Abstract
Zinc is hypothesized to be co-released with glutamate at synapses of the central nervous system. Zinc binds to NR1/NR2A N-methyl-d-aspartate (NMDA) receptors with high affinity and inhibits NMDAR function in a voltage-independent manner. The serine protease plasmin can cleave a number of substrates, including protease-activated receptors, and may play an important role in several disorders of the central nervous system, including ischemia and spinal cord injury. Here, we demonstrate that plasmin can cleave the native NR2A amino-terminal domain (NR2AATD), removing the functional high affinity Zn2+ binding site. Plasmin also cleaves recombinant NR2AATD at lysine 317 (Lys317), thereby producing a ∼40-kDa fragment, consistent with plasmin-induced NR2A cleavage fragments observed in rat brain membrane preparations. A homology model of the NR2AATD predicts that Lys317 is near the surface of the protein and is accessible to plasmin. Recombinant expression of NR2A with an amino-terminal deletion at Lys317 is functional and Zn2+ insensitive. Whole cell voltage-clamp recordings show that Zn2+ inhibition of agonist-evoked NMDA receptor currents of NR1/NR2A-transfected HEK 293 cells and cultured cortical neurons is significantly reduced by plasmin treatment. Mutating the plasmin cleavage site Lys317 on NR2A to alanine blocks the effect of plasmin on Zn2+ inhibition. The relief of Zn2+ inhibition by plasmin occurs in PAR1-/- cortical neurons and thus is independent of interaction with protease-activated receptors. These results suggest that plasmin can directly interact with NMDA receptors, and plasmin may increase NMDA receptor responses through disruption or removal of the amino-terminal domain and relief of Zn2+ inhibition.
N-Methyl-d-aspartate (NMDA)2 receptors are one of three types of ionotropic glutamate receptors that play critical roles in excitatory neurotransmission, synaptic plasticity, and neuronal death (1–3). NMDA receptors are comprised of glycine-binding NR1 subunits in combination with at least one type of glutamate-binding NR2 subunit (1, 4). Each subunit contains three transmembrane domains, one cytoplasmic re-entrant membrane loop, one bi-lobed domain that forms the ligand binding site, and one bi-lobed amino-terminal domain (ATD), thought to share structural homology to periplasmic amino acid-binding proteins (4–6). Activation of NMDA receptors requires combined stimulation by glutamate and the co-agonist glycine in addition to membrane depolarization to overcome voltage-dependent Mg2+ block of the ion channel (7). The activity of NMDA receptors is negatively modulated by a variety of extracellular ions, including Mg2+, polyamines, protons, and Zn2+ ions, which can exert tonic inhibition under physiological conditions (1, 4). Several extracellular modulators such as Zn2+ and ifenprodil are thought to act at the ATD of the NMDA receptor (8–14).
Zinc is a transition metal that plays key roles in both catalytic and structural capacities in all mammalian cells (15). Zinc is required for normal growth and survival of cells. In addition, neuronal death in hypoxia-ischemia and epilepsy has been associated with Zn2+ (16–18). Abnormal metabolism of zinc may contribute to induction of cytotoxicity in neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis (19). Zinc is co-released with glutamate at excitatory presynaptic terminals and inhibits native NMDA receptor activation (20, 21). Zn2+ inhibits NMDA receptor function through a dual mechanism, which includes voltage-dependent block and voltage-independent inhibition (22–24). Voltage-independent Zn2+ inhibition at low nanomolar concentrations (IC50, 20 nm) is observed for NR2A-containing NMDA receptors (25–28). Evidence has accumulated that the amino-terminal domain of the NR2A subunit controls high-affinity Zn2+ inhibition of NMDA receptors, and several histidine residues in this region may constitute part of an NR2A-specific Zn2+ binding site (8, 9, 11, 12). For the NR2A subunit, several lines of evidence suggest that Zn2+ acts by enhancing proton inhibition (8, 11, 29, 30).
Serine proteases present in the circulation, mast cells, and elsewhere signal directly to cells by cleaving protease-activated receptors (PARs), members of a subfamily of G-protein-coupled receptors. Cleavage exposes a tethered ligand domain that binds to and activates the cleaved receptors (31, 32). Protease receptor activation has been studied extensively in relation to coagulation and thrombolysis (33). In addition to their circulation in the bloodstream, some serine proteases and PARs are expressed in the central nervous system, and have been suggested to play roles in physiological conditions (e.g. long-term potentiation or memory) and pathophysiological states such as glial scarring, edema, seizure, and neuronal death (31, 34–36).
Functional interactions between proteases and NMDA receptors have previously been suggested. Earlier studies reported that the blood-derived serine protease thrombin potentiates NMDA receptor response more than 2-fold through activation of PAR1 (37). Plasmin, another serine protease, similarly potentiates NMDA receptor response (38). Tissue-plasminogen activator (tPA), which catalyzes the conversion of the zymogen precursor plasminogen to plasmin and results in PAR1 activation, also interacts with and cleaves the ATD of the NR1 subunit of the NMDA receptor (39, 40). This raises the possibility that plasmin may also interact directly with the NMDA receptor subunits to modulate receptor response. We therefore investigated the ability of plasmin to cleave the NR2A NMDA receptor subunit. We found that nanomolar concentrations of plasmin can cleave within the ATD, a region that mediates tonic voltage-independent Zn2+ inhibition of NR2A-containing NMDA receptors. We hypothesized that plasmin cleavage reduces the Zn2+-mediated inhibition of NMDA receptors by removing the Zn2+ binding domain. In the present study, we have demonstrated that Zn2+ inhibition of agonist-evoked NMDA currents is decreased significantly by plasmin treatment in recombinant NR1/NR2A-transfected HEK 293 cells and cultured cortical neurons. These concentrations of plasmin may be pathophysiologically relevant in situations in which the blood-brain barrier is compromised, which could allow blood-derived plasmin to enter brain parenchyma at concentrations in excess of these that can cleave NR2A. Thus, ability of plasmin to potentiate NMDA function through the relief of the Zn2+ inhibition could exacerbate the harmful actions of NMDA receptor overactivation in pathological situations. In addition, if newly cleaved NR2AATD enters the bloodstream during ischemic injury, it could serve as a biomarker of central nervous system injury.
EXPERIMENTAL PROCEDURES
Immunoblot—Adult rat brain regions were dissected and homogenized in ice-cold RIPA buffer (9.1 mm dibasic sodium phosphate, 1.7 mm monobasic sodium phosphate, 150 mm NaCl, 1% Igepal CA-360, 0.5% sodium deoxycholate, and 0.1% SDS) containing the following protease inhibitors: 0.1 mg/ml phenylmethylsulfonyl fluoride, 30 μl/ml aprotinin, and 1 nm sodium orthovanadate. The membranes were stored at -20 °C and were incubated with increasing concentrations of plasmin (30–500 nm) for 20 min at room temperature. 50–100 μg of protein per sample were incubated with 2% SDS, 62.5 mm Tris, 10% glycerol, 5% β-mercaptoethanol, and 0.05% bromphenol blue for 5 min at 100 °C.
Samples were loaded into 10% Tris glycine pre-cast gels (Invitrogen) and run at 150 V for 1.5 h in running buffer (0.1% SDS, 125 mm Tris base, 1 m glycine). Proteins subsequently were transferred onto polyvinyldine difluoride (PVDF) membranes in transfer buffer (10% methanol, 25 mm Tris, 192 mm glycine) for 2 h at 100 V at 4 °C. Membranes were blocked in 2.5% milk in Tris-buffered saline for 30 min and then incubated with an anti-NR2A primary antibody (1:1000, polyclonal, Upstate Biotech, Lake Placid, NY) on a shaker at 4 °C overnight. Membranes were washed 3 × 10 min in Tris-buffered saline, incubated with goat anti-mouse secondary antibodies (1:1000, Jackson Laboratories, Bar Harbor, ME), washed again, and subsequently detected by enhanced chemiluminescence (Amersham Biosciences).
NR2A Amino-terminal Domain (NR2AATD) Protein Production—The NR2AATD was produced as a thrombin-cleavage glutathione S-transferase (GST) fusion protein in Escherichia coli. NR2AATD (Ala33–Asp421) was generated by polymerase chain reaction with the following primer pairs: sense, 5′-acgattccatggcgctgaacattgcggtgctg-3′ and antisense, 5′-atcgaagctttatcagaggtcctcttcggatatcagcttctgttcgtcctctacgatgacgaagg-3′. The product then was cloned into a pGEX-KG vector. A c-myc tag (underlined in antisense primer; EQKLISEEDL) was incorporated in the COOH terminus of NR2AATD for identification purposes. After transformation into BL21-CodonPlus-RIL cells (Stratagene, La Jolla, CA), cells at OD 0.8 were induced by 0.2 mm isopropyl β-d-thiogalactopyranoside for 3 h at 37 °C. Pelleted cells were lysed using French Press (Thermo Spectronic, Madison, WI), and GST-NR2AATD protein was solubilized with detergents. The fusion protein was purified through batch chromatography using reduced glutathione-Sepharose 4B matrix (Amersham Biosciences). NR2AATD protein was cleaved from the GST by 10 units of thrombin (Calbiochem, San Diego, CA) at room temperature for 1 h.
Zn2+-IDA-Agarose Affinity Binding Assay—Fifty microliters of immobilized iminodiacetic acid (IDA) resin (Pierce) equilibrated in Buffer A (10 mm HEPES, 50 mm NaCl, 0.5% Triton X-100, pH 7.5) were incubated at room temperature for 30 min with a freshly prepared 10 mm ZnCl2 solution. The fully Zn2+-charged IDA-agarose was thoroughly washed with Buffer A. The pelleted Zn2+-IDA-agarose was resuspended in 25 μl of Buffer A and incubated with 25 μl of thrombin-cleaved NR2AATD protein (room temperature for 30 min). The NR2AATD-bound to Zn2+-IDA-agarose was pelleted, and the supernatant was collected (designated as FT). The agarose resin was washed with Buffer A six times, and the first wash solution (W1) was collected for analysis. The bound NR2AATD was eluted from Zn2+-IDA-agarose with several volumes of 50 mm EDTA or 1 mm l-histidine in Buffer A (designated as E1, E2, or E3). Negative controls were performed with IDA-agarose that was not charged with ZnCl2.
Sequencing of NR2AATD Plasmin Cleavage Products—The proteins were separated by gel electrophoresis, blotted onto PVDF membrane, and stained with Coomassie Brilliant Blue R-250. The amino-terminal sequence after plasmin cleavage was determined using Edman degradation, which was performed in the Applied Biosystems model cLC-Procise protein sequencer (41) at the Emory Microchemical Facility.
Molecular Modeling of NR2A Subunit—The homology model of NR2AATD was built using the previously published alignment between NR2AATD and mGluR1 (42). The closed form of mGluR1 found in Protein Data Bank code 1EWK (43) provided the structural template. The mGluR1 structure and alignment were imported into Prime (version 1.5, 2006, Schrodinger, LLC, New York), and the NR2AATD homology model was subsequently built using default settings. The model was refined initially using the Prime loop prediction tool on the two central loops (41–45 and 100–105) of the ATD. The entire structure was then energy minimized, and multiple rounds of energy minimization and side chain optimization were performed on all residues within5Å of the two central loops.
Mutagenesis—Site-directed mutagenesis was performed using the QuikChange kit (Stratagene, La Jolla, CA) as described previously (11). cDNAs for NR1-1a (U11418 and U08261; hereafter NR1) and NR2A (D13211) were provided by Drs. S. Heinemann (Salk Institute) and S. Nakanishi (Kyoto University), respectively. The ATD deletion construct of the NR2A subunit was developed from wild-type NR2A and NR2B-(ΔS28-M394). This construct encoded for the first 28 residues of NR2B, including the signal peptides. The residue following Ser28 was Ala318 of NR2A.
Transfection of HEK 293 Cells and Plasmin Treatment—Human embryonic kidney cells 293 (HEK 293 cells; CRL 1573, ATCC, Rockville, MD) were plated at low confluency on glass coverslips (Warner Instruments, Hamden, CT) coated in 200 μg/ml poly-d-lysine and incubated overnight at 37 °C in humidified 5% CO2 in media (Dulbecco's modified Eagle's medium; Invitrogen) supplemented with 10% fetal bovine serum, 10 units/ml penicillin, and 10 μg/ml streptomycin. Cells then were transiently transfected using the FuGENE transfection reagent (Roche Diagnostics, Basel, Switzerland) with 2.0 μg of plasmid cDNA encoding green fluorescent protein, NR1, and wild-type NR2A, green fluorescent protein, NR1, and mutant NR2A-K317A, or green fluorescent protein, NR1, and mutant NR2A-ΔATD-K317 at a 5:1:1 ratio. After transfection, the cells were incubated overnight at 37 °C in humidified 5% CO2 in media supplemented with 200 μm dl-2-amino-5-phosphonovalerate and 200 μm 7-chlorokynurenic acid. Following transfection and overnight incubation, media was removed and cells were washed in external solution for patch clamp recording. Some washed coverslips then were removed from their wells and gently placed in a new, untreated 24-well plate that contained the external recording solution (500 μl, composition seen below) plus 100 nm plasmin. Clean, untreated plates without cells or bovine serum albumin from the media were used for plasmin treatment to reduce the amount of proteins that could compete with NR2A for cleavage and thus lower the apparent effectiveness of plasmin. The treated coverslips were incubated at 37 °C for 10 min and were transferred to new wells containing only external solution for patch clamp recording. We performed the recordings for both the control and the plasmin-treated groups on the same day.
Neuron Culture—Primary cultures of cortical neurons were prepared from 1- to 3-day postnatal wild type (C57BL/6) or PAR1-/- mice, which were created by breeding PAR1+/- mice, a gift from Dr. Shaun Coughlin (University of California, San Francisco, CA), with C57BL/6 wild-type mice from The Jackson Laboratory. Heterozygous littermates were bred to generate littermate homozygous null mutants and wild-type controls, which were subsequently used to generate homozygous colonies. PAR1-/- mice were >99% C57Bl/6. All procedures involving animals were approved by the Emory University Institutional Animal Care and Use Committee. The cortex was dissected and chopped into small cubes. The tissues were digested by 24 units/ml papain for 1 h in a 37 °C incubator, washed twice with 5–7 ml of inactivation solution (Neurobasal medium with 10% fetal bovine serum and 40 μg/ml DNase), and triturated gently with at least three serial fire-polished Pasteur pipettes. Cells were plated onto glass coverslips coated with 100 μg/ml poly-d-lysine. Cultures were maintained for 5–15 days in Neurobasal medium supplemented with 5% fetal bovine serum, B-27, 1.0 mm sodium pyruvate, 0.5 mm l-glutamine, 0.6% dextrose, 10 units/ml penicillin, and 10 μg/ml streptomycin at 37 °C in humidified 5% CO2. On the recording day, some washed coverslips were transferred to wells of a new plate that contained the plasmin solution (300 nm), incubated at 37 °C for 10 min, and then washed by external recording solution (composition given below).
Two-electrode Voltage-clamp Recordings from Xenopus laevis Oocytes—Preparation and injection of cRNA, as well as two-electrode voltage-clamp recordings from X. laevis oocytes, were performed as previously described (27). Briefly, oocytes were injected with 5–10 ng of cRNAs synthesized in vitro from linearized template cDNA and stored at 15 °C in Barth's solution. The ratio of NR1 to NR2 injected cRNA was 1:2. Two-electrode voltage-clamp recordings were made 2–4 days postinjection at room temperature (23 °C). The recording solution contained (in mm) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2, plus 2–10 μm EDTA. In the experiments for agonist potency, pH was adjusted to 7.6 with NaOH, and EDTA was added to chelate contaminant divalent ions. In the experiments for the concentration-response curve of zinc inhibition, pH was adjusted to 7.2 and Tricine (10 mm) was used to buffer Zn2+ concentrations as described previously (27). Solution exchange was computer-controlled through an 8-modular valve positioner (Digital MVP Valve, Hamilton, CT). Voltage and current electrodes were filled with 0.3–3.0 m KCl, and current responses were recorded at a holding potential of -20 to -40 mV at 23 °C. Data acquisition and voltage control were accomplished with a two-electrode voltage-clamp amplifier (OC-725, Warner Instrument, Hamilton, CT). Only currents greater than 50 nA were included in the analysis. 50 μm Glutamate and 30 μm glycine were used in all oocyte experiments unless otherwise stated.
Whole Cell Voltage-clamp Recording from Transfected HEK 293 Cells—Voltage-clamp recordings (VHOLD -60 mV) were performed on transfected HEK 293 cells using an Axopatch 200B amplifier (Molecular Devices, Union City, CA) at room temperature (23 °C). Recording electrodes (3.5–7 MΩ) were made from thin wall glass pipettes (TW150F-4, World Precision Instruments, Sarasota, FL) pulled using a vertical puller (Narishige PP-830, Tokyo, Japan), and were filled with an internal solution containing (in mm) 110 d-gluconate, 110 CsOH, 30 CsCl, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 NaATP, and 0.3 NaGTP, pH 7.35. Cells were bathed continually at 23 °C in an external solution composed of (in mm) 150 mm NaCl, 10 HEPES, 30 d-mannitol, 3 KCl, 2 CaCl2, and 0.01 EDTA, pH 7.4. Glutamate and glycine agonist solutions were made in external solution that contained 10 μm EDTA or 1 μm Zn2+. Once the cell membrane was ruptured by applying suction, the whole cell was lifted into the path of a 2-barrel fast perfusion system controlled by a piezoelectric translator (Burleigh Instruments, Fishers, NY) with 5–8-ms exchange times for the solutions (29). Cells were exposed first to glycine (50 μm), and subsequently glutamate (50 μm in 50 μm glycine) was applied rapidly for 1 s, after which the cells were again washed in glycine. Steady-state amplitudes and deactivation time constants were measured using ChanneLab (Synaptosoft, Decatur, GA).
Whole Cell Perforated Patch Recording from Cultured Cortical Neurons—Whole cell voltage-clamp current recordings (VHOLD -60 mV) were performed on 5–10-day cultured cortical neurons at room temperature (23 °C). The recording chamber was continually perfused with recording solution composed of (in mm) 150 NaCl, 3 KCl, 2 CaCl2, 5.5 glucose, and 10 HEPES (pH 7.2 by NaOH; osmolality adjusted to 315–320 mosmol with sucrose). 10 μm EDTA was added to chelate contaminant divalent ions except for Zn2+-containing solutions. Thin wall glass pipettes (TW150F-4, World Precision Instrument) were used to make recording electrodes (4–5 mΩ) by a vertical puller (Narishige PP-830, Tokyo, Japan) and filled with (in mm) 150 CsMeSO4, 10 NaCl, 0.5 CaCl2, 10 HEPES, and 25–50 μg/ml gramicidin D (pH adjusted to 7.3 with CsOH and osmolality adjusted to 310 mOsm with sucrose). Fresh stock solution of gramicidin D (1 mg/80 μl in dimethyl sulfoxide) was prepared daily. The final internal solution containing gramicidin D was used within 1 h. It took 15–30 min to achieve acceptable perforation with series resistance ranging from 30 to 60 mΩ. Drugs were applied directly by gravity and controlled by solenoid valves (Lee, Westbrook, CT) with tubes placed above the recorded cell. The solution exchange achieved a complete local perfusion of the recorded cell in ∼400 ms (10–90% rise time), which was measured by applying a high K+ solution onto cultured cortical neurons and measuring the time course of the instantaneous change of the leak current. All drugs were purchased from Sigma except papain and DNase (Worthington, Lakewood, NJ), Neurobasal medium, fetal bovine serum, l-glutamine and sodium pyruvate (Invitrogen), CNQX disodium salt and ifenprodil (Tocris Cookson, Bristol, UK), plasmin (Hematologic Technologies, Inc., Essex Junction, VT), and 2-furoyl-LIGRLO (Emory Microchemical Facility).
Data are expressed as mean ± S.E., and analyzed statistically using unpaired t test or one-way analysis of variance with Tukey's post hoc test. Significance for all tests was set at p < 0.05. Error bars in all figures are S.E.
RESULTS
Plasmin Cleaves the Native NR2A Subunit—To detect whether plasmin can cleave the NR2A subunit in native tissue, membrane preparations from rat cortex were treated with increasing concentrations of plasmin (200–500 nm) for 20 min at room temperature. Membrane proteins were separated using gel electrophoresis and transferred to PVDF membranes, which were probed with a primary antibody that recognizes the NR2A COOH-terminal. These experiments (four experiments from four separate membrane preparations) showed that 200–500 nm plasmin cleaves a ∼40-kDa fragment from the amino-terminal domain, leaving a truncated 140-kDa NR2A subunit (Fig. 1, A and B). Lower plasmin concentrations (30 nm) showed less cleavage of NR2A, suggesting concentration dependence (data not shown). These data indicate that higher concentrations of plasmin can cleave the NR2A subunit of native tissues at the ATD.
FIGURE 1.
Plasmin cleavage of the NR2A subunit in brain tissue. A, Western immunoblot was probed with a COOH-terminal NR2A antibody (Upstate Biotech). Rat brain membranes were incubated with plasmin for 20 min at room temperature prior to loading in a 10% Tris glycine gel. Note that plasmin cleaves a ∼40-kDa fragment from the NR2A amino-terminal domain. B, arrow indicates approximate plasmin cleavage site on NR2A subunit. Line above the COOH-terminal side indicates antibody epitope.
To investigate whether plasmin can cleave the NR1 subunit, Western blots were run on plasmin-treated rat neuronal membranes. Western blots probed with the NR1 monoclonal antibody 54.1 showed several fragments following plasmin treatment (supplemental Fig. S1A). Probing these same Western blots with an anti-COOH-terminal antibody yielded no apparent cleaved fragments but reveals decreasing signal in the original band of the plasmin-treated groups (supplemental Fig. S1B). These data suggest that plasmin cleaves NR1 efficiently at a site near the COOH terminus. In addition, there was very modest cleavage at a site within the S2 domain of the NR1 subunit, which appears less sensitive to plasmin than NR2A. The COOH-terminal site is inaccessible to plasmin in intact cells, and the weakly sensitive NR1-S2 cleavage site is distant from the Zn2+ binding site, which resides within the amino-terminal domain of the NR2 (not NR1) subunit. This result is consistent with the proposed plasmin cleavage sites on NR1 in the recently published report by Samson et al. (44).
Plasmin Cleaves Recombinant NR2AATD Protein—The finding that plasmin cleaves the NR2AATD in neuronal membranes may be significant because this region mediates high affinity voltage-independent Zn2+ inhibition of NR2A-containing receptors. To further investigate plasmin cleavage of the NR2AATD, recombinant NR2AATD protein was expressed in bacteria and purified. To accomplish this, the NR2AATD construct (residues 33–421; Fig. 2A, upper panel, dot boxed) was cloned into pGEX-KG plasmid and COOH-terminal tagged with c-myc. The size of the expressed GST-NR2AATD fusion protein was ∼74.3 kDa (Fig. 2A, middle panel). A thrombin cleavage site was inserted between GST and NR2AATD (Fig. 2A, lower panel). The NR2AATD construct was expressed in E. coli, and the recombinant NR2AATD proteins were purified through both the batch method and column chromatography. The purified recombinant NR2AATD protein was treated with 10 nm plasmin for 20 min at room temperature, and a PVDF membrane containing at least three bands of NR2AATD protein fragments was stained with Coomassie Blue (Fig. 2B). To determine the plasmin cleavage site(s), the cleaved protein fragments were cut, purified, and subjected to amino-terminal peptide sequencing. Sequencing showed that only one isolated fragment (indicated by thick red arrow in Fig. 2B) had a sequence matching the NR2A subunit at residues downstream of Lys317 (Fig. 2C, underlined amino acids); other isolated peptide fragments either did not yield interpretable sequences or were from other unidentified proteins. These results suggest that plasmin can cleave recombinant NR2AATD protein at a site located at Lys317, which is similar to plasmin-cleavage sites on other plasmin substrates (Table 1).
FIGURE 2.
Determination of the cleavage site of the NR2A subunit. A, schematic diagram of NR2A. Upper panel, histidines in the amino-terminal domain are indicated as gray ovals. The four putative membrane-associated domains are indicated as M1–M4. His42, His44, and His128 are critical for high-affinity voltage-independent Zn2+ inhibition of NR1/NR2A receptors and are highlighted by asterisks (11). The NR2AATD construct (residue Ala33–Asp421; dot boxed) was cloned into pGEX-KG vector. Middle panel, the expressed GST-NR2AATD fusion protein was COOH-terminal tagged with c-Myc (∼74.3 kDa). The vector-encoded thrombin cleavage site is indicated by an arrow. Lower panel, thrombin-cleaved NR2AATD (46.3 kDa). B, Coomassie-stained PVDF membrane containing NR2AATD protein fragments. Arrows indicate the potential plasmin (10 nm)-cleaved fragments subjected to amino-terminal sequencing. Only the band indicated by the thick red arrow yielded an interpretable sequence. C, amino acid Lys317 in red indicates the plasmin cleavage site. Note that the underlined amino acids were determined from amino-terminal peptide sequencing on the cleaved protein fragment (the bottom protein fragment in panel B, indicated by the red arrow). D, diagram of the NR2A subunit shows autonomous glutamate and Zn2+ binding domains. E, homology model of the NR2AATD. Left and middle panels, plasmin cleaves (indicated by the red arrow) between Lys317 (CPK colors) and Ala318 (first residue loop of red portion) in the NR2AATD protein (initiating methionine is 1). Zn2+ and its coordinating residues are indicated in the left panel (arrow). In the right panel, the red amino acids are downstream of the plasmin cleavage site and presumably remain attached to the receptor. The light gray portion of the protein harboring the Zn2+ site is free to dissociate from receptor after plasmin treatment.
TABLE 1.
Alignment of plasmin cleavage sites Position 0 (amino acid in bold) is the proposed plasmin cleavage site.
| Proteins/position | –3 | –2 | –1 | 0 | 1 | 2 | 3 |
|---|---|---|---|---|---|---|---|
| PAR-1 (46, 47) | P | E | S | K | A | T | N |
| L | D | P | R | S | F | L | |
| T | E | Y | R | L | V | S | |
| S | I | N | K | S | S | P | |
| P | L | Q | K | Q | L | P | |
| PAR-1 (46) | R | S | S | K | G | R | S |
| S | K | G | R | S | L | I | |
| Fibrinogen α (72) | G | G | V | R | G | P | R |
| R | G | P | R | V | V | E | |
| Fibrinogen β (72) | A | T | Q | K | K | V | E |
| G | G | Y | R | A | R | P | |
| R | P | A | K | A | A | A | |
| P | L | D | K | K | R | E | |
| Fibrinogen γ (72, 73) | T | Y | S | K | A | S | T |
| A | T | W | K | T | R | W | |
| Q | L | I | K | A | I | Q | |
| A | T | L | K | S | R | L | |
| Factor X (74) | I | T | F | R | M | N | V |
| Vitronectin (75) | K | G | Y | R | S | Q | R |
| Osteocalcin (76) | E | A | Y | R | F | Y | G |
| Catestatin (77) | E | D | N | R | D | S | S |
| S | S | M | K | L | S | F | |
| L | S | F | R | A | R | A | |
| F | R | A | R | A | Y | A | |
| Y | G | F | R | G | P | G | |
| Pro-brain-derived neurotropic factor (49) | M | S | M | R | V | R | R |
| Size of side chain (Å) | 51 | 43 | 64 | 86 | 39 | 67 | 52 |
| Basica | 100% | ||||||
| Smallb/non-polarc | 69% | 81% | 88% | ||||
| Small/polard | 85% | 85% | 81% | ||||
| NR2AATD | P | E | A | K | A | S | C |
Basic amino acids: R, K, H
Small amino acids: V, C, P, D, N, A, G, S, T
Non-polar amino acids: P, A, L, V, I, W, M, F, G
Polar amino acids: D, E, C, N, Q, T, Y, S, H, K, R
Homology Model of NR2AATD—To evaluate the accessibility of the proposed cleavage site at the NR2AATD to plasmin (Fig. 2D), a homology model of the NR2AATD was constructed using the alignment between NR2AATD and mGluR1 (43). This model predicts that Lys317 is near the surface of the protein (Fig. 2E) and thus is potentially accessible by plasmin. This model also indicates that the Zn2+-binding residues and binding pocket (magenta arrow in Fig. 2E) are upstream of the plasmin cleavage site (red arrow in Fig. 2E). This implies that the fragment containing the Zn2+-binding pocket within the ATD (gray color in Fig. 2E, right panel) may dissociate from the receptor after plasmin treatment. Therefore, plasmin treatment may relieve Zn2+ inhibition on NR2A-containing NMDA receptors.
Zinc Ions Bind to Recombinant NR2AATD Protein—There is considerable evidence showing that the region containing the amino-terminal domain of the NR2A subunit controls high-affinity Zn2+ inhibition of NMDA receptors, and several histidine residues in this region may constitute part of the NR2A-specific Zn2+ binding site (8, 11). To validate that the recombinant protein was in the correct conformation, we assessed its ability to bind Zn2+. Recombinant NR2AATD protein binds to Zn2+-charged IDA-agarose but not to uncharged IDA-agarose (Fig. 3A). The bound NR2AATD protein could be eluted using the metal-chelating agent EDTA (50 mm), but not low affinity Zn2+ binding agents imidazole and histidine. Several histidine residues in the ATD may constitute part of the NR2A-specific Zn2+ binding site (8, 11, 12). When H44G and H128A mutations were inserted concurrently into the recombinant NR2AATD protein, the binding of the NR2AATD(H44G/H128A) protein to Zn2+-IDA-agarose was weaker than the NR2AATD protein. NR2AATD(H44G/H128A) could be fully displaced by low affinity Zn2+ binding agents imidazole and l-histidine, which we interpreted as evidence that Zn2+ is coordinated by His44 and His128 (Fig. 3B). These data suggest that recombinant NR2AATD might exist in a conformation that resembles the native NR2A subunit and thus is a suitable substrate for analyses of plasmin proteolysis.
FIGURE 3.
A, recombinant NR2AATD proteins bind to Zn2+-charged IDA-agarose (lanes FT1 and FT2 under + zinc), and can be eluted in the flow-through in uncharged IDA-agarose (lanes FT1 and FT2 under - zinc). When the NR2AATD-bound Zn2+-IDA-agarose was eluted with 50 mm EDTA, all bound NR2AATD protein appeared in the elutes (lanes E1 and E2 under + zinc). NR2AATD proteins were probed with an anti-c-Myc antibody (1:10,000, Sigma) against the COOH-terminal c-myc inserted into the NR2AATD construct. All experiments (n = 2) were performed in duplicate (e.g. FT1 and FT2, E1 and E2). The right panel shows a shorter exposure of the same Western blot in the left panel to highlight that NR2AATD proteins that were eluted as a clear single band. M, protein standards (kDa). W1, first wash after incubation of NR2AATD and Zn2+-IDA-agarose. B, a competitive Zn2+-IDA-agarose binding assay showed NR2AATD proteins containing H44G and H128A double mutations bound weaker than the wild-type NR2AATD when challenged with 2 low affinity divalent chelating agents, imidazole and l-histidine. The immunoblot was labeled with the anti-c-Myc antibody (1:10,000, Sigma). L, WT, NR2AATD. FT, flow-through. MT, NR2AATD-(H44G/H128A). E, elute with consecutive agent. Concentrations of each agent are indicated.
Deletion of NR2AATD (ΔATD) at Lys317 Is Functional and Zn2+-insensitive—These biochemical data suggest that plasmin can cleave the Zn2+ binding domain of NR2A, raising the possibility that the remaining receptor may be functional but resistant to Zn2+ inhibition. To evaluate the functional change by deletion of the NR2AATD following plasmin cleavage, we mutated NR2A to delete all of the ATD up to Lys317 (NR2A-ΔATD-K317) (see “Experimental Procedures”; Fig. 4A). We expressed this deletion construct in Xenopus oocytes and evaluated properties of the functional NMDA receptor responses. The EC50 of glutamate measured in the presence of a maximally effective concentration of glycine (100 μm) at NR2A-ΔATD-K317 (5.2 ± 0.3 μm, n = 6) was similar to that of wild-type NR2A (3.3 ± 0.2 μm, n = 6). The EC50 of glycine in the presence of a maximally effective concentration of glutamate (100 μm) was reduced modestly (1.3 ± 0.1 μm, n = 14) compared with wild-type NR1/NR2A (2.5 ± 0.2 μm, n = 12) (Fig. 4B). Zn2+ sensitivity was assessed by measuring the percentage of inhibition of NMDA receptor agonist-evoked steady-state currents in the presence of 1 μm Zn2+, a concentration that saturates the high affinity site. Deletion of the amino-terminal domain of NR2A at Lys317 significantly reduced Zn2+ inhibition (12 ± 1.4%, n = 16) compared with wild-type NR2A (47 ± 3.0%, n = 15; p < 0.001, unpaired t test) (Fig. 4C). To further investigate the effects of ATD deletion from the plasmin cleavage site on zinc inhibition, the concentration-response curve for zinc inhibition was generated from recombinant NMDA receptors expressed in oocytes. High affinity Zn2+ inhibition was present in wild-type NR1/NR2A receptors, but was absent in the ATD deletion construct NR2A-ΔATD-K317. The low affinity site for Zn2+ inhibition was present in both wild-type NR2A and NR2A-ΔATD-K317, and presumably reflects voltage-dependent channel block by Zn2+ (Fig. 4D). These data demonstrate that recombinant NR1/NR2A receptors lacking the residues upstream of the plasmin cleavage site are functional. These data also are consistent with a previous report (9) that NR1/NR2A receptors lacking the ATD are less sensitive to Zn2+ inhibition.
FIGURE 4.
Recombinant NR1/NR2A-ΔATD-K317 is functional and Zn2+ insensitive. A, NR2A-ΔATD-K317 was constructed by inserting an NruI site into wild-type NR2A and subcloning the downstream portion (NruI-NotI) into the vector of NR2B-ΔATD-M394. B, mean normalized concentration-response curves for glutamate (left panel) and glycine (right panel) in oocytes expressing wild-type NR1/NR2A and NR1/NR2A-ΔATD-K317. Note that the EC50 of glutamate of NR2A-ΔATD-K317 (dashed line, 3.3 ± 1.4 μm, n = 6) is similar to the EC50 of glutamate of wild-type NR2A (solid line, 5.2 ± 0.3 μm, n = 6), whereas the EC50 of glycine of NR2A-ΔATD-K317 (dashed line, 2.5 ± 0.2 μm, n = 12) is slightly shifted to the right compared with wild-type NR2A (solid line, 1.3 ± 0.1 μm, n = 14). C, two-electrode voltage-clamp recordings of oocytes injected with wild-type NR1/NR2A and NR1/NR2A-ΔATD-K317 cDNA are shown. Left, a representative two-electrode voltage-clamp current recording obtained from wild-type NR1/NR2A showed significant inhibition by 1 μm Zn2+. Right, a representative recording of currents from NR2A-ΔATD-K317 exhibited less inhibition by 1 μm Zn2+. D, mean normalized concentration-response curves for zinc inhibition in oocytes expressing wild-type NR1/NR2A and NR1/NR2A-ΔATD-K317. Note that the Zn2+ concentration-inhibition curve at wild-type NR1/NR2A receptors (n = 8) was biphasic, with high (IC50, 46.5 nm; Hill slope, 0.9) and low (IC50, 10.5 μm; Hill slope, 2.4) affinity components, whereas NR1/NR2A-ΔATD-K317 receptors (n = 8) only showed a low affinity component (IC50, 13.5 μm, Hill slope, 1.7).
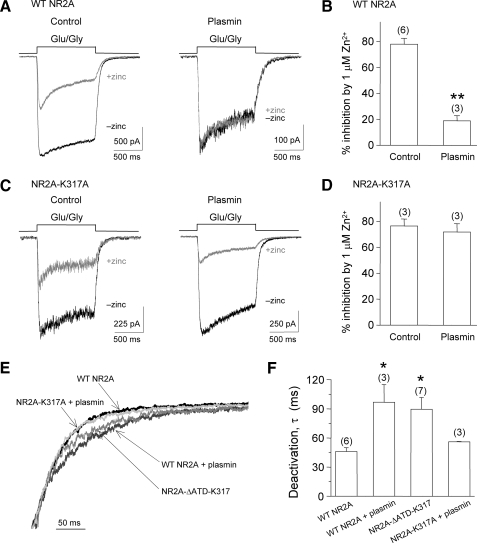
Plasmin Cleavage of NMDA Receptors Relieves Zn2+ Inhibition in Recombinant NR1/NR2A-transfected HEK 293 Cells— To test the hypothesis that plasmin cleavage relieves Zn2+ inhibition, agonist-evoked currents were recorded from recombinant NR1/NR2A-transfected HEK 293 cells by whole cell voltage-clamp recordings in the absence and presence of 1 μm Zn2+ with and without plasmin treatment. Zn2+ inhibition was evaluated as the percentage of inhibition produced by 1 μm Zn2+ on steady-state currents evoked by glutamate/glycine (50 μm). We found that 1 μm Zn2+ inhibited agonist-evoked currents by 78.0 ± 4.6% (n = 6, Fig. 5A, left panel), but only inhibited current responses in HEK 293 cells by 19 ± 13% following a 10-min treatment with 100 nm plasmin (n = 3, Fig. 5A, right panel). There is a significant difference (p < 0.001, unpaired t test; Fig. 5B) between these two groups, suggesting that Zn2+ sensitivity is reduced by plasmin treatment.
FIGURE 5.
Plasmin treatment relieves Zn2+ inhibition in whole cell voltage-clamp current recordings from HEK 293 cells expressing recombinant NR1/NR2A NMDA receptors. A, whole cell voltage-clamp recording of HEK 293 cells transfected with wild-type NR1/NR2A. Left, representative traces of currents elicited by 50μm glycine and 50μm glutamate in 10μm EDTA (black, -zinc) superimposed on currents evoked by 50μm glycine and 50 μm glutamate in 1 μm Zn2+ (gray, +zinc). Right, current evoked by 50 μm glycine and 50 μm glutamate in 10 μm EDTA (black) and 50 μm glycine and 50 μm glutamate in 1 μm Zn2+ (red) following 100 nm plasmin treatment. B, 1 μm Zn2+ inhibited HEK 293 cells expressing wild-type NR1/NR2A by 78 ± 4.6% (n = 6) but only inhibited HEK 293 cells by 19 ± 13% following a 10-min treatment with 100 nm plasmin (n = 3). **, p < 0.001; unpaired t test. C, whole cell voltage-clamp current recordings from HEK 293 cells transfected with NR1/NR2A-K317A. Left, representative traces of currents evoked by 50 μm glycine and 50 μm glutamate in 10 μm EDTA (black) and 50 μm glycine and 50 μm glutamate in 1 μm Zn2+ (gray). Right, representative traces of currents evoked by 50 μm glycine and 50 μm glutamate in 10 μm EDTA (black) and 50 μm glycine and 50 μm glutamate in 1 μm Zn2+ (gray) following 10 min treatment with 100 nm plasmin. D, the summary shows that 1 μm Zn2+ inhibited HEK 293 cells expressing NR1/NR2A-K317A by 77 ± 5.3% (n = 3) and caused 72 ± 6.5% inhibition following 10 min of 100 nm plasmin treatment (n = 3). E, representative traces of the deactivation of normalized current responses evoked by removal of glutamate from HEK 293 cells expressing NR1/NR2A (black), plasmin-treated NR1/NR2A (dark gray), NR1/NR2A-ΔATD-K317 (gray), and plasmin-treated NR1/NR2A-K317A (light gray). F, the deactivation time constants for NR1/NR2A following plasmin treatment (τ = 96.8 ± 18.1 ms; n = 3) and the deletion mutant NR1/NR2A-ΔATD-K317 (τ = 89.5 ± 12.1 ms; n = 7) were similar and statistically longer than untreated NR1/NR2A (τ = 46.2 ± 3.9 ms; n = 6). Plasmin-treated NR1/NR2A-K317A (τ = 56.0 ± 0.35 ms; n = 3) also deactivated rapidly. *, p < 0.05, one-way analysis of variance with Tukey's post-hoc test.
To further confirm the hypothesis that the relief of Zn2+ inhibition is due to plasmin cleavage at Lys317 on the NR2A amino-terminal, Lys317 was mutated to alanine (NR2A-K317A), and Zn2+ inhibition was measured on current responses in recombinant NR1/NR2A-K317A-transfected HEK 293 cells in the control and plasmin treatment groups. Consistent with our previous data, we found that 1 μm Zn2+ inhibited agonist-evoked currents on NR1/NR2A-K317A-transfected HEK 293 cells by 72 ± 6.5% after 10 min of plasmin treatment, similar to 77 ± 5.3% (n = 3) inhibition without plasmin treatment (Fig. 5C). There is no significant difference in Zn2+ sensitivity between the control and plasmin-treated HEK 293 cells transfected with NR1/NR2A-K317A (p = 0.64, unpaired t test). These data suggest that plasmin-induced removal of Zn2+ sensitivity requires cleavage of the NR2A amino-terminal at Lys317.
In addition to examining the relief of Zn2+ inhibition following plasmin treatment, we also measured the deactivation time constant, τ, of evoked currents following the removal of agonist for untreated and plasmin-treated NMDA receptors (Fig. 5, E and F). Untreated NR1/NR2A receptors deactivated rapidly (τ = 46.2 ± 3.9 ms; n = 6), whereas plasmin-treated NR1/NR2A receptors exhibited a significantly prolonged decay (τ = 96.8 ± 18.1 ms; n = 3; p < 0.05). The deactivation time course of NR1/NR2A-ΔATD-K317 was similar to plasmin-treated NR1/NR2A and was significantly slower (τ = 89.5 ± 12.1 ms) than untreated NR1/NR2A (n = 7; p < 0.05). However, the deactivation time course of plasmin-treated NR1/NR2A-K317A was rapid (τ = 56.0 ± 0.35 ms; n = 3) and similar to untreated NR1/NR2A, indicating that the mutated NR2AATD may not have been cleaved by plasmin treatment.
Plasmin Cleavage of NMDA Receptors Relieve Zn2+ Inhibition in Cultured Cortical Neurons—To investigate whether the relief of Zn2+ inhibition by plasmin cleavage also occurs in native NMDA receptors, whole cell perforated patch voltage-clamp recordings were performed on mouse cortical neurons maintained in culture for 5–10 days (Fig. 6A). Neurons were distinguished by inward sodium currents evoked by depolarizing voltage steps (Fig. 6B) that were reversibly abolished by 0.5 μm tetrodotoxin (data not shown). Zn2+ inhibition was evaluated as the percentage of inhibition of 1 μm Zn2+ observed at steady-state on NMDA-evoked whole cell currents from control and the plasmin treatment groups. CNQX and ifenprodil were used to block non-NMDA receptors and NR2B-containing NMDA receptors.
FIGURE 6.
Plasmin treatment relieves the Zn2+ inhibition on NMDA receptors in cultured cortical neurons. A, the figure shows a recording pipette on a representative cultured cortical neuron. B, a representative inward Na+ current is shown in whole cell mode in response to voltage steps from -90 to +50 mV. C, extracellular 1 μm Zn2+ inhibited agonist-evoked currents in wild-type (WT) neurons (50 μm NMDA and 50 μm glycine) by 53 ± 2.6% (n = 6) in the presence of 20 μm CNQX and 3 μm ifenprodil (upper panel), but had less effect (29 ± 4.2%, n = 7) after treatment with 300 nm plasmin (lower panel). D, summary of the Zn2+ inhibition on the wild-type control and plasmin treatment group. *, p < 0.05, unpaired t test.
Plasmin treatment had no significant effect on the steady-state amplitude of NMDA receptor currents in cultured cortical neurons (data not shown; control group: 124 ± 17 pA, n = 15 versus plasmin-treated group: 143 ± 20 pA, n = 14; p = 0.48; unpaired t test). This result suggests that plasmin itself has no direct effect on NMDA receptor function and is consistent with our previous studies (38, 45). We subsequently evaluated the effect of plasmin on Zn2+ sensitivity of NMDA receptor currents from cultured cortical neurons by recording the NMDA current response in the presence and absence of Zn2+. In these experiments, extracellular Zn2+ (1 μm) inhibited NMDA-evoked whole cell currents by 53 ± 2.6% (n = 6) before and 29 ± 4.2% (n = 7) after treatment with 300 nm plasmin (Fig. 6, C and D). These data show that Zn2+ causes significantly less inhibition following treatment with the serine protease plasmin (p < 0.05, unpaired t test). These data suggest that voltage-independent Zn2+ inhibition is relieved by submicromolar concentrations of plasmin, consistent with plasmin-mediated removal of the Zn2+ binding site within the amino-terminal domain.
It is known that proteases, including plasmin, potentiate the NMDA receptor response through activation of PAR1 (37, 38). To evaluate whether relief of Zn2+ inhibition by plasmin involves PAR1 activation, we prepared cultured cortical neurons from PAR1-/- mice. Similar to wild-type neurons, extracellular Zn2+ (1 μm) inhibited agonist-evoked NMDA receptor currents by 40 ± 4.4% (n = 10), whereas it had less effect after treatment with 300 nm plasmin (18 ± 4.0%, n = 6; p < 0.001, unpaired t test; Fig. 7, A and B). These results indicate that the relief of Zn2+ inhibition by plasmin is independent of PAR1 activation. In addition to activating PAR1, plasmin is also capable of activating other PARs, such as PAR2 (46). Therefore, we characterized the effect of PAR2 activation on Zn2+ inhibition. Cultured cortical neurons were pretreated with the PAR2 agonist peptide 2-furoyl-LIGRLO for 30 min at room temperature, and Zn2+ inhibition was evaluated. We found that 1 μm Zn2+ inhibited agonist-evoked NMDA receptor currents by 50 ± 2.2% (n = 3). There is no significant difference (p = 0.54; unpaired t test; Fig. 7C) between the wild-type control and the 2-furoyl-LIGRLO treatment. These data indicate that activation of PAR2 receptors does not influence Zn2+ inhibition. In addition, the relief of Zn2+ inhibition by plasmin treatment is independent of PAR2 activation.
FIGURE 7.
The reduction of Zn2+ inhibition on NMDA receptors of cultured cortical neurons by plasmin is independent of protease-activated receptors. A, extracellular 1 μm Zn2+ inhibited NMDA-evoked whole cell currents in PAR1-/- neurons by 40 ± 4.3% (n = 10) in the presence of CNQX and ifenprodil (upper panel), but had less effect after treatment with 300 nm plasmin (18 ± 4.0%, n = 6; lower panel; **, p < 0.001, unpaired t test, compared with the PAR1-/- control). B, summary of the Zn2+ inhibition of NMDA current responses in PAR1-/- control neurons and the plasmin treatment group of PAR1-/- neurons. C, summary of the Zn2+ inhibition of NMDA current responses on the wild-type control neurons (53 ± 2.6%, n = 6) and the 2-f-LIGRLO-treated neurons (50 ± 2.2%, n = 3).
DISCUSSION
Serine proteases play important roles not only in coagulation and thrombolysis, but also in inflammatory and proliferative responses triggered by tissue injury, as well as in signal pathways in the central nervous system (31, 33). Recent studies indicate serine proteases modulate the function of NMDA receptors by indirect or direct mechanisms (37, 38, 40). The present study demonstrates that nanomolar concentrations of plasmin cleave the NR2A subunit of native and recombinant NMDA receptors, and the cleavage occurs after Lys317 on the NR2A amino-terminal domain. Plasmin cleavage significantly reduced the Zn2+ inhibition on agonist-evoked currents of NMDA receptors from both recombinant transfected HEK 293 cells and cultured mouse cortical neurons. The relief of Zn2+ inhibition by plasmin treatment was rescued by exchange of Lys317 to alanine. These data suggest that plasmin cleavage at Lys317 removes the amino-terminal domain that harbors the high affinity Zn2+ binding site. This finding holds implications for NMDA receptor-mediated toxicity, tPA treatment of stroke, and potential identification of biomarkers of central nervous system injury.
Plasmin Cleavage of NMDA Receptors—Plasmin can cleave numerous proteins, including fibrinogens, PARs, as well as pro-brain-derived neurotrophic factor (47–49). In our current study, plasmin can cleave NR2AATD recombinant proteins and the NR2A amino-terminal in membrane preparations from rat cortex. A ∼40-kDa fragment is removed by plasmin that encodes the NR2AATD (Fig. 1A). Following plasmin treatment, sequencing the cleavage products of the NR2A amino-terminal revealed only one fragment with readable sequence that matched NR2A. Lys317 was identified as the plasmin cleavage site by sequencing the cleavage fragments of NR2AATD recombinant proteins. This cleavage site is similar to the cleavage site of other plasmin substrates (Table 1). The ability of a point mutation to block the effects of plasmin suggests that plasmin cleaves the NR2A amino-terminal at only one site, although we cannot exclude the possibility that there are other plasmin cleavage sites on the NR2A.
Some residual inhibition by 1 μm Zn2+ is still observed after plasmin treatment in both recombinant NR1/NR2A-transfected HEK 293 cells and cultured cortical neurons by 20–30%. We interpret this remaining inhibition to be a combination of incomplete cleavage of the ATD by plasmin and the presence of other NR2B, -2C, or -2D subunit-containing NMDA receptors in cultured cortical neurons. For example, Zn2+ inhibits NR2B with an IC50 of 800 nm, suggesting some inhibition might be caused by NR1/NR2B (27, 50). In addition, a small amount of inhibition caused by voltage-dependent Zn2+ inhibition may contribute to the residual inhibition.
Interaction between Proteases, PARs, and NMDA Receptors— Recent studies (45) suggest that activation of astrocytic PAR1 triggers G-protein-mediated release of glutamate, which causes the activation of neuronal NMDA receptors and potentiation of synaptic NMDA receptor function by a Mg2+-dependent mechanism that appears secondary to depolarization. The protease thrombin potentiates NMDA receptor currents in rat hippocampal neurons (37). Plasmin also potentiates the NMDA receptor-dependent component of miniature excitatory postsynaptic currents and increases NMDA-induced whole cell receptor currents recorded from CA1 pyramidal cells (38). These studies showed that the protease modulates NMDA receptor function through an indirect mechanism, such as PAR1 activation. Comparison of the level of NMDA receptor potentiation produced in thrombin-treated hippocampal slices, TFLLR-treated hippocampal slices, and plasmin-treated hippocampal slices shows striking differences. The peptide TFLLR robustly activates PAR1 without cleaving NR2A, and potentiates NMDA receptor function 1.4-fold. By contrast, treatment of hippocampal slices by plasmin, a weak activator of PAR1, leads to an enhancement of NMDA responses of 2.4-fold, considerably more than observed with more robust PAR1 activation by TFLLR. Our interpretation of this difference in the levels of potentiation is that plasmin treatment both activates PAR1 and cleaves the Zn2+ binding domain, removing tonic high affinity inhibition by ambient concentrations of Zn2+ present. Thus, the two different mechanisms engaged by plasmin enhance NMDA receptor function considerably more than activation by PAR1 alone.
In addition, functionally relevant cleavage of the extracellular portion of NMDA receptors by proteases has been reported by several laboratories. tPA appears to enhance NMDA receptor function and has been suggested to cleave the ATD of the NR1 subunit at arginine 260 (39, 40). Thrombin and plasmin also have been suggested to cleave the NR1 subunit (37, 44). Calpain proteolysis of the COOH termini of NR2A, -2B, and -2C subunits results in NMDA receptor degradation and reduced receptor activity (51–53). Therefore, in addition to indirectly mediating the functional response of NMDA receptors, proteases also can directly modulate NMDA receptor responses through cleavage of the receptors' subunits.
We do not yet fully understand the relative contribution of PAR1 activation and plasmin cleavage of NMDA receptors in normal and pathological situations. For thrombin, it is clear that PAR1 activation occurs at a lower concentration than NR1 cleavage (37). Plasmin appears to activate PAR1 receptors (38) in the same concentration range as it cleaves NR2A. The tPA/plasminogen/plasmin system is thought to play important roles in normal brain functions, including synaptic plasticity, learning, and memory, as well as in a number of pathophysiological conditions, including ischemic stroke, neurodegenerative disease, and seizures (54–57). Our study suggests that if tPA can generate a significant amount of plasmin, this could lead to the cleavage of NR2A to reduce Zn2+ inhibition, which can occur either through tonic levels of extracellular Zn2+ or activity-dependent release of Zn2+ (58).
The Significance of Plasmin Cleavage of the NR2AATD—tPA converts inactive plasminogen into the active protease plasmin, which in turn catalyzes cleavage of fibrin into soluble degraded fragments to facilitate clot dissolution. A successful study by the NINDS, National Institutes of Health, in 1995 using recombinant tPA within 3 hours of acute stroke (59) led to the approval of recombinant tPA (Alteplase) by the FDA as a thrombolytic therapy in acute stroke to reperfuse ischemic tissue. However, the use of tPA is limited by the narrow time window of application, high risk of hemorrhagic transformation, as well as the potential harmful extravascular side effects (60, 61). Plasmin, the catalyzed end product of tPA that facilitates blood clot dissolution, might play important roles in excitotoxic neurodegeneration by proteolytic degradation of the extracellular matrix and immune activation (62, 63). Injection of plasmin or plasminogen into the brain significantly increased the number of apoptotic neurons and the injury area of the intracortical hemorrhage model in rats (63). Harmful consequences of tPA and plasmin in ischemic animal models have been reported (Refs. 37 and 64–66, but see Ref. 67). Our results add an additional potential mechanism that could mediate the harmful effects of tPA/plasmin in the brain. Although tPA can cross the intact blood-brain barrier (68), high concentrations of blood-derived proteases such as tPA and plasmin are unlikely to develop in brain tissue while the blood-brain barrier is intact. Blood-derived tPA can enter in brain tissue in situations in which the blood-brain barrier is disrupted (such as hemorrhagic stroke, aneurysm rupture, penetrating head wound, infection, inflammation, status epileptics, or tPA treatment of stroke) (35). Increased levels of plasmin may cleave the NR2AATD, and the cleaved NR2A fragments may enter into the peripheral circulatory system. Detecting these NR2AATD fragments may offer the possibility to evaluate some features of ischemia. It has previously been proposed that NMDA receptor NR2 subunits are degraded, and the proteolytic fragments enter into the peripheral blood of patients with transient ischemic attack and stroke, resulting in the formation of autoantibodies to the extracellular ligand binding domains of NR2A and NR2B that can be detected by enzyme-linked immunosorbent assay (United State patent 6896872) (69–71). Because these fragments are detectable, the plasmin-cleaved NR2AATD fragments may be capable of entering the bloodstream. Our findings showing that plasmin cleavage of the NR2AATD provide one potential mechanism whereby NMDA receptor fragments could be generated in ischemic stroke. These data may also provide a rationale for monitoring the serum levels of extracellular NR2A fragments in patients receiving tPA.
In conclusion, these data together suggest that plasmin can cleave the amino-terminal domain of the NR2A subunit in recombinant receptors and native tissues and subsequently relieve Zn2+ inhibition. Our results indicate that the protease plasmin may play a role in the modulation of NMDA receptor function and may provide a possible mechanism underlying the harmful effects of tPA and plasmin in the brain. In situations where plasmin may enter brain, such as during tPA treatment for ischemic stroke or break-down of the blood-brain barrier, removal of Zn2+ inhibition could exacerbate NMDA receptor-mediated neuronal damage, as well as provide a biomarker (NR2AATD) that might enter into blood.
Supplementary Material
Acknowledgments
We thank Drs. Kasper B. Hansen and Anders Kristensen for help with the mutagenesis of NR2A-ΔATD-K317 and NR2A-K317A. We also thank Dr. Shaun Coughlin for generously providing PAR1+/- animals, Dr. Jan Pohl and the Emory Microchemical Facility for sequencing the NR2A plasmin cleavage fragments, and Phuong Le and Kimberly Haustein for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health NINDS Grants NS39419 and NS36654 (to S. F. T.). This work was also supported by grants from National Alliance for Research on Schizophrenia and Depression (to S. F. T.), the Michael J. Fox Foundation for Parkinson's Research (to S. F. T.), National Medical Research Council (to C. M. L.), Biomedical Research Council (to C. M. L.), and Academic Research Funds (to C. M. L.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: NMDA, N-methyl-D-aspartate; ATD, amino-terminal domain; PAR, protease-activated receptor; tPA, tissue-plasminogen activator; GST, glutathione S-transferase; PDGF, platelet-derived growth factor; IDA, iminodiacetic acid; HEK, human embryonic kidney; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; BAPTA, 1,2-bis(aminophenoyl)ethane-N,N,N′,N′-tetraacetic acid; CNQX, 6-cyano-7-nitroquinoxaline-2,3-dione.
References
- 1.Dingledine, R., Borges, K., Bowie, D., and Traynelis, S. F. (1999) Pharmacol. Rev. 51 7-61 [PubMed] [Google Scholar]
- 2.Kemp, J. A., and McKernan, R. M. (2002) Nat. Neurosci. 5 (suppl.) 1039-1042 [DOI] [PubMed] [Google Scholar]
- 3.Stys, P. K., and Lipton, S. A. (2007) Trends Pharmacol. Sci. 28 561-566 [DOI] [PubMed] [Google Scholar]
- 4.Erreger, K., Chen, P. E., Wyllie, D. J., and Traynelis, S. F. (2004) Crit. Rev. Neurobiol. 16 187-224 [DOI] [PubMed] [Google Scholar]
- 5.Paoletti, P., and Neyton, J. (2007) Curr. Opin. Pharmacol. 7 39-47 [DOI] [PubMed] [Google Scholar]
- 6.Williams, K. (2001) Curr. Drug Targets. 2 285-298 [DOI] [PubMed] [Google Scholar]
- 7.Woodburn, V. L., and Woodruff, G. N. (1994) Adv. Pharmacol. 30 1-33 [DOI] [PubMed] [Google Scholar]
- 8.Choi, Y. B., and Lipton, S. A. (1999) Neuron 23 171-180 [DOI] [PubMed] [Google Scholar]
- 9.Fayyazuddin, A., Villarroel, A., Le, G. A., Lerma, J., and Neyton, J. (2000) Neuron 25 683-694 [DOI] [PubMed] [Google Scholar]
- 10.Hatton, C. J., and Paoletti, P. (2005) Neuron 46 261-274 [DOI] [PubMed] [Google Scholar]
- 11.Low, C. M., Zheng, F., Lyuboslavsky, P., and Traynelis, S. F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 11062-11067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paoletti, P., Perin-Dureau, F., Fayyazuddin, A., Le, G. A., Callebaut, I., and Neyton, J. (2000) Neuron 28 911-925 [DOI] [PubMed] [Google Scholar]
- 13.Perin-Dureau, F., Rachline, J., Neyton, J., and Paoletti, P. (2002) J. Neurosci. 22 5955-5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong, E., Ng, F. M., Yu, C. Y., Lim, P., Lim, L. H., Traynelis, S. F., and Low, C. M. (2005) Protein Sci. 14 2275-2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallee, B. L., and Falchuk, K. H. (1993) Physiol. Rev. 73 79-118 [DOI] [PubMed] [Google Scholar]
- 16.Aizenman, E., Stout, A. K., Hartnett, K. A., Dineley, K. E., McLaughlin, B., and Reynolds, I. J. (2000) J. Neurochem. 75 1878-1888 [DOI] [PubMed] [Google Scholar]
- 17.Choi, D. W., and Koh, J. Y. (1998) Annu. Rev. Neurosci. 21 347-375 [DOI] [PubMed] [Google Scholar]
- 18.Frederickson, C. J., Koh, J. Y., and Bush, A. I. (2005) Nat. Rev. Neurosci. 6 449-462 [DOI] [PubMed] [Google Scholar]
- 19.Cuajungco, M. P., and Lees, G. J. (1997) Neurobiol. Dis. 4 137-169 [DOI] [PubMed] [Google Scholar]
- 20.Vogt, K., Mellor, J., Tong, G., and Nicoll, R. (2000) Neuron 26 187-196 [DOI] [PubMed] [Google Scholar]
- 21.Westbrook, G. L., and Mayer, M. L. (1987) Nature 328 640-643 [DOI] [PubMed] [Google Scholar]
- 22.Christine, C. W., and Choi, D. W. (1990) J. Neurosci. 10 108-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legendre, P., and Westbrook, G. L. (1990) J. Physiol. 429 429-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer, M. L., and Vyklicky, L., Jr. (1989) J. Physiol. 415 351-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, N., Moshaver, A., and Raymond, L. A. (1997) Mol. Pharmacol. 51 1015-1023 [DOI] [PubMed] [Google Scholar]
- 26.Paoletti, P., Ascher, P., and Neyton, J. (1997) J. Neurosci. 17 5711-5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traynelis, S. F., Burgess, M. F., Zheng, F., Lyuboslavsky, P., and Powers, J. L. (1998) J. Neurosci. 18 6163-6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams, K. (1996) Neurosci. Lett. 215 9-12 [DOI] [PubMed] [Google Scholar]
- 29.Erreger, K., and Traynelis, S. F. (2005) J. Physiol. 569 381-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erreger, K., and Traynelis, S. F. (2008) J. Physiol. 586 763-778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traynelis, S. F., and Trejo, J. (2007) Curr. Opin. Hematol. 14 230-235 [DOI] [PubMed] [Google Scholar]
- 32.Schmidlin, F., and Bunnett, N. W. (2001) Curr. Opin. Pharmacol. 1 575-582 [DOI] [PubMed] [Google Scholar]
- 33.Carmeliet, P., and Collen, D. (1997) Curr. Opin. Lipidol. 8 118-125 [DOI] [PubMed] [Google Scholar]
- 34.Mizutani, A., Saito, H., and Matsuki, N. (1996) Brain Res. 739 276-281 [DOI] [PubMed] [Google Scholar]
- 35.Gingrich, M. B., and Traynelis, S. F. (2000) Trends Neurosci. 23 399-407 [DOI] [PubMed] [Google Scholar]
- 36.Tomimatsu, Y., Idemoto, S., Moriguchi, S., Watanabe, S., and Nakanishi, H. (2002) Life Sci. 72 355-361 [DOI] [PubMed] [Google Scholar]
- 37.Gingrich, M. B., Junge, C. E., Lyuboslavsky, P., and Traynelis, S. F. (2000) J. Neurosci. 20 4582-4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mannaioni, G., Orr, A. G., Hamill, C. E., Yuan, H., Pedone, K. H., McCoy, K. L., Palmini, R. B., Junge, C. E., Lee, C. J., Yepes, M., Hepler, J. R., and Traynelis, S. F. (2008) J. Biol. Chem. 283 20600-20611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Monreal, M., Lopez-Atalaya, J. P., Benchenane, K., Cacquevel, M., Dulin, F., Le Caer, J. P., Rossier, J., Jarrige, A. C., MacKenzie, E. T., Colloc'h, N., Ali, C., and Vivien, D. (2004) J. Biol. Chem. 279 50850-50856 [DOI] [PubMed] [Google Scholar]
- 40.Nicole, O., Docagne, F., Ali, C., Margaill, I., Carmeliet, P., MacKenzie, E. T., Vivien, D., and Buisson, A. (2001) Nat. Med. 7 59-64 [DOI] [PubMed] [Google Scholar]
- 41.Hewick, R. M., Hunkapiller, M. W., Hood, L. E., and Dreyer, W. J. (1981) J. Biol. Chem. 256 7990-7997 [PubMed] [Google Scholar]
- 42.Malherbe, P., Kratochwil, N., Knoflach, F., Zenner, M. T., Kew, J. N., Kratzeisen, C., Maerki, H. P., Adam, G., and Mutel, V. (2003) J. Biol. Chem. 278 8340-8347 [DOI] [PubMed] [Google Scholar]
- 43.Kunishima, N., Shimada, Y., Tsuji, Y., Sato, T., Yamamoto, M., Kumasaka, T., Nakanishi, S., Jingami, H., and Morikawa, K. (2000) Nature 407 971-977 [DOI] [PubMed] [Google Scholar]
- 44.Samson, A. L., Nevin, S. T., Croucher, D., Niego, B., Daniel, P. B., Weiss, T. W., Moreno, E., Monard, D., Lawrence, D. A., and Medcalf, R. L. (2008) J. Neurochem. 107 1091-1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, C. J., Mannaioni, G., Yuan, H., Woo, D. H., Gingrich, M. B., and Traynelis, S. F. (2007) J. Physiol. 581 1057-1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loew, D., Perrault, C., Morales, M., Moog, S., Ravanat, C., Schuhler, S., Arcone, R., Pietropaolo, C., Cazenave, J. P., van Dorsselaer, A., and Lanza, F. (2000) Biochemistry 39 10812-10822 [DOI] [PubMed] [Google Scholar]
- 47.Kuliopulos, A., Covic, L., Seeley, S. K., Sheridan, P. J., Helin, J., and Costello, C. E. (1999) Biochemistry 38 4572-4585 [DOI] [PubMed] [Google Scholar]
- 48.Pang, P. T., Teng, H. K., Zaitsev, E., Woo, N. T., Sakata, K., Zhen, S., Teng, K. K., Yung, W. H., Hempstead, B. L., and Lu, B. (2004) Science 306 487-491 [DOI] [PubMed] [Google Scholar]
- 49.Gray, K., and Ellis, V. (2008) FEBS Lett. 582 907-910 [DOI] [PubMed] [Google Scholar]
- 50.Rachline, J., Perin-Dureau, F., Le, G. A., Neyton, J., and Paoletti, P. (2005) J. Neurosci. 25 308-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bi, X., Rong, Y., Chen, J., Dang, S., Wang, Z., and Baudry, M. (1998) Brain Res. 790 245-253 [DOI] [PubMed] [Google Scholar]
- 52.Guttmann, R. P., Sokol, S., Baker, D. L., Simpkins, K. L., Dong, Y., and Lynch, D. R. (2002) J. Pharmacol. Exp. Ther. 302 1023-1030 [DOI] [PubMed] [Google Scholar]
- 53.Guttmann, R. P., Baker, D. L., Seifert, K. M., Cohen, A. S., Coulter, D. A., and Lynch, D. R. (2001) J. Neurochem. 78 1083-1093 [DOI] [PubMed] [Google Scholar]
- 54.Tsirka, S. E. (1997) J. Mol. Med. 75 341-347 [DOI] [PubMed] [Google Scholar]
- 55.Nagai, T., Yamada, K., Yoshimura, M., Ishikawa, K., Miyamoto, Y., Hashimoto, K., Noda, Y., Nitta, A., and Nabeshima, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3650-3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawlak, R., Melchor, J. P., Matys, T., Skrzypiec, A. E., and Strickland, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 443-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matys, T., Pawlak, R., Matys, E., Pavlides, C., McEwen, B. S., and Strickland, S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 16345-16350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie, X. M., and Smart, T. G. (1991) Nature 349 521-524 [DOI] [PubMed] [Google Scholar]
- 59.Qureshi, N. (1996) N. Engl. J. Med. 334 1406. [PubMed] [Google Scholar]
- 60.Sheehan, J. J., and Tsirka, S. E. (2005) Glia 50 340-350 [DOI] [PubMed] [Google Scholar]
- 61.Kidwell, C. S., Latour, L., Saver, J. L., Alger, J. R., Starkman, S., Duckwiler, G., Jahan, R., Vinuela, F., Kang, D. W., and Warach, S. (2008) Cerebrovasc. Dis. 25 338-343 [DOI] [PubMed] [Google Scholar]
- 62.Liotta, L. A., Goldfarb, R. H., Brundage, R., Siegal, G. P., Terranova, V., and Garbisa, S. (1981) Cancer Res. 41 4629-4636 [PubMed] [Google Scholar]
- 63.Xue, M., and Del Bigio, M. R. (2001) Stroke 32 2164-2169 [DOI] [PubMed] [Google Scholar]
- 64.Tsirka, S. E., Gualandris, A., Amaral, D. G., and Strickland, S. (1995) Nature 377 340-344 [DOI] [PubMed] [Google Scholar]
- 65.Junge, C. E., Sugawara, T., Mannaioni, G., Alagarsamy, S., Conn, P. J., Brat, D. J., Chan, P. H., and Traynelis, S. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13019-13024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Y. F., Tsirka, S. E., Strickland, S., Stieg, P. E., Soriano, S. G., and Lipton, S. A. (1998) Nat. Med. 4 228-231 [DOI] [PubMed] [Google Scholar]
- 67.Nagai, N., De Mol, M., Lijnen, H. R., Carmeliet, P., and Collen, D. (1999) Circulation 99 2440-2444 [DOI] [PubMed] [Google Scholar]
- 68.Benchenane, K., Berezowski, V., Ali, C., Fernandez-Monreal, M., Lopez-Atalaya, J. P., Brillault, J., Chuquet, J., Nouvelot, A., MacKenzie, E. T., Bu, G., Cecchelli, R., Touzani, O., and Vivien, D. (2005) Circulation 111 2241-2249 [DOI] [PubMed] [Google Scholar]
- 69.Bokesch, P. M., Izykenova, G. A., Justice, J. B., Easley, K. A., and Dambinova, S. A. (2006) Stroke 37 1432-1436 [DOI] [PubMed] [Google Scholar]
- 70.Dambinova, S. A., Khounteev, G. A., Izykenova, G. A., Zavolokov, I. G., Ilyukhina, A. Y., and Skoromets, A. A. (2003) Clin. Chem. 49 1752-1762 [DOI] [PubMed] [Google Scholar]
- 71.Dambinova, S. A., Khounteev, G. A., and Skoromets, A. A. (2002) Stroke 33 1181-1182 [DOI] [PubMed] [Google Scholar]
- 72.Takagi, T., and Doolittle, R. F. (1975) Biochemistry 14 5149-5156 [DOI] [PubMed] [Google Scholar]
- 73.Southan, C., Thompson, E., Panico, M., Etienne, T., Morris, H. R., and Lane, D. A. (1985) J. Biol. Chem. 260 13095-13101 [PubMed] [Google Scholar]
- 74.Pryzdial, E. L., Lavigne, N., Dupuis, N., and Kessler, G. E. (1999) J. Biol. Chem. 274 8500-8505 [DOI] [PubMed] [Google Scholar]
- 75.Chain, D., Kreizman, T., Shapira, H., and Shaltiel, S. (1991) FEBS Lett. 285 251-256 [DOI] [PubMed] [Google Scholar]
- 76.Novak, J. F., Hayes, J. D., and Nishimoto, S. K. (1997) J. Bone Miner. Res. 12 1035-1042 [DOI] [PubMed] [Google Scholar]
- 77.Biswas, N., Vaingankar, S. M., Mahata, M., Das, M., Gayen, J. R., Taupenot, L., Torpey, J. W., O'Connor, D. T., and Mahata, S. K. (2008) Endocrinology 149 749-757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.