Abstract
c-Jun N-terminal kinase 3α1 (JNK3α1) is a mitogen-activated protein kinase family member expressed primarily in the brain that phosphorylates protein transcription factors, including c-Jun and activating transcription factor-2 (ATF-2) upon activation by a variety of stress-based stimuli. In this study, we set out to design JNK3-selective inhibitors that had >1000-fold selectivity over p38, another closely related mitogen-activated protein kinase family member. To do this we employed traditional medicinal chemistry principles coupled with structure-based drug design. Inhibitors from the aminopyrazole class, such as SR-3576, were found to be very potent JNK3 inhibitors (IC50 = 7 nm) with >2800-fold selectivity over p38 (p38 IC50 > 20 μm) and had cell-based potency of ∼1 μm. In contrast, indazole-based inhibitors exemplified by SR-3737 were potent inhibitors of both JNK3 (IC50 = 12 nm) and p38 (IC50 = 3 nm). These selectivity differences between the indazole class and the aminopyrazole class came despite nearly identical binding (root mean square deviation = 0.33 Å) of these two compound classes to JNK3. The structural features within the compounds giving rise to the selectivity in the aminopyrazole class include the highly planar nature of the pyrazole, N-linked phenyl structures, which better occupied the smaller active site of JNK3 compared with the larger active site of p38.
Because the initial reports on the discovery of p38 (1) and c-Jun N-terminal kinase (JNK)2 (2–6) these mitogen-activated protein kinase family members have generated great interest as drug targets. p38 especially has garnered considerable interest, particularly for the treatment of rheumatoid arthritis and Crohn disease, and numerous compounds have entered clinical trials for these indications (7–11). Because most of the p38 inhibitors are competitive versus ATP (12–17), and there are 518 kinases in the genome, it was crucial to develop compounds that are selective against a broad panel of kinases so that compounds could be advanced to clinical development. The molecular basis that gives rise to selective p38 inhibitors from numerous structural classes has been reported (18–20) and is centered on amino acid differences at the so-called “gate-keeper” Thr-106 residue in p38 (Met in all of the JNK isoforms and Gln in extracellular regulated kinase, the other mitogen-activated protein kinase family member). Many compounds have been synthesized that take advantage of this deeper hydrophobic pocket in p38, compared with JNK3, and the structures of the compounds have included trifluoromethyl and other large moieties, which all contribute to p38 selectivity (21).
In contrast to p38, there have been fewer reports for selective JNK inhibitors, and the clinical development of JNK inhibitors also lags that of p38. Despite the paucity of highly selective JNK inhibitors that have advanced to clinical development, numerous recent reports have begun to emerge that show compounds from various structural classes (benzothiazole pyrimidines, aminopyridines, benzothien-2-yl amides, aminopyrimidines, and quinolines) having selectivity for JNK over p38 (5, 15, 17, 22–24). The well described toxicity of p38 inhibition (7) necessitates this desired selectivity in any JNK inhibitor program. Interestingly, in 2005 Swahn et al. reported a class of indazole JNK3 inhibitors that were equipotent against p38 with the exception of one compound that showed a 300-fold selectivity for JNK3 over p38 (25). The publications on selective JNK inhibitors came soon after compelling validation studies utilizing either knockout mice or an 11-mer JNK-interacting protein peptide, which indicted JNK as an attractive drug target for stroke (26), Parkinson disease (27, 28), and type II diabetes mellitus (29).
Many high resolution crystal structures for p38α, JNK3α1, and JNK1α1 have been solved (18, 30–34). In 1997, Tong et al. (33) reported the 2.0-Å resolution crystal structure of SB-203580, the original pyridinyl imidazole, with p38α highlighting the hydrogen bonding interaction of the pyridinyl nitrogen of the inhibitor with the main-chain amido nitrogen of Met-109, a key residue within the ATP binding domain of p38. In 1998, Lisnock et al. (19) reported that single digit nanomolar biarylimidazole p38 inhibitors could be rendered inactive (up to 3 μm) on p38 if the Thr-106 residue in p38 was mutated to the corresponding Met residue found in the JNK isoforms. Wilson et al. (20) also showed this gatekeeper residue to be essential for p38 inhibitor selectivity over JNK. In 2003 Scapin et al. (32) published the first crystal structure of JNK3, which demonstrated that the glycine-rich loop of JNK3 undergoes a conformational change that locks the small, flat, planar di-aza-phenanthroline inhibitor into the smaller binding pocket of JNK3 making this compound highly selective over p38, which has a larger ATP binding pocket. This was the first report demonstrating a structural basis for selective JNK3 inhibition (32).
Given the compelling validation data for JNK as a drug target and the emergence of a few structural classes of JNK selective compounds that have been reported (15, 17, 22–24), we set out to investigate whether JNK inhibitor selectivity over p38 could be established. We started our efforts with an indazole class of compounds and were not able to establish significant selectivity for JNK3 over p38. This roadblock led us to an aminopyrazole class of compounds that did indeed afford compounds with >2800-fold selectivity for JNK3 over p38. To do this we utilized traditional structure-activity relationship principals supported by homogeneous time-resolved fluorescence-based biochemical assays and cell-based assays, combined with structure-based drug design. The principal findings of this study were: despite nearly identical binding (r.m.s.d., 0.33 Å) and enzyme affinity (IC50), for the indazole-based JNK3 inhibitor (SR-3737, IC50 = 12 nm) compared with the aminopyrazole-based JNK3 inhibitor (SR-3451, IC50 = 25 nm), the two classes of compounds had significantly different binding affinity to p38 (IC50 = 3.2 nm (SR-3737) and 3.6 μm (SR-3451)). We attribute the weak p38 inhibition potency of SR-3451 and inhibition selectivity for JNK3α1 to the highly planar nature of the pyrazole, N-linked phenyl structures. SR-3451 and its analogs better occupy the smaller active site of JNK3 compared with the larger active site of p38.
EXPERIMENTAL PROCEDURES
Expression and Purification of JNK3—The expression construct pDest_JNK339–402 was expressed in Escherichia coli strain BL21(DE3) (Invitrogen) under the following conditions. 5 ml of a log phase culture grown in Luria broth (Invitrogen) supplemented with 50 μg/ml ampicillin was transferred to 500 ml of the same medium and grown with shaking at 240 rpm and 37 °C. The culture was grown to A600 = 0.4, and the temperature was reduced to 27 °C and incubated with shaking for 60 min. The culture was then induced with 1.0 mm (final concentration) isopropyl 1-thio-β-d-galactopyranoside (Invitrogen) and grown with shaking at 240 rpm for a further 5 h and harvested by centrifugation 3,000 × g at 4 °C. The cell pellet from 1 liter of culture was resuspended in 20 ml of cell lysis buffer (50 mm HEPES, pH 7.0, 100 mm NaCl, 10% (v/v) glycerol, and 5 mm tris(2-carboxyethyl)phosphine) and sonicated in three 20-s bursts on ice. The lysate was clarified by centrifugation at 14,000 rpm for 60 min. A 5-ml SP Fastflow column (Amersham Biosciences) was equilibrated with 5 column volumes of buffer A (50 mm HEPES, pH 7.0, and 10 mm β-mercaptoethanol). The clarified lysate was diluted 10-fold in dilution buffer (50 mm HEPES, pH 7.0, 10% (v/v) glycerol, and 10 mm β-mercaptoethanol) and loaded onto the SP Fastflow column at 4 ml/min. After loading the sample the sample was washed with 5 column volumes of buffer A. The sample was eluted with a 10-column volume gradient of 0–100% buffer B (50 mm HEPES, pH 7.0, 10 mm β-mercaptoethanol, and 1 m NaCl), and 1-ml fractions were collected and analyzed by SDS-PAGE. Fractions containing JNK3 were pooled, diluted 10-fold with dilution buffer, and loaded onto the Mono S column (Amersham Biosciences) at 5 ml/min. The column was washed with 5 column volumes of buffer A and eluted with a 10-column volume gradient of 0–100% buffer B. 0.25-ml fractions were collected and analyzed by SDS-PAGE. Fractions containing JNK3 were pooled and loaded onto a pre-equilibrated Sephacryl S200 (Amersham Biosciences, 50 mm HEPES, pH 7.0, 100 mm NaCl, and 10 mm β-mercaptoethanol). Fractions containing JNK3 were visualized by SDS-PAGE and estimated to be >95% pure. Peak fractions were pooled and concentrated using an Amicon ultracentrifugation filter device, 10K pore size (Millipore) to 10 mg/ml, and stored at 4 °C.
Generation of ATF-21–115 Construct—The pDest_ATF-21–115 was constructed from the full-length ATF-2 cDNA (purchased by Scripps Florida from Open Biosystems as part of the MGC collection). Nucleotides 1–345 of ATF-2, corresponding to amino acids 1–115, were subcloned into pEntrSD/Dtopo (Invitrogen) using the primer sequences 5′-CACC GAT TAC AAG GAT GAC GAC GAT AAG AAA TTC AAG TTA CAT GTG AAT TCT GCC AGG-3′ and 5′-TTA TGC AAG AGG GGA TAA ATC-3′. Standard PCR conditions using Pfu DNA polymerase (Stratagene) and cloning techniques were used. Next, the fusion construct was subcloned into pET104Dest (Invitrogen) using Clonase II according to the manufacturer's instructions (Invitrogen).
Expression and Purification of ATF-2—The expression construct pDest_ATF-2bio1–115flag together with a construct containing the birA gene (Avidity) was expressed in Escherichia coli strain BL21(DE3) (Invitrogen) in Luria broth supplemented with 50 μg/ml ampicillin and 50 μg/ml chloramphenicol (Sigma) at 30 °C. At A600 = 0.6 D-biotin was added to the culture (50 μm final concentration), and the culture was induced with 0.5 mm (final concentration) isopropyl 1-thio-β-d-galactopyranoside (Invitrogen) and grown at 30 °C for a further 2 h. The cell pellet was lysed, centrifuged, and diluted 10-fold with buffer B (20 mm HEPES, pH 7.0, 150 mm NaCl) and applied to a 5-ml monomeric avidin column (Pierce). The column was washed with 10 column volumes of buffer B and eluted via competition with biotin. Purified protein was loaded onto a 2-ml FLAG M2 column (Sigma). The column was washed with 10 column volumes of buffer B and eluted via competition with the FLAG peptide (100 μg/ml). Fractions containing ATF-2 were visualized by SDS-PAGE and estimated to be >90% pure. Further details provided on request.
Homogeneous Time-resolved Fluorescence Assay—Enzyme inhibition studies were performed in 384-well polystyrene homogeneous time-resolved fluorescence plates (Grainier) for 15 min at ambient temperature (∼22 °C) with 0.2 μm biotinylated FL-ATF-2, 1 μm ATP, 0.3 nm activated JNK3α1 (with a control in the absence of kinase for determining the basal signal) in 10-μl volumes containing the final concentrations of the following: 50 mm Hepes, pH 7.0, 2.5 mm MgCl2, 0.1 mg/ml bovine serum albumin, 1 mm dl-dithiothreitol, 0.01% Triton X-100 (all from Sigma-Aldrich), and 5% DMSO (with or without compound). A 10-point titration of all compounds was carried out in 3-fold dilutions from 10 pm to 2000 nm. After 15 min, the kinase reaction was terminated by addition of 10 μl of quenching solution (50 mm Hepes, pH 7.0, with 14 mm EDTA, 0.01% Triton X-100, 400 mm KF (all from Sigma-Aldrich)). The detection reagents, streptavidin-xlAPC (200 nm) and europium cryptate-labeled rabbit polyclonal anti-phospho-ATF-2 (0.43 ng/well), were from Cis-Bio. The homogeneous time-resolved fluorescence signal was detected using a viewlux plate reader (PerkinElmer Life Sciences) 1 h post-quenching. The data from four different experiments were averaged and presented as the mean ± S.D. IC50 values were determined by fitting the data to the equation for a four-parameter logistic. p38 enzyme inhibition assays were performed identically to the JNK3 assays with the exception that the reaction time was 30 min, ATP = 11 μm, and p38 (Millipore) = 0.625 nm.
Cell-based Assays Measuring JNK Activity—INS-1 β-pancreatic cells were plated in a 96-well tissue culture plate at 3.5 × 104 cells/well (Corning) in a media containing RPMI 1640 (± glutamine (2 mm)) and 10% fetal bovine serum (Invitrogen) and incubated overnight at 37 °C in 5% CO2. An assay plate was prepared by coating a 96-half-well plate (Costar) with 50 μl/well p-c-Jun capture antibody (Cell Signaling). Cells were incubated with 4 mm streptozotocin containing various concentrations of potential inhibitor dissolved in DMSO for 3 h at 37 °C in 5% CO2. After treatment the media was removed, and the cells were washed in ice-cold phosphate-buffered saline. The phosphate-buffered saline was removed, and the cells were lysed in ice-cold lysis buffer (100 μl/well) containing 1× protease (Roche Applied Science) and 1× phosphatase inhibitors (Sigma). Lysates were transferred to the corresponding well of the blocked assay plate, covered tightly, and incubated 16 h at 4 °C. The c-Jun detection antibody (100× dilution), and the secondary anti-mouse coupled horseradish peroxidase (1000× dilution) were purchased from Cell Signaling. Inhibition of signal was quantified using TMB substrate (BioFX Laboratories) and read on a microplate reader at an absorbance of 450 nm. IC50 values were determined using a four-parameter logistic and a 10-point dilution curve for each of the inhibitors covering four orders of magnitude of inhibitor concentration.
Synthesis of Indazoles and Aminopyrazoles—The synthesis of SR-3737 and its analogs followed the same general protocol. A full experimental protocol for the synthesis of SR-3737 can be found in the supplemental materials. Starting with commercially available 5-nitroindazole (Sigma-Aldrich), the indazole was N-arylated with ethyl 3-bromobenzoate (Sigma-Aldrich) using standard Ullman procedures (35, 36). Reduction of the nitro group via hydrogenation, followed by Buchwald arylation of the 5-amino group (37, 38) afforded the bis-aryl indazole. Finally, ester hydrolysis and amide coupling afforded the final products.
The compounds in the pyrazole series were made in a similar fashion. A detailed experimental protocol for SR-3451 can be found in the supplemental materials, but other analogs were made following the general protocol herein. Pyrazole (Sigma-Aldrich) was nitrated in fuming sulfuric acid to give the known 4-nitropyrazole after re-crystallization (39). Ullman coupling with ethyl 3-bromobenzoate, followed by ester hydrolysis and amide coupling, afforded the 4-nitro-N-arylated pyrazole amide. Hydrogenation of the nitro group to the amine, followed by reaction with the appropriate aryl isocyanate, gave the final urea product. All final products were purified either by silica gel chromatography, reversed-phase preparative high-performance liquid chromatography, or re-crystallization and were >95% pure as judged by analytical high-performance liquid chromatography analysis.
Crystallization of JNK3—The 10 mg/ml JNK3 sample was mixed with 1 mm AMP-PCP, 2 mm MgCl2, 0.4 mm Zwittergent 3-14, and 10% ethylene glycol prior to crystallization and incubated on ice for 30 min. JNK3 was screened against commercially available sparse matrix screens using a vapor diffusion experiment with the best crystals seen within 1 week in JCSG+ Suite (Qiagen) condition #92 (0.2 m NaCl, 0.1 m Bis-Tris, pH 5.5, and 25% polyethylene glycol 3350). Grid screen optimization using a 0.5-μl protein plus 0.5-μl precipitating solution microbatch drop produced diffraction quality crystals in 0.2 m NaCl, 0.1 m Bis-Tris, pH 5.5, and 28–31% polyethylene glycol 3350. Crystals were seen within 24 h and reached sufficient size by days 3–7. 100 mm stock solutions of SR-3451 and SR-3737 were made in 100% DMSO, and 0.1 μl was injected into the microbatch drop using a Hamilton syringe where the compounds were soaked into the crystals over 36 h. Crystals were mounted in loops on copper-based Hampton Research style pins and plunged into liquid nitrogen for storage.
Data Collection and Structure Solution of JNK3—Liquid nitrogen-cooled crystals were loaded into an SSRL cassette for shipping. Data were collected remotely at SSRK on beamline 9-1 (40). Datasets were indexed and scaled to the orthorhombic space group C2221 using HKL2000 (41). Molecular replacement and subsequent refinement was carried out using PHENIX (42) while using COOT (43) for visualization and model building. The MOLPROBITY web server was used for geometric analysis and clash monitoring (44). Structure files for SR-3737 and SR-3451 ligands were generated using the Dundee PRODRG server (45) and placed into difference density. The final refined structures were deposited into the Protein Data Bank (PDB ID 3FI3 and 3FI2 for SR-3737 and SR-3451, respectively).
RESULTS
JNK3/p38 SAR for Indazole-based Inhibitors—Supplemental Tables S1 and S2 present the IC50 data for the indazole-based inhibitors of JNK3, JNK1, and p38. The parent compound from this class, SR-3737, was a potent JNK3 and p38 inhibitor showing 4-fold greater potency for p38 over JNK3. Of the 15 indazoles synthesized, only one, SR-4186, had >20-fold selectivity for JNK3 over p38, and thus this series was not pursued further.
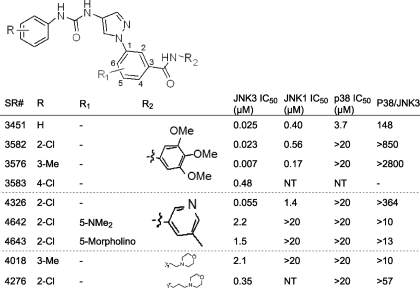
JNK3/p38 SAR for Aminopyrazole-based Inhibitors—Because we were unable to establish a structure-activity relationship (SAR) for the indazole series that maintained JNK3 potency and simultaneously achieved selectivity over p38, we investigated the potential selectivity of aminopyrazole urea-based inhibitors. The aminopyrazole urea-based inhibitors described in Table 1 are similar in structure to the indazole-based inhibitors of supplemental Tables S1 and S2. The side chains are basically the same, but they bear different cores (bicyclic for the indazoles and monocyclic for the pyrazoles). Five of the nine compounds presented in Table 1 showed >50-fold selectivity for JNK3 over p38. This is a lower limit estimate for the selectivity, because we did not measure p38 inhibition at concentrations >20 μm. SR-3576 in particular was a 7 nm JNK3 inhibitor having >2800-fold selectivity over p38 (Table 1). Although 2- and 3-substitution on the N-phenyl-linked urea had little effect on JNK3 inhibition potency (compare SR-3451 with SR-3582 and SR-3576), 4-substitution (SR-3583) decreased JNK3 potency by 19-fold (Table 1). Like the indazoles, many of the more potent aminopyrazoles (e.g. SR-3582 and SR-3576) showed 24-fold selectivity for JNK3 over JNK1 (Table 1).
TABLE 1.
SAR for aminopyrazole inhibitors n ≥ 4 for JNK3 and n ≥ 2 for p38 and JNK1. All standard deviation ≤ 20%.
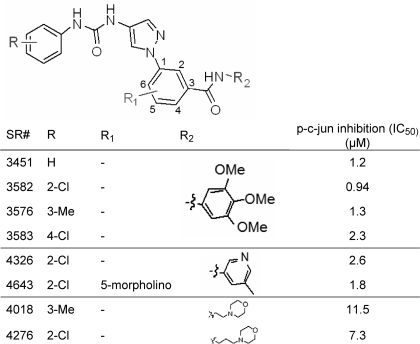
Cell-based JNK Inhibition for Aminopyrazole-based Inhibitors—Given the >2800-fold biochemical selectivity of compounds such as SR-3576 for JNK3 over p38, we tested the cell-based activity of these compounds in INS-1 cells. In this assay several compounds showed IC50 values of near 1 μm for the inhibition of c-Jun phosphorylation (Table 2). The assay was robust with signal-to-background in the range of 6- to 8-fold with Z′ values = 0.77.
TABLE 2.
Cell-based inhibition for aminopyrazole inhibitors n = 2 for p-c-Jun inhibition. All standard deviation ≤ 50%.
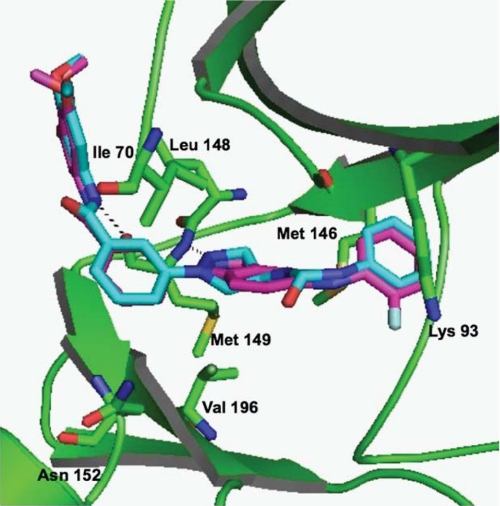
Crystallography of JNK-Inhibitor Complexes—The structures of SR-3737- and SR-3451-soaked JNK3 crystals were solved at 2.20 Å and 2.29 Å, respectively, and refined to final Rwork/Rfree of 18.03/24.88 for SR-3737 and 18.05/26.71 for SR-3451 with further refinement statistics found in Table 3. In the SR-3451 structure, five amino acids between the Tyr-363 and Asp-381 break were built in as polyalanines, because the sequence could not be determined due to lack of side-chain density. Those five amino acids were assigned as unknown residues and numbered 500 to 505. Because SR-3451 differed by only ∼3-fold in potency from SR-3576, and the SR-3451 structure was solved before the SAR evolution generated SR-3576, we felt SR-3451 was a good structural representation for the class. Fig. 1 presents the overlay of SR-3737 (magenta) and SR-3451 (cyan) in JNK3 (green). The r.m.s.d. between the two structures is 0.33 Å. The hydrogen bond formed between the pyrazole N2 of SR-3451 and main-chain atoms of Met-149 in JNK3 and the hydrogen bond formed between the indazole N2 of SR-3737 and main-chain atoms of Met-149 in JNK3 are shown as black dotted lines. Similarly, the hydrogen bond between the main-chain atoms of Met-149 in JNK3 and the amide nitrogen atom of SR-3451 or SR-3737 is also shown as a black dotted line. Other key active site residues are shown, including the gatekeeper Met-146 residue, Val-196, Asn-152, and Ile-70. The distance between C5 on the indazole- and pyrazole-linked phenyl groups Val-196 in JNK3 was 3.79 Å. Similarly the distance between this position in SR-3451 or SR-3737 and Ile-70 of JNK3 was 4.01 Å. The trimethoxyphenyl ring of both compounds stacked against the side chain of Ile-147, whereas the other face of the ring was exposed to solvent. Due to the nature of the compound wrapping itself around the hinge region, the limited contacts allowed the P-loop to be flexible in the crystal to the point that residues 71–75 in beta strand 1 were untraceable at 1 sigma electron density contour level. This flexibility and break in the chain have been seen in previous JNK3 structures (15, 22, 25, 32, 34, 46).
TABLE 3.
Crystallographic data and refinement statistics
| SR-3737 | SR-3451 | |
|---|---|---|
| Space group | C2221 | C2221 |
| Unit cell | a = 82.86 b = 124.11 c = 69.52 | a = 82.69 b = 124.53 c = 69.01 |
| Resolution range (Å) | 48.94–2.20 | 46.23–2.29 |
| No. of unique observations | 17,506 | 14,193 |
| Completeness (%) | 93.79 | 85.86 |
| Rsym (%)a | 4.9 | 5.7 |
| I/σ (I) | 31.15 (2.62) | 26.24 (2.03) |
| Refinement | ||
| Average Wilson B (Å2) | 38.66 | 38.55 |
| Rcryst (%)b | 18.03 | 18.05 |
| Rfree (%)b | 24.88 | 26.71 |
| r.m.s.d. bonds (Å) | 0.007 | 0.008 |
| r.m.s.d. angles (°) | 1.13 | 1.19 |
| Coordinate error (ML-based) (Å) | 0.33 | 0.38 |
Rsym = Σhkl Σi |Ii(hkl)| – (I(hkl))|/Σhkl Σi(|Ii(hkl)|)
Rcryst and Rfree = Σ(||Fo| – |Fc||)/Σ(|Fo|), where Rfree represents a 10% subset of the total reflections that are excluded from refinement. Values in parentheses are for the highest resolution shell
FIGURE 1.
Overlay of SR-3737 (magenta) and SR-3451 (cyan) structures. JNK3 (green) is shown as a schematic ribbon diagram with important residues highlighted in stick format and labeled. Hydrogen bonds between the compounds and the main-chain atoms of Met-149 are shown as black dotted lines. The figures was generated by using PyMOL.
SR-3737 and SR-3451 bound in the ATP pocket of JNK3 in a very similar and well defined fashion. Both compounds caused movement of the “gatekeeper” Met-146 side chain, maintained the conserved hydrogen bond with the main-chain nitrogen of Met-149, and formed a new electrostatic interaction with the main-chain oxygen of Met-149. The three phenyl rings in both compounds overlaid very well in terms of position and angle with the greatest deviation being 0.66 Å in the nitrogens forming the conserved hydrogen bond with Met-149. This was accomplished by a greater dihedral angle between the indazole N-linked phenyl ring and the indazole ring of SR-3737 (36.50°) compared with the angle formed with the pyrazole and the N-linked phenyl ring of SR-3451 (27.95°). The other structural differences between the two scaffolds were facing into the open space of the ATP binding pocket and did not form any specific interactions explaining their similar level of JNK3 inhibition.
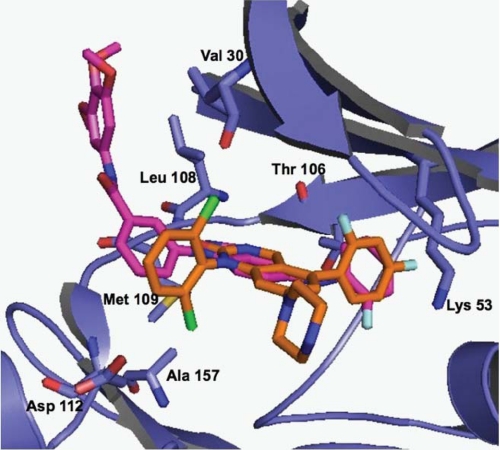
In an attempt to understand the binding mode of SR-3737 to p38, we overlaid it onto the structure for p38 (r.m.s.d. = 2.33 Å). Fig. 2 shows SR-3737 overlaid onto p38 and superimposed upon the crystal structure of a dihydroquinazolinone (compound 14e) inhibitor complexed with p38 taken from Stelmach et al. (18). Compound 14e, like SR-3737, is a 3 nm inhibitor of p38. The figure showed significant overlap for many of the structural features between the two small molecule inhibitors. For example, the difluorophenyl group of compound 14e (orange) overlapped nicely with the 2-fluorophenyl moiety in SR-3737 (magenta). Similarly, the indazole ring of SR-3737 (magenta) overlaid nicely with the 3,4-dihydropyrido[3,2-d]pyrimidin-2(1H)-one of compound 14e (orange) (Fig. 2). While distorted, the N-linked phenyl group of SR-3737 overlapped reasonably well (1.35 Å apart) with the N-linked dichlorophenyl group of compound 14e. The unique trimethoxyl phenyl group of SR-3737 did not appear to contribute any binding affinity to p38 compared with compound 14e given that their IC50 values were the same at 3 nm. Key residues for the p38 protein are given.
FIGURE 2.
Overlay of SR-3737 (magenta) with compound 14e (orange) from PDB ID 1M7Q (32). p38 (blue) is shown as a schematic ribbon diagram with homologous JNK3 highlighted residues from Fig. 1 in stick format and labeled using p38 numbering. The figure was generated by using PyMOL.
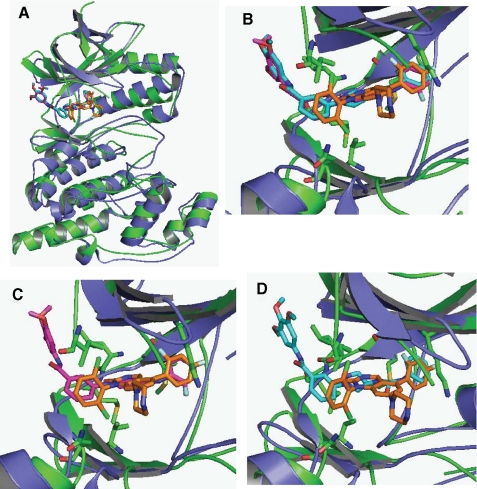
To better compare the structural similarities and differences between JNK3 and p38 with the previously described compounds bound we overlaid JNK3, p38, and the three compounds (Fig. 3). Fig. 3A shows the total protein structure of JNK3 (green), p38 (blue), along with SR-3737 (magenta), SR-3451 (cyan), and compound 14e (orange) from Stelmach et al. (18). Fig. 3B shows the close-up view of the inhibitor binding pocket with all three inhibitors. Fig. 3C shows the close-up view of the inhibitor binding pocket with SR3737 and compound 14e (orange) from Stelmach et al. (18). Fig. 3D shows the close-up view of the inhibitor binding pocket with SR-3451 and compound 14e (orange) from Stelmach et al. (18).
FIGURE 3.
Overlay of SR-3737, SR-3451, and p38 with compound 14e. A, JNK3 (green) and p38 (blue) superimposed in schematic ribbon format (only the SR-3737 JNK3 is shown). Ligands for SR-3737 (magenta), SR-3451 (cyan), and compound 14e (orange) (32) are shown in stick format. B, close-up of the ATP binding pocket showing the overlapping ligands with the highlighted JNK3 amino acids from Fig. 1 in stick format. C, close-up of the ATP binding pocket showing the overlap of SR-3737 and compound 14e with the highlighted JNK3 amino acids from Fig. 1 in stick format. D, close-up of the ATP binding pocket showing the overlap of SR-3451 and compound14e with the highlighted JNK3 amino acids from Fig. 1 in stick format. The figure was generated by using PyMOL.
DISCUSSION
The overarching goal of this work was to design JNK inhibitors that were selective over p38 and to understand what the structure-activity relationships were within the JNK inhibitors that described this selectivity. To do this we employed traditional medicinal chemistry principles and supplemented this approach with structure-based drug design. Structure-based drug design has been extremely helpful in the development of a number of FDA-approved drugs such as the carbonic anhydrase inhibitor, dorzolamide (47), and many HIV protease inhibitors, including indinavir (48, 49) and nelfinavir (50, 51). X-ray crystallography also proved invaluable in explaining the mechanism of action for imatinib, the first FDA-approved kinase inhibitor by illuminating the critical observation that imatinib bound to the inactive conformation of the enzyme (52). Conversely, numerous crystal structures of p38 complexed with various inhibitors have been published (18, 30, 33), but the SAR for p38 inhibitor selectivity (18, 21) and the molecular basis for this selectivity based on site-directed mutagenesis studies (19, 20) had already been established prior to the publication of these structures leaving the crystallography data only to confirm what the SAR and mutagenesis studies had already elucidated. The latter set of observations could also be stated about our current study for JNK selectivity.
In this particular study we had to rely slightly more on traditional medicinal chemistry principles to optimize JNK selective compounds than on insights from the structural data. As an example, much data are not presented surrounding SAR on either ends of the molecules (R or R2). Although modifications to either ends of the molecules (R or R2) in either series (indazoles or pyrazoles) did affect JNK3/1 and p38 activity, these changes had little effect on JNK3/p38 selectivity. Thus, the only modifications that had any implications on JNK3/p38 selectivity came from modifications at R1 on the central phenyl ring attached to either the indazole or pyrazole nitrogen atom. This will be the basis of the discussion below.
JNK3/p38 SAR for Indazole-based Inhibitors—It is well established in the literature that the inhibitor binding pocket in p38 is larger and more accommodating than that in JNK3 (18, 32–34). Although this will be the basis for much of the discussion concerning selectivity, it does not account for all the data in Table 1 supplemental Tables S1 and S2. Most of the indazoles did not show any selectivity for JNK3 over p38 likely due to the orientation of the central indazole-linked N-phenyl ring. Indeed, twists in the indazole-linked N-phenyl ring increased the three-dimensional space occupied by these compounds thereby favoring the larger p38 active site.
One compound that showed more selectivity for JNK3 over p38 was SR-4186, which had a trifluoromethyl group that was quite large and may have non-bonded interactions within the active sites that could explain the drop-off in p38 activity. Indeed, the C5 position of the N-linked phenyl ring of the indazoles was only 3.79 from Val-196 in JNK3. This residue is Ala in p38. The findings of this study showing that SR-4186 was completely impotent as a p38 inhibitor were likely due to the smaller Ala residue present at this position. JNK3, on the other hand has Val at this position and can likely pack tighter with the 5-trifluoromethyl substitution. Lisnock et al. (19) showed that, when the Ala residue in p38 is mutated to the larger Val residue found in JNK3, triarylimidazole inhibitors of p38 become 10-fold more potent against p38. Thus it is likely that the larger Val residue in JNK3 coupled with the 5-trifluoromethyl substitution allowed for remaining JNK3 inhibition, with concomitant loss of all p38 inhibition due to the smaller Ala.
JNK3/p38 SAR for Aminopyrazole-based Inhibitors—A strategy used to eliminate the offending in-plane interactions observed in the indazole series would dictate removing the indazole phenyl ring altogether. This approach led us to a series of N-phenyl-linked pyrazoles (Table 1). To maintain the same position of the indazole 5-anilino group in this series and its occupation of hydrophobic pocket I, we used a urea group to link the pyrazole and aryl ring. We postulated that the conjugated sp-2-like nature of the urea would maintain a planer arrangement, much like the indazoles, and limit free rotation. Fortunately, this approach proved viable, because SR-3451 showed only a 2-fold drop in JNK3 potency yet a 1000-fold drop in p38 activity relative to SR-3737. Although SR-3451 was itself only 150-fold selective for JNK3/p38, other analogs showed even more selectivity against p38 with SR-3576 nearly 3000-fold selective. Now that the indazole phenyl ring had been removed, there were no in-plane steric interactions with the pyrazole N-linked phenyl ring. We hypothesize that, in this series, the pyrazole, its N-linked phenyl ring, as well as the urea side chain can all achieve co-planarity minimizing its three-dimensional requirement for binding space. This arrangement more favorably binds the smaller JNK3 active site, hence, the observed selectivity. Although the crystal structure of SR-3451 in JNK3 showed a slight deviation from planarity between the pyrazole and its N-linked phenyl ring, the urea certainly does maintain a rigid, planer orientation placing the urea phenyl ring into hydrophobic pocket I as anticipated.
In this particular study the x-ray crystallography data (see Figs. 2 and 3) did not provide great insight as to why the indazoles as represented by SR-3737 did not have selectivity for JNK3 where the aminopyrazoles, as represented by SR-3451 did. One possible explanation is that all of the crystal structures for JNK3 and p38 with compounds bound come from the unphosphorylated, inactive state of the enzyme. The enzyme inhibition data of course use the active, phosphorylated enzyme. Perhaps differences in structure between the active and inactive forms of a particular enzyme (say JNK3 or p38) can be attributed to these differences. Alternatively, despite the fact that p38 and JNK3 have very similar structures in the inactive form, perhaps p38 and JNK assume different active conformations, and these different active conformations contribute to the selectivity. Solution of active conformation structures would be needed to test these hypotheses.
JNK3/JNK1 SAR for Aminopyrazole- and Indazole-based Inhibitors—Both the indazoles and pyrazole series displayed a roughly 10-fold to 20-fold selectivity for JNK3 versus JNK1. From the crystal structures of SR-3737 and SR-3451, it was clear that both structural series bound in an induced fit manner within JNK3. In the indazoles, it was the 5-anilinophenyl substituent, and the N-phenyl urea group in the pyrazoles that bound into hydrophobic pocket I, the so-called selectivity pocket. The gatekeeper methionine residue moved to accommodate the ligand in both instances. Given that the selectivity pocket is lined with different amino acids in JNK3 versus JNK1, the induced fit binding appeared to be more favorable for JNK3 and hence the observed selectivity. This observation is in accord with several literature accounts that also note the movement of the gatekeeper residue and binding to the hydrophobic pocket results in modest JNK3/JNK1 selectivity (22, 25, 32, 34).
Cell-based JNK Inhibition for Aminopyrazole-based Inhibitors—The purpose of this cell-based assay is to determine if the compounds we designed were cell permeable and to see what the cell-based potency was. Unlike a biochemical assay, which measures individual JNK isoform activity, the cell-based IC50 values reported for JNK activity in the INS-1 cells is reflective of total JNK activity in the cell. Most of this JNK activity is comprised from JNK1 and JNK2, because INS-1 cells are derived from rat pancreas. It is unknown if this transformed cell line contains JNK3, because there are no studies that have looked for JNK3 expression in this cell line, but, given the expression patterns in humans for JNK3, it is likely that most of the activity measured in this cell line will be from JNK1 and JNK2. For most of the aminopyrazoles presented in Table 2, the cell-based IC50 was within 2- to 8-fold compared with that for the JNK1 biochemical IC50. These results suggest that the aminopyrazoles are cell-permeable JNK inhibitors and that the potency is not greatly shifted in cells. Given this observation, one can speculate that, if there was a cell-based assay for neuronal cells that could monitor only JNK3 activity in the cell, and there were no other impacting factors, the IC50 in cells for JNK3 would be <100 nm for compounds such as SR-3451 and SR-3576.
In summary, through a series of SAR modifications we have been able to develop a class of aminopyrazole inhibitors that are very potent JNK3 inhibitors with >2800-fold selectivity over p38 and have cell-based potencies within 2- to 8-fold of the biochemical JNK1 potency. The structural features within the compounds giving rise to the selectivity in the aminopyrazole class include the highly planar nature of the pyrazole, N-linked phenyl structures, which better occupy the smaller active site of JNK3 compared with the larger active site of p38. Modest selectivity (∼24-fold) for the aminopyrazoles was also found for JNK3 over JNK1. Further SAR studies along with detailed site-directed mutagenesis and x-ray crystallography experiments (particularly with complexes of p38 with these inhibitors) may elucidate structural features that may help enable the design of highly selective JNK isoform-selective inhibitors.
Supplementary Material
Acknowledgments
We are grateful to Yamille Del Rosario for administrative assistance in preparing the manuscript.
The atomic coordinates and structure factors (codes 3FI2 and 3FI3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant U01-NS057153 (to P. L.). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the U. S. Dept. of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Dept. of Energy, Office of Biological and Environmental Research, and by the National Center for Research Resources, NIH, Biomedical Technology Program, and NIGMS, NIH.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, schemes, and Tables S1 and S2.
Footnotes
The abbreviations used are: JNK, c-Jun N-terminal kinase; ATF-2, activating transcription factor-2; AMP-PCP, adenosine 5′-(β,γ-methylenetriphosphate); Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; SAR, structure-activity relationship; r.m.s.d., root mean square deviation.
References
- 1.Lee, J. C., Laydon, J. T., McDonnell, P. C., Gallagher, T. F., Kumar, S., Green, D., McNulty, D., Blumenthal, M. J., Keys, J. R., Landvatter, S. W., Strickler, J. E., McLaughlin, M. M., Siemens, I. R., Fisher, S. M., Livi, G. P., White, J. R., Adams, J. L., and Young, P. R. (1994) Nature 372 739-746 [DOI] [PubMed] [Google Scholar]
- 2.Derijard, B., Hibi, M., Wu, I. H., Barrett, T., Su, B., Deng, T., Karin, M., and Davis, R. J. (1994) Cell 76 1025-1037 [DOI] [PubMed] [Google Scholar]
- 3.Hibi, M., Lin, A., Smeal, T., Minden, A., and Karin, M. (1993) Genes Dev. 7 2135-2148 [DOI] [PubMed] [Google Scholar]
- 4.Kyriakis, J. M., Banerjee, P., Nikolakaki, E., Dai, T., Rubie, E. A., Ahmad, M. F., Avruch, J., and Woodgett, J. R. (1994) Nature 369 156-160 [DOI] [PubMed] [Google Scholar]
- 5.Minden, A., Lin, A., Smeal, T., Derijard, B., Cobb, M., Davis, R., and Karin, M. (1994) Mol. Cell. Biol. 14 6683-6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sluss, H. K., Barrett, T., Derijard, B., and Davis, R. J. (1994) Mol. Cell. Biol. 14 8376-8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez, C., Powers, D. A., and Tamayo, N. (2005) Curr. Opin. Drug Discov. Dev. 8 421-430 [PubMed] [Google Scholar]
- 8.Haddad, J. J. (2001) Curr. Opin. Investig. Drugs 2 1070-1076 [PubMed] [Google Scholar]
- 9.Lee, M. R., and Dominguez, C. (2005) Curr. Med. Chem. 12 2979-2994 [DOI] [PubMed] [Google Scholar]
- 10.Regan, J., Breitfelder, S., Cirillo, P., Gilmore, T., Graham, A. G., Hickey, E., Klaus, B., Madwed, J., Moriak, M., Moss, N., Pargellis, C., Pav, S., Proto, A., Swinamer, A., Tong, L., and Torcellini, C. (2002) J. Med. Chem. 45 2994-3008 [DOI] [PubMed] [Google Scholar]
- 11.Schreiber, S., Feagan, B., D'Haens, G., Colombel, J. F., Geboes, K., Yurcov, M., Isakov, V., Golovenko, O., Bernstein, C. N., Ludwig, D., Winter, T., Meier, U., Yong, C., and Steffgen, J. (2006) Clin. Gastroenterol. Hepatol. 4 325-334 [DOI] [PubMed] [Google Scholar]
- 12.de Dios, A., Shih, C., Lopez de Uralde, B., Sanchez, C., del Prado, M., Martin Cabrejas, L. M., Pleite, S., Blanco-Urgoiti, J., Lorite, M. J., Nevill, C. R., Jr., Bonjouklian, R., York, J., Vieth, M., Wang, Y., Magnus, N., Campbell, R. M., Anderson, B. D., McCann, D. J., Giera, D. D., Lee, P. A., Schultz, R. M., Li, L. C., Johnson, L. M., and Wolos, J. A. (2005) J. Med. Chem. 48 2270-2273 [DOI] [PubMed] [Google Scholar]
- 13.Goldstein, D. M., Alfredson, T., Bertrand, J., Browner, M. F., Clifford, K., Dalrymple, S. A., Dunn, J., Freire-Moar, J., Harris, S., Labadie, S. S., La Fargue, J., Lapierre, J. M., Larrabee, S., Li, F., Papp, E., McWeeney, D., Ramesha, C., Roberts, R., Rotstein, D., San Pablo, B., Sjogren, E. B., So, O. Y., Talamas, F. X., Tao, W., Trejo, A., Villasenor, A., Welch, M., Welch, T., Weller, P., Whiteley, P. E., Young, K., and Zipfel, S. (2006) J. Med. Chem. 49 1562-1575 [DOI] [PubMed] [Google Scholar]
- 14.LoGrasso, P. V., Frantz, B., Rolando, A. M., O'Keefe, S. J., Hermes, J. D., and O'Neill, E. A. (1997) Biochemistry 36 10422-10427 [DOI] [PubMed] [Google Scholar]
- 15.Swahn, B. M., Xue, Y., Arzel, E., Kallin, E., Magnus, A., Plobeck, N., and Viklund, J. (2006) Bioorg. Med. Chem. Lett. 16 1397-1401 [DOI] [PubMed] [Google Scholar]
- 16.Young, P. R., McLaughlin, M. M., Kumar, S., Kassis, S., Doyle, M. L., McNulty, D., Gallagher, T. F., Fisher, S., McDonnell, P. C., Carr, S. A., Huddleston, M. J., Seibel, G., Porter, T. G., Livi, G. P., Adams, J. L., and Lee, J. C. (1997) J. Biol. Chem. 272 12116-12121 [DOI] [PubMed] [Google Scholar]
- 17.Zhao, H., Serby, M. D., Xin, Z., Szczepankiewicz, B. G., Liu, M., Kosogof, C., Liu, B., Nelson, L. T., Johnson, E. F., Wang, S., Pederson, T., Gum, R. J., Clampit, J. E., Haasch, D. L., Abad-Zapatero, C., Fry, E. H., Rondinone, C., Trevillyan, J. M., Sham, H. L., and Liu, G. (2006) J. Med. Chem. 49 4455-4458 [DOI] [PubMed] [Google Scholar]
- 18.Stelmach, J. E., Liu, L., Patel, S. B., Pivnichny, J. V., Scapin, G., Singh, S., Hop, C. E., Wang, Z., Strauss, J. R., Cameron, P. M., Nichols, E. A., O'Keefe, S. J., O'Neill, E. A., Schmatz, D. M., Schwartz, C. D., Thompson, C. M., Zaller, D. M., and Doherty, J. B. (2003) Bioorg. Med .Chem. Lett. 13 277-280 [DOI] [PubMed] [Google Scholar]
- 19.Lisnock, J., Tebben, A., Frantz, B., O'Neill, E. A., Croft, G., O'Keefe, S. J., Li, B., Hacker, C., de Laszlo, S., Smith, A., Libby, B., Liverton, N., Hermes, J., and LoGrasso, P. (1998) Biochemistry 37 16573-16581 [DOI] [PubMed] [Google Scholar]
- 20.Wilson, K. P., McCaffrey, P. G., Hsiao, K., Pazhanisamy, S., Galullo, V., Bemis, G. W., Fitzgibbon, M. J., Caron, P. R., Murcko, M. A., and Su, M. S. (1997) Chem. Biol. 4 423-431 [DOI] [PubMed] [Google Scholar]
- 21.Liverton, N. J., Butcher, J. W., Claiborne, C. F., Claremon, D. A., Libby, B. E., Nguyen, K. T., Pitzenberger, S. M., Selnick, H. G., Smith, G. R., Tebben, A., Vacca, J. P., Varga, S. L., Agarwal, L., Dancheck, K., Forsyth, A. J., Fletcher, D. S., Frantz, B., Hanlon, W. A., Harper, C. F., Hofsess, S. J., Kostura, M., Lin, J., Luell, S., O'Neill, E. A., Orevillo, C. J., Pang, M., Parsons, J., Rolando, A., Sahly, Y., Visco, D. M., and O'Keefe, S. J. (1999) J. Med. Chem. 42 2180-2190 [DOI] [PubMed] [Google Scholar]
- 22.Angell, R. M., Atkinson, F. L., Brown, M. J., Chuang, T. T., Christopher, J. A., Cichy-Knight, M., Dunn, A. K., Hightower, K. E., Malkakorpi, S., Musgrave, J. R., Neu, M., Rowland, P., Shea, R. L., Smith, J. L., Somers, D. O., Thomas, S. A., Thompson, G., and Wang, R. (2007) Bioorg. Med. Chem. Lett. 17 1296-1301 [DOI] [PubMed] [Google Scholar]
- 23.Gaillard, P., Jeanclaude-Etter, I., Ardissone, V., Arkinstall, S., Cambet, Y., Camps, M., Chabert, C., Church, D., Cirillo, R., Gretener, D., Halazy, S., Nichols, A., Szyndralewiez, C., Vitte, P. A., and Gotteland, J. P. (2005) J. Med. Chem. 48 4596-4607 [DOI] [PubMed] [Google Scholar]
- 24.Szczepankiewicz, B. G., Kosogof, C., Nelson, L. T., Liu, G., Liu, B., Zhao, H., Serby, M. D., Xin, Z., Liu, M., Gum, R. J., Haasch, D. L., Wang, S., Clampit, J. E., Johnson, E. F., Lubben, T. H., Stashko, M. A., Olejniczak, E. T., Sun, C., Dorwin, S. A., Haskins, K., Abad-Zapatero, C., Fry, E. H., Hutchins, C. W., Sham, H. L., Rondinone, C. M., and Trevillyan, J. M. (2006) J. Med. Chem. 49 3563-3580 [DOI] [PubMed] [Google Scholar]
- 25.Swahn, B. M., Huerta, F., Kallin, E., Malmstrom, J., Weigelt, T., Viklund, J., Womack, P., Xue, Y., and Ohberg, L. (2005) Bioorg. Med. Chem. Lett. 15 5095-5099 [DOI] [PubMed] [Google Scholar]
- 26.Borsello, T., Clarke, P. G., Hirt, L., Vercelli, A., Repici, M., Schorderet, D. F., Bogousslavsky, J., and Bonny, C. (2003) Nat. Med. 9 1180-1186 [DOI] [PubMed] [Google Scholar]
- 27.Hunot, S., Vila, M., Teismann, P., Davis, R. J., Hirsch, E. C., Przedborski, S., Rakic, P., and Flavell, R. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 665-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia, X. G., Harding, T., Weller, M., Bieneman, A., Uney, J. B., and Schulz, J. B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10433-10438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneto, H., Nakatani, Y., Miyatsuka, T., Kawamori, D., Matsuoka, T. A., Matsuhisa, M., Kajimoto, Y., Ichijo, H., Yamasaki, Y., and Hori, M. (2004) Nat. Med. 10 1128-1132 [DOI] [PubMed] [Google Scholar]
- 30.Fitzgerald, C. E., Patel, S. B., Becker, J. W., Cameron, P. M., Zaller, D., Pikounis, V. B., O'Keefe, S. J., and Scapin, G. (2003) Nat. Struct. Biol. 10 764-769 [DOI] [PubMed] [Google Scholar]
- 31.Heo, Y. S., Kim, S. K., Seo, C. I., Kim, Y. K., Sung, B. J., Lee, H. S., Lee, J. I., Park, S. Y., Kim, J. H., Hwang, K. Y., Hyun, Y. L., Jeon, Y. H., Ro, S., Cho, J. M., Lee, T. G., and Yang, C. H. (2004) EMBO J. 23 2185-2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scapin, G., Patel, S. B., Lisnock, J., Becker, J. W., and LoGrasso, P. V. (2003) Chem. Biol. 10 705-712 [DOI] [PubMed] [Google Scholar]
- 33.Tong, L., Pav, S., White, D. M., Rogers, S., Crane, K. M., Cywin, C. L., Brown, M. L., and Pargellis, C. A. (1997) Nat. Struct. Biol. 4 311-316 [DOI] [PubMed] [Google Scholar]
- 34.Xie, X., Gu, Y., Fox, T., Coll, J. T., Fleming, M. A., Markland, W., Caron, P. R., Wilson, K. P., and Su, M. S. (1998) Structure 6 983-991 [DOI] [PubMed] [Google Scholar]
- 35.Huang, X., Anderson, K. W., Zim, D., Jiang, L., Klapars, A., and Buchwald, S. L. (2003) J. Am. Chem. Soc. 125 6653-6655 [DOI] [PubMed] [Google Scholar]
- 36.Lindley, J. (1984) Tetrahedron 40 1435-1456 [Google Scholar]
- 37.Wolfe, J. P., Tomori, H., Sadighi, J. P., Yin, J., and Buchwald, S. L. (2000) J. Org. Chem. 65 1158-1174 [DOI] [PubMed] [Google Scholar]
- 38.Yang, B. H., and Buchwald, S. L. (1999) J. Organomet. Chem. 576 125-146 [Google Scholar]
- 39.Gross, R. S., Guo, Z., Dyck, B., Coon, T., Huang, C. Q., Lowe, R. F., Marinkovic, D., Moorjani, M., Nelson, J., Zamani-Kord, S., Grigoriadis, D. E., Hoare, S. R., Crowe, P. D., Bu, J. H., Haddach, M., McCarthy, J., Saunders, J., Sullivan, R., Chen, T., and Williams, J. P. (2005) J. Med. Chem. 48 5780-5793 [DOI] [PubMed] [Google Scholar]
- 40.Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M., and Phizackerley, R. P. (2002) J. Appl. Crystallogr. 35 720-726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 42.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Suater, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948-1954 [DOI] [PubMed] [Google Scholar]
- 43.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 44.Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B., 3rd, Snoeyink, J., Richardson, J. S., and Richardson, D. C. (2007) Nucleic Acids Res. 35 W375-W383, web server issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuttelkopf, A. W., and van Aalten, D. M. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1355-1363 [DOI] [PubMed] [Google Scholar]
- 46.Aronov, A. M., Baker, C., Bemis, G. W., Cao, J., Chen, G., Ford, P. J., Germann, U. A., Green, J., Hale, M. R., Jacobs, M., Janetka, J. W., Maltais, F., Martinez-Botella, G., Namchuk, M. N., Straub, J., Tang, Q., and Xie, X. (2007) J. Med. Chem. 50 1280-1287 [DOI] [PubMed] [Google Scholar]
- 47.Smith, G. M., Alexander, R. S., Christianson, D. W., McKeever, B. M., Ponticello, G. S., Springer, J. P., Randall, W. C., Baldwin, J. J., and Habecker, C. N. (1994) Protein Sci. 3 118-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen, Z., Li, Y., Chen, E., Hall, D. L., Darke, P. L., Culberson, C., Shafer, J. A., and Kuo, L. C. (1994) J. Biol. Chem. 269 26344-26348 [PubMed] [Google Scholar]
- 49.Dorsey, B. D., Levin, R. B., McDaniel, S. L., Vacca, J. P., Guare, J. P., Darke, P. L., Zugay, J. A., Emini, E. A., Schleif, W. A., Quintero, J. C., Lin, J. H., Chen, I. W., Holloway, K. M., Fitzgerald, P. M. D., Axel, M. G., Ostovic, D., Anderson, P. S., and Huff, J. R. (1994) J. Med. Chem. 37 3443-3451 [DOI] [PubMed] [Google Scholar]
- 50.Gehlhaar, D. K., Verkhivker, G. M., Rejto, P. A., Sherman, C. J., Fogel, D. B., Fogel, L. J., and Freer, S. T. (1995) Chem. Biol. 2 317-324 [DOI] [PubMed] [Google Scholar]
- 51.Kaldor, S. W., Kalish, V. J., Davies, J. F., 2nd, Shetty, B. V., Fritz, J. E., Appelt, K., Burgess, J. A., Campanale, K. M., Chirgadze, N. Y., Clawson, D. K., Dressman, B. A., Hatch, S. D., Khalil, D. A., Kosa, M. B., Lubbehusen, P. P., Muesing, M. A., Patick, A. K., Reich, S. H., Su, K. S., and Tatlock, J. H. (1997) J. Med. Chem. 40 3979-3985 [DOI] [PubMed] [Google Scholar]
- 52.Schindler, T., Bornmann, W., Pellicena, P., Miller, W. T., Clarkson, B., and Kuriyan, J. (2000) Science 289 1938-1942 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.