Abstract
Arsenicals are both environmental carcinogens as well as therapeutic agents for the treatment of trypanosomiasis and more recently cancer. Arsenic trioxide (ATO) has been successfully used for the treatment of acute promyelocytic leukemia (APL) and has activity in multiple myeloma (MM). While signaling events associated with carcinogenesis have been well studied, it still remains to be determined which of these events are involved in anti-cancer signaling. To better define this response, gene expression profiling following ATO treatment of four MM cell lines was performed. The pattern was consistent with a strong antioxidative response, particularly of genes activated by Nrf2. While Nrf2 is expressed constitutively at the mRNA level, the protein is not detected in untreated cells. Consistent with inactivation of Keap1, Nrf2 protein is stabilized and present in the nucleus within 6 h of ATO treatment. Despite the activation of this antioxidative response, ROS may not be important in ATO-induced death. Inhibition of ATO-induced ROS with butylated hydroxyanisole (BHA) does not affect Nrf2 activation or cell death. Moreover, silencing Nrf2 had no effect on ATO-induced apoptosis. Together these data suggest that ROS is not important in the induction of the antioxidative response or cellular death by ATO.
Arsenic trioxide (ATO)2 is an effective treatment for patients with relapsed or refractory acute promyelocytic leukemia (APL) (1). ATO induces the degradation of the PML-RARα fusion protein through interactions with cysteines in the PML portion, as well as activation of MAPK pathway (1). PML-RARα degradation can result in either terminal differentiation and/or induction of apoptosis (1). ATO has also been studied, as a single agent or in combination with other drugs, in other hematological malignancies, including multiple myeloma (MM) (2–8). Several preclinical studies demonstrated that ATO induces growth inhibition and apoptosis in different lymphoid and myeloid malignant cells but the exact mechanism(s) for cells that do not express the PML-RARα fusion protein are not known (4, 9). The in vitro sensitivity of cultured MM cell lines to clinically achievable concentrations (1–2 μm) led to the investigation of its mechanism in MM (10). Several studies have suggested that induction of oxidative stress as a result of mitochondrial dysfunction leads to cell death and consistent with this possibility GSH depletion sensitizes cells to ATO (5, 11–14). Moreover heme oxygenase-1 (HO-1) expression was observed in myeloma cells that survived a long term treatment with a low concentration of ATO (15). HO-1 is a transcriptional target of the nuclear factor erythroid-derived 2-like 2 (Nrf2) (16, 17).
Nrf2 is a member of the leucine zipper family of transcription factors. Numerous studies have demonstrated an essential function of Nrf2 in the transcriptional regulation of antioxidant response element (ARE)-controlled genes (16–19). Nrf2 is not only required for the basal expression but also for the inducible expression of a number of ARE-controlled genes. Inactive Nrf2 resides in the cytoplasm and it is rapidly degraded through the ubiquitin-26S proteasome pathway. The cytoplasmic Nrf2-binding protein Keap1 targets the ubiquitination of Nrf2 through its association with Cullin-3 and Rbx1 (17). Activated Nrf2 translocates into the nucleus, binds to the ARE of target genes, and induces the transcription of these genes. The activation of Nrf2 is not completely understood, but two mechanisms are supported by the available evidence. In the first mechanism, modification of the sulfydryl groups present in Keap1 by chemical inducers dissociates Nrf2 from Keap1 and the subsequent translocation into the nucleus, thereby activating ARE (20). Alternatively, several upstream signaling kinases, including protein kinase C (PKC), phosphoinositol 3-kinase (PI3K), and mitogen-activated protein kinases (p38, JNK, and ERK1/2) have been reported to regulate Nrf2/ARE activity (21–23). Together with the ARE-regulated target genes, the Nrf2-Keap1 pathway is a sensing/transcriptional gene regulation mechanism critical in defense against endogenous and environmental toxic compounds. Loss of Nrf2 transcriptional activity has been associated with augmented sensitivity to carcinogens and certain diseases (24–26). Consistent with this role, activation of Nrf2 is a common target of many chemopreventive agents and is activated by antioxidants (27). In this study we found that ATO activates Nrf2 and induces apoptosis in MM cell lines, through a mechanism that is independent of ROS.
MATERIALS AND METHODS
Cell Lines—U266 and 8226/S were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The MM.1s cell line was obtained from Dr. Steven Rosen (Northwestern University, Chicago, IL), and the KMS11 cell line was provided by Dr. P. Leif Bergsagel (Mayo Clinic, Scottsdale, AZ). Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2, in RPMI 1640 medium, supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 2 mmol/liter of l-glutamine (all from Cellgro, Mediatech, Herndon, VA).
Reagents—Zinc chloride (ZnCl2), hemin, propidium iodide (PI), diphenyleneiodonium chloride (DPI), ebselen (Ebs), butylated hydroxyanisole (BHA), hydrogen peroxide (H2O2), melphalan, buthionine sulfoximine (BSO), 2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPO), 4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPOL), α-lipoic acid (LA), and cycloheximide (CHX) were purchased from Sigma-Aldrich. N-Acetylcysteine (NAC) was purchased from Bedford Laboratories (Bedford, OH). Mn(III)tetrakis(4-benzoic acid) porphyrin chloride (MnTBAP) was purchase from Oxis International, Inc. (Foster City, CA). EUK-134 was provided by Eukarion Inc. (Bedford, MA). ATO was provided by Cell Therapeutics Inc. (Seattle, WA). Bortezomib (PS-341, Velcade®) was provided by Millennium Pharmaceuticals (Cambridge, MA). G418 was purchased from Cellgro. Dihydroethidium (DHE) was purchased from Invitrogen (Carlsbad, CA). Staurosporine (STS) was obtained from EMD Biosciences (La Jolla, CA).
Antibodies—The following primary antibodies were used: rabbit anti-HO-1 polyclonal antibody (pAb) (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-NQO1 monoclonal antibody (mAb) (Cell Signaling, Danvers, MA); rabbit anti-actin pAb (Sigma-Aldrich); rabbit anti-Nrf2 pAb (Santa Cruz Biotechnology); rabbit anti-Keap1 pAb (Proteintech Group Inc., Chicago, IL); rabbit anti-Lamin A/C pAb (Abcam, Cambridge, MA), and the mouse anti-Noxa mAb (Abcam). The ECL Rabbit IgG, horseradish peroxidase-linked whole Ab (from donkey) (GE HealthCare, NJ) and the anti-mouse IgG1-HRP conjugate (Roche Applied Science, Indianapolis, IN) were used as secondary antibody for Western blot.
Vectors and Stable Expression of HO-1—HO-1 cDNA was obtained from ATCC (10589047, in pExpress1 vector, ATCC) and amplified by PCR with specific primers to introduce the KpnI and XbaI restriction enzyme sites at the 5′- and 3′-ends, respectively. Amplified HO-1 cDNA was cloned into the KpnI-XbaI sites of pcDNA3.1(+) vector (Invitrogen). HO-1 expression vector was introduced into U266 cells by nucleofection (Amaxa, Gaithersburg, MD) following the manufacturer's instructions. Briefly, 5 × 106 cells were electroporated in 100 μl of cell line nucleofector solution (Amaxa Reagent C) with 2 μg of DNA, using preselected Amaxa Program X005. Electroporated cells were plated with supplemented RPMI 1640 medium and 0.5 μg/ml G418 were used for selection of HO-1-expressing U266 cells.
Cell Viability by Annexin V-Fluorescein Isothiocyanate (FITC) and PI Staining—Cell viability was measured by Annexin V-FITC (Biovision, Palo Alto, CA) and PI staining, following manufacturer's instructions as previously described (13). Samples were acquired on a FACScan flow cytometer (Becton Dickinson, San Jose, CA) and analyzed with CellQuest software (Becton Dickinson). Viability or percent (%) of control viability was determined as percent of Annexin-V-negative cells.
Cellular Assays—All cells were incubated at 2.5 × 105 cells/ml with the indicated concentrations of ATO and/or BSO (100 μm), NAC (10 mm), hemin, ZnCl2, or PS-341. Hemin and ZnCl2 pretreatment: cells were pretreated with indicated concentrations of hemin or ZnCl2, for 24 h prior to ATO addition, to induce HO-1 and metallothionein-1 (MT1), respectively. Annexin-V-FITC/PI staining was used to determine ATO-induced apoptosis. ATO plus Bortezomib treatment: cells were treated with ATO (2 μm), bortezomib (40 nm), or the combination, for 6 h. Samples were analyzed by annexin-V-FITC/PI staining for cell viability, and cell pellets were frozen for protein analysis by Western blot. Cytosolic and nuclear fractions were also obtained for nuclear translocation analysis by Western blot (see “Subcellular Fractionation”).
Affymetrix Array—Four cell lines were treated for 0, 6, 24, and 48 h with 2 μm ATO. Total RNA was isolated using the RNeasy® Mini Kit (Qiagen, Valencia, CA) and the hybridization and initial data analysis performed by Expression Analysis Inc. (Durham, NC). Affymetrix Hu133 2.0 Plus Chips (Affymetrix, Santa Clara, CA), containing over 50,000 (54,675 including controls) probe sets were used as previously described (28).
RT-PCR for MT1—To assess mRNA expression, semiquantitative RT-PCR was performed using the GeneAmp RNA PCR Kit (Applied Biosystems, Foster City, CA). Briefly, 1 μg of total RNA was reverse-transcribed by the MuLV Reverse Transcriptase in a 20-μl reaction and the generated cDNA (1 μl) was amplified by the AmpliTaq DNA Polymerase, LD, using the following specific mixture of primers for MT1: MT1-Forward Primer: 5′-ATG GAC CCC AAC TGC TCC TG-3′ (for all the MT1 forms), MT1XH-Reverse Primer: 5′-TCA GGC ACA GCA GCT GCA CTT-3′(for MT1X and MT1H forms) and MT1FG-Reverse Primer: 5′-TCA GGC GCA GCA GCT GCA CTT-3′ (for MT1F and MT1G forms).
Real-time PCR—cDNA was amplified using TaqMan® Gene Expression Assay (Applied Biosystems) on the 7700 Sequence Detection System following manufacturer's protocol and the 20× human MT1H and MT2A TaqMan® assays (Applied Biosystems). TaqMan® human actin was used as endogenous control. MT1H and MT2A mRNA expression was calculated as relative quantification (RQ), normalized using actin mRNA expression.
Western Blot Analysis—Western blotting was performed using standard techniques as previously described (28). 10–30 μg of total proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Amersham Biosciences, Buckinghamshire, UK).
Subcellular Fractionation—Nuclear fractions were obtained as follows. Cells were harvested, washed twice with PBS, and centrifuged at 500 × g for 5 min at room temperature. The cell pellets were resuspended in lysis buffer (10 mm HEPES, pH 7.9, 1.5 mm MgCl2,10 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, 0.2% Nonidet P-40), plus protease inhibitor mixture, and incubated on ice for 15 min. After centrifugation at 10,000 × g for 10 min at 4 °C, supernatants (cytosolic fractions) were collected and stored at -80 °C, whereas the pellets were further processed to obtain nuclear extracts. Pellets were resuspended in radioimmune precipitation assay buffer plus protease inhibitors and incubated for 10 min on ice. Nuclear extracts were isolated by centrifugation at 10,000 × g for 10 min at 4 °C (supernatant). Protein concentration was determined using a BCA Protein Assay kit (Pierce Biotechnology).
Silencing Studies using Small Interfering RNAs (siRNA)—siRNAs were obtained from DHARMACON RNA Technologies Inc. (Chicago, IL) selecting the ON-TARGET-plus SMARTpool duplexes as the RNAi-specific technology platform. siRNA against human Nrf2 (NFE2L2, L-003755), Keap1 (KEAP1, L-012453), and the siCONTROL non-targeting siRNA [siRNA(-)] were used. siRNAs were introduced into cells by nucleofection (Amaxa) following the manufacturer's instructions as previously described (28). Then, cells were treated with ATO, and samples were harvested at 24 and 48 h, for ATO-induced apoptosis determination by Annexin-V-FITC/PI staining and at 6 and 24 h, for protein expression analysis by Western blot.
Intracellular ROS Detection—DHE was used as fluorescent probe to detect intracellular ROS as superoxide. Briefly, 2 × 105 cells (control or treated cells) were washed twice with PBS and stained for 30 min at 37 °C with 10 μm DHE in PBS. Samples were washed once with PBS and then were acquired on a FACScan flow cytometer (Becton Dickinson) and analyzed with CellQuest software (Becton Dickinson).
RESULTS
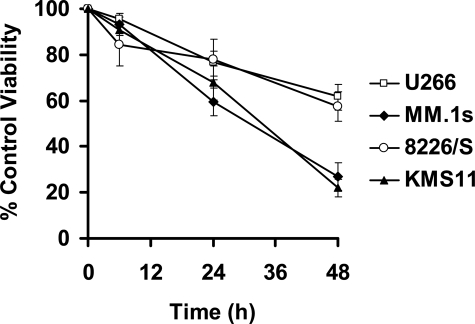
Gene Expression Profiling of the Response to ATO Treatment in MM Cell Lines—We and others have been studying the induction of apoptosis by ATO in MM cell lines and while most studies clearly point to induction of caspase activation, it is not clear what the initiating events that ultimately result in caspase activation are or what factors may regulate this signaling (5, 13, 29, 30–32). To this end, we performed an analysis of the gene expression profile in response to ATO. We elected to do this analysis in four MM cell lines that display two different sensitivities to ATO. All four cell lines were sensitive to ATO in a time (Fig. 1) and dose-dependent manner (28). However, two cell lines (U266 and 8226/S) were less sensitive than the other two cell lines. This is exemplified at 48 h where the percent of control viability for U266, MM.1s, 8226/S and KMS11 were 61.9 ± 5.1, 26.9 ± 6.0, 57.5 ± 6.4 and 21.9 ± 3.7%, respectively.
FIGURE 1.
ATO-induced apoptosis in four human myeloma cell lines. U266, MM.1s, 8226/S, and KMS11 were treated for 6, 24, and 48 h with 2μm ATO. Viability was measured by Annexin-V-FITC/PI staining. Percent (%) of control viability, as percent of Annexin-V-negative cells, was plotted versus time (h). The data are presented as the mean ± S.D. of eight independent experiments.
To study the ATO response, cells were treated with 2 μm ATO for 0, 6, 24, and 48 h, and total RNA was obtained for probing Affymetrix Hu133 2.0 Plus Arrays. Ratios at 6, 24, and 48 h versus baseline gene expression were calculated and up- and down-regulation was initially considered as a 2-fold change from the baseline expression. Interestingly, MM.1s and KMS11, the more sensitive cell lines, displayed a larger number of genes up- or down-regulated compared with U266 and 8226/S. However, given the number of changes observed in 3 of 4 individual cell lines, the data sets were too complex to determine if a pattern of expression consistent with an ATO myeloma response exists (Table 1). Therefore, to develop a more manageable data set, we elected to initially focus on genes that were regulated in a similar fashion in all four cell lines. However, this number appeared to be restricted by the relatively low number of genes that changed in 8226/S cells. Therefore, we expanded this data set to include all genes that changed (I or D call) regardless of the magnitude of the change. This resulted in a data set that contained no more than 343 up-regulated genes and 318 down-regulated genes (Table 1). The complete list of these up- and down-regulated genes can be found in the supplemental Table S1 and S2. Interestingly, the peak of up-regulated genes occurred early. This was in part due to a subset of genes that were up-regulated at 6 h and then returned to baseline expression. This group consisted primarily of heat shock proteins (HSPs, see supplemental Table S1). Many of the other genes that were up-regulated were consistent with an antioxidant response, including NAD(P)H dehydrogenase, quinone 1 and 2 (NQO1 and NQO2), HO-1, and metallothionein-1 and 2A (MT1 and MT2A).
TABLE 1.
Number of up- and down-regulated probes during ATO-induced apoptosis in U266, MM.1s, 8226/S, and KMS11 cell lines at 6, 24, and 48 h
| Changes | U266 | MM.1s | 8226/S | KMS11 | ALL |
|---|---|---|---|---|---|
| Up | |||||
| 6 h | 1471 (245)a | 3120 (730) | 1646 (262) | 3263 (961) | 343 (55) |
| 24 h | 2644 (396) | 2763 (641) | 722 (81) | 2514 (602) | 176 (35) |
| 48 h | 2824 (606) | 3201 (823) | 1333 (204) | 3002 (1076) | 272 (46) |
| Down | |||||
| 6 h | 2048 (243) | 3001 (648) | 1798 (143) | 2933 (544) | 282 (10) |
| 24 h | 2740 (555) | 2893 (648) | 1217 (92) | 3093 (601) | 286 (6) |
| 48 h | 3059 (791) | 3228 (1120) | 1817 (186) | 3263 (1145) | 318 (7) |
The numbers of probe sets that were increased or decreased by 2-fold or greater are shown in parentheses
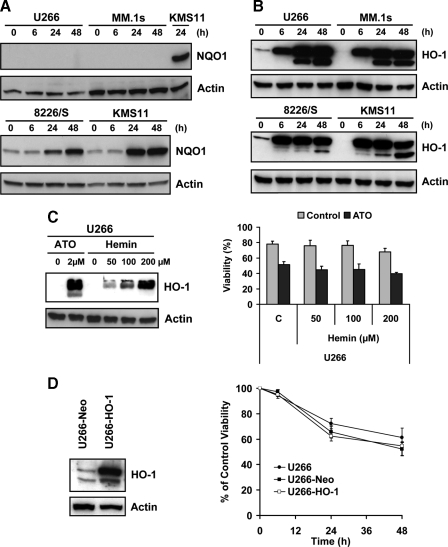
NQO1 was up-regulated at the mRNA level in all four cell lines as early as in 6 h (supplemental Table S1). Interestingly, in U266 and MM.1s, NQO1 protein was not present or up-regulated after ATO treatment and in 8226/S and KMS11 was up-regulated only after 24 h ATO treatment (Fig. 2A). These results suggest that NQO1 is unlikely to be related with any protection from this antioxidant response. Therefore, we initially focused on the two most up-regulated genes HO-1 and metallothioneins (MTs).
FIGURE 2.
Induction of HO-1 is not protective against ATO-induced apoptosis in MM cell lines. U266, MM.1s, 8226/S and KMS11 were treated for 0, 6, 24, and 48 h with 2 μm ATO. Total protein lysates were obtained and NQO1 (A) and HO-1 (B) protein expression was determined by Western blot. KMS11 treated with ATO for 24 h was used as a positive control for the U266 and MM.1s NQO1 blot. C, U266 cell line was pretreated for 24 h with 50, 100, and 200 μm of hemin and HO-1 protein up-regulation was demonstrated by Western blot. Cells treated with ATO for 24 h were used as a positive control. Hemin-pretreated U266 cells were treated with 2 μm ATO for 24 h and viability was evaluated by Annexin-V-FITC/PI staining. Data are presented as mean ± S.D. of three independent experiments. D, U266 cell line was stably transfected with pcDNA3-HO-1 or an empty vector control (Neo). Expression of HO-1 was confirmed by Western blot. U266, U266-Neo, and U266-HO-1 were treated with 2 μm ATO for the indicated time and viability was evaluated by annexin-V-FITC/PI staining. Percent (%) of control viability was plotted versus time (h). Data are presented as mean ± S.D. of three independent experiments.
ATO-induced Up-regulation of HO-1 Is Not Protective against Cell Death—HO-1 was strongly up-regulated in all four cell lines, not only at the mRNA level (Table S1) but also at protein level (Fig. 2B). A second band of ∼25 kDa was obtained at 24 and 48 h, coinciding with caspase activation. Inhibition of caspases with BocD-Fmk, prevented the appearance of this band and suggests that it is a result of caspase-dependent cleavage of HO-1 (not shown). Interestingly, the HO-1 baseline expression was higher in U266 and 8226/S than in MM.1s and KMS11, suggesting that HO-1 could be protective against ATO. This is consistent with a previous report demonstrating HO-1 expression in the surviving fraction of cells treated for 7 days with ATO (15). We tested this possibility by determining the effect of pretreating cells with hemin to induce HO-1 protein expression. Pretreatment of U266 (Fig. 2C) and MM.1s (not shown) cell lines, with hemin at 50, 100, and 200 μm for 24 h, induced up-regulation of HO-1 at the protein level, in a dose-dependent manner. Regardless, hemin-pretreated U266 cell line remained sensitive to ATO (Fig. 2C). Similar findings were observed with MM.1s (not shown). We next directly tested the effect of HO-1 on ATO-induced death by stably transfecting cells with an HO-1 expression vector. Western blot analysis demonstrated overexpression of HO-1 protein (Fig. 2D) in the U266-HO-1 cell line compared with the U266-Neo cell line. Consistent with induction of HO-1 by hemin, overexpression of HO-1 was not protective against ATO-induced cell death (Fig. 2D).
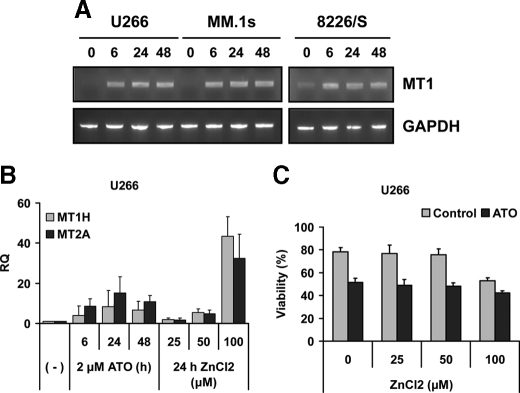
ATO-induced Up-regulation of MTs Is Also Not Protective— MT1 gene expression was measured by RT-PCR at 0, 6, 24, and 48 h after ATO treatment, for U266, MM.1s, and 8226/S. Up-regulation of the MT1 gene was observed in the three cell lines tested, as early as 6 h (Fig. 3A), confirming the microarray data obtained for all four cell lines (supplemental Table S1). To test if MT1 and/or MT2A are protective, the mRNA was up-regulated using ZnCl2 pretreatment for 24 h, and gene expression was determined by real-time PCR. As seen in Fig. 3B both MT1H and MT2A are induced by ATO and ZnCl2. Similar results were obtained for MM.1s (not shown). However, when ZnCl2-pretreated cells were tested for ATO-induced apoptosis and no differences were found compared with untreated cells as control (Fig. 3C and not shown).
FIGURE 3.
MT induction does not protect cells from ATO-induced cell death. A, U266, MM.1s, and 8226/S were treated for 0, 6, 24, and 48 h with 2 μm ATO, and total RNA was obtained. RT-PCR for MT1 and GAPDH was performed as described under “Materials and Methods.” B, U266 was treated with 25, 50, and 100 μm of ZnCl2, and MT1H and MT2A gene expression was measured by real-time PCR. C, untreated and ZnCl2-pretreated U266 cells were treated with 2 μm ATO for 24 h, and viability was evaluated by annexin-V-FITC/PI staining. Data are presented as mean ± S.D. of three independent experiments.
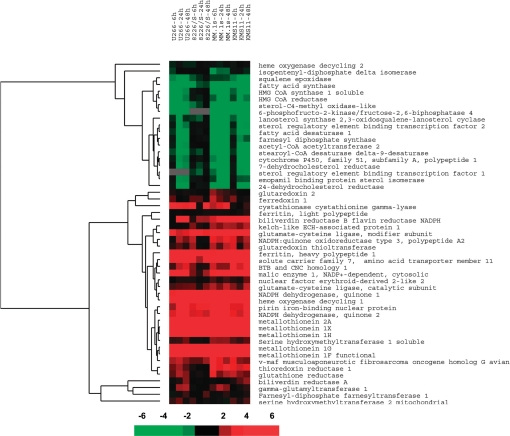
ATO Induces an Antioxidant Response Related to the Activation of Nrf2—A closer analysis of the array data results revealed the common up- and down-regulation of a subset of genes related to the Nrf2-Keap1 pathway. A supervised clustering of known target and related genes indicated activation of this pathway (Fig. 4). Down-regulated genes are consistent with transcriptional repression of genes involved in the cholesterol and lipid biosynthetic pathways. Up-regulated genes included: MTs, HO-1, and related enzymes (biliverdin reductase B, ferritin, and pirin iron-binding nuclear protein), NQO1, and NQO2, NADPH-generating enzymes (malic enzyme), and genes related to the GSH de novo synthesis and salvage pathways as well as enzymes and carriers that would facilitate synthesis and transport of GSH building block amino acids (solute carrier family 7, glutamate-cysteine ligase, serine hydroxylmethyl-transferase-1 soluble, γ-glutamyl-transferase-1, cystathionase, and glutathione reductase).
FIGURE 4.
ATO induced activation of the Nrf2-Keap1 pathway. Genes related to the Nrf2-Keap1 signaling pathway, included in the array, were clustered using Cluster and TreeView. The clustering of these genes was supervised to maintain the time course of the arrays. Ratios versus baseline expression at 6, 24, and 48 h were included in the analysis for U266, MM.1s, 8226/S, and KMS11. The scale represents the magnitude of indicated changes. A black square indicates no change in expression at that time point compared with control, while a gray box indicates that no expression was observed in that cell line.
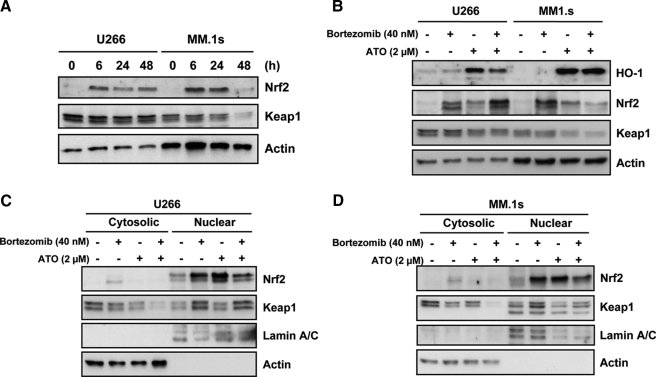
Activation of Nrf2 by ATO—Nrf2 is regulated primarily at the protein level through stabilization and nuclear translocation. The protein is targeted for degradation by a cullin ring ligase complex containing Cul3, Rbx1, and Keap1 (21–23). Oxidative stress stabilizes Nrf2 by either inhibiting the ubiquitination of Nrf2 or causing its dissociation from Keap1 (17). Consistent with these possibilities, Nrf2 mRNA is present in all cells and is not up-regulated in response to ATO (supplemental Table S1). In contrast, the protein is absent and rapidly up-regulated in response to ATO (Fig. 5A). We also tested the effect of melphalan and staurosporine on Nrf2 protein, in U266 and MM.1s. These two drugs have been reported to induce caspase activation and downstream ROS production (33–36). Both drugs induced apoptosis in myeloma cells however Nrf2 up-regulation was not observed (not shown). The rapid up-regulation of Nrf2 protein expression by ATO is not associated with loss of Keap1 protein which suggests that inhibition of Cul3-Rbx1-Keap1 complex by ATO is not due to Keap1 degradation. To confirm that protein stabilization of Nrf2 occurs following ATO treatment, cells were treated with ATO, the proteasome inhibitor Bortezomib or the combination of both for 6 h, and accumulation of Nrf2 monitored. As seen in Fig. 5B, both ATO and Bortezomib resulted in the accumulation of Nrf2, while having little effect on Keap1 expression. Additionally, cells treated with 100 nm of Bortezomib for 2 h, washed and then treated with 10 μg/ml of CHX or the combination of CHX plus ATO (2 μm), demonstrated that ATO could maintain Nrf2 protein in the absence of de novo protein synthesis (supplemental Fig. S1). Taken together, these data demonstrate that ATO induces Nrf2 by stabilizing the protein presumably through inhibition of the Cul3·Rbx1·Keap1 complex. Surprisingly, while proteasome inhibition is sufficient to induce Nrf2 accumulation, it does not result in the activation of a target gene (Fig. 5B). HO-1 expression is not induced by Bortezomib in either cell line tested despite the accumulation of Nrf2 protein, similar to that observed with ATO. This is not due to differences in localization as Bortezomib-induced Nrf2 accumulates in the nucleus in a similar fashion as Nrf2 induced by ATO (Fig. 5C). Together these data suggest that Nrf2 accumulation and nuclear localization are not necessarily sufficient for target gene expression, and that ATO addition likely results in other changes that allow for HO-1 expression.
FIGURE 5.
ATO induces up-regulation of Nrf2 protein, translocation to the nucleus and target gene expression. A, Western blot analysis of Nrf2 and Keap1 protein expression at 0, 6, 24, and 48 h after 2 μm ATO treatment, for U266 and MM.1s cell lines. B, cells were treated with 40 nm of Bortezomib and/or 2 μm ATO for 6 h, lysed, and subjected to Western blot analysis for HO-1, Nrf2, Keap1, and actin protein expression. C, U266 and D, MM.1s cells were treated as in B and fractionated into cytosolic and nuclear protein fractions. Nrf2 and Keap1 protein expression was analyzed by Western blot. Actin and Lamin A/C were used as cytosolic and nuclear loading controls, respectively.
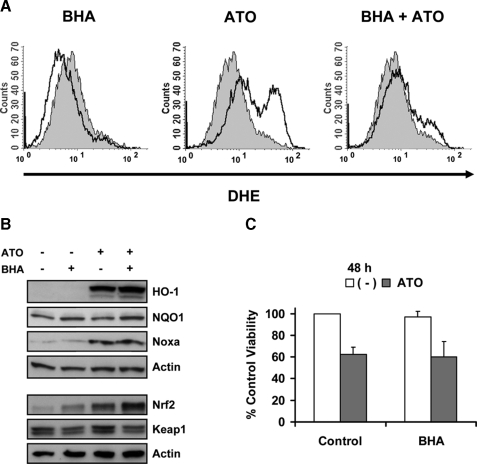
Activation of the Nrf2-Keap1 Pathway Is Not Mediated by ATO-induced ROS—We next determined the role of ROS in Nrf2 activation and ATO-induced cell death. KMS11 cells were treated with 1 μm ATO for 6 h and DHE staining revealed an increased superoxide production in ATO-treated cells compared with control (Fig. 6A). We next pretreated cells for 16 h with several different antioxidants and/or ROS scavengers (including NAC, DPI, ebselen, BHA, MnTBAP, TEMPO, TEMPOL, α-lipoic acid, and EUK-134) to block superoxide production induced by ATO. Interestingly, of all the antioxidant compounds tested, only NAC, α-lipoic acid, and BHA were able to inhibit ATO-induced ROS production (Fig. 6A and not shown). We and others (5, 13, 15, 37) have reported that NAC can protect cells from ATO-induced apoptosis due to an increase in intracellular glutathione, which in addition to its antioxidant properties, is the primary means of arsenic inactivation via conjugation (38). Similar to NAC, α-lipoic acid was able to block ROS production and protect cells from ATO-induced apoptosis (not shown). However for both NAC and α-lipoic acid, the protective effects were dependent on GSH synthesis (not shown). These data are consistent with reports demonstrating that α-lipoic acid can increase GSH synthesis (39). BHA treatment was able to lower endogenous ROS production as well as inhibits ATO-induced ROS (Fig. 6A). These changes in ROS were associated with BHA-induced increases in Nrf2 expression (Fig. 6B). However, the mechanism and consequences of this induction appear to be different than that observed with ATO. 16-h treatment of cells with BHA resulted in a decrease in Keap1 expression, no HO-1 expression and only a modest change in NQO1 expression. This resembles the effect of stabilizing Nrf2 with Bortezomib rather than activation with ATO. More importantly, the induction of Nrf2 (Fig. 6B) and inhibition of ROS (Fig. 6A) had no effect on ATO-induced changes in HO-1, Noxa (Fig. 6B), or cell death (Fig. 6C). Together these data bring into question the role of ROS in ATO apoptotic signaling as well as whether the Nrf2 response confers any protection against ATO-induced apoptosis.
FIGURE 6.
Activation of Nrf2 and Noxa up-regulation by ATO are not mediated by ATO-induced ROS. A, KMS11 was incubated with 100 μm BHA for 16 h. Control and BHA-treated cells were then treated with 1 μm ATO for 6 h. DHE was used for ROS determination. The shaded histogram represents untreated cells while the bold-lined open histograms represent the indicated treatment. B, KMS11 was treated as in A and protein lysates were subjected to Western blot analysis of HO-1, NQO1, Nrf2, Keap1, Noxa, and actin. C, KMS11 was treated as in A, and cell viability was measured by Annexin-V-FITC/PI staining after 48 h of ATO treatment. Percent (%) of control viability was calculated, and data are presented as mean ± S.D. of four independent experiments.
Inhibition of Nrf2 Does Not Affect Sensitivity to ATO-induced Apoptosis—To test the role of Nrf2 signaling in ATO-induced apoptosis we determined the effect of silencing Nrf2 and Keap1. Knockdown of Nrf2 and Keap1 proteins was confirmed by Western blot analysis in MM.1s and KMS11 cell lines (Fig. 7A). Up-regulation of Nrf2 protein by ATO was almost completely inhibited by siNrf2 in both cell lines. HO-1 up-regulation, as a target of this pathway, was only partially inhibited after Nrf2 silencing. Residual Nrf2 activation or up-regulation of HO-1 by ATO through a different mechanism cannot be excluded. Keap1 silencing did not induce any change on HO-1 up-regulation by ATO (Fig. 7A). Consistent with the ROS data, Nrf2 silencing had a statistically significant albeit biologically modest effect at 24 h in KMS11 cells. However this effect was not observed at 48 h and no effect on ATO-induced apoptosis was seen in MM.1s cells (Fig. 7B). Surprisingly Keap1 silencing sensitized cells to ATO-induced death (Fig. 7B).
FIGURE 7.
Nrf2 signaling is not protective during ATO-induced apoptosis. A, MM.1s and KMS11 were electroporated with si(-) (negative control), siNrf2, and siKeap1. After 16 h, cells were treated with 2 μm ATO. Silencing of Nrf2 and Keap1 proteins was determined by Western blot at 6 and 24 h after ATO treatment. B, viability was evaluated by Annexin-V-FITC/PI staining. Percent (%) of control (untreated, transfected, UT) viability was plotted versus time (h). Student's t test was used to compare differences between samples, si(-) and experimental samples unless otherwise indicated, with confidence intervals of 95%. ND, no difference; *, p < 0.05.
DISCUSSION
Arsenic has been extensively studied as an environmental carcinogen, however in the last 15 years it has re-emerged as an anti-cancer agent (7, 12). While many studies have examined how arsenic kills cells under these conditions, few have examined the role of the response observed during carcinogenesis. In this study we examined the gene expression profile using clinically relevant concentrations of arsenic trioxide in a cell model of a disease, where therapeutic responses have been reported (5, 10, 13). Despite the fact that the concentration of ATO used kills 40–60% of cells in 48 h, the primary response observed appeared to be protective in nature. Therefore, we determined the nature and role of this response.
Sensitivity to ATO inversely correlated with the number of up- or down-regulated genes. In U266 and 8226/S, only one-third of the total number of genes displayed a 2-fold up- or down-regulation compared with the more sensitive MM.1s and KMS11 cell lines at 6 h (Table 1). This indicates that the difference in sensitivity is most likely to be early in the process of detecting the presence of ATO and implies that the less sensitive lines are effectively exposed to less ATO. This could reflect differences in uptake or metabolism and experiments are in progress to determine if such differences exist. Regardless, when we analyzed the data in a fashion that excluded these differences by not taking into account the magnitude of the changes, most of the up-regulated genes observed were associated with antioxidative stress response signaling. HSPs, MT1, HO-1, and NQO1 were the most up-regulated genes in all cell lines.
HSPs are a conserved family of stress response proteins with chaperone functions playing a key role in the correct folding of cellular proteins (40). They are overexpressed in numerous cancers and are implicated in cell proliferation, differentiation, and metastasis with inhibitory effects on apoptosis (40). Interestingly, they have been found as target genes of the Nrf2-Keap1 pathway (41); however, the transient nature of the HSP gene signature suggests that their regulation by ATO is distinct from the other genes that we observed. Indeed we have found that ATO can activate transcription from heat shock elements (HSE) as well as AREs.3 Regardless, because HSPs expression peaks at 6 h and return to baseline within 24 h, it is unlikely to play a significant protective role in response to ATO. MT1 and HO-1 were particularly interesting because a previous report has suggested that these two proteins could be related with ATO resistance after 7 days exposure to arsenic (15).
HO-1, a microsomal enzyme, has been shown to protect cells in different stress models (42–44). HO-1 is induced by heme, but also by cytokines, heat shock, metals, and other cellular stress agents that generate ROS. The precise mechanism, by which HO-1 protects cells, is unclear. HO-1 catalyzes the degradation of heme to produce biliverdin, carbon monoxide (CO) and free iron (45–47). Biliverdin is rapidly converted to bilirubin by biliverdin reductase, another gene that is up-regulated in all the cell lines by ATO. Both biliverdin and bilirubin possess antioxidant activity (48, 49). Additionally it has been shown that CO may also initiate intracellular signaling cascades related to cell survival (50, 51). However our results did not demonstrate any protection to the cells by HO-1 up-regulation. While HO-1 expression is higher in the two less sensitive cells, HO-1 was strongly up-regulated within 6 h for all cell lines and cells were still sensitive to ATO-induced apoptosis. Moreover, HO-1 up-regulation was actually higher for the more sensitive MM.1s and KMS11 cell lines; therefore there was no correlation with ATO-sensitivity and HO-1 induction. Furthermore, pretreatment with hemin, a well documented inducer and a substrate of HO-1, did not protected cell lines from ATO-induced apoptosis. Finally, stable overexpression of HO-1 did not protect U266 from ATO-induced cell death.
MTs are cysteine-rich, low molecular weight proteins with a high affinity for metals including arsenicals (52–54). MTs not only bind metals but also scavenge ROS and may play a role in drug resistance (55). MT gene expression levels are increased in MM cells exposed to ATO (15), suggesting a possible role in the ATO resistance phenotype. Consistent with that possibility, MT2A can chelate ATO and possibly sequester ATO intracellularly (53). Both MT1 and MT2A were up-regulated in all cell lines within 6 h, displaying the strongest overall up-regulation during ATO-induced apoptosis. However, pretreatment with ZnCl2 to increase MT1/MT2A expression in U266 and MM.1s did not confer protection against ATO-induced cell death.
HO-1 and MT1 up-regulation, as individual elements, were not enough to protect cells from ATO-induced apoptosis. However, the antioxidative stress response after ATO treatment is not limited to these two proteins. Myeloma cells elicited a complex protective response driven by the Nrf2-Keap1 pathway that also induced up-regulation of the NQO1 and the GSH synthesis pathway as well as down-regulation of the lipid/cholesterol pathways. Nrf2 protein was strongly up-regulated by ATO as early as 6 h in all the cell lines, while gene expression was only slightly up-regulated in two of the four cell lines. Consistent with reported post-translational regulation (20, 56), proteasome inhibition with Bortezomib resulted in protein accumulation; however, this did not enhance ATO-induced Nrf2, suggesting that ATO also inhibits the degradation of Nrf2. ATO stabilized Nrf2 accumulated in the nucleus and could activate HO-1 expression. Interestingly, Bortezomib induced only accumulation but not the activation of Nrf2 (as measured by HO-1 up-regulation). Together these data suggest that nuclear accumulation of Nrf2 is not sufficient for HO-1 induction. Therefore, ATO is likely to also influence Nrf2 function independent of stability. Several studies suggest that phosphorylation could also be important in Nrf2 activation (57, 58). ATO also activates members of the MAPK pathway, some of which have protective roles (59–61). These could also play a role in HO-1 induction.
These data are consistent with the idea that arsenic induces ROS and that ROS could be a mediator of ATO-induced cell death. Indeed, as we and others have previously demonstrated, ATO induces superoxide production in myeloma cells (13, 29, 62, 63). However, ATO-induced ROS does not play a key role in either activation of Nrf2 or the induction of cell death by ATO. BHA was able to block ATO-induced ROS, but did not inhibit activation of Nrf2 or the up-regulation of Noxa, a BH3-only protein required for ATO-induced cell death (28). This is consistent with our previous findings that in some myeloma cell lines, ROS production occurs downstream of caspase activation and is therefore a late event (29). Thus, much of the antioxidant response induced by ATO may not be useful as it functions primarily to protect the cells from a component of the arsenic response that is not required for cell death.
To directly determine the role of this antioxidative stress response, the effect of Nrf2-Keap1 silencing on ATO sensitivity was measured. Keap1 silencing has been reported to up-regulate ARE-containing genes. Transfection of human HaCaT cells with Keap1 siRNA markedly enhanced endogenous levels of Nrf2 protein and increased transcription of an ARE-driven reporter gene by 2.3-fold (64). Consistent with our findings with Bortezomib, Keap1 silencing experiments did not induce any additional accumulation or activation of Nrf2 protein or HO-1 up-regulation. Contrary to expected results, Keap1 silencing sensitized the cells to ATO-induced apoptosis (Fig. 6B) and even induced more death in untreated transfected cells compare with si(-) transfected cells (data not shown). These data suggest that Keap1 may regulate cell survival independent of Nrf2. Interestingly the only other reported substrate recognized by Keap1 is a Bcl-xL-binding protein (65).
Nrf2 silencing was able to inhibit Nrf2 protein accumulation after ATO treatment and partially inhibited the HO-1 up-regulation in myeloma cell lines. However, little effect on ATO sensitivity was observed. This lack of effect of a diminished antioxidant response is consistent with ROS not being an important mediator of ATO-induced cell death. However, the silencing of Nrf2 and its effect on HO-1 induction were not complete therefore it is possible that the protective function of this pathway remained intact. At least three transcription factors have been reported to target the HO-1 gene (66–68): Nrf2, activator protein-1 (AP-1), and nuclear factor-κB (NFκB). Nrf2 and AP-1 can have independent roles in the up-regulation of HO-1 by arsenite in murine embryonic fibroblasts (67). Alternatively, it is also possible that the apoptotic signaling induced by ATO is dominant to the protective pathway. This is the favorable scenario with all chemotherapeutic agents.
Finally in thinking about arsenic as a therapeutic agent, the data suggest that combinations with ATO should be carefully considered. It may not be advisable to combine ATO with agents that function primarily through the induction of oxidative stress. The protective response induced by ATO may prevent the desired synergistic effects in a combination therapy.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA129968, CA97243, and CA127910. This work was also supported by a Senior Research Award from the Multiple Myeloma Foundation.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.
Footnotes
The abbreviations used are: ATO, arsenic trioxide; PBS, phosphate-buffered saline; HSP, heat shock protein; ROS, reactive oxygen species; MM, multiple myeloma; BHA, butylated hydroxyanisole; HO-1, heme oxygenase-1; ARE, antioxidant response element; FITC, fluorescein isothiocyanate; NAC, N-acetylcysteine; CHX, cycloheximide.
L. H. Boise, submitted manuscript.
References
- 1.Wang, Z. Y., and Chen, Z. (2006) Blood 111 2505-2515 [DOI] [PubMed] [Google Scholar]
- 2.Berenson, J. R., and Yeh, H. S. (2006) Clin. Lymphoma Myeloma. 7 192-198 [DOI] [PubMed] [Google Scholar]
- 3.Hussein, M. A. (2001) Med. Oncol. 18 239-242 [DOI] [PubMed] [Google Scholar]
- 4.Kalmadi, S. R., and Hussein, M. A. (2006) Acta Haematol. 116 1-7 [DOI] [PubMed] [Google Scholar]
- 5.Bahlis, N. J., McCafferty-Grad, J., Jordan-McMurry, I., Neil, J., Reis, I., Kharfan-Dabaja, M., Eckman, J., Goodman, M., Fernandez, H. F., Boise, L. H., and Lee, K. P. (2002) Clin. Cancer Res. 8 3658-3668 [PubMed] [Google Scholar]
- 6.Munshi, N. C., Tricot, G., Desikan, R., Badros, A., Zangari, M., Toor, A., Morris, C., Anaissie, E., and Barlogie, B. (2002) Leukemia 16 1835-1837 [DOI] [PubMed] [Google Scholar]
- 7.Bonati, A., Rizzoli, V., and Lunghi, P. (2006) Curr. Pharm. Biotechnol. 7 397-405 [DOI] [PubMed] [Google Scholar]
- 8.Rousselot, P., Larghero, J., Arnulf, B., Poupon, J., Royer, B., Tibi, A., Madelaine-Chambrin, I., Cimerman, P., Chevret, S., Hermine, O., Dombret, H., Claude Brouet, J., and Paul Fermand, J. (2004) Leukemia 18 1518-1521 [DOI] [PubMed] [Google Scholar]
- 9.Miller, W. H., Schipper, H. M., Lee, J. S., Singer, J., and Waxman, S. (2002) Cancer Res. 62 3893-3903 [PubMed] [Google Scholar]
- 10.Gallagher, R. E. (2003) Leuk. Res. 25 237-239 [DOI] [PubMed] [Google Scholar]
- 11.Dai, J., Weinberg, R. S., Waxman, S., and Jing, Y. (1999) Blood 93 268-277 [PubMed] [Google Scholar]
- 12.Dilda, P. J., and Hogg, P. J. (2007) Cancer Treat. Rev. 33 542-564 [DOI] [PubMed] [Google Scholar]
- 13.Grad, J. M., Bahlis, N. J., Reis, I., Oshiro, M. M., Dalton, W. S., and Boise, L. H. (2001) Blood 98 805-813 [DOI] [PubMed] [Google Scholar]
- 14.Chen, D., Chan, R., Waxman, S., and Jing, Y. (2006) Cancer Res. 66 11416-11423 [DOI] [PubMed] [Google Scholar]
- 15.Zhou, P., Kalakonda, N., and Comenzo, R. L. (2005) Br. J. Haematol. 128 636-644 [DOI] [PubMed] [Google Scholar]
- 16.Nguyen, T., Yang, C. S., and Pickett, C. B. (2004) Free Radic. Biol. Med. 37 433-441 [DOI] [PubMed] [Google Scholar]
- 17.Kensler, T. W., Wakabayashi, N., and Biswal, S. (2007) Annu. Rev. Pharmacol. Toxicol. 47 89-116 [DOI] [PubMed] [Google Scholar]
- 18.Ma, Q., Kinneer, K., Bi, Y., Chan, J. Y., and Kan, Y. W. (2004) Biochem. J. 377 205-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., Oyake, T., Hayashi, N., Satoh, K., Hatayama, I., Yamamoto, M., and Nabeshima, Y. (1997) Biochem. Biophys. Res. Commun. 36 313-322 [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, A., Kang, M. I., Watai, Y., Tong, K. I., Shibata, T., and Yamamoto, M. (2006) Mol. Cell Biol. 26 221-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. S., and Surh, Y. J. (2005) Cancer Lett. 224 171-184 [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, M., and Yamamoto, M. (2005) Antioxid. Redox. Signal. 7 385-394 [DOI] [PubMed] [Google Scholar]
- 23.Martin, D., Rojo, A. I., Salinas, M., Diaz, R., Gallardo, G., Alam, J., De Galarreta, C. M., and Cuadrado, A. (2004) J. Biol. Chem. 279 8919-8929 [DOI] [PubMed] [Google Scholar]
- 24.Hu, X., Roberts, J. R., Apopa, P. L., Kan, Y. W., and Ma, Q. (2006) Mol. Cell Biol. 26 940-954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos-Gomez, M., Kwak, M. K., Dolan, P. M., Itoh, K., Yamamoto, M., Talalay, P., and Kensler, T. W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3410-3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma, Q., Battelli, L., and Hubbs, A. F. (2006) Am. J. Pathol. 168 1960-1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, X., and Talalay, P. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10446-10451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales, A. A., Gutman, D., Lee, K. P., and Boise, L. H. (2008) Blood 111 5152-5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCafferty-Grad, J., Bahlis, N. J., Krett, N., Aguilar, T. M., Reis, I., Lee, K. P., and Boise, L. H. (2003) Mol. Cancer Ther. 2 1155-1164 [PubMed] [Google Scholar]
- 30.Liu, Q., Hilsenbeck, S., and Gazitt, Y. (2003) Blood 101 4078-4087 [DOI] [PubMed] [Google Scholar]
- 31.Hayashi, T., Hideshima, T., Akiyama, M., Richardson, P., Schlossman, R. L., Chauhan, D., Munshi, N. C., Waxman, S., and Anderson, K. C. (2002) Mol. Cancer Ther. 1 851-860 [PubMed] [Google Scholar]
- 32.Park, W. H., Seol, J. G., Kim, E. S., Hyun, J. M., Jung, C. W., Lee, C. C., Kim, B. K., and Lee, Y. Y. (2000) Cancer Res. 60 3065-3071 [PubMed] [Google Scholar]
- 33.Min, J. Y., Park, M. H., Park, M. K., Park, K. W., Lee, N. W., Kim, T., Kim, H. J., and Lee, D. H. (2006) J. Neural Transm. 113 1821-1826 [DOI] [PubMed] [Google Scholar]
- 34.Circu, M. L., Stringer, S., Rhoads, C. A., Moyer, M. P., and Aw, T. Y. (2009) Biochem. Pharmacol. 77 76-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krause, C., Klüttermann, K., and Mauz-Körholz, C. (2008) Anticancer Res. 28 2585-2593 [PubMed] [Google Scholar]
- 36.Gouill, S., Bataille, R., and Amiot, M. (2005) Oncogene 24 8076-8079 [DOI] [PubMed] [Google Scholar]
- 37.Hu, X. M., Hirano, T., and Oka, K. (2003) Cancer Chemother. Pharmacol. 52 47-58 [DOI] [PubMed] [Google Scholar]
- 38.Kumagai, Y., and Sumi, D. (2007) Annu. Rev. Pharmacol. Toxicol. 47 243-262 [DOI] [PubMed] [Google Scholar]
- 39.Fujita, H., Shiosaka, M., Ogino, T., Okimura, Y., Utsumi, T., Sato, E. F., Akagi, R., Inoue, M., Utsumi, K., and Sasaki, J. (2008) Brain Res. 1206 1-12 [DOI] [PubMed] [Google Scholar]
- 40.Calderwood, S. K., Khaleque1, M. A., Sawyer, D. B., and Ciocca, D. R. (2006) Trends Biochem. Sci. 31 164-172 [DOI] [PubMed] [Google Scholar]
- 41.Kwak, M. K., Wakabayashi, N., Itoh, K., Motohashi, H., Yamamoto, M., and Kensler, T. W. J. (2003) Biol. Chem. 278 8135-8145 [DOI] [PubMed] [Google Scholar]
- 42.Nowis, D., Legat, M., Grzela, T., Niderla, J., Wilczek, E., Wilczynski, G. M., Głodkowska, E., Mrówka, P., Issat, T., Dulak, J., Józkowicz, A., Waś, H., Adamek, M., Wrzosek, A., Nazarewski, S., Makowski, M., Stokłosa, T., Jakóbisiak, M., and Gołab, J. (2006) Oncogene 25 3365-3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki, T., Yoshida, K., Kondo, H., Ohmori, H., and Kuniyasu, H. (2005) Virchows Arch. 446 525-531 [DOI] [PubMed] [Google Scholar]
- 44.Liu, Z. M., Chen, G. G., Ng, E. K., Leung, W. K., Sung, J. J., and Chung, S. C. (2004) Oncogene 23 503-513 [DOI] [PubMed] [Google Scholar]
- 45.Abraham, N. G., Asija, A., Drummond, G., and Peterson, S. (2007) Curr. Gene Ther. 7 89-108 [DOI] [PubMed] [Google Scholar]
- 46.Ryter, S. W., and Choi, A. M. (2002) Antioxid. Redox. Signal. 4 625-632 [DOI] [PubMed] [Google Scholar]
- 47.Fang, J., Akaike, T., and Maeda, H. (2004) Apoptosis 9 27-35 [DOI] [PubMed] [Google Scholar]
- 48.Baranano, D. E., Rao, M., Ferris, C. D., and Snyder, S. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16093-16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stocker, R., Yamamoto, Y., McDonagh, A. F., Glazer, A. N., and Ames, B. N. (1987) Science 235 1043-1046 [DOI] [PubMed] [Google Scholar]
- 50.Brouard, S., Otterbein, L. E., Anrather, J., Tobiasch, E., Bach, F. H., Choi, A. M., and Soares, M. P. (2000) J. Exp. Med. 192 1015-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato, K., Balla, J., Otterbein, L., Smith, R. N., Brouard, S., Lin, Y., Csizmadia, E., Sevigny, J., Robson, S. C., Vercellotti, G., Choi, A. M., Bach, F. H., and Soares, M. P. (2001) J. Immunol. 166 4185-4194 [DOI] [PubMed] [Google Scholar]
- 52.Ngu, T. T., and Stillman, M. J. (2006) J. Am. Chem. Soc. 128 12473-12483 [DOI] [PubMed] [Google Scholar]
- 53.Toyama, M., Yamashita, M., Hirayama, N., and Murooka, Y. (2002) J. Biochem. 132 217-221 [DOI] [PubMed] [Google Scholar]
- 54.Albores, A., Koropatnick, J., Cherian, M. G., and Zelazowski, A. J. (1992) Chem. Biol. Interact. 85 127-140 [DOI] [PubMed] [Google Scholar]
- 55.Volm, M., Koomagi, R., Mattern, J., and Efferth, T. (2002) Br. J. Cancer 87 251-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto, N., Sawada, H., Izumi, Y., Kume, T., Katsuki, H., Shimohama, S., and Akaike, A. (2007) J. Biol. Chem. 282 4364-4372 [DOI] [PubMed] [Google Scholar]
- 57.Huang, H. C., Nguyen, T., and Pickett, C. B. (2002) J. Biol. Chem. 277 42769-42774 [DOI] [PubMed] [Google Scholar]
- 58.Chen, C. Y., Jang, J. H., Li, M. H., and Surh, Y. J. (2005) Biochem. Biophys. Res. Commun. 331 993-1000 [DOI] [PubMed] [Google Scholar]
- 59.Yan, W., Arai, A., Aoki, M., Ichijo, H., and Miura, O. (2007) Biochem. Biophys. Res. Commun. 355 1038-1044 [DOI] [PubMed] [Google Scholar]
- 60.Kajiguchi, T., Yamamoto, K., Iida, S., Ueda, R., Emi, N., and Naoe, T. (2006) Cancer Sci. 97 540-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kannan-Thulasiraman, P., Katsoulidis, E., Tallman, M. S., Arthur, J. S., and Platanias, L. C. (2006) J. Biol. Chem. 281 22446-22452 [DOI] [PubMed] [Google Scholar]
- 62.Woo, S. H., Park, I. C., Park, M. J., An, S., Lee, H. C., Jin, H. O., Park, S. A., Cho, H., Lee, S. J., Gwak, H. S., Hong, Y. J., Hong, S. I., and Rhee, C. H. (2004) Int. J. Cancer 112 596-606 [DOI] [PubMed] [Google Scholar]
- 63.Shen, Z. Y., Shen, W. Y., Chen, M. H., Shen, J., and Zeng, Y. (2003) Int. J. Mol. Med. 11 479-484 [PubMed] [Google Scholar]
- 64.Devling, T. W. P., Lindsay, C. D., McLellan, L. I., McMahon, M., and Hayes, J. D. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 7280-7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lo, S. C., and Hannink, M. (2006) J. Biol. Chem. 281 37893-37903 [DOI] [PubMed] [Google Scholar]
- 66.Ferrandiz, M. L., and Devesa, I. (2008) Curr. Pharm. Design. 14 473-486 [DOI] [PubMed] [Google Scholar]
- 67.Harada, H., Sugimoto, R., Watanabe, A., Taketani, S., Okada, K., Warabi, E., Siow, R., Itoh, K., Yamamoto, M., and Ishii, T. (2008) Free Radic. Res. 42 297-304 [DOI] [PubMed] [Google Scholar]
- 68.Alam, J., and Cook, J. L. (2007) Am. J. Respir. Cell Mol. Biol. 36 166-174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.