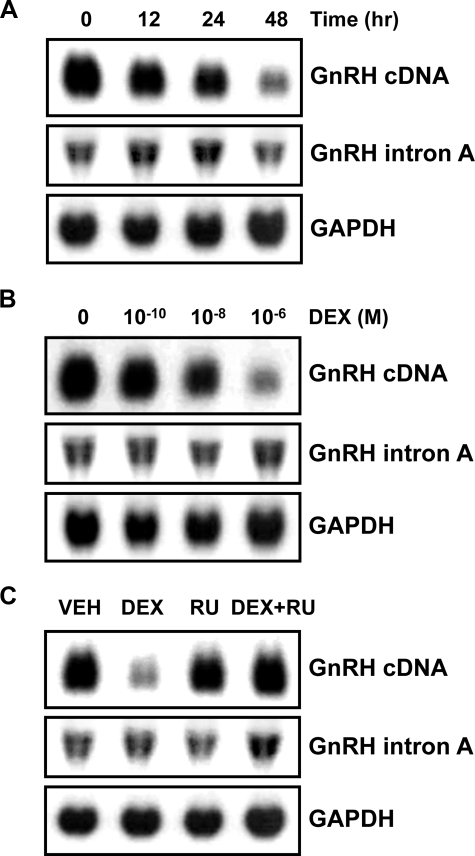
Production of GnRH mRNA and Retention of Intron A in Response to Treatment with DEX—GC was shown to down-regulate GnRH gene expression at the level of transcription in the rat hypothalamus and in GnRH-producing GT1 cells (9, 10). The possible involvement of post-transcriptional processes in the down-regulation is unknown. To address this question, we determined levels of both mature GnRH mRNA and its intron A-containing variant in GT1-1 cells treated with DEX. Northern blot analysis using a GnRH cDNA probe revealed that DEX dramatically reduced GnRH mRNA levels in time- and dose-dependent manners (Fig. 1, A and B, top panels). The DEX-evoked decrease in GnRH mRNA level was reversed by RU486, a GR antagonist (Fig. 1C, top). Since excision of intron A from the GnRH pre-mRNA is a rate-limiting step in GnRH production (11, 16), we determined intron A-retained transcript levels using an intron A probe. In contrast to the drastic reduction in mature mRNA level, the level of intron A-retained transcript was altered very slightly by DEX (Fig. 1, A–C, middle panels). Although it appeared that the intron A-retained transcript was increased slightly at the DEX + RU486 point (Fig. 1C, middle), the expression level of it was not changed significantly when it was analyzed by quantitative real time RT-PCR (data not shown). The constant level of intron A-retained transcript together with the decreased level of mature mRNA suggested that DEX reduces the processing of GnRH pre-mRNA into mature mRNA in addition to transcriptional repression. The exon 2-skipped transcript, another variant form of GnRH gene transcripts (16), was barely detectable in the GT1-1 cells and was not influenced by DEX treatment (Fig. S2).
FIGURE 1.
Dexamethasone down-regulates GnRH mRNA but not intron A-containing transcript levels in GT1-1 cells. GT1-1 cells were grown to confluence and treated with 10–6 m DEX for 12, 24, and 48 h (A) and with vehicle (VEH; 0.1% ethanol) or various doses of DEX, as indicated, for 24 h (B). C, the GT1-1 cells were treated with vehicle, 10–6 m DEX, and/or 5 × 10–6 m RU486, a GC receptor antagonist, for 24 h. 30-μg samples of total RNA were electrophoresed on 1.2% formaldehyde denaturing agarose gels and transferred to nylon membrane. The membranes were probed with a 32P-labeled mouse GnRH cDNA or an intron A probe and then reprobed with a 32P-labeled GAPDH cDNA probe.
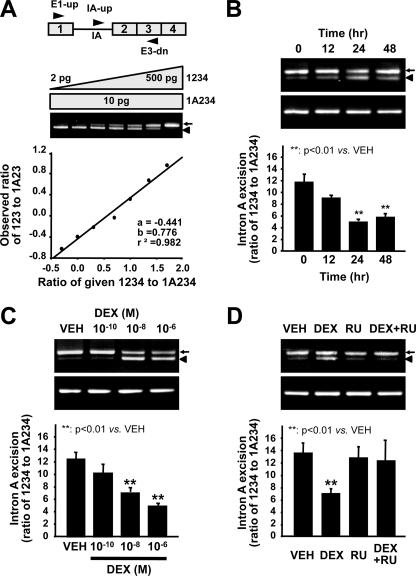
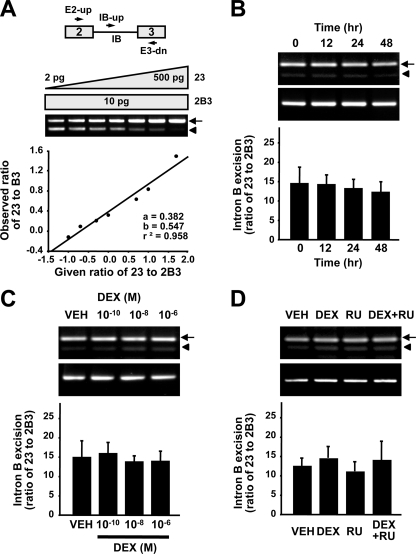
DEX Inhibits Excision of GnRH Intron A—To quantify the effect of GC on the rate of intron A excision, we employed a competitive RT-PCR analysis involving three primers, targeting both GnRH mature mRNA, 123, and intron A-retained transcript, 1A23, as described previously (12). The competitive RT-PCR data were derived from a standard curve of the ratio of PCR product 123 to A23 as a function of the ratio of cDNA fragments 1234 to 1A234, which gave a linear regression coefficient of 0.982 (Fig. 2A). In agreement with the Northern blot results, DEX decreased the ratio of 123 to 1A23 in a time- and dose-dependent manner (Fig. 2, B and C), and this effect was reversed by RU486 (Fig. 2D). By contrast, the excision rate of intron B, a strong intron from the GnRH pre-mRNA, as determined by competitive RT-PCR analysis using three primers encompassing exons 2 and 3 (23), intron B, and exon 3 (B3) was not affected by DEX (Fig. 3). These results suggest that GC specifically inhibits excision of the weak GnRH intron A.
FIGURE 2.
Dexamethasone attenuates the excision of GnRH intron A. A, the structure of the GnRH gene and positions of the primers (top). A standard curve to measure 1234 and 1A234 cDNAs was constructed by competitive PCR using 10 pg of the 1A234 cDNA fragment and serial dilutions of the 1234 cDNA fragment (bottom). B–D, total RNA was extracted from GT1-1 cells treated with vehicle (VEH) or DEX, as indicated, and RT-PCR was performed. Intron A excision was calculated from the ratio of 123 to 1A23 on the basis of a standard curve. B, time course of the effect of DEX. C, effect of vehicle or various doses of DEX for 24 h on the 123/1A23 ratio. D, the effect of RU486 on GnRH intron A excision. Bars, means ± S.E. (n = 6). **, p < 0.01 versus vehicle-treated cells. Arrows and arrowheads, mature mRNA and intron A-retained transcript, respectively.
FIGURE 3.
Effect of dexamethasone on the excision of GnRH intron B. A, GnRH gene structure and locations of primers (top). A standard curve to measure 23 and 2B3 cDNAs was constructed by competitive PCR using 10 pg of the 2B3 cDNA fragment and serial dilutions of the 23 cDNA fragment (bottom). B, time course of the effect of 10–6 m DEX on intron B excision (n = 6). C, effect of 24-h treatment with the indicated doses of DEX (n = 6). D, effect of DEX and/or RU486 on the excision of GnRH intron B (n = 6). The arrows and arrowheads indicate transcripts lacking intron B and containing intron B, respectively.
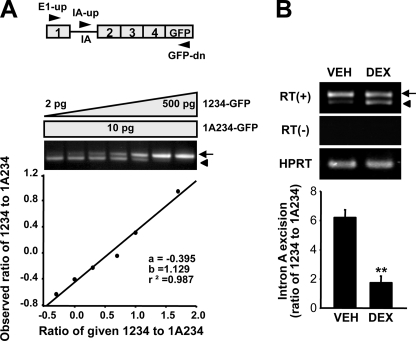
To verify the impaired excision of the intron A by DEX, we used a splicing reporter (named p1A234-GFP) mimicking the intron A-containing GnRH gene transcript under the control of the GC-insensitive cytomegalovirus promoter (Fig. 4A) and a strong SV40 poly(A) signal as previously described (13). GT1-1 cells were transfected with p1A234-GFP and treated with DEX (10–6 m) for 24 h. Total RNA extracted from VEH- or DEX-treated cells were used in competitive RT-PCR with the E1-up, IA-up, and GFP-dn primers to determine the intron A excision rate by measuring 1234-GFP/1A234-GFP ratios with a standard curve (Fig. 4A). Like the endogenous GnRH gene transcript, intron A excision from the p1A234-GFP reporter was down-regulated by DEX (Fig. 4B), implying DEX-mediated inhibition of GnRH pre-mRNA splicing.
FIGURE 4.
The splicing efficiency of GnRH-GFP reporter by treatment with dexamethasone. A, schematic diagram of the 1A234-GFP reporter construct containing the cytomegalovirus promoter and SV40 poly(A) signal (top). A standard curve for quantification of 1234-GFP and 1A234-GFP RNA species was constructed (bottom). B, GT1-1 cells were transiently transfected with 100 ng of the p1A234-GFP reporter plasmid expressing pre-mRNA substrates and treated with 10–6 m DEX or vehicle (VEH) for 24 h. RT-PCR was performed with three primers, E1-up, IA-up, and GFP-dn, allowing simultaneous and competitive amplification of the mature mRNA (1234-GFP) and intron A-containing RNA species (1A234-GFP). Bars, means ± S.E. (n = 6). **, p < 0.01 versus vehicle-treated cells. The arrows and arrowheads indicate mature mRNA and intron A-retained transcript, respectively.
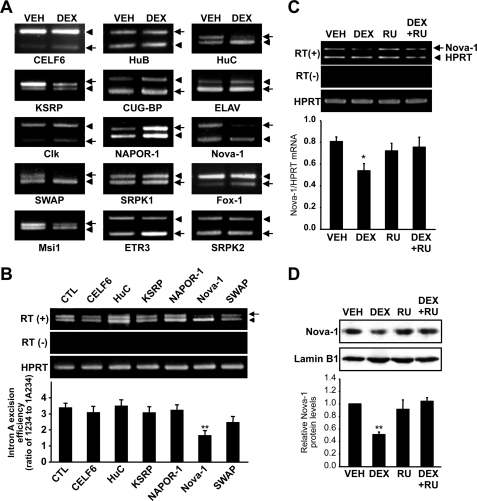
Nova-1 Is Involved in the DEX-evoked Inhibition of Intron A Removal—Since intron A excision is largely dependent on regulatory splicing factors, we hypothesized that an effect on one or another trans-acting splicing factor could be responsible for the DEX-induced attenuation of intron A excision. Although a subset of SR and SR-related proteins, such as Tra2α, 9G8, and SRp30c, is known to cooperate in the excision of GnRH intron A (13, 14), DEX did not have any significant effect on the expression of any of these factors (Fig. S3). Therefore, we hypothesized that DEX may affect the GnRH pre-mRNA splicing by an additional splicing regulator(s), whose actions on GnRH were not yet characterized. To identify possible candidate(s) whose expression is influenced by DEX, we screened the transcript levels of 15 brain-enriched RNA-binding splicing regulators in DEX-treated GT1-1 cells (Table S1). The expression of six splicing regulators, CELF-6, HuC, KSRP, NAPOR-1, nova-1, and SWAP, was altered by 24-h DEX treatment (Fig. 5A). We then measured intron A excision rates after overexpression of antisense cDNAs against each of these six splicing factors. Intron A excision was only affected by the Nova-1 antisense vector (Fig. 5B and Fig. S4). In accordance with the effect of DEX on the GnRH pre-mRNA splicing, decreased nova-1 mRNA and protein level by DEX was reversed by RU486, indicating GR-mediated regulation (Fig. 5, C and D).
FIGURE 5.
Expression patterns of RNA binding splicing regulators implicated in GnRH pre-mRNA splicing. A, differential mRNA expression of 15 brain-enriched splicing regulators determined by semiquantitative RT-PCR. To compare the mRNA expression levels of the target genes in GT1-1 cells treated with 10–6 m DEX for 24 h, the non-SR splicing regulators and HPRT cDNAs were co-amplified in the same tubes. B, effect of knockdown of the indicated splicing regulator gene on GnRH pre-mRNA splicing. Empty vector (pcDNA3.1) or individual antisense overexpression constructs were cotransfected with the p1A234-GFP reporter into GT1-1 cells. After 2 days, total RNA was isolated, and RT-PCR was performed to measure GnRH splicing efficiency. (**, p < 0.01 versus CTL; n = 5). The arrow and arrowhead indicate mature GnRH mRNA and intron A-retained transcript, respectively. C, nova-1 mRNA expression was calculated from the ratio of nova-1 to HPRT mRNA, and the effect of RU486 on nova-1 mRNA expression was analyzed. Bars, mean ± S.E. (n = 3). *, p < 0.05 versus vehicle-treated cells. D, GT1-1 cells were treated with vehicle (VEH), 10–6 m DEX, and/or RU486 for 24 h. The nuclear extract was isolated from GT1-1 cells, and then 15 μg of proteins were analyzed by immunoblotting with anti-Nova-1 (top) and anti-lamin B1 (bottom) antibodies. Bars, mean ± S.E. (n = 3). **, p < 0.01 versus vehicle-treated cells.
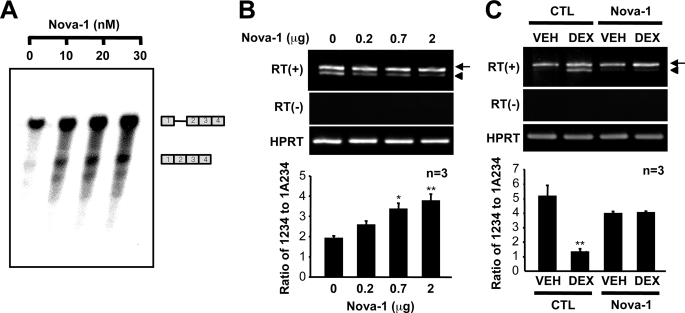
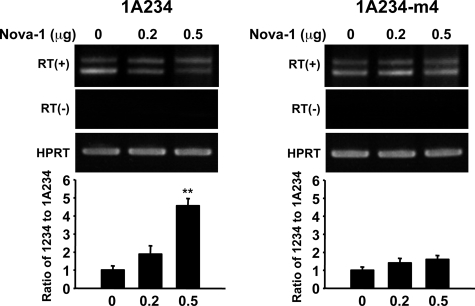
To address the role of Nova-1 in GnRH pre-mRNA splicing, we employed an in vitro splicing assay using a 1ΔA234 minigene construct and a splicing-deficient cytoplasmic S100 extract as mentioned previously (13, 14) in the presence of increasing concentrations of purified GST-Nova-1 protein. Nova-1 augmented intron A excision in a dose-dependant manner (Fig. 6A). In agreement with the in vitro splicing assay, Nova-1 overexpression in NIH3T3 mouse fibroblast cells also enhanced intron A excision from the transfected 1A234-GFP construct (Fig. 6B). Furthermore, Nova-1 overexpression in GT1-1 cells abolished the DEX-induced suppression of intron A excision (Fig. 6C). Collectively, these results indicate that the down-regulation of Nova-1 can reside under the attenuated intron A excision by DEX.
FIGURE 6.
Nova-1 blocks dexamethasone-suppressed GnRH intron A splicing. A, in vitro splicing was assayed in the presence of a cytoplasmic S100 extract, and purified GST-Nova-1 was added to the splicing reactions at the indicated concentrations. Unspliced and spliced RNA species are indicated diagrammatically on the right. B, Myc-tagged Nova-1 was overexpressed in NIH3T3 cells with the p1A234-GFP splicing reporter, and intron A excision was examined by competitive RT-PCR. *, p < 0.05; **, p < 0.01 versus control vector-transfected cells. C, empty vector (CTL; pCMV-Tag3) or a Nova-1 expression vector was cotransfected with a p1A234-GFP splicing reporter into GT1-1 cells. The cells were exposed to vehicle (VEH) or DEX (10–6 m) for 24 h, and intron A excision was examined by competitive RT-PCR. The arrows and arrowheads indicate GnRH mRNA and intron A-retained transcript, respectively.
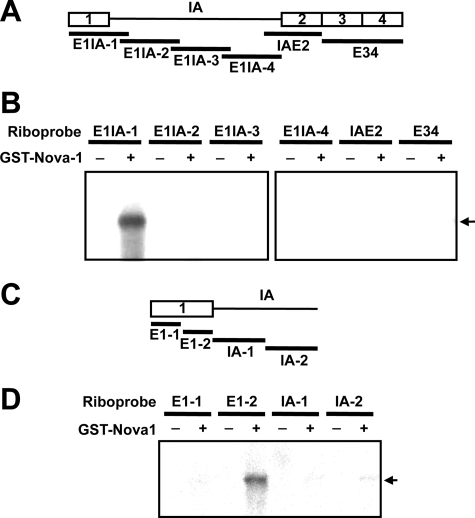
Nova-1 Recognizes ACAU Sequences on Exon 1 of the GnRH Pre-mRNA—Recently, Nova-1 was shown to bind to the primary transcripts of many target genes (17, 18). To identify a Nova-1 binding site in GnRH pre-mRNA, we purified GST-tagged Nova-1 using a baculovirus expression system and employed it in UV cross-linking assays with a series of fragments of the pre-mRNA (Fig. 7A). Only E1IA-1 consisting of exon 1 and the proximal region of intron A interacted with Nova-1 (Fig. 7B). When E1IA-1 was further dissected into four regions (designated E1-1, E1-2, IA-1, and IA-2) (Fig. 7C), only the E1–2 RNA construct bound to Nova-1, indicating that Nova-1 interacts with the distal region of exon 1 (Fig. 7D).
FIGURE 7.
Binding of Nova-1 to a discrete region of GnRH exon 1. A, schematic diagram of the RNA probes used for UV cross-linking. B, UV cross-linking of purified GST-tagged Nova-1 (300 nm per 15-μl reaction mixture) with various radiolabeled RNA probes (E1IA-1, E1IA-2, E1IA-3, E1IA-4, IAE2, and E34). C, diagram of the RNA probes used for UV cross-linking. D, UV cross-linking of Nova-1 (300 nm) with several RNA probes (E1–1, E1–2, IA-1, and IA-2). The arrows indicate the specific binding of Nova-1 to the probes.
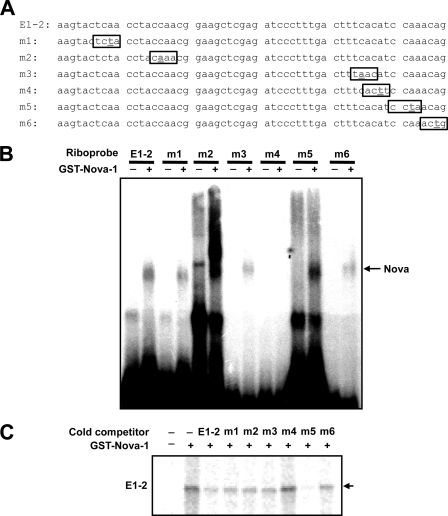
It is known that Nova-1 recognizes YCAY sequences in pre-mRNA and that the CA is critical for binding (19–21). To identify the RNA motif in GnRH pre-mRNA recognized by Nova-1, we mutated the CA-containing regions, designated m1, m2, m3, m4, m5, and m6, in E1-2 (Fig. 8A) and performed electrophoretic mobility shift assays in the presence or absence of Nova-1. All of the wild type and mutant E1–2 RNA probes except m4 bound Nova-1, as shown by the shifted bands (Fig. 8B), indicating that the CA motif mutated in m4 is responsible for Nova-1 binding. This observation was supported by UV cross-linking assays in which only excess unlabeled m4 competitor failed to block the binding between Nova-1 and the E1-2 riboprobe (Fig. 8C).
FIGURE 8.
Nova-1 interacts with the distal region of GnRH exon 1. A, sequences of the point-mutated RNA probes used for UV cross-linking. B, electrophoretic mobility shift assays of Nova-1 (300 nm) with various point-mutated E1-2 RNA probes (E1-2, m1, m2, m3, m4, m5, and m6). C, UV cross-linking of Nova-1 (300 nm) with radiolabeled E1-2 RNA probes alone or with excess unlabeled E1-2, m1, m2, m3, m4, m5, or m6 RNA. The arrows indicate specific binding of Nova-1 to the probes.
To confirm the role of the ACAU motif in the distal region of GnRH exon 1 in the activity of Nova-1 in GnRH pre-mRNA splicing, we mutated the ACAU motif to AUUU in p1A234-GFP, yielding p1A234-GFP-m4. Cotransfection of this mutant reporter plasmid with the Nova-1 expression vector into NIH3T3 cells resulted in reduced intron A excision (Fig. 9). Clearly the ACAU motif within the distal exon 1 is necessary for the regulation of GnRH pre-mRNA splicing by Nova-1.
FIGURE 9.
Nova-1 enhances GnRH intron A excision efficiency by binding to ACAU within exon 1. A, NIH3T3 cells were cotransfected with the indicated amount of Nova-1 expression construct plus a wild type or mutant (ACAT to ATTT) p1A234-GFP-m4 construct. Ratios of mRNA to intron A-containing transcript were determined by RT-PCR. Bars, means ± S.E. (n = 3). **, p < 0.01 versus control vector-transfected cells. The arrows and arrowheads indicate GnRH mRNA and intron A-retained transcript, respectively.
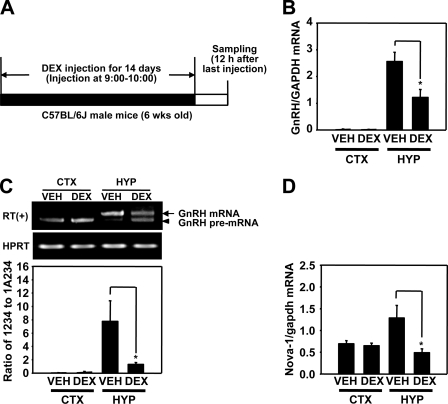
DEX Down-regulates Nova-1 Expression with Reduced GnRH pre-mRNA Splicing in Vivo—To determine whether DEX suppresses GnRH pre-mRNA splicing in vivo, as well as reducing nova-1 gene expression, we administered DEX to male C57BL/6J mice for 14 days at a dose of 4 mg/kg/day and determined intron A excision rates and nova-1 mRNA levels by RT-PCR using total RNAs derived from the hypothalamus and cerebral cortex (Fig. 10A). Consistent with previous reports (9), chronic injection of DEX reduced GnRH mRNA expression in the hypothalamus, as measured by real time RT-PCR analysis (Fig. 10B) and drastically decreased excision of intron A in the hypothalamus but not in the cortex (Fig. 10C). In addition, the nova-1 mRNA level was specifically reduced in the hypothalamus (Fig. 10D). These results suggest that chronic DEX administration lowers the intron A excision rate as well as GnRH mRNA levels, presumably by inhibiting Nova-1 expression.
FIGURE 10.
Effect of dexamethasone on GnRH pre-mRNA splicing in adult male mice. A, schematic diagram of the experiments. Male mice (6-week-old, 4 mice/group) were injected subcutaneously with DEX 21-phosphate (4 mg/kg) or vehicle (VEH) for 14 days and sacrificed 12 h after the last injection. Total RNA from cerebral cortex (CTX) and hypothalamus (HYP) was analyzed for GnRH mRNA level by real time RT-PCR (B), for GnRH pre-mRNA splicing level by competitive RT-PCR (C), and for nova-1 mRNA level by real time RT-PCR (D). Data shown are means ± S.E. (n = 4). *, p < 0.05 versus vehicle-injected mice.
DISCUSSION
In the present study, we addressed a question regarding how stress can influence reproductive functions and sexual maturation by interfering with the hypothalamic-pituitary-gonadal neuroendocrine axis (2, 22, 23). Indeed, it has been well established that the activation of the hypothalamic-pituitary-adrenal axis can impair the mammalian reproductive system at multiple levels (24, 25), and especially stressful stimuli, such as repeated cold stresses, directly suppressed hypothalamic GnRH gene expression (26). In this context, GC was proposed as a key mediator for stress-evoked inhibition of GnRH expression and secretion (9). GR in the hypothalamic GnRH neurons and GT1-1 cells participates in suppression of the transcriptional activity of the GnRH promoter (8, 27). However, the fact that the inhibitory effect of GC on the GnRH promoter activity in GT1-1 cells was much smaller than its effect on GnRH mRNA (Fig. 1 and Fig. S1) (8), suggested that GC might also regulate GnRH gene expression by a post-transcriptional mechanism, such as pre-mRNA splicing and/or mRNA stability.
Our findings clearly proved this hypothesis; DEX significantly lowered the ratio of mature GnRH mRNA to its intron A-retained variant in both GT1-1 cells and the mouse hypothalamus, as shown in Figs. 1, 2, and 10. This is evidently due to the attenuated intron A excision rate, because similar impairment of intron A excision efficiency by DEX was observed in a splicing reporter construct bearing a strong viral promoter and a poly(A) signal (Fig. 4). It should be noted that the excision of the intron A serves as a rate-limiting step to produce a functional GnRH mRNA and was proposed to contribute to developmental stage- and cell type-specific expression of GnRH (12, 15). Considering the crucial role of the intron A excision in the translation initiation (15), the present study strongly suggests that this post-transcriptional mechanism can also participate in the temporary regulation of GnRH gene expression in response to extracellular stimuli like GC.
It is often observed that GC can affect RNA splicing. For instance, alternative RNA splicing of slo, a gene which encodes a calcium- and voltage-activated potassium channel and contains a stress axis-regulated exon, is influenced by injection of ACTH into hypophysectomized rats and induces the excitable behavior of Slo channels (28). Similarly, GC modulates the splicing of slo genes in cultured pituitary and adrenal cells (29). Also, GC has been implicated in the alternative splicing of acetylcholinestrase and insulin receptor genes (30, 31). Therefore, it is plausible that a subset of splicing regulators resides under the control of GC signaling. However, GC does not appear to significantly alter general splicing machinery, since DEX treatment shows no effect on the intron B removal, which is a consensus intron easily excised by general splicing apparatus (Fig. 3) (14).
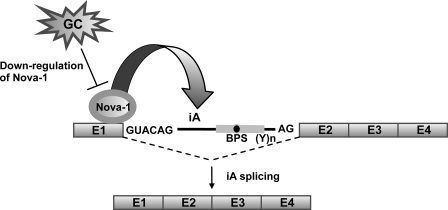
Considering that DEX specifically suppresses GnRH intron A excision, there might be a crucial factor(s) that can be directly regulated by DEX and mediate DEX-induced down-regulation of GnRH intron A splicing. In the next set of experiments, we thus attempted to find such splicing regulator(s). Previously, we demonstrated that excision of intron A required GnRH neuron-specific splicing machinery and a subset of SR and SR-like proteins, such as Tra2α, 9G8, and SRp30c (13, 14). However, these SR splicing regulators were not affected by DEX treatment of GT1-1 cells (Fig. S3). Since a number of tissue-specific non-SR RNA binding proteins can mediate neuron-specific alternative RNA splicing in the brain (32, 33), we screened neuron-specific non-SR RNA-binding proteins influenced by DEX and found that Nova-1 mRNA and protein were reduced by DEX treatment and that blocking of its expression reduced the rate of intron A excision. The promoter region of nova-1 contains several predictive GC response elements and negative GC response elements. However, further examination is required to determine whether these elements can mediate effects of DEX on nova-1 gene expression at the level of transcription. Nova is a neuron-specific KH-type RNA-binding protein (21, 34) that affects a large number of pre-mRNA splicing processes (17, 18, 20). Furthermore, several KH-type splicing factors were shown to regulate neuron-specific splicing, such as c-src exon N1 in association with general RNA-binding proteins (35–37). Nova-1 enhanced intron A removal in a dose-dependent manner in the in vitro splicing assay system and in NIH3T3 cells (Fig. 6). Since the S100 contains minimal components required for splicing pre-mRNA, our in vitro splicing assay clearly demonstrated that Nova-1 can stimulate the intron A excision (38). More importantly, overexpression of Nova-1 overcame the DEX-evoked inhibition of intron A excision in GT1-1 cells (Fig. 6C). Based on these findings, we propose that Nova-1 is a key splicing regulator mediating the effect of DEX on GnRH pre-mRNA splicing (Fig. 11).
FIGURE 11.
A model of glucocorticoid-regulated GnRH RNA splicing by Nova-1. GnRH intron A excision is enhanced by binding of Nova-1 to the distal region of exon 1. Attenuation of GnRH pre-mRNA splicing by GC is mediated by down-regulation of Nova-1 gene expression.
Most RNA-binding proteins recognize specific target-binding motifs, and precise binding of splicing factors to these sequences is important for RNA processing. Thus, for instance, Nova-1 failed to enhance E9 inclusion during splicing of GABAA receptor γ2 subunit pre-mRNA, when its target sequence was disrupted by mutation (19). We showed above that Nova-1 recognized the ACAU motif in the distal region of GnRH exon 1 and that mutation of this binding site completely abolished intron A excision by Nova-1 (Figs. 8B and 9). Interestingly, nova-1 binding close to the splice donor site was proposed to be inhibitory for intron removal by blocking U1 small nuclear ribonucleoprotein binding. By contrast, the intronic binding site of Nova-1 could enhance the excision of the bound intron (39). However, there was no Nova-1 binding on intron A, and the exonic binding site was required for its intron A-removing activity. Therefore, Nova-1 seems to enhance the excision of GnRH intron A by a mode of action distinct from that previously suggested.
Nova-1 has been detected in the neurons of a variety of brain regions, including the preoptic area of the hypothalamus, which contains a large population of GnRH-producing neurons (12, 34). This spatial expression of Nova-1 also suggested its involvement in GnRH gene regulation in GnRH-producing neurons in vivo. To confirm this, we performed in vivo experiments similar to those reported elsewhere (9), and the outcome was that intron A excision was inhibited in adult mice chronically treated with DEX and that they contained reduced GnRH mRNA levels. Interestingly, Nova-1 mRNA expression was also down-regulated in the hypothalamus but not in the cortex (Fig. 10). These results imply that down-regulation of Nova-1 expression and subsequent impairment of GnRH pre-mRNA splicing may function as a significant mechanism underlying suppressive effects of hyperactivated GC signaling on the mammalian reproductive functions.
In conclusion, the present study provides novel insights into molecular regulation of GnRH gene expression as well as a well recognized cross-talk between the hypothalamic-pituitary-adrenal and the hypothalamic-pituitary-gonadal axes. Our findings clearly demonstrate that the excision of GnRH intron A can be used in the temporary regulation of this neurohormone production in addition to its highly cell type-restricted expression. More importantly, the present study identified a novel function of Nova-1 to enhance GnRH pre-mRNA splicing. This expands our understanding of the strictly regulated splicing of the GnRH intron A, because Nova-1 is obviously distinct, in its neuron-enriched expression, from the SR and SR-related proteins, which were previously characterized to be responsible for GnRH pre-mRNA splicing. The exact molecular mechanism underlying GC-evoked down-regulation of Nova-1 still needs to be further delineated. Considering the versatility of Nova-1 in neural functions (18), this will also be greatly helpful to understand stress- or GC-evoked changes in alternative RNA splicing in the central nervous system.
Supplementary Material
This work was supported by a grant from the Brain Research Center of the 21st Century Frontier Research Program in Neuroscience funded by the Ministry of Science and Technology, Republic of Korea.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, and Figs. S1–S4.
Footnotes
The abbreviations used are: ACTH, adrenocorticotropic hormone; GnRH, gonadotropin-releasing hormone; GC, glucocorticoid; DEX, dexamethasone; GR, GC receptor; RT, reverse transcription; GFP, green fluorescent protein; EGFP, enhanced GFP; GST, glutathione S-transferase; Nova, neuro-oncological ventral antigen.
References
- 1.Mastorakos, G., Pavlatou, M. G., and Mizamtsidi, M. (2006) Pediatr. Endocrinol. Rev. 3 Suppl. 1, 172–181 [PubMed] [Google Scholar]
- 2.Breen, K. M., and Karsch, F. J. (2004) Endocrinology 145 692–698 [DOI] [PubMed] [Google Scholar]
- 3.Chatterton, R. T. (1990) Int. J. Fertil. 35 8–13 [PubMed] [Google Scholar]
- 4.Genazzani, A. D. (2005) Pediatr. Endocrinol. Rev. 2 661–668 [PubMed] [Google Scholar]
- 5.Wray, S., Gahwiler, B. H., and Gainer, H. (1988) Peptides 9 1151–1175 [DOI] [PubMed] [Google Scholar]
- 6.Dobson, H., Ghuman, S., Prabhakar, S., and Smith, R. (2003) Reproduction (Camb.) 125 151–163 [DOI] [PubMed] [Google Scholar]
- 7.Ferin, M. (1999) J. Clin. Endocrinol. Metab. 84 1768–1774 [DOI] [PubMed] [Google Scholar]
- 8.Chandran, U. R., Attardi, B., Friedman, R., Dong, K. W., Roberts, J. L., and DeFranco, D. B. (1994) Endocrinology 134 1467–1474 [DOI] [PubMed] [Google Scholar]
- 9.Gore, A. C., Attardi, B., and DeFranco, D. B. (2006) Mol. Cell. Endocrinol. 256 40–48 [DOI] [PubMed] [Google Scholar]
- 10.Chandran, U. R., Attardi, B., Friedman, R., Zheng, Z., Roberts, J. L., and DeFranco, D. B. (1996) J. Biol. Chem. 271 20412–20420 [DOI] [PubMed] [Google Scholar]
- 11.Seong, J. Y., Park, S., and Kim, K. (1999) Mol. Endocrinol. 13 1882–1895 [DOI] [PubMed] [Google Scholar]
- 12.Seong, J. Y., Kim, B. W., Park, S., Son, G. H., and Kim, K. (2001) Endocrinology 142 4454–4461 [DOI] [PubMed] [Google Scholar]
- 13.Seong, J. Y., Han, J., Park, S., Wuttke, W., Jarry, H., and Kim, K. (2002) Mol. Endocrinol. 16 2426–2438 [DOI] [PubMed] [Google Scholar]
- 14.Park, E., Han, J., Son, G. H., Lee, M. S., Chung, S., Park, S. H., Park, K., Lee, K. H., Choi, S., Seong, J. Y., and Kim, K. (2006) J. Biol. Chem. 281 401–409 [DOI] [PubMed] [Google Scholar]
- 15.Son, G. H., Jung, H., Seong, J. Y., Choe, Y., Geum, D., and Kim, K. (2003) J. Biol. Chem. 278 18037–18044 [DOI] [PubMed] [Google Scholar]
- 16.Zhen, S., Dunn, I. C., Wray, S., Liu, Y., Chappell, P. E., Levine, J. E., and Radovick, S. (1997) J. Biol. Chem. 272 12620–12625 [DOI] [PubMed] [Google Scholar]
- 17.Jensen, K. B., Dredge, B. K., Stefani, G., Zhong, R., Buckanovich, R. J., Okano, H. J., Yang, Y. Y., and Darnell, R. B. (2000) Neuron 25 359–371 [DOI] [PubMed] [Google Scholar]
- 18.Ule, J., Ule, A., Spencer, J., Williams, A., Hu, J. S., Cline, M., Wang, H., Clark, T., Fraser, C., Ruggiu, M., Zeeberg, B. R., Kane, D., Weinstein, J. N., Blume, J., and Darnell, R. B. (2005) Nat. Genet. 37 844–852 [DOI] [PubMed] [Google Scholar]
- 19.Dredge, B. K., and Darnell, R. B. (2003) Mol. Cell. Biol. 23 4687–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ule, J., Jensen, K. B., Ruggiu, M., Mele, A., Ule, A., and Darnell, R. B. (2003) Science 302 1212–1215 [DOI] [PubMed] [Google Scholar]
- 21.Jensen, K. B., Musunuru, K., Lewis, H. A., Burley, S. K., and Darnell, R. B. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5740–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEwen, B. S. (1994) Ann. N. Y. Acad. Sci. 743 1–18 [DOI] [PubMed] [Google Scholar]
- 23.Handa, R. J., Burgess, L. H., Kerr, J. E., and O'Keefe, J. A. (1994) Horm. Behav. 28 464–476 [DOI] [PubMed] [Google Scholar]
- 24.Dobson, H., and Smith, R. F. (2000) Anim. Reprod. Sci. 60–61, 743–752 [DOI] [PubMed] [Google Scholar]
- 25.Rivier, J. E., Porter, J., Rivier, C. L., Perrin, M., Corrigan, A., Hook, W. A., Siraganian, R. P., and Vale, W. W. (1986) J. Med. Chem. 29 1846–1851 [DOI] [PubMed] [Google Scholar]
- 26.Tanebe, K., Nishijo, H., Muraguchi, A., and Ono, T. (2000) J. Neuroendocrinol. 12 13–21 [DOI] [PubMed] [Google Scholar]
- 27.Ahima, R. S., and Harlan, R. E. (1992) Neuroendocrinology 56 845–850 [DOI] [PubMed] [Google Scholar]
- 28.Xie, J., and McCobb, D. P. (1998) Science 280 443–446 [DOI] [PubMed] [Google Scholar]
- 29.Lai, G. J., and McCobb, D. P. (2006) Endocrinology 147 3961–3967 [DOI] [PubMed] [Google Scholar]
- 30.Kosaki, A., and Webster, N. J. (1993) J. Biol. Chem. 268 21990–21996 [PubMed] [Google Scholar]
- 31.Nijholt, I., Farchi, N., Kye, M., Sklan, E. H., Shoham, S., Verbeure, B., Owen, D., Hochner, B., Spiess, J., Soreq, H., and Blank, T. (2004) Mol. Psychiatry 9 174–183 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, W., Liu, H., Han, K., and Grabowski, P. J. (2002) RNA (N.Y.) 8 671–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Q., Lee, J. A., and Black, D. L. (2007) Nat. Rev. 8 819–831 [DOI] [PubMed] [Google Scholar]
- 34.Buckanovich, R. J., Yang, Y. Y., and Darnell, R. B. (1996) J. Neurosci. 16 1114–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan, R. C., and Black, D. L. (1997) Mol. Cell. Biol. 17 4667–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou, M. Y., Rooke, N., Turck, C. W., and Black, D. L. (1999) Mol. Cell. Biol. 19 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min, H., Turck, C. W., Nikolic, J. M., and Black, D. L. (1997) Genes Dev. 11 1023–1036 [DOI] [PubMed] [Google Scholar]
- 38.Krainer, A. R., and Maniatis, T. (1985) Cell 42 725–736 [DOI] [PubMed] [Google Scholar]
- 39.Ule, J., Stefani, G., Mele, A., Ruggiu, M., Wang, X., Taneri, B., Gaasterland, T., Blencowe, B. J., and Darnell, R. B. (2006) Nature 444 580–586 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.