Abstract
Mycobacterium tuberculosis is thought to undergo transformation into its non-replicating persistence state under the influence of hypoxia or nitric oxide (NO). This transformation is thought to be mediated via two sensor histidine kinases, DosS and DosT, each of which contains two GAF domains that are responsible for detecting oxygen tension. In this study we determined the crystal structures of the first GAF domain (GAF-A) of DosS, which shows an interaction with a heme. A b-type heme was embedded in a hydrophobic cavity of the GAF-A domain and was roughly perpendicular to the β-sheet of the GAF domain. The heme iron was liganded by His-149 at the proximal heme axial position. The iron, in the oxidized form, was six-coordinated with a water molecule at the distal position. Upon reduction, the iron, in ferrous form, was five-coordinated, and when the GAF domain was exposed to atmospheric O2, the ferrous form was oxidized to generate the Met form rather than a ferrous O2-bound form. Because the heme is isolated inside the GAF domain, its accessibility is restricted. However, a defined hydrogen bond network found at the heme site could accelerate the electron transferability and would explain why DosS was unable to bind O2. Flavin nucleotides were shown to reduce the heme iron of DosS while NADH was unable to do so. These results suggest that DosS is a redox sensor and detects hypoxic conditions by its reduction.
Mycobacterium tuberculosis is still one of the most dreaded pathogens in existence, and one of the reasons for its success as a pathogen lies in its ability to persist for years within its host. One-third of the world population is estimated to carry M. tuberculosis in the dormant form (1), and while in this state, the pathogen is insensitive to most available chemotherapy. M. tuberculosis has been shown to undergo a metabolic transformation to its non-replicating persistence state under the influence of environmental stimuli such as hypoxia or nitric oxide (2). Recent studies have implied that CO is also an environmental trigger of mycobacterial persistence (3, 4).
A two-component regulatory system mediates the genetic response to oxygen limitation and NO exposure in M. tuberculosis (5). The regulatory system consists of two sensor proteins, DosS and DosT and the cognate response regulator DosR (6, 7). DosS and DosR are also known as DevS and DevR (6). DosS and DosT are histidine kinases that undergo autophosphorylation in response to an environmental change, and subsequently transduce the signal to DosR (a transcriptional regulator). DosS and DosT each contain two GAF domains at the N-terminal sensory domain and an HATPase (histidine kinase-like ATPase) domain at its C terminus (8). Hypoxia sensing by DosS or DosT is presumably carried out through the GAF domains.
GAF domains are small molecule binding domains found in many proteins from various organisms and are known to play important roles as regulatory elements. Many GAF-containing proteins have two GAF domains in tandem and the two domains have separate functions, binding a cyclic nucleotide and dimerization (9, 10). The two GAF domains in the M. tuberculosis DosS and DosT proteins are also arranged in tandem. The first GAF domain (GAF-A) is expected to contain a heme (8) while the second GAF domain (GAF-B) is not suitable to bind a small ligand such as cyclic nucleotides (11). Thus it is possible that DosS and DosT might be controlled through the binding of oxygen or NO at the GAF-A domain.
Heme-based sensors are a diverse group of signal transduction proteins that respond to gases like O2 and CO (12), and four distinct families of heme-based sensor proteins have been characterized. The bacterial oxygen sensors, FixL and HemAT have a PAS fold and a globin fold domain, respectively, to hold the heme in place (13, 14). The CO sensor CooA, belongs to the cAMP receptor protein family (15) while the NO sensor soluble guanylate cyclase, contains an H-NOX domain (16). These proteins have quite distinct heme-binding protein scaffolds. Although the GAF domain is structurally similar to the PAS domain, its heme-binding mode differs from that of the PAS domain (17).
Analyses using electron paramagnetic resonance (EPR)2 and UV-visible spectroscopy suggested that DosS and DosT recognize oxygen from O2, NO, or CO through its binding to the heme (18–21). Under low O2 concentrations, the heme exists in the deoxy form and DosS becomes active while atmospheric O2 concentrations inactivate DosS (20, 21). Recently the crystal structure of DosT GAF-A showed its direct interaction to O2 through the heme (17). However, a detailed sensing mechanism for DosS based on a molecular structure has not yet been reported. There is controversy over whether DosS senses O2 directly or not. As a heme-based O2 sensor with a Kd value of 3 μm, DosS is active when it exists in an unliganded ferrous form, while its conversion to the oxy form results in its inactivation (20). Alternatively, DosS can be thought of as a redox sensor (21). During aerobic respiration, DosS exists as the Met (Fe3+) form and under hypoxic conditions a reductant converts DosS to its reduced active form to induce the Dos regulon.
In this study, the crystal structures of the GAF-A domain of DosS were determined showing a heme-binding structure. Structural and spectroscopic analyses of GAF-A upon reduction and oxidation suggested that DosS is a redox sensor, which undergoes conversion to an oxidized Met form with a six-coordinated iron upon exposure to atmospheric O2. Finally, it was found that flavin nucleotides were capable of reducing DosS. The results imply that DosS recognizes hypoxic conditions in a cell via the accumulation of a reducing agent.
EXPERIMENTAL PROCEDURES
Gene Cloning and Protein Purification—The DosS gene (Rv3132c) was cloned from the chromosomal DNA of the M. tuberculosis H37Rv strain by polymerization chain reaction (PCR). The region encoding the GAF-A domain of DosS (Asp-63 to Lys-210) was amplified by PCR using primer sets carrying BamHI and EcoRI restriction sites (5′-CGGGATCCGGACCTGGAGGC-3′ and 5′-CCGGAATTCACTTAGCCTGCTGG-3′) and was cloned into the pHIS-parallel vector (22), a His-fusion protein expression vector containing a recombinant TEV protease cleavage site. The integrity of the insertion was verified by direct DNA sequencing.
The expression of selenomethionine (SeMet)-labeled DosS GAF-A was induced by 0.2 mm isopropyl-1-thio-β-d-galactopyranoside in the E. coli strain B834 (Novagen) grown at the log phase (A600 = 0.6) with M9 medium containing 50 μg/ml SeMet at 37 °C. The cells were grown for an additional 24 h at 25 °C. For the native protein, the E. coli strain BL21 (DE3), carrying the recombinant plasmid, was grown in LB medium. The cells were grown for an additional 12 h at 18 °C after induction. The expressed protein was purified by affinity chromatography using an Ni-NTA column (Qiagen). The recombinant protein was digested using rTEV at 10 °C in the presence of 0.5 mm EDTA and 1 mm dithiothreitol. After complete digestion, the hexa-His tag was removed using an Ni-NTA column. Gel filtration was performed with a Superdex G75 column (GE Healthcare) equilibrated with 20 mm Tris-HCl, pH 7.5, and the fractions containing GAF-A protein were collected and concentrated using Centriprep YM10 (Millipore) for crystallization screening and spectroscopic analysis. The purified protein contained an additional five amino acids (GAMDP) at the N terminus arising from the cloning procedure.
Crystallization and Data Collection—Crystallization of the purified protein was initially performed with commercially available sparse-matrix screens (Hampton Research and Emerald Biostructures) using the sitting-drop vapor diffusion method at 21 °C. After optimization, the best crystals of SeMet-substituted DosS GAF-A were obtained under conditions of 0.1 m Tris-HCl, pH 7.0 containing 17% (w/v) PEG 6000 and 0.2 m calcium acetate, with a protein concentration of 33 mg/ml. The crystal of the native protein was obtained from the drop containing 1.5 μl of 33 mg/ml protein and 1.5 μl of a reservoir solution, which contained 20% PEG4000 and 0.2 m calcium acetate in Tris-HCl, pH 7.0. The crystals were transferred to a cryoprotectant solution containing 19% (w/v) PEG 6000 for the SeMet crystal, or 22% (w/v) PEG4000 for the native crystal, in addition to 0.2 m calcium acetate and 16% glycerol, prior to x-ray data diffraction collection.
For the reduced form of the GAF-A, the crystals were obtained by the addition of 50 mm sodium dithionite to the crystallization conditions for the SeMet-substituted protein. The color of the crystal was red while that of the native crystal was dark brown. The crystal was soaked in the cryoprotectant containing 50 mm sodium dithionite before mounting on the beam. The air-oxidized crystal was obtained from the drop of the reduced form after maintenance under aerobic conditions. The color of the crystal turned to dark brown again. The crystal of the cyanide complex was obtained with a crystallization solution containing 20 mm potassium cyanide.
After a fluorescence scan, single anomalous x-ray dispersion data for a SeMet crystal were collected at a wavelength corresponding to the Se absorption peak (0.9793 Å) using a Quantum 210 CCD detector on the beam line 4A at the Pohang Accelerator Laboratory. The data were indexed, integrated, and scaled using the HKL2000 package (23). The crystal belongs to the space group P212121 with the unit cell parameters of a = 35.8 Å, b = 87.0 Å, and c = 101.2 Å. The diffraction data for the native, reduced, air-oxidized, and cyanide complex forms were collected at a wavelength of 1.0000 Å. The unit cell parameters for the crystals are similar to those for the SeMet crystal. The statistics of the crystals are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data set | SeMet | Native | Reduced | Air-oxidized | Cyanide-bound |
|---|---|---|---|---|---|
| Experimental Data | |||||
| X-ray source | PAL 4A | PAL 4A | PAL 4A | PAL 4A | PAL 4A |
| Wavelength (Å) | 0.9792 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit cell parameters (Å) | 35.7 × 87.0 × 101.2 | 36.0 × 86.9 × 100.4 | 36.7 × 86.2 × 101.9 | 36.4 × 86.8 × 100.8 | 36.6 × 84.4 × 100.5 |
| Resolution limit (Å) | 50–2.0 | 50–1.6 | 40–1.6 | 50–1.4 | 40–1.8 |
| Total reflections | 218,305 | 277,449 | 491,300 | 649,385 | 170,746 |
| Unique reflections | 40,262 | 41,345 | 40,766 | 55,575 | 29,219 |
| Redundancy | 5.4 | 6.7 | 12.1 | 11.7 | 5.8 |
| Completeness (%) | 98.3 | 96.8 | 93.2 | 87.1 | 98.3 |
| Rsymm (%)a | 11.8 | 7.3 | 6.9 | 7.3 | 6.2 |
| Average I/σ (I) | 16.2 | 26.0 | 50.3 | 43.9 | 33.1 |
| Refinement Details | |||||
| Resolutions (Å) | 2.0 | 1.6 | 1.6 | 1.4 | 1.8 |
| Refractions (working) | 18,107 | 39,191 | 38,636 | 52,671 | 27,686 |
| Refractions (test) | 991 | 2,088 | 2,076 | 2,821 | 1,485 |
| Rworkb | 0.213 | 0.200 | 0.210 | 0.192 | 0.215 |
| Rfreeb | 0.278 | 0.234 | 0.234 | 0.217 | 0.260 |
| Number of waters | 260 | 171 | 291 | 249 | |
| R.m.s. deviation from ideal geometry | |||||
| Bond length (Å) | 0.008 | 0.012 | 0.011 | 0.013 | 0.010 |
| Bond angle (°) | 1.175 | 1.427 | 1.445 | 1.450 | 1.324 |
| Average B factors (Å) | |||||
| Molecule A (main/side chain) | 15.1 (13.9/16.4) | 17.5 (15.9/19.1) | 32.6 (31.1/34.1) | 22.3 (20.2/24.5) | 30.9 (28.8/33.0) |
| Molecule B (main/side chain) | 11.9 (10.8/13.1) | 15.7 (14.0/17.3) | 29.5 (27.9/31.1) | 21.5 (19.5/23.5) | 30.1 (28.7/31.6) |
| Waters | 17.6 | 25.7 | 39.3 | 31.3 | 38.0 |
Rsym = Σ|Ii – <I>|/ΣI where Ii is the intensity of the ith observation and <I> is the mean intensity of the reflections
Rwork = Σ||Fobs| – |Fcalc||/Σ Fobs|, crystallographic R factor, and Rfree = Σ||Fobs| – |Fcalc||/Σ|Fobs| when all reflections belong to a test set of randomly selected data
Structural Determination—The structure of SeMet-substituted GAF-A was determined by the single anomalous dispersion (SAD) method at a resolution of 2.0 Å. Six Se atoms were identified in the asymmetric unit using SOLVE (24) and the initial phase had a figure of merit (FOM) of 0.279. Density modification and subsequent automated model building were done with RESOLVE (25), increasing FOM to 0.635 with 61% (187 amino acid residues) of the residues built. The RESOLVE-built partial model was used as a guide to build the remainder of the protein manually into density-modified electron density maps with the program COOT (26). Refinement with isotropic displacement parameters was performed with Refmac5 (27) in the CCP4 suite (28). The Rwork and Rfree values of the refined structure were 0.213 and 0.278, respectively. The structures of the native, reduced, air-oxidized, and cyanide complexes were determined by the Molecular Replacement method with AMORE (29) using the SeMet-substituted crystal structure as the template at resolutions of 1.6 Å, 1.6 Å, 1.4 Å, and 1.8 Å, respectively. The crystallographic data statistics are summarized in Table 1. The final models have been deposited in the worldwide Protein Data Bank (PDB) (30) under the PDB ID code 2W3D, 2W3E, 2W3F, 2W3G, and 2W3H for the SeMet-substituted, oxidized native, reduced and its air-oxidized-, and cyanide-bound forms, respectively.
Reduction and Oxidation—All of the absorption spectra were measured on a UV-visible spectrophotometer (Optizen 3220UV, Mecasys). 10 μm of DosS GAF-A in Tris-HCl, pH 7.5 buffer was subjected to wavelength scanning. Oxidation and reduction of the GAF-A domain was performed by the addition of 0.1 mm potassium ferricyanide and 1 mm sodium dithionite, respectively. For atmospheric O2 exposure, the cuvette, filled to two-thirds its volume with the reduced GAF-A solution, was inverted three times to ensure a thorough mixing with air bubbles. Reduction of GAF-A by NADH and FMN was examined by the addition of 100 μm of NADH or 8 μm of FMN in the presence of 1.5 units of Clostridium kluyveri diaphorase (Sigma), 10 mm KCl, and 100 μm of NADH for FMN reduction. The effects of electron carriers on the reduction of GAF-A in the presence of sodium dithionite were performed by addition of 2 μm of FMN, 2 μm of FAD, or 2 μm of menaquinone with 1 mm fresh sodium dithionite solution. A little excess dithionite was necessary to keep the reduced mixture from re-oxidizing because of the presence of diffused oxygen in the atmosphere.
RESULTS
The Protein Fold—The crystal structure of the heme-bound GAF-A domain (residue Asp-63 to Arg-210) in M. tuberculosis DosS has been determined using single-wavelength anomalous diffraction of the SeMet-substituted protein for experimental phase determination. The crystal structures of the protein in its native met, reduced ferrous, air-oxidized, and cyanide-bound forms have also been solved (Table 1).
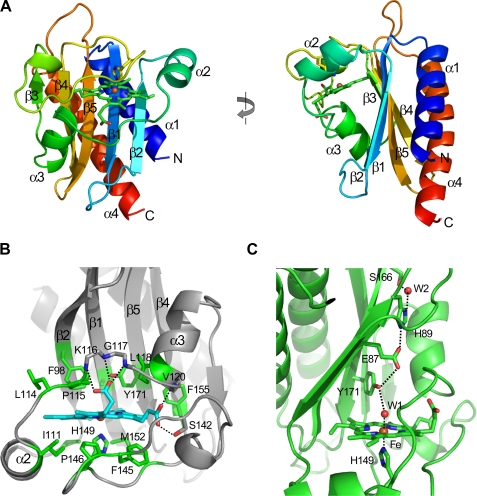
GAF-A consists of one five-stranded antiparallel β-sheet and four α-helices (Fig. 1A). The strand order of the sheet is 3-4-5-1-2 and two helices (α1 and α4) at the N terminus and C terminus are located on one side of the sheet. A peptide region connecting strands β2 to β3 at each end of the sheet contains two helices (α2 and α3) and crosses over to the other side of the sheet with a space existing between the sheet and a loop connecting the α2 and α3 helices. Two loops connecting strands β1 to β2, and strands β3 to β4, cover the space at the top and bottom positions completing the inside cavity. A heme group is tightly packed into the cavity. In the crystal structure, two GAF-A molecules (Mol-A and Mol-B) were found in an asymmetric unit. Two conformations of the molecules were almost identical except for a slight shift of helix α3 and the loop region between strands β1 and β2, indicating possible movement of these regions (supplemental Fig. S1A).
FIGURE 1.
DosS GAF-A structure and its heme interaction. A, ribbon diagram of DosS GAF-A showing a β-α-β structure with five-stranded antiparallel β-sheet. The left figure was rotated by 90°. The plane of the heme is perpendicular to the sheet. B, heme is surrounded by hydrophobic residues, and the propionic groups interact with main chain amide groups. C, heme iron has bonded with His-149 and a water molecule, which interacts to His-89 through Tyr-171 and Glu-87.
The overall folding of the GAF-A domain of DosS is similar to the GAF-B domain of DevS from M. smegmatis, except for the absence of a strand in front of the α3-helix completing the six-stranded β-sheet, which is a key feature of a GAF-domain (supplemental Fig. S2, A and B). The canonical GAF domain has a curved six-stranded β-sheet forming a half-barrel structure containing a cyclic nucleotide; however, the GAF-A sheet is five-stranded and rather flat (see Fig. 1A). The GAF-A domain consists of the sheet and two α-helices on each side of the sheet with the secondary structure order of α-β(2)-α(2)-β(3)-α. It is topologically similar to the PAS domain found in the E. coli protein Dos. Both GAF and PAS domains belong to the Profilin-like domain family, an α+β protein with α-β-α layers. The canonical GAF domain consists of α(2)-β(3)-α-β(3)-α while PAS has α-β(2)-α(2)-β(3) structure (supplemental Fig. S2, C and D). The GAF-A domain has a relatively long loop between strands β3 and β4 compared with the PAS domain while the canonical GAF domain has an α-helix in the corresponding loop. Although, topologically, the GAF-A domain of DosS is similar to a PAS domain, its overall folding is that of the canonical GAF domain.
Heme Environment—In the crystal structure of the GAF-A domain, a b-type heme was found in the cavity between the β-sheet and the loop region covering the sheet, with liganding of the iron by H149 from a long loop connecting the β3- and β4-strands at the proximal position of the heme. The plane of the porphyrin rings is roughly perpendicular to the sheet (Fig. 1A). The heme binding in the GAF-A domain differs from that in the PAS domain, in which a heme is inserted into a crevice between the β-sheet and an α-helix. The plane of the porphyrin ring is parallel to the β-sheet (supplemental Fig. S2D). In the GAF-A domain of DosS, the heme group is embedded in a defined hydrophobic space surrounded by Ile-103, Ile-111, Phe-145, Pro-146, and Met-152 at the proximal position (at a distance of 3.5–3.6 Å to the heme) and residues Ala-85, Phe-98, Ile-121, Leu-114, Pro-115, Phe-155, and Tyr-171 at the distal position (Fig. 1B). Two heme propionate groups have hydrogen bonds to a loop connected to the α3 helix of GAF-A. One binds the main chain amide groups of Lys-116, Gly-117, and Leu-118 and the other bonds with the amide groups of Val-120 and the hydroxyl group of Ser-142.
In the native crystal structure, the iron is six-coordinated with five nitrogen atoms from the porphyrin rings and the imidazole ring of His-149, and one oxygen atom from a water molecule at the distal position (Fig. 2A). The iron atoms are essentially coplanar with the four pyrrole nitrogen atoms, as expected for a six-coordinated low-spin iron. The bond distances from the iron to the imidazole ring of His-149, and the water molecule are 2.1–2.2 Å and 2.2–2.3 Å, respectively (Fig. 3A). Interestingly, in the SeMet-substituted GAF-A structure, the distance from the water molecule at the distal position to the iron is 2.8 Å (Fig. 3B). This rather long Fe-H2O distance has been observed in horseradish peroxidase isozyme C, with its Resonance Raman spectra showing a six-coordinated high-spin heme (31). Thus two different heme iron environments would be available in the six-coordinated state of GAF-A. In the distal position of the heme, Tyr-171 from the β5 strand is located close to the iron, and the hydroxyl group of Tyr-171 has a hydrogen bond to the water molecule coordinating the iron (Fig. 1C).
FIGURE 2.
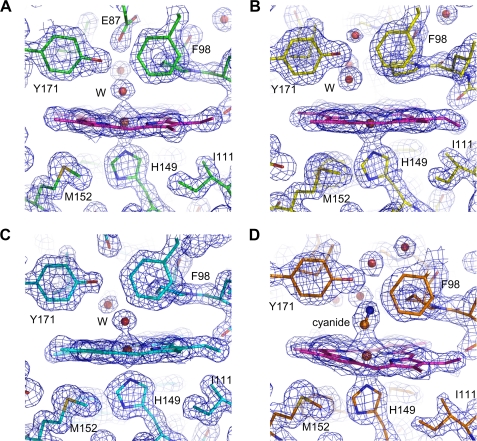
Electron density maps around the heme at DosS GAF-A. A, water molecule interacts with the heme iron at the distal position in the native structure. B, ferrous iron is five coordinated in the reduced form of GAF-A. C, upon air exposure, a water molecule ligands the heme iron at the distal position. D, cyanide interacts with the heme at the distal position, and Tyr-171 guides the cyanide interaction. The 2Fo-Fc electron density maps were contoured at the 1.5 σ level.
FIGURE 3.
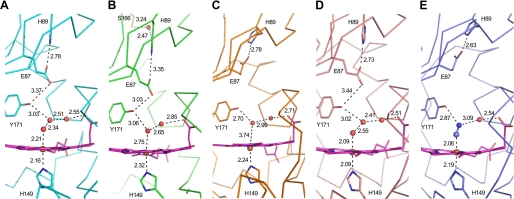
Interactions of ligand and residues at the heme sites. The numbers next to the dashed lines indicate the distances (Å) between two atoms. The residues are water molecules at the heme sites of the native (A), selenomethionine-substituted (B), reduced by sodium dithionite (C) air-oxidized (D), and cyanide complex (E) forms of DosS GAF-A. Mol-A is shown in each of the asymmetric units of the five crystals.
Because the heme group is buried in the hydrophobic cavity, the binding of a ligand at the heme would cause a rearrangement of the hydrogen bond network between water molecules and a few hydrophilic residues at the distal position. In the crystal structures, two or three water molecules are employed to connect the iron and a propionate group of the heme (Fig. 3). When two water molecules connect the iron and propionate group, the hydroxyl group of Tyr-171 has a hydrogen bond with the water molecule coordinating the iron (Fig. 3B). When three water molecules bridge the iron and the propionate group, Tyr-171 has a hydrogen bond with the second water molecule from the iron (Fig. 3A). It is worth paying attention to the two residues, Glu-87 and His-89 in the β1 strand. It is possible that the carboxyl group of Glu-87 could form a hydrogen bond with the hydroxyl group of Tyr-171 and the imidazole group of His-89 (Fig. 1C). Therefore, a hydrogen bonding network from the iron, in a hydrophobic environment, to the protein surface is completed via the water molecule coordinating the iron, Tyr-171, Glu-87, and His-89. When the iron transits between the ferric and ferrous states, this hydrogen bond network could be one possible pathway for electron transport.
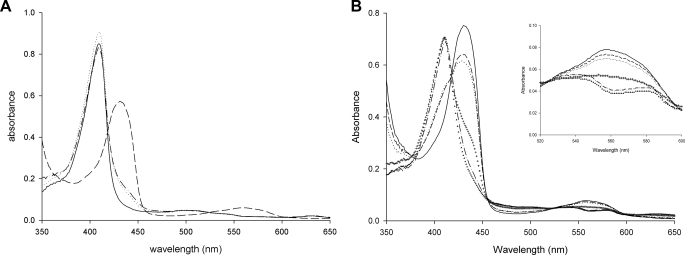
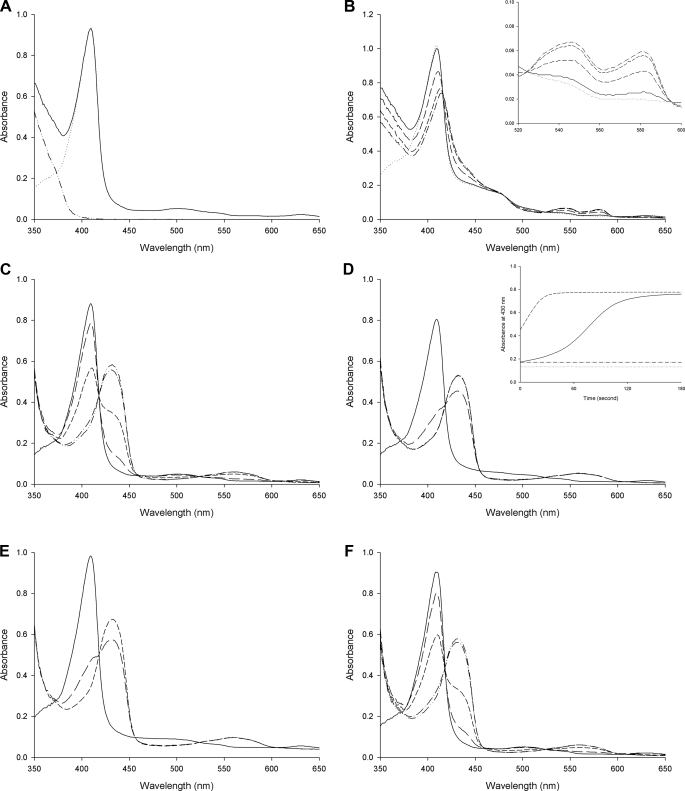
DosS Is Oxidized Rather Than Oxygenated upon Air Exposure— The oxidation status of DosS has been studied using UV-visible spectrum, EPR spectroscopy, and Resonance Raman spectroscopy (18–21). When UV-visible absorption spectra of the purified DosS GAF-A domain were measured, a typical Soret peak at 409 nm was observed with additional weak bands at 500 nm and 630 nm but no distinctive α and β bands (around 540 nm and 580 nm) (Fig. 4A). When GAF-A was treated with an excess of the oxidant Fe(CN6)3-, no changes were observed, thus confirming that the protein was in its ferric form. The reduction of GAF-A by sodium dithionite shifted the Soret peak to 430 nm and a new peak at 560 nm was observed (Fig. 4A). The ferrous form spectrum returned to that of the oxidized form upon treatment with ferricyanide. GAF-A was purified under aerobic conditions as an oxidized met form and did not bind an oxygen molecule as shown in the native crystal structure. The possibility that GAF-A could not bind an oxygen molecule because the heme was already in its ferric form cannot be ruled out.
FIGURE 4.
UV-visible spectra of the oxidized and reduced forms of DosS GAF-A. A, in the presence of ferricyanide (dotted line) as an oxidant, the spectrum of the purified DosS GAF-A (solid line) is not changed while the Soret peaks are shifted in the presence of sodium dithionite (dashed line) as a reductant. The reduced DosS is re-oxidized by ferricyanide (dashed line with a dot). B, oxidation of DosS GAF-A upon atmospheric O2 exposure is observed. Spectra of reduced GAF-A (solid line) and after mixing with air bubbles (dashed line) are shown with at 1, 2, 3, and 4 min (dots, crosses, dashed line with one dot, and triangles), respectively. The inset shows an expansion of the region around 560 nm.
A reduced DosS protein would either become oxidized or exist in its oxygen-bound form in the presence of O2. When the reduced DosS was exposed to atmospheric O2, the Soret peak returned to the wavelength of a native met form (409 nm) while the peak at 560 nm disappeared (Fig. 4B). This indicates that DosS has been oxidized to its ferric state. During the oxidation of DosS, peaks at 540 nm and 580 nm were observed (Fig. 4B). These peaks have been suggested as characteristic peaks of an oxygen-bound ferrous form of DosS (20). The crystal structures of the reduced form and the atmospheric O2-exposed form were determined after reduction. A heme-containing ferrous iron was obtained by soaking a crystal in a cryosolution containing dithionite. In the reduced structure, the heme iron is five-coordinated with an absence of water molecule interaction at the distal position (Fig. 2B). The iron atom is not coplanar with the four pyrrole nitrogen atoms and shifts to a proximal imidazole ring of His-149, as expected, with other reduced five-coordinated irons. The distance of the nearest water molecule to the iron is 3.7–3.8 Å, while that between the iron and imidazole nitrogen is unchanged (Fig. 3C). This water molecule has hydrogen bonds directly to the hydroxyl group of Tyr-171 and to the propionate group via another water molecule. The side chain of Glu-87 does not interact with Tyr-171 and is rotated to face His-89.
Upon reduction of the crystal by sodium dithionite under aerobic conditions, O2 does not bind at the heme. It may be due to slow, delayed O2 binding to the GAF-A heme or to low O2 concentrations because the dithionite consumed most of the O2 around DosS. A reduced crystal was kept under aerobic conditions and the color of the crystal changed from red to dark brown. The air-oxidized crystal structure shows a water molecule coordinating the iron at a distal position (Fig. 2C). The internal arrangement of water molecules and residues is similar to that of the native structure (Fig. 3D). The distance between the water molecule and iron is 2.1–2.2 Å and Tyr-171 has a hydrogen bond with the second water molecule from the iron. The side chain of Glu-87 is directed toward Tyr-171 with the result that the imidazole ring of His-87 rotates toward the heme side. Despite exposition to atmospheric O2, DosS did not bind an oxygen to the heme iron and re-oxidized to its ferric form. This unambiguously confirms that DosS is indeed a redox sensor and not a hypoxia sensor.
DosS was reported to bind CO and NO using its heme iron when in its reduced state (21). An attempt was also made to obtain a crystal structure bound to a diatomic ligand such as CO or NO, using a CO-saturated buffer and a solution containing an NO generating agent with DosS reduced by dithionite. However, the attempt was unsuccessful (data not shown). To determine possible changes of the heme environment for the DosS GAF-A domain upon ligand binding, the structure of a cyanide complex was determined. Cyanide coordinated the heme iron at the distal position (Fig. 2D). The cyanide bound directly to the iron and Tyr-171 with distances of ∼2.2 Å and 2.8 Å, respectively (Fig. 3E). It also interacted with the propionate group through a water molecule. The internal arrangement of atoms between the iron and the propionate group is similar to that of the oxidized structure containing three water molecules. The distance from the iron to the hydroxyl group of Tyr-171 is rather similar to their reduced form due to the short distance between the two cyanide atoms. The carboxyl group of Glu-87 rotates toward His-89 and forms a hydrogen bond with the imidazole group of His-89 (Fig. 3E). GAF-A employs Tyr-171 near the heme distal position to guide the diatomic ligand binding with a hydrogen bond. A cyanide complex could represent the binding of a diatomic ligand in the heme pocket.
Flavin Nucleotide Reduces DosS—Under hypoxic conditions, the electron transfer system of M. tuberculosis growing by aerobic respiration would cease to function due to the limitation O2, the final electron acceptor. Then electron carriers accumulated in this status could reduce the DosS kinase to its active form. This raises the question of a possible electron donor for DosS in a cell. NADH and reduced flavin mononucleotide (FMNred) were investigated as possible reductants for DosS. Upon reduction of DosS by dithionite, the Soret peak was shifted and a new peak at around 560 nm was observed. When NADH was added to GAF-A, there were no changes to the absorbance spectra (Fig. 5A). FMN can be reduced using diaphorase with NADH. When the GAF-A domain was treated with a mixture of FMN, diaphorase and NADH, the Soret peak shifted to 420 nm and peaks at 540 nm and 580 nm were observed with the consumption of NADH (Fig. 5B). This implies that FMNred can reduce DosS while NADH cannot. The differences in the characteristic peaks of DosS reduced by FMNred to that of DosS reduced by dithionite may be due to the low activity of diaphorase in producing FMNred. The reduction rate of DosS under aerobic conditions using diaphorase was slower than that using dithionite. Peaks at 540 nm and 580 nm during the oxidization of reduced DosS were observed (see Fig. 4B).
FIGURE 5.
Absorption spectra of DosS GAF-A upon reduction. A, spectrum of GAF-A (dotted line) is unchanged after addition of NADH (solid line). An increase of absorbance in the UV-region is due to NADH itself (dashed line with two dots). B, DosS GAF-A reduced by FMNred. Spectra for DosS GAF-A at 1, 2, and 3 min (long-dash, short-dashed, and dashed with one dot line, respectively) after addition of NADH (solid line) to the mixture of GAF-A, diaphorase, and FMN (dotted line) are shown. The inset shows an increase in the α and β peaks. C, reduction of DosS GAF-A by sodium dithionite. D, FMN accelerates the reduction of DosS GAF-A by dithionite. Spectra before and after addition of sodium dithionite in the present of FMN are shown. The inset shows an increase in absorbance at 430 nm by reduction of GAF-A due to the addition of FMN and dithionite (dashed line) compared with that with dithionite alone (solid line). The dotted line and dashed line with one dot show the absorbance values without dithionite. E, FAD accelerates the reduction of DosS GAF-A by dithionite. F, menaquinone did not accelerate the reduction of DosS GAF-A by dithionite. Spectra before (solid line) and at 1, 2, and 3 min (short-dashed and dashed lines with one and two dots) respectively, after addition of sodium dithionite are shown for C, D, E, and F.
Because dithionite can reduce FMN, DosS reduction was examined with FMN and dithionite. Although dithionite can reduce GAF-A directly, when FMN and dithionite were used together, a much faster reduction of GAF-A was observed compared with that using dithionite alone (Fig. 5, C and D). This suggests that the reduction of DosS via FMNred is more efficient than direct reduction by dithionite. Another flavin nucleotide, flavin adenine dinucleotide (FAD), was tested for DosS reduction. FAD can also accelerate the reduction of GAF-A by dithionite, suggesting that FAD can also reduce DosS (Fig. 5E). This implies that flavin nucleotides would be electron donors for DosS in the cell. To test the possibility of menaquinol as an electron donor, menaquinone was treated with dithionite for DosS reduction, however it did not accelerate the reduction of DosS GAF-A (Fig. 5F). Considering the location of menaquinol in a membrane, it is unlikely to act as an electron donor for DosS. Although early sequence analysis suggested the location of DosS with three putative transmembrane helices (6), which were all subsequently revealed as belonging to helices in GAF domains (11), DosS is likely a soluble protein.
DISCUSSION
The DosS and DosT proteins of M. tuberculosis are sensor histidine kinases that recognize environmental changes in diatomic gases such as O2, NO, or CO (20, 21). The GAF-A domains of these sensory kinases contain a heme at their N-terminal sensory domain and are responsible for the recognition of diatomic ligands. Analysis of the crystal structures of GAF-A revealed that the GAF domain binds heme differently, not only to PAS, but also to the other heme binding proteins such as globin and the H-NOX domain. In the other heme-binding proteins, the heme was found in a crevice between secondary structural elements with the result that the heme was almost parallel to the secondary structure. The amino acid residue liganding the heme iron at the proximal position, originated from a β-strand or α-helix. In GAF-A, the heme was embedded in the domain hydrophobic pocket and was roughly perpendicular to the β-sheet. The His-149 residue liganding the heme at the proximal position originated from one of the long loop regions surrounding the heme.
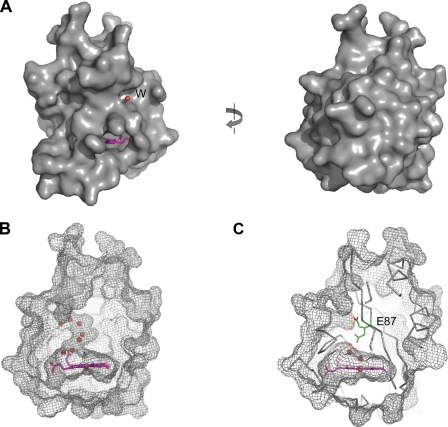
Unlike other heme-binding proteins, DosS GAF-A domain encloses its heme within the domain, making it difficult for a ligand molecule to access the heme (Fig. 6A). This might be one of the reasons why DosS is most likely not a direct oxygen sensor. In the hydrophobic cavity of GAF-A, the small amount of room left after heme binding was occupied by two or three water molecules. A defined water channel connecting the isolated iron in the cavity to an outside solvent was found between the α3 helix and a loop between strands β1 and β2 (Fig. 6B). Small ligands such as a cyanide ion or NO might gain access to the heme iron through this channel. Two different rotameric conformations for Glu-87 near the channel were found in the crystal structure, and it was noted that one alternative position of the Glu-87 side chain could block the channel (Fig. 6C).
FIGURE 6.
Heme is embedded in an inside cavity of DosS GAF-A. A, molecular surface of DosS GAF-A in the cyanide complex form. A water molecule (red circle) in the water channel and a part of the heme group (pink) are shown. The left figure was rotated by 180°. B, water molecules are in a channel connecting the heme site inside the GAF-A (mesh) and outside. C, side chain of Glu-87 (stick) has two alternative positions as found in the native conformation, and when the side chain is directed toward the heme, the channel could be blocked.
Because an oxygen molecule can only bind to the ferrous form of iron, it must access the heme before the iron oxidizes to its ferric form. DosS has a well-established hydrogen bond network, connecting the heme iron to the surface of the protein through Tyr-171, Glu-87, and His-89, which would accelerate the oxidation of the heme iron. Because FMN and FAD are much larger molecules than dithionite, these molecules could not reduce the DosS heme iron by direct interaction since accessibility to the heme is restricted. However, the iron was rapidly reduced by dithionite when FMN or FAD was present. This implies that there is an effective way of transferring an electron to the iron. One possible way is through Glu-87 and His-89. Reduced accessibility and faster oxidation could be the reasons why the heme in the DosS GAF-A domain was unable to bind an oxygen molecule directly.
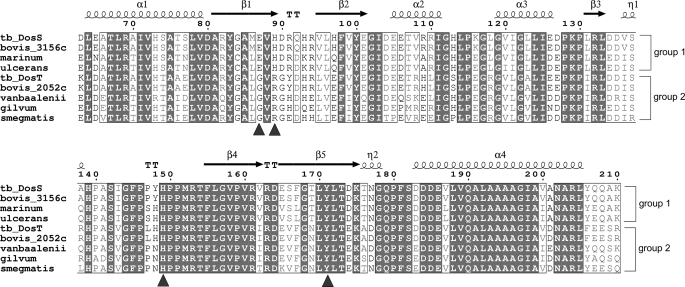
DosT, another hypoxia sensor in M. tuberculosis, has high sequence homology with DosS (61% identity) and both DosS and DosT share a response regulator, DosR. Because GAF-A contains a heme thought to be capable of sensing hypoxia or NO, the sequence differences between the two GAF-A domains of DosS and DosT may confer a different sensing feature. It has been suggested that DosT may act as an oxygen sensor while DosS is a redox sensor (21). The amino acid sequences of M. tuberculosis DosS GAF-A were compared with the GAF-A domains of DosT and of DosS homologues in other species of Mycobacterium (Fig. 7). The N- and C-terminal helices and β4 strand of the domain are mostly conserved while major differences were observed in strands of the β1 to β2 region, including Glu-87 and His-89. In DosT, the residues corresponding to Glu-87 and His-89 of DosS, are Gly and Arg, respectively. Although His-149 and Tyr-171 near the heme iron are conserved, the absence of Glu-87 would affect the hydrogen bond network from the heme iron. The oxidation of DosS is known to be faster than that of DosT, although the auto-oxidation rate of DosS in air was estimated as the half-life of oxy-DosS, being 4 h at 37 °C (20).
FIGURE 7.
Sequence alignment of DosS GAF-A and its homologues. Amino acid sequences of DosS (NP_217648) and DosT (NP_216543) GAF-A domains from M. tuberculosis and its corresponding regions in DosS homologues from M. bovis (NP_856801 for 3156c and NP_855702 for 2052c), M. marinum (YP_001849823), M. ulcerans (YP_906245), M. vanbaalenii (YP_952235), M. gilvum (YP_001135104), and M. smegmatis (YP_889487) were compared. Numbering was done using DosS of M. tuberculosis. The arrows and coils above the aligned sequences indicate the secondary structural elements of DosS GAF-A. DosS homologues could be divided into two groups: Group 1, including DosS, has a conserved Glu-86 and His-89, while Group 2, to which DosT belongs, has Gly and Arg conserved at these positions. Residues involved in interactions at the heme site are indicated by blue triangles. Multiple alignment was done using the T-coffee software and visualized using ESPript.
As DosT contains the smallest amino acid, Gly instead of Glu-87 in DosS, this could make the channel to the heme slightly wider than the DosS channel, thus facilitating its accessibility to O2. Consequently, DosT might be capable of sensing oxygen directly, unlike DosS. The nine GAF-A domains in the Mycobacterium DosS homologues could be divided into two groups: one group, including DosS, has a conserved Glu and His at the β1 strand, while the other group, to which DosT belongs, has Gly and Arg conserved at these positions (Fig. 7). This suggests that DosS and DosT are evolutionarily diversified proteins with distinct sensing characteristics.
During aerobic respiration, Mycobacterium utilizes O2 as final electron acceptor. Under hypoxic conditions, the electron transport system would be blocked and reduced electron carriers, such as NADH, would accumulate. As a redox sensor, DosS could recognize this condition by its reduction. Because NADH cannot reduce DosS directly while FMNred can, M. tuberculosis might require a cytosolic enzyme, which reduces FMN using NADH. Recently, it was reported that the M. tuberculosis Rv2540c DNA sequence encodes a bifunctional chorismate synthase capable of accelerating NADH-dependent FMN reduction (32). It suggests that DosS could be reduced by chorismate synthase using NADH as the reducing agent.
The abilities of DosS and DosT to sense O2, NO, and CO through their heme irons were characterized by various spectroscopic studies including UV-visible, EPR, and Resonance Raman spectroscopy. Under anaerobic conditions, the ferrous form of DosS showed a peak at 558 nm, in addition to the Soret peak at 430 nm, and upon treatment of O2 this characteristic single peak changed to a dual peak (542 and 578 nm) with a shift of the Soret peak to 416 nm (20). The ferrous form of the DosS GAF-A domain adopted a pure 5-coordinated high-spin configuration with peaks at 428 and 562 nm (18). Upon CO binding to the reduced GAF-A domain, the Soret peak shifted to 422 nm with the detection of a dual band at 540 and 570 nm. The dual peak at around 560 nm used to be interpreted as indicating the binding of a ligand such as O2 or CO. However, a sharp Soret peak (409 nm), α (575 nm), and β (536 nm) bands, which are indicative of a hexa-coordinated, high-spin heme protein, were observed with the ferric from of DosS, and these peaks then shifted to 430 nm and converged to 557 nm, respectively, upon reduction (21). When DosS GAF-A was reduced by dithionite, the Soret peak at 409 nm shifted to 430 nm and visible bands at 560 nm were observed (Fig. 4A). However, upon air-exposure, DosS converted to its oxidized form, the peak at 560 nm decreased and faint peaks at 540 nm and 580 nm remained (Fig. 4B). It is unlikely that the dual peaks represent an oxygen-bound form. The air-oxidized form was a six-coordinated ferric form binding a water molecule, as shown in the crystal structure, and in addition, the peaks at 540 nm and 580 nm were not increased during air exposure. Reduction of DosS by FMN using diaphorase and NADH also resulted in the increase of absorption peaks at 540 nm and 580 nm (Fig. 5B). This dual peak could be the characteristic peak of a reduced form of DosS. It is possible that two different types of the reduced form might exist as oxidized DosS, because two different distances were observed for the water molecule coordinating the iron in the native and SeMet-substituted structures.
The reduced form of DosS containing a ferrous iron represents the active form while the ferric form is inactive (20, 21). A dramatic conformational change in GAF-A was not found in the crystal structures upon reduction, which may be due to crystallographic packing restricting the possible conformation changes. The β5 strand of Mol-B and β2 strand of Mol-A are in close contact with Tyr-148 from Mol-A and Mol-B, respectively, in the neighboring asymmetric units (supplemental Fig. S1B). The five crystals used in this study showed almost identical crystallographic packing of Mol-A and Mol-B in an asymmetric unit with the same space group of P212121. All five Mol-B structures had a similar arrangement of the residues Tyr-171, Glu-87, and His-89 at the distal position possibly due to the crystallographic packing. Hydrogen bond networks around the heme irons in the Mol-A structures, which were less restricted in the crystal, were compared (Fig. 3C). The arrangement of water molecules and side-chains of the residues was changed upon reduction of GAF-A. The water molecule liganding the heme iron at a distance of 2.1 Å in the six-coordinated ferric form interacted with Tyr-171 through another water molecule and allowed the side chain of Glu-87 to direct toward Tyr-171. In the ferrous form, the water molecule near the iron, at a distance of 3.8 Å, formed a hydrogen bond with the hydroxyl group of Tyr-171, pushing the side chain of Glu-87 toward His-89. Consequently the side chain of His-87, facing the heme in its oxidized form, rotated in the opposite direction upon reduction. His-89 could also have a hydrogen bond with the main chain carbonyl oxygen of Asp-90 or the hydroxyl group of Ser-166 via a water molecule. These interactions maintain the local conformation of the β1-β2 loop region. Thus the internal rearrangement of the hydrogen bond network resulted in the movements of motifs such as the β1-β2 loop. Although dramatic conformational changes upon reduction of the GAF-A domain were not observed, a small change would be enough to control the kinase activity.
In this study, we have determined crystal structures for the DosS GAF-A domain, which showed a novel heme-binding feature. Upon reduction of DosS, the heme iron converted to a five-coordinated ferrous form. Atmospheric O2 exposure oxidized the heme iron of DosS rather than O2 binding at the heme. A flavin nucleotide would be the electron donor for DosS. These results suggest that DosS is a redox sensor and would sense hypoxic conditions through its reduction by a reducing agent such as FMNred, which accumulates due to the blocking of the electron transport system. When the reduced DosS is exposed to O2, it may be difficult for O2 to access the heme iron inside the GAF domain. However, a series of hydrophilic residues, which establish a hydrogen-bond network with water molecules from the iron to a solvent containing the O2, may facilitate oxidation of the heme iron. This could explain why this heme-based protein is not an oxygen sensor but a redox sensor. The M. tuberculosis Rv3132c gene product was named DosS after dormancy survival regulator sensor (33). Although the name is similar to the E. coli heme-containing PAS protein, Dos (direct oxygen sensor) (34), DosS is unlikely to sense oxygen directly.
Supplementary Material
Acknowledgments
We thank the staff members of beamline 4A at Pohang Accelerator Laboratory for assistance.
The atomic coordinates and structure factors (codes 2W3D, 2W3E, 2W3F, 2W3G, and 2W3H) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Education Science, and Technology, Republic of Korea.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: EPR, electron paramagnetic resonance; DosS, dormancy survival regulator sensor; R.m.s., root mean square; NTA, nitrilotriacetic acid.
References
- 1.Parrish, N. M., Dick, J. D., and Bishai, W. R. (1998) Trends Microbiol. 6 107-112 [DOI] [PubMed] [Google Scholar]
- 2.Wayne, L. G., and Sohasjey, C. D. (2001) Annu. Rev. Microbiol. 55 139-163 [DOI] [PubMed] [Google Scholar]
- 3.Kumar, A., Deshane, J. S., Crossman, D. K., Bolisetty, S., Yan, B., Kramnik, I., Agarwal, A., and Steyn, A. J. C. (2008) J. Biol. Chem. 283 18032-18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiloh, M. U., Manzanillo, P., and Cox, J. S. (2008) Cell Host Microbe 3 277-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherman, D. R., Voskuil, M., Schnappinger, D., Liao, R., Harrell, M. I., and Schoolnik, G. K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7534-7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasgupta, N., Kapur, V., Singh, K. K., Das, T. K., Sachdeva, S., Jyothisri, K., and Tyagi, J. S. (2000) Tuber Lung Dis. 80 141-159 [DOI] [PubMed] [Google Scholar]
- 7.Roberts, D. M., Liao, R. P., Wisedchaisri, G., Hol, W. G. J., and Sherman, D. R. (2004) J. Biol. Chem. 279 23082-23087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sardiwal, S., Kendall, S. L., Movahedzadeh, F., Rison, S. C. G., Stoker, N. G., and Djordjevic, S. (2005) J. Mol. Biol. 353 929-936 [DOI] [PubMed] [Google Scholar]
- 9.Martinez, S. E., Wu, A. Y., Glavas, N. A., Tang, X., Turley, S., Hol, W. G. J., and Beavo, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13260-13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez, S. E., Bruder, S., Schultz, A., Zheng, N., Schultz, J. E., Beavo, J. A., and Linder, J. U. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3082-3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, J., Cho, H. Y., Cho, H. J., Ko, I., Park, S. W., Baik, H., Oh, J., Eom, C., Kim, Y. M., Kang, B. S., and Oh, J. (2008) J. Bacteriol. 190 6795-6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilles-Gonzalez, and M., Gonzalez, G. (2005) J. Inorg. Biochem. 99 1-22 [DOI] [PubMed] [Google Scholar]
- 13.Gong, W., Hao, B., Mansy, S. S., Gonzalez, G., Gilles-Gonzalez, M. A., and Chan, M. K. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 15177-15182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, W., and Philips, G. N., Jr. (2003) Structure 11 1097-1110 [DOI] [PubMed] [Google Scholar]
- 15.Lanzilotta, W. N., Schuller, D. J., Thorsteinsson, M. V., Kerby, R. L., Roberts, G. P., and Poulos, T. L. (2000) Nat. Struct. Biol. 7 879-880 [DOI] [PubMed] [Google Scholar]
- 16.Pellicena, P., Karow, D. S., Boon, E. M., Marletta, M. A., and Kuriyan, J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 12854-12859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podust, L. M., Ioanoviciu, A., and Ortiz de Montellano, P. R. (2008) Biochemistry 47 12523-12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yukl, E. T., Ioanoviciu, A., Ortiz de Montellano, P. R., and Moënne-Loccoz, P. (2007) Biochemistry 46 9728-9736 [DOI] [PubMed] [Google Scholar]
- 19.Ioanoviciu, A., Yukl, E. T., Moënne-Loccoz, P., and Ortiz de Montellano, P. R. (2007) Biochemistry 46 4250-4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sousa, E. H., Tuckerman, J. R., Gonzalez, G., and Gilles-Gonzalez, M. (2007) Protein Sci. 16 1708-1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, A., Toledo, J. C., Patel, R. P., Lancaster, J. R., and Steyn, A. J. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11568-11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheffield, P., Garrard, S., and Derewenda, Z. (1999) Protein Expr. Purif. 15 34-39 [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. D 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger, T. C. (2000) Acta Crystallogr. D 56 965-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D 53 240-255 [DOI] [PubMed] [Google Scholar]
- 28.CCP4 (Collaborative Computational Project Number4) (1994) Acta Crystallogr. D 50 760-763 [DOI] [PubMed] [Google Scholar]
- 29.Navaza, J. (1994) Acta Crystallogr. A 50 157-163 [Google Scholar]
- 30.Berman, H. M., Henrick, K., and Nakamura, H. (2003) Nat. Struct. Biol. 10 980. [DOI] [PubMed] [Google Scholar]
- 31.Smulevich, G., Feis, A., Indiani, C., Becucci, M., and Marzocchi, M. P. (1999) J. Biol. Inorg. Chem. 4 39-47 [DOI] [PubMed] [Google Scholar]
- 32.Ely, F., Nunes, J. E., Schroeder, E. K., Frazzon, J., Palma, M. S., Santos, D., and Basso, L. A. (2008) BMC Biochem. 9 13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., and Schoolnik, G. K. (2003) J. Exp. Med. 198 705-713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delgado-Nixon, V. M., Gonzalez, G., and Gilles-Gonzalez, M. A. (2000) Biochemistry 39 2685-2691 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.