Abstract
Ryk is a Wnt receptor that plays an important role in neurogenesis, neurite outgrowth, and axon guidance. We have reported that the Ryk receptor is cleaved by γ-secretase and that its intracellular domain (ICD) translocates to the nucleus upon Wnt stimulation. Cleavage of Ryk and its ICD is important for the function of Ryk in neurogenesis. However, the question of how the Ryk ICD is stabilized and translocated into the nucleus remains unanswered. Here, we show that the Ryk ICD undergoes ubiquitination and proteasomal degradation. We have identified Cdc37, a subunit of the molecular chaperone Hsp90 complex, as a Ryk ICD-interacting protein that inhibits proteasomal degradation of the Ryk ICD. Overexpression of Cdc37 increases Ryk ICD levels and promotes its nuclear localization, whereas Cdc37 knockdown reduces Ryk ICD stability. Furthermore, we have discovered that the Cdc37-Ryk ICD complex is disrupted during neural differentiation of embryonic stem cells, resulting in Ryk ICD degradation. These results suggest that Cdc37 plays an essential role in regulating Ryk ICD stability and therefore in Ryk-mediated signal transduction.
Wnt signaling plays an essential role in several developmental processes, including cell proliferation, cell migration, and cell fate determination (1–3). Wnt signaling is mediated by various receptors that activate different signal transduction pathways. One of these receptor families includes seven-transmembrane Frizzled proteins that, along with their coreceptor low density lipoprotein receptor-related protein (LRP), control β-catenin-dependent signaling through activation of Tcf/Lef transcription factors. Frizzled proteins also activate β-catenin-independent signaling, referred to as the planar cell polarity and calcium pathways (4, 5). Wnt proteins also activate a different type of signaling via the Ryk receptor (6, 7).
Ryk, whose structure is related to that of receptor protein-tyrosine kinases (RTKs)4, consists of a glycosylated extracellular domain, a transmembrane domain, and an intracellular kinase domain. The extracellular domain exhibits sequence homology to Wnt inhibitory factor, suggesting that it binds to secreted Wnt growth factors (8). Indeed, recent studies show that Wnt1, -3, -3a, and -5a bind directly to the Ryk extracellular domain and suggest a possible role for Ryk as a Wnt receptor in several developmental processes, including neurite outgrowth, cell fate determination, organogenesis, and axon guidance (9–13). The Ryk intracellular domain (ICD) contains 11 distinct subdomains that are highly conserved within the kinase domain of RTKs. Despite this, Ryk belongs to a subclass of catalytically inactive RTKs, which includes CCK4, ErbB3, EphB6, and Ror1 (14). A comparison of Ryk intracellular domains from several species with catalytically active RTKs shows amino acid substitutions in subdomains I and II, suggesting a loss of function at the ATP binding site. Subdomain VII also displays a highly unusual amino acid substitution in the catalytic loop, which may also account for loss of catalytic activity (14, 15). Thus, the mechanism by which the Ryk receptor transduces signals is unknown. One possibility is that Ryk signals via heterodimerization with other RTKs. This hypothesis is supported by studies showing that Ryk-deficient mice exhibit a cleft palate and defects in craniofacial morphology, similar to the phenotypes exhibited by EphB2/EphB3-deficient mice, and that Ryk binds to EphB2 and EphB3 (16). Recently, we discovered that the Ryk receptor undergoes intramembrane proteolytic cleavage to directly transduce intracellular signaling and that this event is required for neurogenesis (17).
Ryk cleavage is mediated by γ-secretase (17), releasing the ICD into the cytoplasm where it translocates to the nucleus and likely regulates transcription of target genes in a manner similar to the ICDs of Notch and ErbB4 (18–20). γ-Secretase can also facilitate degradation of other transmembrane proteins (18), and in some cases, the ICDs of some γ-secretase substrates such as syndecan-3, nectin-1α, and p75 are rapidly degraded. For example, in the absence of Notch binding, the cleaved ICD of the Notch ligand Delta is degraded by the proteasomal machinery (18). All of these observations suggest that the Ryk ICD may require stabilization to transmit Ryk signaling.
Cdc37 is an Hsp90 co-chaperone that physically interacts with several signaling protein kinases, including Cdk4 and Raf-1 (21–23). The Cdc37-Hsp90 complex is required for conformational maturation and stabilization of protein kinases (24). Inhibition of Cdc37-Hsp90 function causes proteasome-mediated Raf degradation (25).
Here, we show that Cdc37 binds to the Ryk ICD, promoting stabilization of the ICD fragment and nuclear translocation. We also show that the Ryk ICD is degraded following disruption of the Cdc37-Ryk ICD complex during the course of neural differentiation of embryonic stem cells (ESCs). These results suggest a biological role for Cdc37-mediated stabilization of the Ryk ICD in Ryk signaling.
EXPERIMENTAL PROCEDURES
Derivation of Ryk ESC Lines and Cell Cultures—Ryk+/- mice were kindly provided by Dr. Steven A. Stacker (Ludwig Institute of Royal Melbourne Hospital). To prepare Ryk-/- ESCs, blastocysts were flushed from the uteruses of pregnant females 3.5 days after mating a Ryk+/- breeder pair, and cells were explanted on feeder layers of irradiated mouse embryonic fibroblasts in growth medium containing 80% Knockout™ Dulbecco's modified Eagle's medium (Invitrogen), 15% Knockout™ serum replacement (Invitrogen), 5% fetal bovine serum (ESC-qualified, HyClone), 2 mm l-glutamine, 1× nonessential amino acids, 1× penicillin/streptomycin (Invitrogen), 1000 units/ml mouse leukemia inhibitory factor (LIF; Millipore), and 0.1 mm 2-mercaptoethanol (Invitrogen). After 3 days, most of the blastocysts had attached to the mouse embryonic fibroblasts, and the culture medium was changed; the medium was again changed every other day until cellular outgrowths were removed and dissociated. Between days 7 and 10 of co-culture, inner cell mass outgrowths were plucked from the mouse embryonic fibroblast feeder layer and dissociated with trypsin/EDTA. Dissociated cells were plated on fresh mouse embryonic fibroblasts in 24-well plates. The medium was changed daily, and cultures were carefully monitored. ESC colonies became visible within a week and were passaged. Colonies were further expanded by enzymatic dissociation with trypsin/EDTA and genotyped using PCR. To obtain feeder-free ESCs, all ESC lines were cultured in Glasgow minimum essential medium (Sigma) containing 15% fetal bovine serum (Invitrogen), 0.1 mm 2-mercaptoethanol, 1000 units/ml LIF, and nonessential amino acids on gelatin-coated tissue culture dishes (26). Embryoid bodies in the absence of LIF were grown with or without 1 μm all-trans-retinoic acid (Sigma) to promote differentiation. Differentiation of ESCs into neural progenitors on monolayer cultures was performed as described previously (27).
Construction of Plasmids and Viruses—Plasmids encoding FLAG-Cdc37 and Cdc37ΔCT were kindly provided by Dr. Robert L. Matts (Oklahoma State University). Plasmids encoding the Ryk ICD and deletion mutants Δ319–366, Δ367–438, Δ439–493, and Δ494–579 were generated by PCR using wild-type Ryk as a template, whereas an oligonucleotide was used to create a C-terminal Myc tag. To generate enhanced GFP fusions, a DNA fragment encoding enhanced GFP was inserted at the N terminus into the plasmids encoding Ryk ICDs. To produce the recombinant Ryk ICD protein (amino acids 367–579) fused to glutathione S-transferase in Escherichia coli, the PCR product encoding the Ryk ICD was inserted into pGEX4T. To create a tetracycline-inducible system in a lentiviral expression system, the digested rtTA2S-M2 fragment (28) was inserted into pFUIPW, which contains an internal ribosomal entry site followed by the puromycin resistance gene (FUrt-TAIPW). The ubiquitin promoter of pFUW was replaced with a tetracycline-responsive element containing a cytomegalovirus minimal promoter to construct pFTREW. Green fluorescent protein (GFP) alone, GFP-ICD, and GFP-ICDΔ494–579 were subcloned into pFTREW. To obtain constructs for tamoxifen-inducible Cdc37 shRNA, the lox recombinant plasmid was generated as described previously (29). To express multiple proteins from a single promoter, a cDNA encoding Zeocin, the foot-and-mouth disease virus peptide (30), and mCherry were assembled by PCR and subcloned downstream of the ubiquitin promoter between the loxP sites. To regulate recombination by tamoxifen, a fragment encoding Cre-ERT2 was inserted into pFUW. Five different constructs (Open Biosystems) were tested for Cdc37 shRNA activity, and three positive Cdc37 shRNA were selected and used in experiments. Oligonucleotides encoding this sequence were annealed and inserted into a restriction site downstream of the second loxP site. Lentivirus was generated as described previously (31), and titers were determined by GFP expression after serial dilution.
Cell Cultures—Doxycycline-inducible stable ESCs were generated using lentivirus expressing FUrtTAIPW or FTREW containing GFP, GFP-ICD, and GFP-ICDΔ494–579 and cultivated with 1 μg/ml puromycin. A transduction efficiency of 100% was confirmed by fluorescence microscopy in doxycycline-treated cells. To obtain a tamoxifen-inducible Cdc37-knockdown ESC line, the lox recombinant plasmid containing Cdc37 shRNA and Cre-ERT2 was introduced into Ryk+/+, Ryk-/- GFP, and GFP-ICD ESCs by lentiviral infection, and ESCs were cultured with 20 μg/ml Zeocin for 6 days. To induce Cdc37 shRNA expression, cells were treated with tamoxifen and analyzed by Western blotting with anti-Cdc37 antibodies and fluorescence microscopy after two or three passages. 293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 100 μg/ml penicillin-streptomycin and transiently transfected with the indicated plasmids using the calcium phosphate precipitation method.
Immunoprecipitation, Western Blotting, and Reagents—Cells were lysed in a lysis buffer containing 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 10 mm sodium pyrophosphate, 10 mm β-glycerophosphate, 1 mm sodium orthovanadate, 10% glycerol, and protease inhibitors (Roche Applied Science). Subcellular fractions were extracted using the ProteoExtract™ kit (Calbiochem). For immunoprecipitation, cell lysates were incubated with a specific antibody for 2 h at 4 °C and then with Protein A/G-agarose beads (Pierce) overnight. For detection of Ryk ICD ubiquitination, immunoprecipitation was performed using lysis buffer containing 0.1% SDS. Immunoprecipitates were eluted using SDS sample buffer and separated using 8 or 10% SDS-PAGE. After blocking, the blots were incubated with a primary antibody and then with a peroxidase-conjugated secondary antibody. The bound secondary antibody was then detected using enhanced chemiluminescence (ECL) reagent (Santa Cruz Biotechnology). Antibodies and antisera were obtained from the following manufacturers: monoclonal anti-Myc (9E10), anti-hemagglutinin (HA; F-7), anti-Hsp90 (α + β), anti-ubiquitin, and anti-actin antibodies (Santa Cruz Biotechnology); monoclonal anti-E-cadherin and anti-Cdc37 antibodies (BD Biosciences); monoclonal anti-FLAG tag antibody (Sigma); and monoclonal and polyclonal GFP antibodies (Invitrogen). Ryk antibody has been described previously (17). The corresponding secondary antibodies were from Jackson ImmunoResearch Laboratories. The following reagents were also used: 17-AGG (BIOMOL); MG132 and lactacystin (Calbiochem); and doxycycline, 4-hydroxytamoxifen, and FLAG peptide (Sigma).
RESULTS
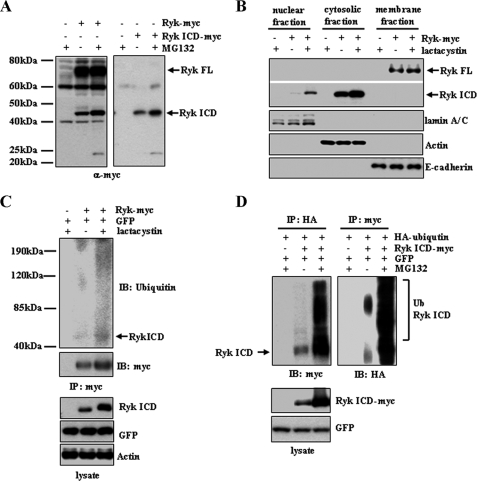
Proteasomal Degradation of the Ryk ICD Prevents Nuclear Localization—We evaluated the stability of the cleaved Ryk ICD using overexpression assays in HEK293T cells. To examine levels of the cleaved C-terminal fragment in cells, a full-length Ryk or the Ryk ICD construct with a C-terminal Myc tag was transiently transfected into HEK293T cells, and cells were treated with or without proteasomal inhibitors MG132 or lactacystin for 6 h. Treatment with MG132 increased levels of the ∼42-kDa form of the Ryk ICD protein (Fig. 1A). Western blot analysis of the cytosolic and nuclear extracts also showed increased levels of the Ryk ICD in cells treated with lactacystin compared with dimethyl sulfoxide-treated control cells (Fig. 1B), suggesting that the Ryk ICD undergoes proteasomal degradation. When proteasomal degradation is inhibited, the Ryk ICD is detected in the nucleus. To determine whether the Ryk ICD is ubiquitinated prior to degradation, the Ryk ICD-Myc immunoprecipitates from cytosolic extracts were subjected to Western blotting with the anti-ubiquitin antibody. A high molecular mass form of the ubiquitinated Ryk ICD was detected in lactacystin-treated 293T cells expressing wild-type Ryk-Myc (Fig. 1C), indicating that cytoplasmic levels of the Ryk ICD may be down-regulated through ubiquitin-dependent proteasome-mediated degradation. To confirm that the smearing high molecular mass bands represented the ubiquitinated Ryk ICD, we transfected 293T cells with constructs encoding HA-ubiquitin and the Ryk ICD-Myc. These cells were treated with or without proteasomal inhibitors MG132 for 6 h. As expected, smearing high molecular mass protein bands containing HA-ubiquitin were detected in the Ryk ICD-Myc immunoprecipitates (Fig. 1D). Treatment with MG132 increased the amount of high molecular mass smearing. In the anti-HA immunoprecipitate, Western blotting using anti-Myc antibody suggests that the smearing high molecular mass proteins are the Myc-tagged Ryk ICD. These results together support the idea that the Ryk ICD is polyubiquitinated.
FIGURE 1.
Proteasomal degradation of the Ryk ICD. A, treatment with the proteasome inhibitor MG132 increases Ryk ICD protein levels. 293T cells were transfected with a plasmid encoding C-terminal Myc-tagged wild-type Ryk or an empty control vector. MG132 was added 6 h prior to lysis, and lysates were analyzed by Western blotting with an anti-Myc antibody. A similar experiment using 293T cells transfected with a Myc-tagged Ryk ICD construct or a control vector was also performed (right panel). B, nuclear localization of the Ryk ICD increases in cells treated with the proteasome inhibitor lactacystin. Nuclear, cytosolic, and membrane extracts of cells transfected with Ryk constructs or control plasmids were subjected to Western blotting using an anti-Myc antibody for full-length (FL) Ryk and Ryk ICD proteins. lamin A/C, actin, and E-cadherin were used as protein markers for nuclear, cytosolic, and membrane fractions, respectively. C, the Ryk ICD is ubiquitinated. Cells transfected with Ryk-Myc and GFP were treated with the proteasome inhibitor lactacystin. Cytosolic extracts were immunoprecipitated (IP) using an anti-Myc antibody and analyzed by Western blotting using antibodies against ubiquitin (Ub) and Myc. Antibodies against GFP and actin served as controls. D, 293T cells were transfected with HA-ubiquitin and Ryk ICD-Myc or empty vector and then treated with MG132 for 6 h prior to lysis. Cell lysates were subjected to immunoprecipitation using anti-HA or anti-Myc antibodies, followed by Western blotting (IB) using anti-Myc or anti-HA antibodies.
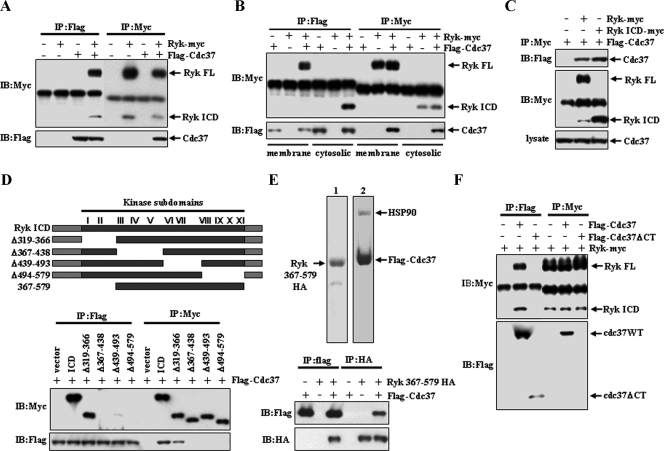
Ryk Associates with Cdc37-Hsp90—Next, we undertook a yeast two-hybrid screen to identify potential interacting proteins with the Ryk ICD and identified the co-chaperone protein Cdc37 (data not shown). This interaction was confirmed in 293T cells transiently transfected with FLAG-Cdc37 and full-length Ryk-Myc (Fig. 2A). Both the full-length and cleaved Ryk ICD were detected in FLAG-Cdc37 immunoprecipitates. Furthermore, immunoprecipitation analysis following subcellular fractionation showed the presence of full-length Ryk in the membrane and the ICD in the cytoplasm. Both of these proteins bind to Cdc37 (Fig. 2B). Detection of the Ryk ICD in FLAG-Cdc37 immunoprecipitates suggests that the Ryk ICD mediates the association with Cdc37. This was confirmed when Cdc37 was detected in the anti-Myc immunoprecipitate from 293T cells transfected with plasmids encoding the C-terminal Myc-tagged ICD of Ryk and FLAG-Cdc37 (Fig. 2C).
FIGURE 2.
Ryk binds to Cdc37. A, FLAG-tagged Cdc37 and Myc-tagged Ryk were transfected into 293T cells. Binding of Cdc37 and Ryk was analyzed by immunoprecipitation (IP) followed by Western blotting (IB) using anti-Myc or anti-FLAG antibodies. FL, full-length. B, a Western blot analysis of anti-FLAG and anti-Myc immunoprecipitates from membrane and cytosolic extracts is shown. C, Cdc37 binds to the ICD region of Ryk. 293T cells expressing Cdc37-FLAG were transfected with the wild-type Ryk, Ryk ICD, or a control plasmid. An anti-Myc immunoprecipitate from whole cell lysates was analyzed by Western blotting using anti-FLAG and anti-Myc antibodies. D, Cdc37 recognizes the tyrosine kinase domain of Ryk. A schematic of the wild-type and mutant of Ryk ICD constructs shows that the Ryk tyrosine kinase domain consists of 11 conserved subdomains (top panel). Each mutant was constructed by deletion of the indicated subdomains. Wild-type and subdomain I/II-deleted mutants, but not other mutants, bind Cdc37 (bottom panel). Plasmids encoding wild-type and mutant ICD proteins or a control vector were cotransfected with FLAG-Cdc37. Cell lysates were analyzed by immunoprecipitation and Western blotting with anti-Myc or anti-FLAG antibodies. E, the fragment encoding amino acids 367–579 of the Ryk kinase domains is sufficient to bind to Cdc37 directly. HA-tagged Ryk-(367–579) was purified from bacteria. Cdc37-FLAG was purified by immunoprecipitation from 293T cells transfected with the construct encoding Cdcd37-FLAG. A Coomassie Blue-stained SDS-polyacrylamide gel containing purified proteins (top panel) confirms the purity of the protein preparations. Arrows indicate purified Ryk-(367–579) (lane 1) and Cdc37-FLAG (lane 2). Purified protein samples were incubated under the indicated conditions, and immunoprecipitation and Western blot analyses were performed using anti-HA or anti-FLAG antibody (lower panel). F, the Cdc37 C terminus is required for binding of Ryk. 293T cells expressing wild-type (WT) Ryk-Myc were transfected with plasmids encoding either wild-type Cdc37-FLAG or C-terminal (CT) deletion mutant (Cdc37ΔCT-FLAG). Anti-FLAG and anti-Myc immunoprecipitates were subjected to Western blotting with anti-Myc and anti-FLAG antibodies.
Cdc37 interacts directly with the kinase domain of several protein kinases, including Raf-1, B-Raf, Akt1, and Cdk4, via its N terminus to induce their activation and stability as well as with the Hsp90 chaperone via its C terminus (21, 25, 32, 33). We used a deletion mutant series to identify the Cdc37-binding site in the Ryk ICD kinase domain. Immunoprecipitation using anti-FLAG and anti-Myc antibodies showed that ICDΔ367–438 (lacking subdomains III–V), Δ439–493 (lacking subdomains VI and VII), and Δ494–579 (lacking subdomains VII–XI) mutants did not bind to FLAG-Cdc37, whereas the wild-type Ryk ICD-Myc and ICDΔ319–366-Myc (lacking subdomains I and II) did (Fig. 2D), demonstrating that Ryk ICD subdomains III–XI (amino acids 367–579) in the kinase domain, but not those in the N terminus, are required for Cdc37 binding. This observation was verified by showing that glutathione S-transferase fusions of HA-tagged subdomains III–XI expressed in E. coli (Fig. 2E, top left panel) are sufficient for Cdc37 binding (Fig. 2E). A FLAG-Cdc37 protein was purified from 293T cells transiently transfected with FLAG-Cdc37 using anti-FLAG immunoprecipitation followed by elution with FLAG peptide (Fig. 2E, top right panel). To map the Cdc37 domain binding to Ryk, a plasmid encoding FLAG-tagged Cdc37 deleted at the C terminus (FLAG-Cdc37ΔCT) was transfected into 293T cells expressing wild-type Ryk-Myc. Ryk was detectable in immunoprecipitates derived from cells transfected with full-length FLAG-Cdc37 but not with FLAG-Cdc37ΔCT (Fig. 2F, IP:FLAG), and the findings were corroborated by an inverse immunoprecipitation using Ryk-Myc (IP: Myc). These results indicate that the C terminus of Cdc37 is required for the Cdc37/Ryk interaction.
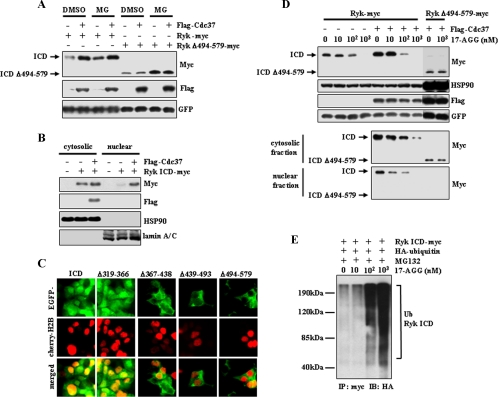
Both Stability and Nuclear Localization of Ryk ICD Require Interaction with Cdc37—The observation that the Ryk ICD binds to Cdc37 in the cytoplasm suggests that Ryk ICD stability is regulated by Cdc37, a hypothesis supported by evidence that proteasomal degradation of Cdk4 is prevented by binding to Cdc37-Hsp90 (24). To determine whether Cdc37 binding alters Ryk ICD stability, we cotransfected 293T cells with plasmids encoding wild-type Ryk-Myc or mutant RykΔ494–579-Myc protein that cannot bind Cdc37 with FLAG-Cdc37 or control vector (Fig. 3A). The level of the wild-type Ryk ICD increased in Cdc37-expressing cells compared with mock-transfected cells. In contrast, expression levels of Ryk Δ494–579 were not altered by Cdc37 overexpression. Treatment of cells with MG132 increased the level of both the wild-type and mutant Ryk ICD but had no effect on increased levels of the wild-type Ryk ICD seen in the presence of overexpressed Cdc37. This data strongly suggests that Cdc37 protects the Ryk ICD from proteasomal degradation. Furthermore, Western analysis of nuclear extracts showed increased nuclear localization of the Ryk ICD in FLAG-Cdc37-transfected cells (Fig. 3B). Consistent with this result, GFP-tagged ICDΔ367–438, ICDΔ439–493, and ICDΔ494–579 mutant proteins were localized to the cytoplasm, whereas GFP-wild-type ICD and GFP-ICDΔ319–366 proteins, which are capable of binding Cdc37, were detected in the nucleus and cytoplasm (Fig. 3C). To determine whether Hsp90 regulates Ryk ICD stability, 17-AGG, which inhibits Hsp90 chaperone function and disrupts formation of the Hsp90-Cdc37 complex (14), was added to 293T cells transiently transfected with wild-type and mutant (Δ494–579) Ryk-Myc plus FLAG-Cdc37 or mock-transfected cells. 17-AGG treatment significantly blocked increases in levels of the wild-type Ryk ICD protein seen following overexpression of FLAG-Cdc37 in a dose-dependent manner (Fig. 3D, top panels). In contrast, no significant change in the level of the Ryk ICDΔ494–579 protein was detected following 17-AGG treatment. These observations suggest that Hsp90 is directly related to the stability of the Ryk ICD. To determine whether 17-AGG-mediated reduction of Ryk ICD levels is caused by ubiquitin-mediated proteasomal degradation, 293T cells cotransfected with HA-ubiquitin and Ryk ICD-Myc were treated with 17-AGG in the presence of MG132. Ubiquitination of the Ryk ICD was determined by immunoprecipitation using anti-Myc antibody and Western blot analysis using anti-HA antibody. Ubiquitination of the Ryk ICD increased in a 17-AGG dosage-dependent manner. This data indicates that the inhibition of Hsp90 induces ubiquitination of the Ryk ICD as well as a decrease in Ryk ICD levels.
FIGURE 3.
Cdc37 increases protein levels and nuclear localization of the Ryk ICD, and an Hsp90 inhibitor inhibits the activity of Cdc37 on the Ryk ICD. A, expression of Cdc37-FLAG increases Ryk ICD protein levels, whereas it has no effect on Ryk ICDΔ494–579, which does not bind to Cdc37. Addition of MG132 (MG) 6 h prior to cell lysis prevents degradation of both wild-type Ryk and Ryk ICDΔ494–579. 293T cells were transfected with FLAG-Cdc37, wild-type Ryk, or Ryk ICDΔ494–579-Myc plus GFP. DMSO, dimethyl sulfoxide. B, Cdc37 enhances nuclear localization of the Ryk ICD. Subcellular fractions from 293T cells transiently expressing Ryk ICD-Myc alone or together with Cdc37-FLAG were analyzed by Western blotting using anti-Myc, anti-FLAG, anti-Hsp90, and anti-lamin A/C antibodies. Detection of lamin A/C confirmed the purity of nuclear fractionation. C, the Ryk ICD and ICDΔ319–366 are localized in the nucleus and cytoplasm, whereas Ryk ICD mutants that do not bind to Cdc37 (see Fig. 2D) remain cytoplasmic. The Ryk ICD and its mutants were fused with GFP. Nuclei were visualized using mCherry-tagged histone H2B. EGFP, enhanced green fluorescent protein. D, treatment with the Hsp90 inhibitor 17-AGG decreases the levels of the wild-type Ryk ICD but not Ryk ICDΔ494–579. Cells were transiently transfected with wild-type Ryk-Myc alone or together with Cdc37-FLAG or with Ryk ICDΔ494–579-Myc plus Cdc37-FLAG. Cells were then treated with the indicated concentration of 17-AGG for 24 h prior to lysis. ICD levels were analyzed by Western blotting of samples from whole cell lysates (top panels) and subcellular fractions (bottom panels). 17-AGG treatment had no effect on expression levels of Cdc37, Hsp90, and GFP. E, treatment with the Hsp90 inhibitor 17-AGG induces ubiquitination of the Ryk ICD. 293T cells transfected with HA-ubiquitin (Ub) and/or Ryk ICD-Myc were treated for 6 h with MG132 and different doses of 17-AGG. Immunoprecipitation (IP) was performed using anti-Myc antibody followed by Western blotting (IB) using anti-HA antibody.
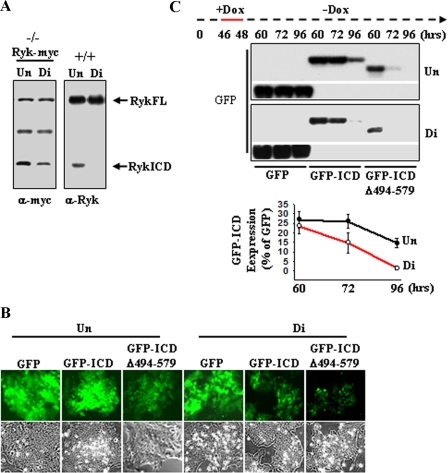
Disruption of Ryk Binding to Cdc37 Reduces Ryk ICD Levels during Neural Differentiation of Embryonic Stem Cells—We demonstrated recently that Ryk ICD levels are higher in differentiating neurons than in neural progenitor cells isolated from mouse embryo cortices (17). Although generation of the Ryk ICD protein likely requires intramembrane cleavage, the observation that the Ryk ICD binds to Cdc37-Hsp90 raises the possibility that the cleaved Ryk ICD may be unstable and rapidly degraded in neural progenitors. To examine this possibility, we used mouse ESCs, which can be differentiated into neural progenitors. To evaluate a functional link between Ryk ICD stability and Cdc37-Hsp90 during neural differentiation, we initially analyzed levels of the Ryk ICD in the Ryk+/+ ESCs and in the Ryk-Myc-expressing Ryk-/- ESCs, which were generated by transduction of lentivirus expressing Ryk-Myc into Ryk-/- ESC lines derived from Ryk-/- blastocysts. We found decreased levels of Ryk ICD when cells were cultured under monolayer neural differentiation conditions for 6 days, compared with those cultured as undifferentiated ESCs in LIF-containing medium (Fig. 4A, Un). In such differentiation conditions, neural progenitor markers Sox1 and nestin were detectable, but the immature neuron markers TUJ1 and mature neuron marker MAP2 (data not shown; supplemental figure) were not, indicating the differentiation of ESCs into neural progenitor cells. To address the stability of the intracellular Ryk ICD, we employed doxycycline-inducible expression of GFP-fused forms of ICD and monitored GFP, GFP-ICD, and GFP-ICDΔ494–579 signals induced by doxycycline treatment in Ryk-/- ESCs during neural differentiation using fluorescence microscopy. Fluorescent signals were noticeably decreased in both Ryk-/- GFP-ICD and GFP-ICDΔ494–579 ESCs undergoing differentiation compared with undifferentiated cells (Fig. 4B, top panels). In contrast, Ryk-/- GFP ESCs showed a similar GFP signal intensity when cultured under stem cell maintenance or differentiation conditions. We next evaluated ICD stability during differentiation using Western blot analysis. Specifically, GFP-ICD or GFP-ICDΔ494–579 was induced with doxycycline for 2 h, and then, following doxycycline withdrawal, GFP-ICD levels were measured at time intervals in cells cultured under either stem cell maintenance or differentiation conditions (Fig. 4C). Peak levels of GFP-ICD, first detected at 60 h, were reduced 18-fold at 96 h in ESCs cultured under differentiation conditions compared with ESCs cultured in the presence of LIF. Furthermore, GFP-ICDΔ494–579 levels were not detectable at 72 or 96 h. These results verify the hypothesis that the Ryk ICD is rapidly degraded during differentiation into neural progenitor cells.
FIGURE 4.
The Ryk ICD is down-regulated during differentiation of ESCs. A, protein levels of the Ryk ICD are decreased during neural differentiation of ESCs. Ryk-/- ESCs transduced by lentivirus expressing wild-type Ryk-Myc (left panel) and Ryk+/+ ESCs (right panel) were cultured under undifferentiation (Un) and neural differentiation conditions (Di). Cellular lysates were blotted with anti-Myc antibody or anti-Ryk antibody, which recognizes the C terminus of Ryk. FL, full-length. B, a visualization of the Ryk ICD during differentiation of ESCs is shown. GFP fusions of the Ryk ICD and its mutant were placed under control of a doxycycline (Dox)-inducible promoter in a lentiviral vector. Constructs were transduced into Ryk-/- ESCs. After doxycycline treatment for 24 h under undifferentiation conditions, Ryk-/- GFP, Ryk-/- GFP-ICD, and Ryk-/- GFP-ICDΔ494–579 ESCs were further cultured under neural differentiation conditions for 4 days. Expression of GFP, GFP-ICD, and GFP-ICDΔ494–579 was analyzed using fluorescence microscopy. Top panels, the fluorescence of GFP-ICD and GFP-ICDΔ494–579 was reduced under neural differentiation culturing conditions. Bottom panels, phase-contrast images are shown. C, the reduction of stability of the Ryk ICD during differentiation is shown. ESCs cultured under undifferentiation and neural differentiation conditions were incubated with doxycycline for 2 h (red line), further cultured without doxycycline, and lysed at the indicated times. Cell lysates were analyzed by Western blotting with anti-GFP antibody (upper panel), and ICD levels were quantified by densitometry (lower panel). Each error bar represents the mean ± S.D. of three independent experiments.
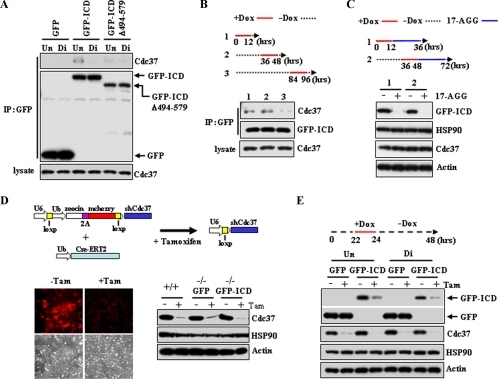
We next examined the Ryk/Cdc37 interaction during neural differentiation of ESCs (Fig. 5A). Cdc37 levels remained constant during the course of differentiation. However, endogenous Cdc37 was detected in GFP-ICD immunoprecipitates only in undifferentiated cultures. Immunoprecipitates of GFP-ICDΔ494–579 revealed no binding to Cdc37 in lysates from cells cultured under either stem cell maintenance or differentiation conditions, consistent with the requirement of amino acids 494–570 of the Ryk ICD for Cdc37 binding. To further analyze the dynamics of the Ryk ICD/Cdc37 interaction during differentiation, Ryk GFP-ICD expression was induced by doxycycline for 12 h at different stages of differentiation, and Cdc37/GFP-ICD association was determined by immunoprecipitation. The GFP-ICD/Cdc37 interaction was reduced during ESC differentiation, as evidenced by reduced Cdc37 levels in immunoprecipitated samples at 84–96 h compared with those at 0–12 and 36–48 h (Fig. 5B, compare lane 3 with lanes 1 and 2). To confirm that reduction in ICD levels is dependent upon Hsp90 during ESC differentiation, 17-AGG was added to cultures after induction of GFP-ICD levels by doxycycline at different times during differentiation. The GFP-ICD protein was undetectable after a 24-h exposure to 17-AGG (Fig. 5C).
FIGURE 5.
Treatment of cells with the Hsp90 inhibitor 17-AGG and knockdown of Cdc37 reduce Ryk ICD levels in ESCs. A, Cdc37 binds to GFP-ICD, but not GFP-ICDΔ494–579 in ESCs. Prior to lysis, cells were treated with doxycycline (Dox) for 12 h on day 5 of culture growth. Mouse monoclonal anti-GFP immunoprecipitates (IP) were blotted with anti-Cdc37 antibodies or rabbit polyclonal anti-GFP antibodies. Un, undifferentiation conditions; Di, neural differentiation conditions. B, binding of Cdc37 to the Ryk ICD decreases during differentiation. Differentiating cells were cultured without doxycycline for various times (dashed line) and then treated with doxycycline for 12 h (red lines) to induce GFP-ICD expression. Each lysate was immunoprecipitated using anti-GFP antibodies followed by Western blotting with anti-Cdc37 and anti-GFP antibodies. C, the Hsp90 inhibitor 17-AGG affects GFP-ICD stability. After doxycycline treatment for the indicated times (red lines), Ryk-/- GFP-ICD ESCs were cultured with 17-AGG (1 μm) for 24 h (blue line). Western analysis with a GFP antibody shows a significant decrease in ICD levels at 36 and 72 h. Antibodies against Hsp90, Cdc37, and actin served as controls. D, a generation of ESCs inducibly expressing shRNA of Cdc37 is shown. A scheme of inducible expression of Cdc37 shRNA via tamoxifen (Tam)-induced recombination is shown in the top panel. Tamoxifen-induced recombination leads to reduction of mCherry signals (bottom left panel). Ryk+/+, Ryk-/- GFP, and Ryk-/- GFP-ICD ESCs infected with constructs expressing Cdc37 shRNA and Cre-ERT were treated with vehicle or tamoxifen and lysed after three passages. Western blot analysis confirms tamoxifen-regulated Cdc37 knockdown (bottom right panel). Ub, ubiquitin. E, GFP-ICD stability is reduced by Cdc37 knockdown. Ryk-/- GFP and Ryk-/- GFP-ICD ESCs expressing the empty construct or encoding Cdc37 shRNA were cultured under undifferentiation or neural differentiation conditions, incubated with doxycycline for the indicated time (red line) to induce GFP or GFP-ICD, and analyzed by Western blotting using antibodies against the indicated proteins.
To determine whether Cdc37 regulates Ryk ICD stability in ESCs, we generated a Cdc37 knockdown in ESCs using tamoxifen-induced recombination. In this modified inducible RNA interference system, a DNA fragment encoding fusions of Zeocin, retroviral 2A protein, and mCherry, flanked by loxP sites, is inserted between the U6 promoter and shRNA targeting Cdc37, and a Cre-estrogen receptor fusion protein is expressed in a separate vector. Both lentiviral vectors are then transduced into ESCs to make a Zeocin-resistant cell line. After tamoxifen-induced recombination, Zeocin-2A-mCherry was floxed out, and expression of Cdc37 shRNA was driven by the U6 promoter (Fig. 5D, top panel). Loss of the mCherry fluorescence signal indicated that a complete recombination event had occurred (Fig. 5D). Levels of Cdc37 were noticeably reduced in Ryk+/+, Ryk-/-GFP, and Ryk-/-GFP-ICD ESC lines (Fig. 5D). When protein expression was induced by doxycycline in Ryk-/- ESC lines, GFP-ICD levels were significantly reduced in a tamoxifen-induced Cdc37 knockdown compared with control ESCs, whereas the level of GFP alone was not affected by Cdc37 knockdown (Fig. 5E). Taken together, these results strongly suggest that Ryk ICD stabilization is regulated through Cdc37-Hsp90 chaperone activity during neural differentiation of ESCs.
DISCUSSION
Intramembrane proteolysis of a single-pass transmembrane receptor can modulate signaling in one of two ways: by regulating the respective signaling pathway via the cleaved ICD or by degradation of cleavage products (18, 34). Our previous data has shown that the receptor Ryk undergoes intramembrane proteolysis, resulting in the generation of a cleavage product, the Ryk ICD (17). The cleaved Ryk ICD exhibits nuclear localization in differentiating premature neurons both in vitro and in vivo. The Ryk-deficient mouse exhibits defects in neuronal differentiation during neurogenesis. These results can be recapitulated in differentiation of ESCs. When Ryk+/+ and Ryk-/- ESCs undergo neural differentiation, no significant difference in the number of nestin-positive cells were observed between the Ryk+/+ and Ryk-/-cell cultures, whereas the number of TUJ1-positive neurons were reduced in Ryk-/- culture (supplemental figure). We have demonstrated that the cleavage of Ryk and nuclear signaling of the Ryk ICD are critical for neurogenesis (17). Therefore, regulation of Ryk ICD stability in Ryk signaling, such as increased stability in differentiating premature neurons and decreased stability in neural stem cells, may be important for neurogenesis.
We showed that the cleaved Ryk ICD undergoes ubiquitination and proteasomal degradation in 293T cells and that the Ryk ICD is rapidly degraded during differentiation of ESCs into neural progenitor cells. Therefore, this implies that the stability of the cleaved ICD is down-regulated to prevent the activation of Ryk signaling in neural progenitor cells. Furthermore, the up-regulated stability of the Ryk ICD may activate Ryk signaling in cells differentiating from neuronal progenitor cells into neurons. This is supported by the observation that overexpression of the Ryk ICD increases the number of neurons differentiating from neural progenitor cells (17).
We identified Cdc37 as a Ryk ICD-interacting protein. Cdc37, a Hsp90 co-chaperone, is implicated in stabilization and/or activation of Hsp90 client kinases, including Cdk4 and Raf, which function in cell cycle control, development, and transcriptional regulation (23, 35, 36). Given that Cdc37 knockdown or treatment with17-AGG, which disrupts Hsp90 association with client kinases, eliminates measurable levels of the Ryk ICD protein and that Ryk is a catalytically inactive protein kinase (14), we conclude that Cdc37-Hsp90 is required for Ryk ICD stabilization rather than for activation of a kinase.
Cdc37 interacts with kinase catalytic domains through a glycine-rich loop (GXGXXG) in the N terminus (37). This loop is highly conserved among protein kinases or RTKs. However, the QXGXXG sequence in Ryk subdomain I contains a glutamine substitution of the first glycine (14). Our Ryk deletion mutants of subdomains I and II and a point mutant with alanine substituted for the remaining glycines (QXAXXA; data not shown) interacted with Cdc37, but C-terminal deletion mutants did not. Thus, the N-terminal lobe of Ryk is not necessary for Cdc37 binding. The requirement for the C-terminal lobe of kinases to bind the Cdc37-Hsp90 complex has been reported previously. V600E B-Raf, which is mutated in the activation segment of the C-terminal portion, shows increased association with Cdc37 compared with wild-type B-Raf. Treatment of cells with 17-AGG also decreased the interaction between V600E B-Raf and Cdc37 (25). Additionally, a recent study showed that the Cdc37-Hsp90 complex binds to the kinase domain of Lck via structures present in both the N- and C-terminal lobes (38).
We observed that association of the Ryk ICD with Cdc37 decreased during neural differentiation of ESCs, consistent with decreasing levels of the Ryk ICD. Treatment with 17-AGG significantly decreased Ryk ICD protein levels during early differentiation, showing the binding of the Ryk ICD and Cdc37 at that time. These results show a positive correlation between Ryk ICD stability and Cdc37-Hsp90 association, although it is not known how binding affinity of Cdc37-Hsp90 to Ryk is regulated during neural differentiation of ESCs.
In conclusion, we identified Cdc37 as a Ryk ICD-interacting protein and demonstrated that stability of the cytoplasmic Ryk ICD is regulated by Cdc37-Hsp90 activity, which then regulates translocation to the nucleus. Our findings suggest that Cdc37-Hsp90-mediated stabilization of the Ryk ICD is a crucial event in the process of Ryk-mediated signal transduction. These results also contribute to understanding a molecular mechanism whereby the receptor Ryk transduces a signal directly from the cell surface to the nucleus via a cleaved ICD during neurogenesis.
Supplementary Material
Acknowledgments
We are grateful for the suggestions and comments from Drs. Martin Pera and Michael Stallcup. We also thank Steve Stacker for providing the Ryk knock-out mice and Dr. Robert L. Matts (Oklahoma State University) for providing Cdc37 plasmids.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
Footnotes
The abbreviations used are: RTK, receptor protein-tyrosine kinase; LIF, leukemia inhibitory factor; ICD, intracellular domain; ESC, embryonic stem cell; shRNA, short hairpin RNA; 17-AGG, 17-(allylamino)-17-demethoxygeldanamycin.
References
- 1.Miller, J. R. (2002) Genome Biol. 3 reviews3001.1-3001.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moon, R. T., Bowerman, B., Boutros, M., and Perrimon, N. (2002) Science 296 1644-1646 [DOI] [PubMed] [Google Scholar]
- 3.Wodarz, A., and Nusse, R. (1998) Annu. Rev. Cell Dev. Biol. 14 59-88 [DOI] [PubMed] [Google Scholar]
- 4.Clevers, H. (2006) Cell 127 469-480 [DOI] [PubMed] [Google Scholar]
- 5.Logan, C. Y., and Nusse, R. (2004) Annu. Rev. Cell Dev. Biol. 20 781-810 [DOI] [PubMed] [Google Scholar]
- 6.Harris, K. E., and Beckendorf, S. K. (2007) Development (Camb.) 134 2017-2025 [DOI] [PubMed] [Google Scholar]
- 7.Yoshikawa, S., McKinnon, R. D., Kokel, M., and Thomas, J. B. (2003) Nature 422 583-588 [DOI] [PubMed] [Google Scholar]
- 8.Patthy, L. (2000) Trends Biochem. Sci. 25 12-13 [DOI] [PubMed] [Google Scholar]
- 9.Green, J. L., Inoue, T., and Sternberg, P. W. (2008) Cell 134 646-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeble, T. R., Halford, M. M., Seaman, C., Kee, N., Macheda, M., Anderson, R. B., Stacker, S, A., and Cooper, H. M. (2006) J. Neurosci. 26 5840-5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, Y., Shi, J., Wu, C., Zhang, H., Zheng, Z., Geiger, T. L., Nuovo, G. L., Liu, Y., and Zheng, P. (2005) Nat. Neurosci. 8 1151-115916116452 [Google Scholar]
- 12.Lu, W., Yamamoto, V., Ortega, B., and Baltimore, D. (2004) Cell 119 97-108 [DOI] [PubMed] [Google Scholar]
- 13.Schmitt, A. M., Shi, J., Wolf, A. M., Lu, C. C., King, L. A., and Zou, Y. (2006) Nature 439 31-37 [DOI] [PubMed] [Google Scholar]
- 14.Halford, M. M., and Stacker, S. A. (2001) BioEssays 23 34-45 [DOI] [PubMed] [Google Scholar]
- 15.Katso, R. M., Russell, R. B., and Ganesan, T. S. (1999) Mol. Cell. Biol. 19 6427-6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halford, M. M., Armes, J., Buchert, M., Meskenaite, V., Grail, D., Hibbs, M. L., Wilks, A. F., Farlie, P. G., Newgreen, D. F., Hovens, C. M., and Stacker, S. A. (2000) Nat. Genet. 25 414-418 [DOI] [PubMed] [Google Scholar]
- 17.Lyu, J., Yamamoto, V., and Lu, W. (2008) Dev. Cell 15 773-780 [DOI] [PubMed] [Google Scholar]
- 18.Kopan, R., and Ilagan, M. X. (2004) Nat. Rev. Mol. Cell Biol. 5 499-504 [DOI] [PubMed] [Google Scholar]
- 19.Ni, C. Y., Murphy, M. P., Golde, T. E., and Carpenter, G. (2001) Science 294 2179-2181 [DOI] [PubMed] [Google Scholar]
- 20.Sardi, S. P., Murtie, J., Koirala, S., Patten, B. A., and Corfas, G. (2006) Cell 127 185-197 [DOI] [PubMed] [Google Scholar]
- 21.Grammatikakis, N., Lin, J. H., Grammatikakis, A., Tsichlis, P. N., and Cochran, B. H. (1999) Mol. Cell. Biol. 19 1661-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter, T., and Poon, R. Y. (1997) Trends Cell Biol. 7 157-161 [DOI] [PubMed] [Google Scholar]
- 23.MacLean, M., and Picard, D. (2003) Cell Stress Chaperones 8 114-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, H., Goode, T., Iakova, P., Albrecht, J. H., and Timchenko, N. A. (2002) EMBO J. 21 930-941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grbovic, O. M., Basso, A. D., Sawai, A., Ye, Q., Friedlander, P., Solit, D., and Rosen, N. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 57-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols, J., Evans, E. P., and Smith, A. G. (1990) Development (Camb.) 110 1341-1348 [DOI] [PubMed] [Google Scholar]
- 27.Ying, Q. L., Nichols, J., Chambers, I., and Smith, A. (2003) Cell 115 281-292 [DOI] [PubMed] [Google Scholar]
- 28.Urlinger, S., Baron, U., Thellmann, M., Hasan, M. T., Bujard, H., and Hillen, W. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7963-7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura, A., Meissner, A., Dillon, C. P., McManus, M., Sharp, P. A., Van Parijs, L., Jaenisch, R., and Jacks, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10380-10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang, J., Qian, J. J., Yi, S., Harding, T. C., Tu, G. H., Van Roey, M., and Jooss, K. (2005) Nat. Biotechnol. 23 584-590 [DOI] [PubMed] [Google Scholar]
- 31.Lois, C., Hong, E. J., Pease, S., Brown, E. J., and Baltimore, D. (2002) Science 295 868-872 [DOI] [PubMed] [Google Scholar]
- 32.Scholz, G., Hartson, S. D., Cartledge, K., Hall, N., Shao, J., Dunn, A. R., and Matts, R. L. (2000) Mol. Cell. Biol. 20 6984-6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terasawa, K., and Minami Y. (2005) FEBS J. 272 4684-4690 [DOI] [PubMed] [Google Scholar]
- 34.Brown, M. S., Ye, J., Rawson, R. B., and Goldstein, J. L. (2000) Cell 100 391-398 [DOI] [PubMed] [Google Scholar]
- 35.Schulte, T. W., Blagosklonny, M. V., Ingui, C., and Neckers, L. (1995) J. Biol. Chem. 270 24585-24588 [DOI] [PubMed] [Google Scholar]
- 36.Stepanova, L., Leng, X., Parker, S. B., and Harper, J. W. (1996) Genes Dev. 10 1491-1502 [DOI] [PubMed] [Google Scholar]
- 37.Terasawa, K., Yoshimatsu, K., Iemura, S., Natsume, T., Tanaka, K., and Minami, Y. (2006) Mol. Cell. Biol. 26 3378-3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prince, T., and Matts, R. L. (2004) J. Biol. Chem. 279 39975-39981 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.