Abstract
Filaggrin is a component of the cornified cell envelope and the precursor of free amino acids acting as a natural moisturizing factor in the stratum corneum. Deimination is critical for the degradation of filaggrin into free amino acids. In this study, we tried to identify the enzyme(s) responsible for the cleavage of deiminated filaggrin in vitro. First, we investigated citrulline aminopeptidase activity in the extract of newborn rat epidermis by double layer fluorescent zymography and detected strong activity at neutral pH. Monitoring the citrulline-releasing activity, we purified an enzyme of 280 kDa, comprised of six identical subunits of 48 kDa. The NH2 terminus of representative tryptic peptides perfectly matched the sequence of rat bleomycin hydrolase (BH). The enzyme released various amino acids except Pro from β-naphthylamide derivatives and hydrolyzed citrulline-β-naphthylamide most effectively. Thus, to break down deiminated filaggrin, another protease would be required. Among proteases tested, calpain I degraded the deiminated filaggrin effectively into many peptides of different mass on the matrix-assisted laser desorption/ionization-time of flight mass spectrum. We confirmed that various amino acids including citrulline were released by BH from those peptides. On the other hand, caspase 14 degraded deiminated filaggrin into a few peptides of limited mass. Immunohistochemical analysis of normal human skin revealed co-localization of BH and filaggrin in the granular layer. Collectively, our results suggest that BH is essential for the synthesis of natural moisturizing factors and that calpain I would play a role as an upstream protease in the degradation of filaggrin.
The mammalian epidermal keratinocytes arise from proliferating basal cells and move outward through a series of distinct differentiation events to form the stratum corneum (1, 2). During this progressive epidermal differentiation, keratinocytes express different proteins such as keratins, profilaggrin/filaggrin, involucrin, small proline-rich proteins, loricrin, cystatin A, and elafin, which form the cornified envelope of mature corneocytes (3–7). Profilaggrin is synthesized as a large, extremely insoluble phosphoprotein that consists of a unique NH2-terminal Ca2+-binding protein of the S-100 family, linked to 10–20 tandem filaggrin monomer repeats (8–10). Each individual filaggrin repeat is completely removed by proteolysis to generate the mature filaggrin monomer (a molecular mass of 37 kDa in human). Then, filaggrin is completely degraded in the uppermost layer of the stratum corneum to produce a mixture of free and modified hygroscopic amino acids that are important for maintaining epidermal hydration (2, 11–13). In addition, a number of proteins are subjected to various post-translational modifications such as disulfide bonding, N-(γ-glutamyl)-lysine isopeptide cross-linking, and deimination during the terminal differentiation of epidermal keratinocytes (4, 6, 14, 15). Deimination is catalyzed by peptidylarginine deiminase (PAD),2 which converts arginine to citrulline in proteins (17–19). The modification seems essential for the processing into free amino acids including citrulline.
Several proteases reportedly participate in the processing of profilaggrin. Furin, a member of the proprotein convertase family, has been proposed to cleave the NH2 terminus of profilaggrin, facilitating the release of the NH2-terminal S-100 protein (20, 21). In contrast, calpain I and profilaggrin endopeptidase I (PEP-I) were implicated in the processing of the linker regions between the filaggrin monomer repeats to generate the filaggrin monomer (22–25). Recently, significant results regarding the conversion of profilaggrin to filaggrin have been obtained with the knock-out of matriptase/MT-SP1, prostasin/channel-activating serine protease 1/Prss 8, and caspase 14 in mice (26–28). These proteases were a key component of the profilaggrin-processing pathway in terminal epidermal differentiation. However, although the signal initiating the degradation of profilaggrin at a defined stage of the maturation of the stratum corneum was found to be the water gradient within the stratum corneum itself (11), the proteases for the processing of filaggrin and/or the deiminated form into peptides following the breakdown of these peptides to amino acids including citrulline remain unknown.
In this study, we have purified a novel aminopeptidase using a deiminated substrate from rat skin homogenate and identified it as a neutral cysteine protease, bleomycin hydrolase (BH). Furthermore, we investigated the processing of the deiminated filaggrin by calpain I or caspase 14. Based on these results, we proposed that calpain I participated preferentially in the processing of deiminated filaggrin into peptides and then BH appeared essential for the breakdown of the peptides into amino acids.
EXPERIMENTAL PROCEDURES
Materials—Recombinant human filaggrin and mouse PAD type 3 (mPAD3) were produced and purified according to the methods reported by Kanno et al. (29) and by Ohsugi et al. (30), respectively. BH was purified from the homogenate of newborn rat epidermis as described previously (31). Human calpain I was purchased from Calbiochem (Darmstadt, Germany). Human caspase 14 was from BIOMOL International LP (Plymouth, PA). Citrulline-β-naphthylamide (Cit-β-NA) and citrulline-4-methylcoumaryl-7-amide (Cit-MCA) were obtained from Bachem Bioscience (Bubendorf, Switzerland). Fluorescence resonance energy transfer substrate (FRETS)-25Cit libraries were commercially synthesized at Peptide Institute Inc. (Osaka, Japan). All other chemicals used were of reagent grade.
Detection of Citrulline Aminopeptidase Activity by Double Layer Fluorescent Zymography—Double layer fluorescent zymography was performed as originally described by Katunuma et al. (32). The rat tissue extract was mixed with an equal volume of a treatment buffer consisting of 0.25 m Tris-HCl (pH 6.8), 20% glycerol, and 0.02% bromphenol blue and then applied to a 5% polyacrylamide gel. Electrophoresis was performed at 4 °C at 13 mA. The gel was then washed twice with distilled water and flooded with 0.1 m phosphate buffer (pH 6.0) or 0.1 m Tris-HCl (pH .5) containing 10 mm DTT and 5 mm EDTA at 37 °C for 30 min to equilibrate the pH. A cellulose acetate membrane was washed with distilled water, immersed in 10% glycerol for 20 min, and air-dried. The membrane was soaked in 5 ml of 0.1 m phosphate buffer (pH 6.0) or Tris-HCl (pH 7.5) containing 0.1 mm Cit-MCA substrate, 0.04% Brij 35, 10 mm DTT, and 5 mm EDTA. Then, the gel was covered with the fluorescent substrate membrane to create a double layer. Following incubation at 37 °C for 30 min, fluorescent proteolytic activity bands in the double layer were observed using a Print-graph (ATTO, Tokyo, Japan). Fluorescent gel images were recorded using a Lumix digital camera (Panasonic, Osaka, Japan).
Purification of an Enzyme with Citrulline Aminopeptidase Activity from Newborn Rat Skin—Newborn rat skin (2 days old, Sprague-Dawley rats) was homogenized first with 10 volumes of 20 mm Tris-HCl (pH 7.5) containing 150 mm NaCl, 10 mm 2-mercaptoethanol, and 5 mm EDTA (buffer A) in a POLY-TRON system PT2100 (KINEMATICA AG, Lucerne, Switzerland) and then a second time in a glass homogenizer. The supernatant was fractionated by precipitation with 45–55% saturated ammonium sulfate. After the removal of insoluble materials, the clear solution was applied to a Sephacryl S-200 column (2.5 × 90 cm). After dialysis of the active fraction from the column developed with 20 mm Tris-HCl (pH 7.5) containing 1 mm 2-mercaptoethanol (buffer B), the sample was applied to a DEAE-cellulose column (1 × 30 cm), and the column was washed with buffer B. The proteins were fractionated with a linear gradient of NaCl from 0 to 350 mm in buffer B. The active fractions were pooled, concentrated by ultrafiltration, and dialyzed against buffer B. The dialysate was applied to a Mono Q column-equipped HPLC system (Amersham Biosciences). The protein was eluted with the same buffer. Enzyme activity was specifically detected by measuring aminopeptidase activity using 2 mm Cit-β-NA during the course of preparation. The purified enzyme showed a single band of molecular mass 48 kDa on SDS-PAGE.
Deimination of Filaggrin by mPAD3—Filaggrin (8.39 μm) was incubated with mPAD3 (0.17 μm) at an enzyme:substrate molar ratio of 1:50 at 37 °C for different periods in a buffer of 100 mm Tris-HCl (pH 7.4), 10 mm CaCl2, and 5 mm DTT. After deimination for different periods, filaggrin (5.95 μm) was incubated at 37 °C for 1 h with trypsin (0.03 μm) in 20 mm Tris-HCl (pH 8.0) containing 20 mm CaCl2. The digestion was performed at a trypsin:filaggrin molar ratio of 1:200. The reaction products were resolved by SDS-PAGE according to the methods of Laemmli (55) using 12.5% gels. The gels were stained with Coomassie Brilliant Blue R-250 (CBB R-250).
Degradation of Deiminated Filaggrin with Calpain I or Caspase 14—Filaggrin (8.39 μm) was deiminated at 37 °C overnight at mPAD3 (0.17 μm):filaggrin molar ratio of 1:50. The degradation of unmodified and deiminated filaggrin was performed at an enzyme:filaggrin molar ratio of 1:20 for all digests. Calpain I (0.32 μm) was incubated at 30 °C with filaggrin (6.31 μm) in 100 mm Tris-HCl (pH 7.5) containing 0.5 mm CaCl2 and 10 mm DTT, whereas caspase 14 (0.32 μm) was incubated at 37 °C with filaggrin (6.31 μm) in 100 mm HEPES (pH 7.0), 60 mm NaCl, 0.01% CHAPS, 5 mm DTT, and 1.1 m sodium citrate as a kosmotropic buffer. After incubation for an optimum period, the degradation product in the reaction mixture was detected by SDS-PAGE. A densitometric analysis of scanned gel images was performed by using the NIH ImageJ program on a Windows XP computer. The digests (1 μl) were mixed with the matrix (α-cyano-4-hydroxycinnamic acid dissolved in 50% acetonitrile/0.1% trifluoroacetic acid) and applied to the sample target for MALDI-TOF MS. After air drying, MALDI-TOF MS was conducted using a Voyager-DE STR (Applied Biosystems, Framingham, MA) in reflector mode.
Analyses of Free Amino Acids Released from Peptides Treated with Calpain I or Caspase 14 by BH—The amounts of newly formed α-amino groups were measured using their reaction with fluorescamine (33). After the digestion of unmodified and deiminated filaggrin (6.31 μm) with calpain I (0.32 μm) or caspase 14 (0.32 μm) for 1 h at an enzyme:filaggrin molar ratio of 1:20, the digests treated with caspase 14 were filtrated by centrifugation with a Microcon YM-3 (Millipore) for the exclusion of kosmotropic salts and then adjusted the volume. The reaction products were incubated with BH (13.0 μm) in 50 mm HEPES-NaOH (pH 7.5), 10 mm DTT, and 5 mm EDTA at 37 °C. Aliquots were removed at different times, and the enzymatic reaction was stopped by boiling. Samples were mixed with 100 μl of 50 mm HEPES-NaOH (pH 8.0) and 50 μl of acetone solution of fluorescamine (0.3 mg/ml). The mixture was vortexed, and 500 μl of 50 mm HEPES-NaOH (pH 8.0) was added. Fluorescence was measured fluorometrically (excitation = 370 nm, emission = 475 nm). The generated amino groups were estimated using several concentrations of l-leucine as a standard.
After a 1-h degradation period with calpain I or caspase 14, enzymes and undegraded filaggrin were removed by filtration (Nanosep-10, Pall Gelman Sciences), and the flow-through fractions were incubated with BH overnight at 37 °C. Free amino acids were analyzed using a Hitachi model L-8800 amino acid analyzer equipped with columns (number 2622: 4.6 × 40 mm + 4.6 × 40 mm, Hitachi, Tokyo, Japan) by gradient elution with MCI buffer L-8500-PF kit (Wako, Osaka, Japan) according to the physiological fluids analysis method. Amino acids post-labeled with ninhydrin were detected by measuring the absorbance at 440 and 570 nm.
Immunohistochemistry—Human specimens were obtained from the informed healthy volunteers. They received detailed information about the study and gave their written informed consent. The study was approved by Sagami Women's University Ethical Committee and by the Shiseido Committee on Human Ethics. Indirect immunofluorescence microscopy was performed with 5-μm-thick cryosections of human skin. After rehydration in PBS at room temperature, sections were incubated in a moist chamber for 30 min with PBS containing 1% bovine serum albumin and 0.05% Tween 20 and then overnight at 4 °C with the anti-rat BH IgG and monoclonal anti-human filaggrin IgG diluted in the same buffer. After four washes in PBS containing 0.05% Tween 20, sections were incubated for 1 h at room temperature with Alexa Fluor 555 goat anti-rabbit IgG (H+L) (1/500) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) (1/500) in PBS containing 1% bovine serum albumin and 0.05% Tween 20. After four washes in PBS containing 0.05% Tween 20, sections were mounted in an antifading solution before observation under a confocal laser microscope (LSM 5 PASCAL, Carl Zeiss, Oberkochen, Germany), with excitation at 488 and 543 nm by argon and He-Ne lasers, respectively. For negative controls, sections were incubated without primary antibodies.
Amino Acid Sequence Analyses of Tryptic Peptides—After the enzyme with citrulline aminopeptidase activity was dipyridylated, it was digested with trypsin (31). The digested peptides were separated by a reverse-phase Shimadzu HPLC on an ODT-120T column (Tosoh, Tokyo, Japan) using a linear gradient of 0–60% acetonitrile in 0.1% trifluoroacetic acid for 60 min at a flow rate of 0.5 ml/min. Peak fractions detected by measuring the absorbance at 215 nm were collected in test tubes and lyophilized. The NH2-terminal amino acid sequences were determined using an Applied Biosystems model 120A PHT analyzer.
RESULTS
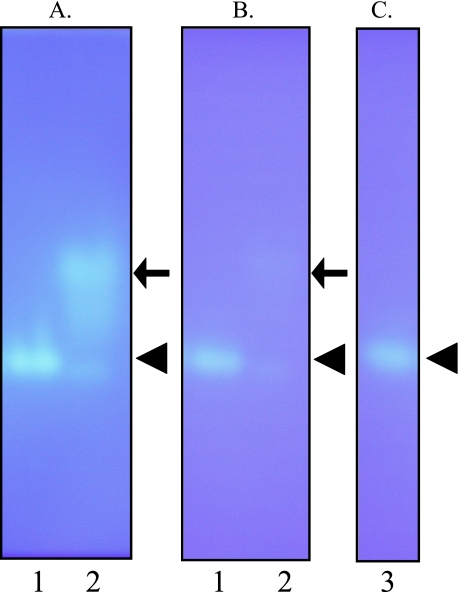
Detection of Citrulline Aminopeptidase Activity with Double Layer Fluorescent Zymography—We analyzed exo- and endopeptidase activities using Cit-MCA and FRETS-25Cit libraries, respectively, in newborn rat epidermal extract by double layer fluorescent zymography. As shown in Fig. 1, strong citrulline aminopeptidase activity was detected in rat epidermal extract at both pH 6.0 and pH 7.5 (Fig. 1, A and B, lane 1, arrowhead). When rat kidney extract was analyzed, a faint band was detected at the same position at both pHs as well as strong activity with low mobility at pH 6.0 but not at pH 7.5 (Fig. 1, A and B, lane 2, arrow). However, no endopeptidase activity against FRETS-25Cit libraries was detected (data not shown). These results suggested the presence of a novel enzyme with strong citrulline aminopeptidase activity at neutral pH in rat epidermal extract.
FIGURE 1.
Analysis of the citrulline aminopeptidase activity by double layer fluorescent zymography. The citrulline aminopeptidase activity in rat epidermis extracts was analyzed by double layer fluorescent zymography. Samples were loaded onto a 5% polyacrylamide gel at 4 °C. Following native PAGE, the gel was washed and immersed in 0.1 m phosphate buffer (pH 6.0) (A) and 0.1 m Tris-HCl (pH 7.5) (B and C) for 30 min and then covered with a 0.1 mm Cit-MCA fluorescent substrate membrane to create a double layer prior to incubation at 37 °C for 30 min. White bands of fluorescent activity generated from the citrulline aminopeptidase located in the double layer were clearly observed on the UV transilluminator. Lane 1, newborn rat epidermis extracts; lane 2, 6-week-old rat kidney extracts; lane 3, rat BH.
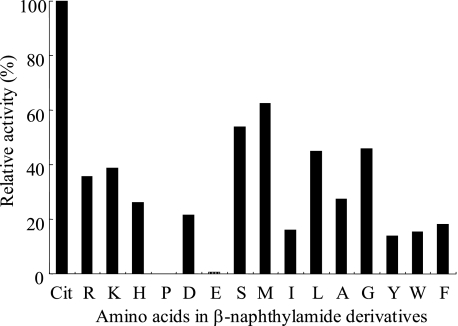
Purification and Characterization of an Enzyme with Citrulline Aminopeptidase Activity from Newborn Rat Epidermis—A three-step protocol was developed using Sephacryl S-200, DEAE-Sepharose, and Mono Q, resulting in the 4,300-fold purification of an enzyme with citrulline aminopeptidase activity from newborn rat epidermal extract. The purified enzyme revealed a single band with a molecular mass of about 48 kDa on SDS-PAGE and resolved as a single symmetrical peak with a molecular mass of about 280 kDa by gel filtration. These results suggested that the enzyme with citrulline aminopeptidase activity is a hexameric protein. We examined the aminopeptidase properties of the enzyme by comparing its activity toward various aminoacyl-β-NA substrates (Fig. 2). The enzyme released different amino acids from the substrates, indicating a broad specificity, but it released no Pro and little Glu. Cit-β-NA was the best substrate for the aminopeptidase function of the enzyme. Lineweaver-Burk plots showed a linear relationship between 1/V (nmol/min/μg) and 1/[S] (mm) and gave a Km value of 1.85 mm and Vmax value of 4.30 nmol/min/μg. However, no activity was found toward any protein substrates including deiminated filaggrin, myelin basic protein, azocasein, bovine serum albumin, myoglobin, and cystatin A. This result suggested that the enzyme probably participates in the degradation of citrulline-containing peptides in vivo.
FIGURE 2.
The properties of the enzyme with citrulline aminopeptidase activity purified from newborn rat epidermis. The purified enzyme (0.1 unit) was assayed for its activity toward 2 mm β-naphthylamide derivatives as described by Takeda et al. (31). Activity toward Cit-β-NA was set at 100%. The data are representative of three independent experiments and are expressed as the percentages of activity toward different substrates relative to activity toward Cit-β-NA.
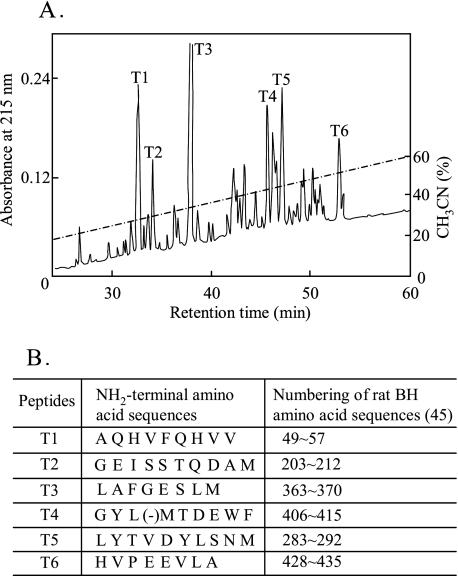
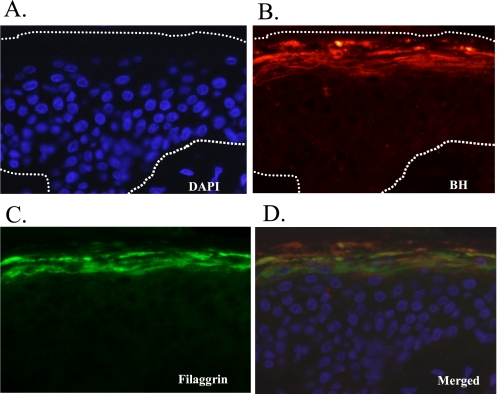
We have analyzed a peptide map and the NH2-terminal amino acid residues of tryptic peptides of the purified enzyme. The digest of the enzyme obtained with trypsin was separated by reverse-phase HPLC as shown in Fig. 3A, in which six major peaks were designated as T1–T6 in order of elution. The NH2-terminal amino acid residues of the six peptides are summarized in Fig. 3B, indicating that each tryptic peptide, T1, T2, T3, T4, T5, and T6, coincided with the amino acid numbers 49–57, 203–212, 363–370, 406–415, 283–292, and 428–434, respectively, of the sequence of rat BH (34). When double layer fluorescent zymography was conducted using rat BH, strong activity was detected at the position where the band was observed in newborn rat epidermal extract at pH 7.5 (Fig. 1C, lane 3, arrowhead). In addition, citrulline aminopeptidase activity was usually coeluted with activity to hydrolyze bleomycin throughout the process of purification from newborn rat epidermis. It was found that the enzyme with citrulline aminopeptidase activity was identical to BH in rat skin, which should show aminopeptidase activity rather than bleomycin hydrolase activity in the normal mammalian epidermis. Furthermore, indirect immunofluorescence microscopy with cryosections of normal human skin allowed us to locate an enzyme with citrulline aminopeptidase activity using anti-BH antibody and filaggrin in the granular layer of human epidermis (Fig. 4, B and C). Double staining using both antibodies revealed a convincing co-localization of BH and filaggrin in the granular layer (Fig. 4D).
FIGURE 3.
Trypsin-digested peptide map and NH2-terminal sequences of tryptic peptides of the enzyme with citrulline aminopeptidase activity purified from newborn rat epidermis. The purified enzyme with citrulline aminopeptidase activity was digested with trypsin as described under “Experimental Procedures.” A, reverse-phase HPLC of the trypsin-digested peptides. Peptides were detected by measuring UV absorption at 215 nm. The label on each peak indicates the main peptides. B, NH2-terminal amino acid sequences of the tryptic peptides (T1–T6).
FIGURE 4.
Immunohistochemical localization of BH and filaggrin. Immunofluorescence staining for BH (red) and filaggrin (green) in paraffin-embedded sections of human skin tissue was performed. A, nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). B and C, paraffin-embedded sections of human skin tissue labeled with dual immunofluorescence were subjected to confocal microscopy. BH and filaggrin co-localized in the granular layer. The upper dotted lines and bottom dotted lines indicate the outer borders of the stratum corneum and the border between the epidermis and dermis. D, merged image of all panels A–C. Magnification of ×40.
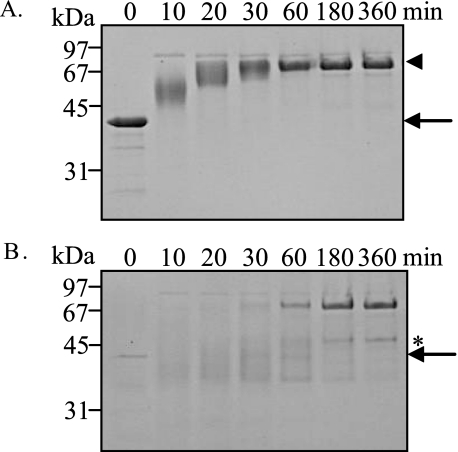
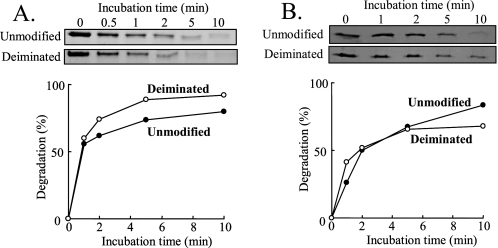
Degradation of Deiminated Filaggrin by Calpain I or Caspase 14—The recombinant filaggrin was purified from bacterial extract of Escherichia coli BL21 carrying pET-filaggrin, which contains 30 arginine residues (29). It was completely deiminated with mPAD3 that converted the guanidino side chain of arginine to the ureido side chain of citrulline, resulting in a change of mobility of the filaggrin band on SDS-PAGE (Fig. 5A, arrow to arrowhead). The deiminated filaggrin after an appropriate incubation was digested by trypsin to confirm the conversion from arginine to citrulline (Fig. 5B). The deiminated filaggrin was resistant to trypsin digestion but was fragmented at two susceptible Lys residues as presumed from the presence of faint bands with a low molecular mass (Fig. 5B, asterisks). Then, we analyzed its degradation by calpain I or caspase 14. Fig. 6, A and B, show the time course of the decrease in the filaggrin and deiminated filaggrin bands on SDS-PAGE, respectively, with calpain I or caspase 14. The initial proteolysis of both substrates was very rapid, and about 70% of the substrate was degraded within 10 min, indicating that the two proteases seem to have a high degree of specificity toward unmodified and deiminated filaggrins. Furthermore, the deiminated filaggrin was more susceptible to calpain I than the unmodified filaggrin. These results suggested calpain I to be the best protease for the degradation of deiminated filaggrin.
FIGURE 5.
Deimination of filaggrin by mPAD3. The human filaggrin repeat unit was deiminated using a mPAD3 (0.17 μm):filaggrin (8.39 μm) molar ratio of 1:50. After incubation for 0, 10, 20, 30, 60, 180, or 360 min, an aliquot of the reaction mixture was incubated at 37 °C for 1 h without (A) or with (B) trypsin, and subjected to SDS-PAGE. Arrows indicate the unmodified filaggrin, arrowheads indicate the deiminated filaggrin, and the asterisk indicates the faint band with a low molecular mass.
FIGURE 6.
Degradation of the deiminated filaggrin by calpain I or caspase 14. The degradation of the unmodified (closed circles) and deiminated (opened circles) filaggrin by calpain I (A) or caspase 14 (B) was analyzed at an enzyme: filaggrin molar ratio of 1:20 for all digests in vitro. Calpain I (0.32 μm) was incubated at 30 °C with filaggrin (6.31 μm) in 100 mm Tris-HCl (pH 7.5) containing 0.5 mm CaCl2 and 10 mm DTT, whereas caspase 14 (0.32 μm) was incubated at 37 °C with it (6.31 μm) in 100 mm HEPES (pH 7.0), 60 mm NaCl, 0.01% CHAPS, 5 mm DTT, and 1.1 m sodium citrate as a kosmotropic buffer. The extent of degradation was determined by SDS-PAGE followed by a densitometric analysis.
To further characterize the degradation of deiminated filaggrin by calpain I, we measured MALDI-TOF MS. Supplemental Fig. S1 shows the MS spectra at m/z 4,000–40,000 of the products from unmodified (A, C, and E) and deiminated (B, D, and F) filaggrin degraded by calpain I. In spectra obtained at the earliest time point, signals for 1+, 2+, and 3+ ions from both filaggrins can be seen (labeled A1+, A2+, and A3+ for unmodified filaggrin and B1+, B2+, and B3+ for deiminated filaggrin) as well as a few other ions (A and B). These signals disappeared, and many new signals appeared in the spectra of the products of both filaggrins digested for 60 min by calpain I (C–F), indicating that calpain I degraded the filaggrins into many peptides of different mass. Furthermore, the spectral pattern showed that deiminated filaggrin was degraded into shorter fragments than unmodified filaggrin by calpain I. Supplemental Fig. S2 shows the MS spectra at m/z 3,500–15,000 of products from unmodified (A) and deiminated (B) filaggrins degraded by caspase 14. Caspase 14 degraded them into a few peptides of limited mass with slightly different patterns. These results indicated that calpain I acts preferentially on deiminated filaggrin when compared with caspase 14.
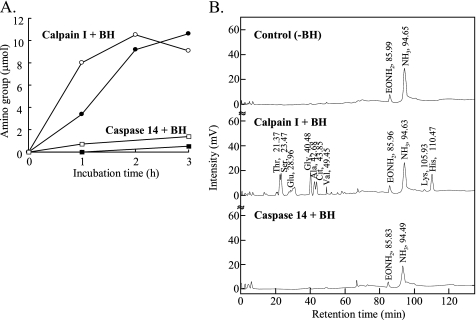
Detection of Free Amino Acids Derived from Peptides of Deiminated Filaggrin Treated with Calpain I or Caspase 14—We investigated the free amino acids released from calpain I- or caspase 14-digested filaggrin peptides by BH using a fluorescamine post-labeling method and amino acid analysis. Free amino groups were released from the calpain I-digested filaggrin and deiminated filaggrin peptides treated with BH in a time-dependent manner (Fig. 7A). The peptides derived from deiminated filaggrin were more susceptible to BH than those from filaggrin. Furthermore, serine, glycine, alanine, citrulline, threonine, and histidine, etc., composing deiminated filaggrin peptides, were released by BH (Fig. 7B). On the other hand, few amounts of free amino groups or amino acids were detected from the caspase 14-digested peptides treated by BH. These results suggest that calpain I and BH are essential for the breakdown of deiminated filaggrin peptides into amino acids.
FIGURE 7.
Free amino acids released by BH from peptides of deiminated filaggrin degraded with calpain I or caspase 14. A, the time course of the released free amino groups from calpain I- or caspase 14-digested filaggrin peptides (closed circles and squares) and deiminated filaggrin peptides (opened circles and squares) by BH in vitro. After the digestion of unmodified and deiminated filaggrin (6.31 μm) with calpain I (0.32 μm) (circles) or caspase 14 (0.32μm) (squares) for 1 h, the digests treated with caspase 14 were filtrated by centrifugation with a Microcon YM-3 (Millipore) for the exclusion of kosmotropic salts, and then the volume was adjusted. The reaction products were incubated with BH in 50 mm HEPES-NaOH (pH 7.5), 10 mm DTT, and 5 mm EDTA at 37 °C. Aliquots were removed at different times, and the enzymatic reaction was stopped by boiling. The free amino groups were measured with fluorescamine. B, the representative elution patterns of amino acid in reaction solutions of calpain I- or caspase 14-digested deiminated filaggrin peptides with BH. Control indicated an elution pattern of amino acids in the reaction solution without BH. Samples were prepared according to the methods as described under “Experimental Procedures.” Free amino acids were analyzed using a Hitachi model L-8800 amino acid analyzer equipped with columns (number 2622: 4.6 × 40 mm + 4.6 × 40 mm) by gradient elution with MCI buffer L-8500-PF kit (Wako) according to the physiological fluids analysis method. Amino acids post-labeled with ninhydrin were detected by measuring the absorbance at 440 and 570 nm. EONH2 indicates ethanolamine as internal marker.
DISCUSSION
We identified and purified a novel enzyme with citrulline aminopeptidase activity in a rat epidermal homogenate. An unexpected result of the present study is that the enzyme is identical with rat BH, a bleomycin-detoxifying enzyme whose natural function and substrate are still unknown. Our finding suggests that BH plays an important role in the synthesis of free amino acids acting as NMFs from deiminated filaggrin peptides in the stratum corneum.
BH is similar to a 20 S proteasome and also belongs to a family of self-compartmentalizing intracellular proteases (34, 35). The evolutionary conservation and wide distribution of BH imply that the enzyme has a conserved cellular function. However, this function has remained enigmatic for many years. Recently, BH was shown to be involved in the post-translational processing of peptides for antigen presentation (36) and a protective mechanism against homocysteine toxicity (37). Furthermore, BH has been shown to interact with several proteins, such as cAMP-binding ectoprotein in yeast (38), the human homologue of ubiquitin-conjugating enzyme 9 (39), human ribosomal proteins (40), and amyloid precursor protein (41). In the last case, we inferred that BH participated in the degradation of amyloid β-peptides derived from amyloid precursor protein (42). Schwartz et al. (43) demonstrated that BH knockout newborn null mice displayed a mild and transient ichthyosis-like phenotype that strikingly resembles the phenotype of the filaggrin-deficient flaky tail ft/ft mouse mutant and also exhibited the tail auto-amputation phenotype (44), indicating that BH has an important role in maintaining epidermal integrity. The novel enzyme exhibited mainly aminopeptidase activity with broad specificity toward aminoacyl-β-naphthylamide derivatives without Pro, among which Cit-β-NA was the most effective substrate at neutral pH. However, it had no activity toward any protein substrates including myelin basic protein, azocasein, bovine serum albumin, myoglobin, and cystatin A. The enzyme was co-localized with filaggrin in the upper layers of the stratum corneum, where it was present in large amounts, in mammalian skin (31, 45–47). These results strongly suggested that BH participated in the release of free amino acids including citrulline in peptides processed from deiminated filaggrin in mammalian skin.
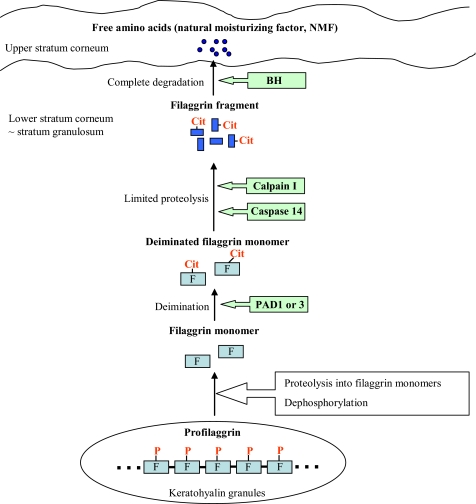
We proposed a reasonable scheme for the degradation of filaggrin as shown in Fig. 8. NMFs are present at high concentrations within corneocytes and represent up to 20–30% of the dry weight of the stratum corneum (48). By absorbing atmospheric water and dissolving in their own water, hygroscopic NMF components act as very efficient humectants (2). NMFs consist primarily of amino acids or their derivatives such as pyrrolidine carboxylic acid and urocanic acid, together with lactic acid and urea, citrate, and sugars (49, 50). The calcium concentration is critical to epidermal differentiation and implicated in the expression and post-transcriptional modification of numerous proteins in the epidermis (51, 52). PAD activity was significantly regulated by the calcium concentration during epidermal differentiation, where PAD1 and/or PAD3 probably participated in the deimination of filaggrin monomers, because of their potential co-localization demonstrated by immunoelectron microscopy (53, 54). However, it is not known which peptidase is involved in the degradation of deiminated filaggrin into its peptides and then into free amino acids. Our findings show that calpain I, a calcium-activated cysteine protease in cytoplasm, preferentially cleaved the deiminated filaggrin to peptides of low molecular masses when compared with filaggrin. Finally, those peptides were degraded into free amino acids by BH. The recent review summarized that caspase 14 is involved in the processing of filaggrin, not in the profilaggrin-processing pathway (16). The present study revealed that caspase 14 processed the deiminated filaggrin into limited fragments because of aspartate residues at a specific cleavage site. However, there was no difference in susceptibility to caspase 14 with deimination, and also, few amounts of amino acids were released from caspase 14-digested peptides by BH. These results suggest that calpain I plays a major role in the processing of deiminated filaggrin into peptides.
FIGURE 8.
Scheme of the degradation of filaggrin in mammalian epidermis. Filaggrin (F) is a component of the cornified envelope in the outer layer of the epidermis. It accumulates in the keratohyalin granules as a high molecular weight, extremely insoluble phosphorylated precursor protein (10). During terminal differentiation, the phosphorylated filaggrin (P-F) is dephosphorylated and then proteolytically cleaved to polypeptides (each ∼35 kDa). The enzymatic properties and epidermal localization of PAD1 and/or PAD3 suggest that they are responsible for the deimination of filaggrin (54). As shown in the present study, caspase 14 partially participates in the processing of the deiminated filaggrin (Cit-F) into limited fragments as illustrated by Denecker et al. (16). On the other hand, calpain I preferentially fragments deiminated filaggrin into the smaller peptides, and then these peptides are degraded into free amino acids acting as NMFs by BH in the lower stratum corneum.
In conclusion, the results of these experiments have identified BH as essential for the degradation of the peptides derived from deiminated filaggrin into amino acids, although more work is needed to reveal the precise functional role of BH during terminal epidermal differentiation.
Supplementary Material
This work was supported by research grants from Sagami Women's University.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental figures.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: PAD, peptidylarginine deiminase; NMF, natural moisturizing factor; Cit-MCA, citrulline-4-methylcoumaryl-7-amide; Cit-β-NA, citrulline-β-naphthylamide; BH, bleomycin hydrolase; DTT, dithiothreitol; PBS, phosphate-buffered saline; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; FRETS, fluorescence resonance energy transfer substrate; MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight; MS, mass spectrometry; HPLC, high pressure liquid chromatography.
References
- 1.Watt, F. M. (1989) Curr. Opin. Cell Biol. 1 1107–1115 [DOI] [PubMed] [Google Scholar]
- 2.Rawlings, A. V., and Harding, C. R. (2004) Dermatol. Ther. 17 Suppl. 1, 43–48 [DOI] [PubMed] [Google Scholar]
- 3.Wiedow, O., Schroder, J. M., Gregory, H., Young, J. A., and Christophers, E. (1990) J. Biol. Chem. 265 14791–14795 [PubMed] [Google Scholar]
- 4.Hohl, D., Mehrel, T., Lichti, U., Turner, M. L., Roop, D. R., and Steinert, P. M. (1991) J. Biol. Chem. 266 6626–6636 [PubMed] [Google Scholar]
- 5.Eckert, R. L., Yaffe, M. B., Crish, J. F., Murthy, S., Rorke, E., and Welter, J. F. (1993) J. Investig. Dermatol. 100 613–617 [DOI] [PubMed] [Google Scholar]
- 6.Steinert, P. M., and Marekov, L. N. (1995) J. Biol. Chem. 270 17702–17711 [DOI] [PubMed] [Google Scholar]
- 7.Takahashi, M, Tezuka, T., and Katunuma, N. (1996) Arch. Biochem. Biophys. 329 123–126 [DOI] [PubMed] [Google Scholar]
- 8.Harding, C. R., and Scott, I. R. (1983) J. Mol. Biol. 170 651–673 [DOI] [PubMed] [Google Scholar]
- 9.Dale, B. A., Resing, K. A., and Lonsdale-Eccles, J. D. (1985) Ann. N. Y. Acad. Sci. 455 330–342 [DOI] [PubMed] [Google Scholar]
- 10.McGrath, J. A., and Uitto, J. (2008) Trends Mol. Med. 14 20–27 [DOI] [PubMed] [Google Scholar]
- 11.Scott, I. R., and Harding, C. R. (1986) Dev. Biol. 115 84–92 [DOI] [PubMed] [Google Scholar]
- 12.Rawlings, A. V., Scott, I. R., Harding, C. R., and Bowser, P. A. (1994) J. Investig. Dermatol. 103 731–741 [DOI] [PubMed] [Google Scholar]
- 13.Verdier-Sevrain, S., and Bonte, F. (2007) J. Cosmet. Dermatol. 6 75–82 [DOI] [PubMed] [Google Scholar]
- 14.Eckert, R. L., and Rorke, E. A. (1989) Environ. Health Perspect. 80 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, S. C., Kim, I. G., Marekov, L. N., O'Keefe, E. J., Parry, D. A., and Steinert, P. M. (1993) J. Biol. Chem. 268 12164–12176 [PubMed] [Google Scholar]
- 16.Denecker, G., Ovaere, P., Vandenabeele, P., and Declercq, W. (2008) J. Cell Biol. 180 451–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarcsa, E., Marekov, L. N., Mei, G., Melino, G., Lee, S. C., and Steinert, P. M. (1996) J. Biol. Chem. 271 30709–30716 [DOI] [PubMed] [Google Scholar]
- 18.Vossenaar, E. R., Zendman, A. J., Van Venrooji, W. J., and Pruijin, G. J. (2003) BioEssays 25 1106–1118 [DOI] [PubMed] [Google Scholar]
- 19.Chavanas, S., Mechin, M. C., Nachat, R., Adoue, V., Coudane, F., Serre, G., and Simon, M. (2006) J. Dermatol. Sci. 44 63–72 [DOI] [PubMed] [Google Scholar]
- 20.Presland, R. B., Kimball, J. R., Kautsky, M. B., Lewis, S. P., Lo, C. Y., and Dale, B. A. (1997) J. Investig. Dermatol. 108 170–178 [DOI] [PubMed] [Google Scholar]
- 21.Pearton, D. J., Nirunsuksiri, W., Rehemtulla, A., Lewis, S. P., Presland, R. B., and Dale, B. A. (2001) Exp. Dermatol. 10 193–203 [DOI] [PubMed] [Google Scholar]
- 22.Resing, K. A., Walsh, K. A., Haugen-Scofield, J., and Dale, B. A. (1989) J. Biol. Chem. 264 1837–1845 [PubMed] [Google Scholar]
- 23.Resing, K. A., al-Alawi, N., Blomquist, C., Fleckman, P., and Dale, B. A. (1993) J. Biol. Chem. 268 25139–25145 [PubMed] [Google Scholar]
- 24.Resing, K. A., Thulin, C., Whiting, K., al-Alawi, N., and Mostad, S. (1995) J. Biol. Chem. 270 28193–28198 [DOI] [PubMed] [Google Scholar]
- 25.Yamazaki, M., Ishidoh, K., Suga, Y., Saido, T. C., Kawashima, S., Suzuki, K., Kominami, E., and Ogawa, H. (1997) Biochem. Biophys. Res. Com. 235 652–656 [DOI] [PubMed] [Google Scholar]
- 26.List, K., Szabo, R., Wertz, P. W., Segre, J., Haudenschild, C. C., Kim, S. Y., and Bugge, T. H (2003) J. Cell Biol. 163 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leyvraz, C., Charles, R. P., Rubera, I., Guitard, M., Rotman, S., Breiden, B., Sandhoff, K., and Hummler, E. (2005) J. Cell Biol. 170 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denecker, G., Hoste, E., Gilbert, B., Hochepied, T., Ovaere, P., Lippens, S., Van Den Broecke, C., Van Damme, P., D'Herde, K., Hachem, J. P., Borgonie, G., Presland, R. B., Schoonjans, L., Libert, C., Vandekerckhove, J., Gevaert, K., Vandenabeele, P., and Declercq, W. (2007) Nat. Cell Biol. 9 666–674 [DOI] [PubMed] [Google Scholar]
- 29.Kanno, T., Kawada, A., Yamanouchi, J., Yoshida-Noro, C., Yoshiki, A., Shiraiwa, M., Kusakabe, M., Manabe, M., Tezuka, T., and Takahara, H. (2000) J. Investig. Dermatol. 115 813–823 [DOI] [PubMed] [Google Scholar]
- 30.Ohsugi, I., Takahara, H., Shiraiwa, M., and Sugawara, K. (1995) Arch. Biochem. Biophys. 317 62–68 [DOI] [PubMed] [Google Scholar]
- 31.Takeda, A., Higuchi, D., Yamamoto, T., Nakamura, Y., Masuda, Y., Hirabayashi, T., and Nakaya, K. (1996) J. Biochem. (Tokyo) 119 29–36 [DOI] [PubMed] [Google Scholar]
- 32.Katunuma, N., Le, Q. T., Miyauchi, R., and Hirose, S. (2005) Anal. Biochem. 347 208–212 [DOI] [PubMed] [Google Scholar]
- 33.Tamura, N., Lottspeich, F., Baumeister, W., and Tamura, T. (1998) Cell 95 637–648 [DOI] [PubMed] [Google Scholar]
- 34.Joshua-Tor, L., and Johnston, S. A. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D., and Woessner, J. F., eds) 2nd Ed., pp. 1197–1201, Elsevier Ltd., New York and London
- 35.Berti, P., and Storer, A. C. (1995) J. Mol. Biol. 246 273–283 [DOI] [PubMed] [Google Scholar]
- 36.Stoltze, L., Schirle, M., Schwarz, G., Schroter, C., Thompson, M. W., Hersh, L. B., Kalbacher, H., Stevanovic, S., Rammensee, H. G., and Schild, H. (2000) Nat. Immunol. 1 413–418 [DOI] [PubMed] [Google Scholar]
- 37.Zimny, J., Sikora, M., Guranowski, A., and Lakubowski, H. (2006) J. Biol. Chem. 281 22485–22492 [DOI] [PubMed] [Google Scholar]
- 38.Niemer, I., Muller, G., Strobel, G., and Bandlow, W. (1997) Curr. Genet. 32 41–51 [DOI] [PubMed] [Google Scholar]
- 39.Koldamova, R. P., Lefterov, I. M., Di Sabella, M. T., and Lazo, J. S. (1998) Mol. Pharmacol. 54 954–961 [DOI] [PubMed] [Google Scholar]
- 40.Koldamova, R. P., Lefterov, I. M., Di Sabella, M. T., Almonte, C., Watkins, S. C., and Lazo, J. S. (1999) Biochemistry 38 7111–7117 [DOI] [PubMed] [Google Scholar]
- 41.Lefterov, I. M., Koldamova, R. P., and Lazo, J. S. (2000) FASEB J. 14 1837–1847 [DOI] [PubMed] [Google Scholar]
- 42.Kajiya, A., Kaji, H., Isobe, T., and Takeda, A. (2006) Protein Pept. Lett. 13 119–123 [DOI] [PubMed] [Google Scholar]
- 43.Schwartz, D. R., Homanics, G. E., Hoyt, D. G., Klein, E., Abernethy, J., and Lazo, J. S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4680–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Presland, R. B., Boggess, D., Lewis, S. P., Hull, C., Fleckman, P., and Sundberg, J. P. (2000) J. Investig. Dermatol. 115 1072–1081 [DOI] [PubMed] [Google Scholar]
- 45.Takeda, A., Masuda, Y., Yamamoto, T., Hirabayashi, T., Nakamura, Y., and Nakaya, K. (1996) J. Biochem. (Tokyo) 120 353–359 [DOI] [PubMed] [Google Scholar]
- 46.Takeda, A., Nonaka, M., Ishikawa, A., and Higuchi, D. (1999) Arch. Dermatol. Res. 291 238–240 [DOI] [PubMed] [Google Scholar]
- 47.Kamata, Y., Itoh, Y., Kajiya, A., Karasawa, S., Sakatani, C., Takekoshi, S., Osamura, R. Y., and Takeda, A. (2007) J. Biochem. (Tokyo) 141 69–76 [DOI] [PubMed] [Google Scholar]
- 48.Trianse, S. J. (1974) Cosmetics Perfumery 89 57 [Google Scholar]
- 49.Cler, E. J., and Fourtanier, A. (1981) Int. J. Cosmet. Sci. 3 101–113 [DOI] [PubMed] [Google Scholar]
- 50.Scott, I. R., Harding, C. R., and Barrett, J. G. (1982) Biochim. Biophys. Acta 719 110–117 [DOI] [PubMed] [Google Scholar]
- 51.Bikle, D. D., and Pillai, S. (1993) Endocr. Rev. 14 3–19 [DOI] [PubMed] [Google Scholar]
- 52.Elias, P. M., Nau, P., Hanley, K., Cullander, C., Crumrine, D., Bench, G., Sideras-Haddad, E., Mauro, T., Williams, M. L., and Feingold, K. R. (1998) J. Investig. Dermatol. 110 399–404 [DOI] [PubMed] [Google Scholar]
- 53.Senshu, T., Kan, S., Ogawa, H., Manabe, M., and Asaga, H. (1996) Biochem. Biophys. Res. Com. 225 712–719 [DOI] [PubMed] [Google Scholar]
- 54.Mechin, M. C., Enji, M., Nachat, R., Chavanas, M., Ishida-Yamamoto, A., Serre, G., Takahara, H., and Simon, M. (2005) CMLS Cell Mol. Life Sci. 62 1984–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.