Abstract
Nucleotide excision repair is distinguished from other DNA repair pathways by its ability to process a wide range of structurally unrelated DNA lesions. In bacteria, damage recognition is achieved by the UvrA·UvrB ensemble. Here, we report the structure of the complex between the interaction domains of UvrA and UvrB. These domains are necessary and sufficient for full-length UvrA and UvrB to associate and thereby form the DNA damage-sensing complex of bacterial nucleotide excision repair. The crystal structure and accompanying biochemical analyses suggest a model for the complete damage-sensing complex.
Nucleotide excision repair is distinguished from other DNA repair pathways by its ability to process a diverse set of lesions. In bacteria, the initial steps are carried out by three proteins: UvrA, UvrB, and UvrC. The UvrA·UvrB complex conducts surveillance of DNA and recognizes damage. Having located a lesion, UvrA “loads” UvrB onto the DNA at the damaged sites and then dissociates. Damage searching, formation of the UvrB·DNA “preincision” complex, and dissociation of UvrA are regulated by ATP (1). UvrB subsequently recruits the endonuclease UvrC, which catalyzes incisions on either side of the lesion (2, 3). Following incision, UvrC and the damage-containing oligonucleotide are removed by UvrD (helicase II), whereas UvrB remains bound to the gapped DNA and recruits DNA polymerase I for repair synthesis. Sealing of the single-stranded nick completes the repair process and restores the original DNA sequence (4).
Since its discovery more than 40 years ago, bacterial nucleotide excision repair has been extensively studied, resulting in a large body of work that describes the protein components and the details of how they operate. Notwithstanding the trove of genetic and biochemical data, several key questions remain unanswered. For example, how does the same set of proteins handle a diverse set of lesions while maintaining specificity? How do UvrA and UvrB cooperate during damage recognition, and what is the precise role of ATP? Ongoing studies in the field, including those described below, aim to address these issues.
Recently, we reported the structure of Geobacillus stearothermophilus UvrA and the identification of binding sites for DNA and UvrB (5). We also established that the identified UvrB-binding domain is necessary and sufficient to mediate the UvrA-UvrB interaction and that the isolated interaction domains of UvrA (5) and UvrB (6) bind to each other in solution.
To understand the interaction between UvrA and UvrB, we have determined the crystal structure of the complex between the two isolated interaction domains. The structure revealed that UvrA-UvrB interaction interface is largely polar, mediated by several highly conserved charged residues. Site-directed mutagenesis and biochemical characterization of the mutant proteins confirmed the importance of the observed interactions. Based on the interaction domain complex structure, we have constructed a structural model for the full-length UvrA·UvrB ensemble and propose two models for lesion recognition that will serve as a basis for future experiments.
EXPERIMENTAL PROCEDURES
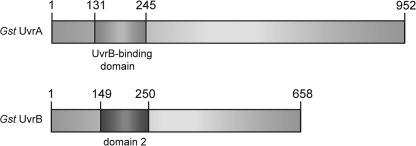
Expression and Purification of G. stearothermophilus UvrA and UvrB Interaction Domain Complex—The DNA sequences encoding the interaction domains (Fig. 1) were amplified from the plasmids containing the genes for full-length UvrA and UvrB (5), cloned into pET-28a (+) (Novagen; see Table 3), and confirmed by sequencing. The UvrA and UvrB domain expression constructs contained residues 131–245 of UvrA and residues 149–250 of UvrB, respectively, with an N-terminal His6 tag and a thrombin cleavage site. The proteins were expressed in Escherichia coli BL21(DE3) pLysS. The cells were grown in LB broth at 37 °C until A600 reached 0.5–0.6, at which point expression was induced by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside. The cells were allowed to grow at 30 °C for 4 h and harvested by centrifugation. The cell pellet was resuspended in lysis buffer (50 mm NaPO4, pH 8.0, 500 mm NaCl, 10 mm imidazole, 5 mm β-ME),4 flash frozen in liquid nitrogen, and stored at -80 °C.
FIGURE 1.
The location of the interaction domains in the primary sequence of G. stearothermophilus UvrA and UvrB.
TABLE 3.
Sequences of primers used in the amplification of G. stearothermophilus UvrA and UvrB interaction domains and the construction of mutants The recognition sequences of the restriction enzymes used for cloning of the PCR products are underlined (NdeI and HindIII). The positions of Arg → Glu, Asp → Arg, and Glu → Arg mutations are in bold type. fwd, forward, rev, reverse.
| Primers | Sequences (5′ → 3′) |
|---|---|
| UvrA 131–245 fwd | CGCGGCAGCCATATGCCCATTTGCCCGACGCAC |
| UvrA 131–245 rev | GGCCGCAAGCTTTTACGAAAAGCCGCAGTACGGAC |
| UvrB 149–250 fwd | CGCGGCAGCCATATGGGGTCGCCGGAAGAATATCGG |
| UvrB 149–250 rev | GGCCGCAAGCTTTTACACGAAGTGCGACGCCGG |
| UvrA-R176E fwd | ATTCGCAAACAAGGGTATGTGGAAGTCCGTATTGACCGCGAGATG |
| UvrA-R176E rev | CATCTCGCGGTCAATACGGACTTCCACATACCCTTGTTTGCGAAT |
| UvrA-E185R fwd | CGTATTGACCGCGAGATGCGCCGTTTGACGGGGGACATTGAGCTT |
| UvrA-E185R rev | AAGCTCAATGTCCCCCGTCAAACGGCGCATCTCGCGGTCAATACG |
| UvrA-D219R fwd | GGCATCGCCGCCAGGCTTGCCCGTTCGCTTGAGACGGCGCTGAAG |
| UvrA-D219R rev | CTTCAGCGCCGTCTCAAGCGAACGGGCAAGCCTGGCGGCGATGCC |
| UvrA-R206E fwd | CATTCGATTGATGTCGTCGTCGACGAAATCATCATCAAAGACGGCATCGCC |
| UvrA-R206E rev | GGCGATGCCGTCTTTGATGATGATTTCGTCGACGACGACATCAATCGAATG |
| UvrB-R183E fwd | CTCGTTGACATCCAATACGACGAAAATGACATCGATTTTCGCCGCGG |
| UvrB-R183E rev | CCGCGGCGAAAATCGATGTCATTTTCGTCGTATTGGATGTCAACGAG |
| UvrB-D198R fwd | GGCACGTTCCGCGTCCGCGGCCGTGTTGTCGAAATTTTCCCCGCG |
| UvrB-D198R rev | CGCGGGGAAAATTTCGACAACACGGCCGCGGACGCGGAACGTGCC |
| UvrB-E215R fwd | GTCGCGCGATGAACATTGCATTCGCGTCCGCTTTTTCGGCGATGAAATCGAG |
| UvrB-E215R rev | CTCGATTTCATCGCCGAAAAAGCGGACGCGAATGCAATGTTCATCGCGCGAC |
| UvrB-E222R fwd | GAATTTTTCGGCGATGAAATCCGTCGCATCCGCGAGGTGGACGCC |
| UvrB-E222R rev | GGCGTCCACCTCGCGGATGCGACGGATTTCATCGCCGAAAAATTC |
| UvrB-R223E fwd | TTTTTCGGCGATGAAATCGAGGAAATCCGCGAGGTGGACGCCTTA |
| UvrB-R223E rev | TAAGGCGTCCACCTCGCGGATTTCCTCGATTTCATCGCCGAAAAA |
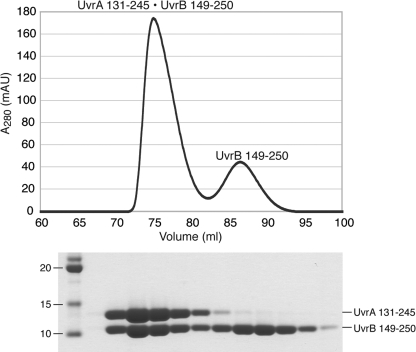
UvrA 131–245 and UvrB 149–250 were separately purified using nickel-nitrilotriacetic acid-agarose (Qiagen), after which the His6 tag was removed by thrombin cleavage. The resulting proteins were further purified by size exclusion chromatography (Superdex 75; GE Healthcare; 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm β-ME). The interaction domain complex was made by mixing UvrA 131–245 with molar excess of UvrB 149–250. The complex was then purified from excess UvrB 149–250 by size exclusion chromatography (Superdex 75; GE Healthcare; 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm β-ME; Fig. 2).
FIGURE 2.
Purification of the UvrA·UvrB interaction domain complex for structural studies. The complex was formed by mixing UvrA 131–245 with molar excess of UvrB 149–250 and purified by size exclusion chromatography.
Crystallization of UvrA·UvrB Interaction Domain Complex— The UvrA·UvrB interaction domain complex was crystallized using the hanging drop vapor diffusion method. The crystallization drop consisted of a 1:1 ratio of protein solution (20–30 mg/ml complex in 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm β-ME) and reservoir solution (12–18% polyethylene glycol 4000, 0.1 m sodium acetate, pH 5.0, 0.2 m NaCl, or ammonium sulfate). The crystals grew to ∼200 × 200 × 200 μm in 1 week. For x-ray diffraction, the crystals were transferred to a 10-μl drop of crystallization buffer containing 15% (±) 1, 2-propanediol for cryoprotection and flash frozen in liquid nitrogen.
Structure Determination—X-ray diffraction data were collected on the NE-CAT beamline 24ID-C (λ = 0.97949 Å) at the Advanced Photon Source, Argonne National Laboratory. The processed data (HKL2000 (7)) revealed that the crystal belonged to the tetragonal space group P41212 with cell parameters a = b = 84.49 Å, c = 60.87 Å, α = β = γ = 90.0°, and contained one complex in the asymmetric unit. The structure was solved at 1.8 Å by molecular replacement (PHASER (8)) using the structures of the corresponding domains in the full-length proteins (residues 131–153 and 200–245 of UvrA from 2R6F (5) and residues 157–250 of UvrB from 1T5L (6)) as search models. Automated model building (9) followed by manual building (10) and crystallographic refinement (11–13) resulted in a model with residues 131–245 of UvrA, residues 157–250 of UvrB, and 53 water molecules. The model displays good geometry (91.1 and 8.9% of the residues in the most favored and additional allowed regions of Ramachandran space, PROCHECK (14)) with a crystallographic R factor of 22.99% and Rfree of 24.75%. The accuracy of the model was confirmed by the positions of selenium atoms determined using anomalous diffraction data collected on a selenomethionine-substituted crystal (λ = 0.97926 Å). Data collection and refinement statistics are in Table 1. The coordinates and structure factors have been deposited to the Protein Data Bank with the accession code 3FPN.
TABLE 1.
Data collection and refinement statistics The data were collected from a single crystal. The values in parentheses are for the highest resolution shell. Rsym = Σ|I – <I>|/ΣI where I is the integrated intensity of a given reflection. Rwork = Σ|F(obs) – F(calc)|/ΣF(obs), where F(obs) and F(calc) are the observed and calculated structure factor amplitudes, respectively. Rfree = Σ|F(obs) – F(calc)|/ΣF(obs), calculated using 5% of the data omitted from the refinement.
| UvrA 131–245·UvrB 149–250 | SeMet UvrA 131–245·UvrB 149–250 | |
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Cell dimensions | ||
| a, b, c (Å) | 84.49, 84.49, 60.87 | 84.79, 84.79, 61.00 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0-1.80 (1.83-1.80) | 50.0-2.20 (2.24-2.20) |
| Rsym (%) | 0.043 (0.443) | 0.120 (0.440) |
| I/σI | 70.7 (6.0) | 70.4 (10.5) |
| Unique reflections | 20904 (1012) | 11885 (569) |
| Completeness (%) | 99.9 (100.0) | 100.0 (100.0) |
| Redundancy | 14.2 (14.5) | 26.5 (23.9) |
| Refinement | ||
| Resolution (Å) | 50.0-1.80 | |
| No. reflections | 19,555 | |
| Rwork/Rfree (%) | 22.99/24.75 | |
| No. atoms | ||
| Protein | 1704 | |
| Water | 53 | |
| B factors | ||
| Protein | 35.45 | |
| Water | 32.95 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.0118 | |
| Bond angles (°) | 1.367 | |
| Ramachandran statistics | ||
| Most favored (%) | 91.1 | |
| Additional allowed (%) | 8.9 | |
| Generously allowed (%) | 0.0 | |
| Disallowed (%) | 0.0 |
Construction of UvrA and UvrB Mutants—Point mutants of full-length UvrA and UvrB were constructed using QuikChange II XL site-directed mutagenesis kit (Stratagene), and the mutations were confirmed by sequencing (see Table 3). The mutant proteins were purified using the wild-type protocol (5).
Biochemical Characterization of UvrA and UvrB Mutants— Interaction between UvrA and UvrB (8 nmol each) was analyzed by size exclusion chromatography (Superdex 200; GE Healthcare) at 4 °C in UvrAB complex buffer (20 mm Tris-HCl, pH 7.5, 150 mm KCl, 5% (v/v) glycerol, 5 mm β-ME, 5 mm MgCl2, 2 mm ATP). The presence of proteins in each fraction was determined by SDS-PAGE.
Multi-angle Laser Light Scattering—UvrA·UvrB complexes were formed in the following ratios (2A:1B, 1A:1B, and 1A:2B) in 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5% (v/v) glycerol, 5 mm β-ME, 5 mm MgCl2, and 2 mm ATP. The samples were then applied to a Shodex KW-804 column equilibrated with the same buffer. Light scattering and refractive index signals were measured using a Wyatt Optilab and Dawn EOS system. Scattering curves were processed using the provided Astra software package (15, 16).
RESULTS
Structure of the UvrA·UvrB Interaction Domain Complex— The G. stearothermophilus UvrA·UvrB interaction domain complex crystallized in the tetragonal space group P41212 with one molecule of each protein in the asymmetric unit. The structure was solved by molecular replacement at 1.8 Å using the relevant domains in the full-length protein structures as search models. The final model consists of residues 131–245 of UvrA, residues 157–250 of UvrB, and 53 water molecules, with a crystallographic R factor of 22.99% and Rfree of 24.75%. The accuracy of the model was confirmed by the positions of selenium atoms determined using anomalous diffraction recorded on a selenomethionine-substituted crystal (Table 1).
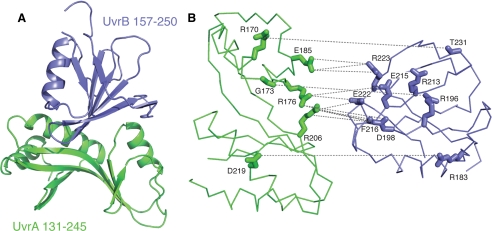
The overall structure of the complex is shown in Fig. 3A. Residues 154–199 of UvrA that were disordered in our previous structure (Protein Data Bank code 2R6F) (5) are now ordered and form a part of the interface. Residues 157–250 of UvrB are identical to the corresponding region in the full-length Bacillus caldotenax UvrB (Protein Data Bank code 1T5L) (6). Interestingly, residues 157–250 of UvrB are disordered in every structure in the data base except for this one, in which a point mutation Y96A led to a favorable crystal packing interaction. These observations suggest that the substructures that are involved in the UvrA-UvrB interaction are largely flexible, both internally and with respect to the rest of the protein, and only adopt a single conformation upon binding to its interaction partner.
FIGURE 3.
Structural basis for UvrA-UvrB interaction. UvrA 131–245 and UvrB 157–250 are shown in green and blue, respectively. A, overall structure of the complex between the interaction domains of UvrA (residues 131–245) and UvrB (residues 157–250) shown as a ribbon diagram. B, exploded view of the interaction interface. The interface is largely polar, consisting of a large number of direct and water-mediated hydrogen bonds, as well as electrostatic interactions between conserved residues. UvrA and UvrB interaction domains are shown as Cα trace. Residues that are involved in direct contacts across the interface are shown as sticks. The interactions are drawn as dashed lines. This view was generated by separating the two proteins by 10 Å and rotating them by 65° away from each other. This orientation was chosen to most clearly depict the interactions (see Table 2 for a complete list of interactions).
The interface between UvrA and UvrB is largely polar. Contacts consist of both direct and water-mediated hydrogen bonds, as well as electrostatic interactions between charged residues (Fig. 3B and Tables 2 and 3). For example, Asp219 of UvrA makes a direct contact with Arg183, and water-mediated contacts Arg169 and Gly197 of UvrB. The amino acids that participate in the UvrA-UvrB interaction are highly conserved (17). The composition of the interface is consistent with experiments suggesting that the A-B interaction is considerably weakened in high ionic strength buffers (6, 18).
TABLE 2.
Interactions observed at the interface between UvrA and UvrB interaction domains
| UvrA residues | UvrB residues | Distancea | Water-mediated | ||
|---|---|---|---|---|---|
| Å | |||||
| Arg170 | NH1 | Thr231 | O | 3.6 | No |
| Lys171 | O | Arg194 | NH1 | 2.8, 3.5 | Yes |
| Gly173 | O | Arg196 | NH2 | 2.8 | No |
| Arg176 | NH1 | Glu215 | OE1 | 3.4 | No |
| Glu222 | OE2 | 3.2 | No | ||
| Asp198 | O | 3.1, 3.2 | Yes | ||
| Glu215 | OE1 | 3.2, 2.6 | Yes | ||
| Phe216 | N | 3.1, 3.1 | Yes | ||
| Phe216 | O | 3.1, 2.8 | Yes | ||
| Glu222 | OE2 | 3.2, 2.7 | Yes | ||
| NH2 | Asp198 | OD1 | 3.0 | No | |
| Asp198 | OD2 | 3.4 | No | ||
| Asp198 | OD1 | 2.9, 2.7 | Yes | ||
| Asp198 | OD2 | 2.9, 3.3 | Yes | ||
| Asp198 | O | 2.7, 3.2 | Yes | ||
| Phe216 | N | 2.7, 3.1 | Yes | ||
| Phe216 | O | 2.7, 2.8 | Yes | ||
| Glu185 | OE1 | Arg223 | NH2 | 3.4 | No |
| OE2 | Arg223 | NH1 | 2.8 | No | |
| Arg223 | NH2 | 3.4 | No | ||
| Val204 | O | Arg196 | O | 2.7, 2.8 | Yes |
| Arg206 | NE | Asp198 | OD2 | 2.8 | No |
| NH1 | Glu222 | OE1 | 2.9, 2.6 | Yes | |
| Glu222 | OE2 | 2.9, 3.2 | Yes | ||
| NH2 | Asp198 | OD2 | 3.4 | No | |
| Phe216 | O | 3.0 | No | ||
| Glu222 | OE2 | 2.7 | No | ||
| Asp198 | O | 3.5, 3.2 | Yes | ||
| Phe216 | N | 3.5, 3.1 | Yes | ||
| Phe216 | O | 3.5, 2.8 | Yes | ||
| O | Asp198 | OD2 | 2.6, 2.8 | Yes | |
| Phe216 | O | 2.6, 3.4 | Yes | ||
| Arg216 | Stacking | Arg169 | Stacking | No | |
| NH1 | Arg169 | N | 3.1, 2.9 | Yes | |
| Asp219 | OD2 | Arg183 | NH2 | 2.9 | No |
| Arg169 | NH2 | 2.8, 3.0 | Yes | ||
| Gly197 | N | 2.7, 3.1 | Yes | ||
| Ser220 | OG | Arg169 | NH2 | 2.8, 3.0 | Yes |
For water-mediated contacts, the two numbers listed are the distances from the water to UvrA and UvrB, respectively
Comparison of the complete UvrB-binding domain of UvrA to other proteins in the Protein Data Bank (19, 20) revealed that the closest structural neighbors are a region of the small subunit of ribulose-1,5-bisphosphate carboxylase (Protein Data Bank code 1BXN; Z score of 6.2), and domain I of the ribosomal protein L1 (Protein Data Bank code 2OV7; Z score of 5.4). A detailed examination confirmed the structural similarity. In both cases, the domain in question makes protein-protein contacts (intermolecular in the case of ribulose-1,5-bisphosphate carboxylase, and intramolecular in the case of ribosomal protein L1). The interactions, however, are not identical to those observed at the UvrA-UvrB interface. Therefore, the functional significance of this similarity is not established.
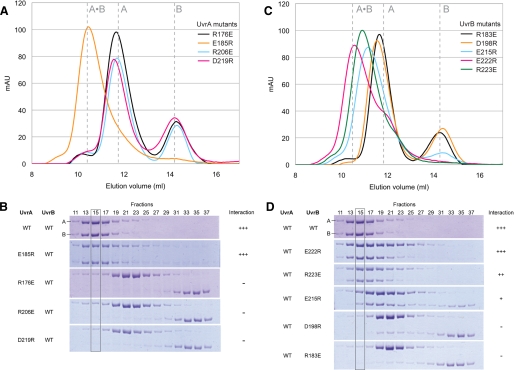
The Observed Interface Is Authentic—To determine whether the structure observed in the crystal represents the physiological UvrA-UvrB interface, we studied the behavior of a series of point mutant proteins. For this purpose, conserved residues in UvrA and UvrB whose side chains participate in direct contacts at the interaction interface were targeted. Negatively charged residues were replaced with arginine, whereas positively charged residues were substituted by glutamate. Nine mutant proteins were prepared (four UvrA mutants: R176E, E185R, R206E, and D219R; and five UvrB mutants: R183E, D198R, E215R, E222R, and R223E) in the context of the complete proteins. The mutants were purified using protocols identical to those used with the wild-type proteins. The ability of the mutants to bind the wild-type interaction partner was measured using size exclusion chromatography (Fig. 4), which resolves the UvrA·UvrB complex from free UvrA and UvrB. Chromatography was carried out in the presence of MgCl2 and ATP because these are crucial for complex formation (21).5 The presence of UvrA and/or UvrB in each fraction was analyzed by SDS-PAGE.
FIGURE 4.
Site-directed mutagenesis in the context of full-length proteins confirmed the importance of the observed interactions. Interaction between UvrA and UvrB (8 nmol each) was analyzed by size exclusion chromatography (Superdex 200; GE Healthcare) at 4 °C in UvrAB complex buffer (20 mm Tris-HCl, pH 7.5, 150 mm KCl, 5% (v/v) glycerol, 5 mm β-ME, 5 mm MgCl2, 2 mm ATP). A and B, elution profile and SDS-PAGE analysis of the samples containing mutant UvrA and wild-type UvrB. C and D, elution profile and SDS-PAGE analysis of the samples containing wild-type UvrA and mutant UvrB. Disruption of UvrA-UvrB interaction can be clearly seen in fraction 15 (gray box) in C and D.
Three of the four UvrA mutations (R176E, R206E, and D219R) completely abolished interaction with UvrB (Fig. 4, A and B), confirming the importance of these positions in complex formation. The E185R mutant, on the other hand, retained wild-type activity (Fig. 4, A and B), indicating that perturbation of Glu185 of UvrA is not sufficient to abolish interaction. Two of the five UvrB mutants studied (D198R and R183E) completely abrogated UvrA-UvrB interaction, and two others (E215R and R223E) showed milder effects (Fig. 4, C and D). The E222R mutant of UvrB displayed wild-type level of UvrA binding activity (Fig. 4, C and D). Taken together, these data suggest that the structure seen in the crystal represents the authentic interface between UvrA and UvrB.
DISCUSSION
We have determined the crystal structure of the complex formed between the interaction domains of G. stearothermophilus UvrA and UvrB. These domains are necessary and sufficient to mediate UvrA-UvrB interaction. The structure showed that interface between UvrA and UvrB is largely polar, consisting of a large number of direct and water-mediated hydrogen bonds, as well as electrostatic interactions between conserved residues.
Interaction across the UvrA-UvrB Interface—We have performed site-directed mutagenesis and biochemical experiments to establish that the direct contacts seen in our structure are important for the formation of the UvrA·UvrB ensemble in solution. The observation that UvrA mutants either retained full activity or became completely defective in complex formation, whereas the UvrB mutants displayed a range of intermediate phenotypes (Fig. 4), could be attributed to the fact that one residue from UvrA often interacts with multiple residues from UvrB (Fig. 3B and Table 2). For example, Arg176 of UvrA forms direct contacts with Glu215, Glu222, and Asp198 of UvrB, and Arg206 of UvrA directly interacts with Asp198, Glu222, and Phe216 of UvrB. The only exception to this is the interaction between Glu185 of UvrA and Arg223 of UvrB, which does not appear to significantly contribute to the UvrA·UvrB complex formation.
Besides direct interactions between side chains, the UvrA-UvrB interaction interface also involves several side chain-main chain interactions and water-mediated contacts (Table 2). Additionally, several of the highly conserved residues form intramolecular interactions with residues that participate in the contacts at the interface. For example, Arg213 of UvrB, which is one of the most highly conserved residues in the interaction domain, does not participate in the UvrA-UvrB interaction. Instead, it makes a water-mediated contact with Glu215, which in turn makes both direct and water-mediated contacts with UvrA. Such buttressing interactions, observed at several positions around the A-B interface, could help stabilize the substructures of the interaction domains that are important for UvrA-UvrB binding.
We note that Truglio et al. (6) reported a biochemical study of B. caldotenax UvrB mutants (R183E, R194A/R916A, R194E/R916E, and R213A/E215A) with substitutions in the UvrA-binding region (domain 2). Our study confirms the accuracy of this earlier analysis and adds additional residues to the interface, providing a more complete inventory of residues that are important for UvrA-UvrB interaction.
Based on biochemical studies of various E. coli UvrB constructs, Hsu et al. (18) proposed additional contacts between UvrA and UvrB involving residues 547–673, which are located at the C terminus of UvrB. If these contacts do form, we do not believe that they are energetically significant because several UvrB point mutants in the interface captured by our structure completely abolish interaction between the full-length proteins (Fig. 4). Additionally, UvrB*, a known proteolytic fragment lacking the C-terminal 43 amino acids, has been documented to form complexes with UvrA and get “loaded” at the site of damage (22). Lastly, examination of the structure of B. caldotenax UvrB revealed that only a small portion of the region in question was modeled (residues 547–595), and the resulting structure, we believe, would not form a stable entity that could productively interact with UvrA. We are thus confident that we have identified the energetically significant contacts between UvrA and UvrB.
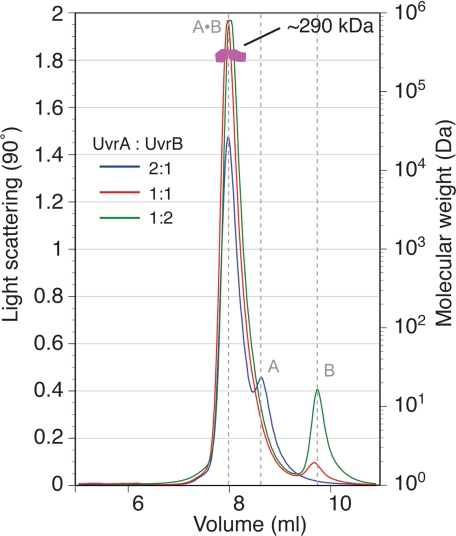
Structural Model for the Complete UvrA·UvrB Damage Sensor—Despite having been extensively studied, the structure of the UvrA·UvrB ensemble has not been elucidated; even its stoichiometry remains controversial. Hydrodynamic studies suggest the stoichiometry of A2B1 (21), whereas atomic force microscopy (23) and fluorescence resonance energy transfer (24) measurements imply A2B2. We have used multi-angle laser light scattering measurements to establish that the molecular mass of the G. stearothermophilus complex is ∼290 kDa. The only combination of UvrA (molecular mass = 107 kDa) and UvrB (molecular mass = 78 kDa) that gives this molecular mass is A2B1 (Fig. 5). Although dimeric UvrA contains two sites that could bind UvrB, and nothing in our structure (5) prohibits a larger ensemble, light scattering analysis of samples containing a large excess of UvrB failed to show any sign of an A2B2 species. We thus combined our analysis of the A-B interaction domains and our molecular mass measurements to construct a model of the intact UvrA2·UvrB1 ensemble. The model was built by superimposing the corresponding domains of full-length UvrA (Protein Data Bank code 2R6F) and UvrB (Protein Data Bank code 1T5L) onto our structure (Fig. 6A).
FIGURE 5.
Multi-angle laser light scattering suggests a 2:1 stoichiometry for the full-length UvrA·UvrB complex. UvrA·UvrB complex was formed using different UvrA:UvrB ratios and subjected to size exclusion chromatography (20 mm Tris-HCl, pH 7.5, 150 mm KCl, 5% (v/v) glycerol, 5 mm β-ME, 5 mm MgCl2, 2 mm ATP), and multi-angle laser light scattering. The complex appeared monodisperse with an apparent molecular mass of ∼290 kDa, approximating that of UvrA2·UvrB1.
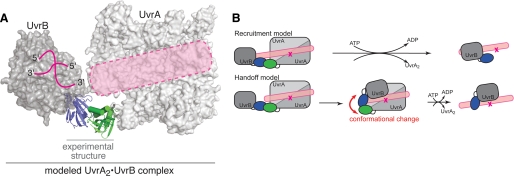
FIGURE 6.
Model of the nucleotide excision repair damage sensor. A, modeled UvrA·UvrB complex based on superposition of the corresponding domains onto the experimentally determined structure. Note that the proposed DNA-binding path on UvrA (5) and the DNA-binding site on UvrB (Protein Data Bank code 2FDC (25)) are aligned (pink). B, the models for lesion recognition by the UvrA·UvrB complex.
In our model, the A-B sensor adopts a flat and open structure with overall dimensions of 160 × 80 × 60 Å. Strikingly, the expected path of DNA over UvrA, as determined by site-directed mutagenesis (5), neatly aligns with the crystallographically established position of DNA on UvrB (25). Moreover, the approximate length of DNA that would be associated with the A2B1 complex is ∼43 bp, a value in remarkable agreement with estimates from DNase I foot-printing (26). We note that the approximate length of DNA occupied by an A2B2 ensemble would be considerably larger, ∼58 bp, and would be in poor agreement with the experimentally determined value.
Models for Lesion Recognition by the UvrA·UvrB Complex—We envision two limiting models for lesion recognition and UvrB loading (Fig. 6B). In the first model, termed the recruitment model, damaged DNA binds along the sensor in the conformation suggested by our structure. This model predicts that contacts to the damaged moiety are made exclusively by UvrA, consistent with photocross-linking experiments (27). Using geometric considerations, UvrB would have to bind at an adjacent site without directly contacting the lesion. Upon ATP hydrolysis, UvrA exits the complex, leaving UvrB stably bound. UvrB could move closer to the lesion, and there is some evidence suggesting that this type of subtle conformational rearrangement is possible (28, 29). Departure of UvrA would leave the damaged site accessible to UvrC. In this model, the UvrA·UvrB complex does not undergo large structural changes upon damage detection and loading of UvrB.
A second model, termed the handoff model, envisages the lesion being recognized most likely by UvrA within the complex in the conformation suggested by our structure. Successful lesion detection would lead to handoff of the damaged site to UvrB. Such a handoff would clearly require major structural changes in the damage sensor that alter the relative orientation between UvrA and UvrB. The presence of flexible linkers between the interaction domains and the remainder of the full-length proteins could enable dramatic changes of the scale suggested in this model. For example, these changes could reorient UvrB from the conformation in our structural model to a location on the double helix opposite the lesion. Available data do not, at present, permit these two models to be distinguished. These models make specific predictions that can serve as a basis for future experiments. Understanding the mechanism of damage recognition must await future studies to more precisely delineate the structure of the damage sensor and its interaction with damaged DNA.
Interactions between UvrA and Transcription-Repair Coupling Factor—In addition to global genome repair, which involves damage recognition by the UvrA·UvrB ensemble, nucleotide excision repair also has another subpathway, termed transcription-coupled repair. Transcription-coupled repair preferentially removes lesions from the DNA strand that is being transcribed (30), thus ensuring that the RNA transcript contains the correct information. This process is initiated by the protein transcription-repair coupling factor (TRCF), also known as Mfd (31, 32). TRCF recognizes RNA polymerase stalled at a lesion, terminates transcription, releases the transcript, and recruits UvrA to the site of damage so that nucleotide excision repair can take place (32). Amino acid sequence analysis of TRCF revealed that the N-terminal region of the protein is similar to a portion of UvrB (32); this region of TRCF was later shown to be important for the TRCF-UvrA interaction (33). The crystal structure of TRCF revealed a surface corresponding to the UvrA-binding region of UvrB, supporting the hypothesis that TRCF and UvrB would interact with UvrA in a similar manner (34). Superposition of the portion of UvrB in our structure on TRCF (Protein Data Bank code 2EYQ) reveals a likely correspondence between residues that participate at the A-B interface and the A-TRCF interface. The more complete inventory of contacts between UvrA and UvrB provided by our structure enables a more precise definition of the TRCF-UvrA interface.
Acknowledgments
We thank Marie Spong, Alexander Ruthenburg, and members of the Verdine laboratory for useful discussions and critical reading of the manuscript and Lauren Verdine for assistance with protein purification.
The atomic coordinates and structure factors (code 3FPN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant CA100742 (to G. L. V.). This work was also supported by Award RR-15301 from the National Center for Research Resources at the National Institutes of Health.
Footnotes
The abbreviations used are: β-ME, β-mercaptoethanol; TRCF, transcription-repair coupling factor.
D. Pakotiprapha, G. L. Verdine, and D. Jeruzalmi, unpublished observation.
References
- 1.Truglio, J. J., Croteau, D. L., Van Houten, B., and Kisker, C. (2006) Chem. Rev. 106 233-252 [DOI] [PubMed] [Google Scholar]
- 2.Lin, J. J., and Sancar, A. (1992) J. Biol. Chem. 267 17688-17692 [PubMed] [Google Scholar]
- 3.Verhoeven, E. E., van Kesteren, M., Moolenaar, G. F., Visse, R., and Goosen, N. (2000) J. Biol. Chem. 275 5120-5123 [DOI] [PubMed] [Google Scholar]
- 4.Orren, D. K., Selby, C. P., Hearst, J. E., and Sancar, A. (1992) J. Biol. Chem. 267 780-788 [PubMed] [Google Scholar]
- 5.Pakotiprapha, D., Inuzuka, Y., Bowman, B. R., Moolenaar, G. F., Goosen, N., Jeruzalmi, D., and Verdine, G. L. (2008) Mol. Cell 29 122-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truglio, J. J., Croteau, D. L., Skorvaga, M., DellaVecchia, M. J., Theis, K., Mandavilli, B. S., Van Houten, B., and Kisker, C. (2004) EMBO J. 23 2498-2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 8.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrakis, A., Harkiolaki, M., Wilson, K. S., and Lamzin, V. S. (2001) Acta Crystallogr. D Biol. Crystallogr. 57 1445-1450 [DOI] [PubMed] [Google Scholar]
- 10.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 11.Brunger, A. T. (2007) Nat. Protoc. 2 2728-2733 [DOI] [PubMed] [Google Scholar]
- 12.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 13.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 14.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 15.Wyatt, P. J. (1991) Biochem. Soc. Trans. 19 485. [DOI] [PubMed] [Google Scholar]
- 16.Folta-Stogniew, E. (2006) Methods Mol. Biol. 328 97-112 [DOI] [PubMed] [Google Scholar]
- 17.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu, D. S., Kim, S. T., Sun, Q., and Sancar, A. (1995) J. Biol. Chem. 270 8319-8327 [DOI] [PubMed] [Google Scholar]
- 19.Holm, L., Kaariainen, S., Rosenstrom, P., and Schenkel, A. (2008) Bioinformatics 24 2780-2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berman, H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., Fagan, P., Marvin, J., Padilla, D., Ravichandran, V., Schneider, B., Thanki, N., Weissig, H., Westbrook, J. D., and Zardecki, C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58 899-907 [DOI] [PubMed] [Google Scholar]
- 21.Orren, D. K., and Sancar, A. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 5237-5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, J. J., Phillips, A. M., Hearst, J. E., and Sancar, A. (1992) J. Biol. Chem. 267 17693-17700 [PubMed] [Google Scholar]
- 23.Verhoeven, E. E., Wyman, C., Moolenaar, G. F., and Goosen, N. (2002) EMBO J. 21 4196-4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malta, E., Moolenaar, G. F., and Goosen, N. (2007) Biochemistry 46 9080-9088 [DOI] [PubMed] [Google Scholar]
- 25.Truglio, J. J., Karakas, E., Rhau, B., Wang, H., DellaVecchia, M. J., Van Houten, B., and Kisker, C. (2006) Nat. Struct. Mol. Biol. 13 360-364 [DOI] [PubMed] [Google Scholar]
- 26.Bertrand-Burggraf, E., Selby, C. P., Hearst, J. E., and Sancar, A. (1991) J. Mol. Biol. 219 27-36 [DOI] [PubMed] [Google Scholar]
- 27.DellaVecchia, M. J., Croteau, D. L., Skorvaga, M., Dezhurov, S. V., Lavrik, O. I., and Van Houten, B. (2004) J. Biol. Chem. 279 45245-45256 [DOI] [PubMed] [Google Scholar]
- 28.Moolenaar, G. F., Herron, M. F., Monaco, V., van der Marel, G. A., van Boom, J. H., Visse, R., and Goosen, N. (2000) J. Biol. Chem. 275 8044-8050 [DOI] [PubMed] [Google Scholar]
- 29.Delagoutte, E., Fuchs, R. P., and Bertrand-Burggraf, E. (2002) J. Mol. Biol. 320 73-84 [DOI] [PubMed] [Google Scholar]
- 30.Mellon, I., and Hanawalt, P. C. (1989) Nature 342 95-98 [DOI] [PubMed] [Google Scholar]
- 31.Selby, C. P., Witkin, E. M., and Sancar, A. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 11574-11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selby, C. P., and Sancar, A. (1993) Science 260 53-58 [DOI] [PubMed] [Google Scholar]
- 33.Selby, C. P., and Sancar, A. (1995) J. Biol. Chem. 270 4882-4889 [DOI] [PubMed] [Google Scholar]
- 34.Deaconescu, A. M., Chambers, A. L., Smith, A. J., Nickels, B. E., Hochschild, A., Savery, N. J., and Darst, S. A. (2006) Cell 124 507-520 [DOI] [PubMed] [Google Scholar]