Abstract
The transcription factors GATA-1 and GATA-2 have been implicated in promoting differentiation of eosinophilic leukocytes. In this study, we examined the roles of GATA-1 and GATA-2 in activating transcription of the secretory ribonuclease, the eosinophil-derived neurotoxin (EDN/RNase 2). Augmented expression of both GATA-1 and GATA-2 was detected in eosinophil promyelocyte HL-60 clone 15 cells in response to biochemical differentiation with butyric acid. Deletion or mutation of one or both of the two consensus GATA-binding sites in the extended 1000-bp 5′ promoter of the EDN gene resulted in profound reduction in reporter gene activity. Antibody-augmented electrophoretic mobility shift and chromatin immunoprecipitation analyses indicate that GATA-1 and GATA-2 proteins bind to both functional GATA consensus sequences in the EDN promoter. Interestingly, RNA silencing of GATA-1 alone had no impact on EDN expression; silencing of GATA-2 resulted in diminished expression of EDN, and also diminished expression of GATA-1 in both butyric acid-induced HL-60 clone 15 cells and in differentiating human eosinophils derived from CD34+ hematopoietic progenitors. Likewise, overexpression of GATA-2 in uninduced HL-60 clone 15 cells resulted in augmented transcription of both EDN and GATA-1. Taken together, our data suggest that GATA-2 functions directly via interactions with the EDN promoter and also indirectly, via its ability to regulate the expression of GATA-1 in differentiating eosinophils and eosinophil cell lines.
Eosinophils remain among the most enigmatic of the mammalian leukocytes, although consensus opinion is that they contribute in some fashion to the pathophysiology of allergic asthma and gastrointestinal dysfunction (1). Despite disagreement as to their function, eosinophils are clearly identified in peripheral blood and tissues by their unique morphology and staining characteristics, including their characteristic bilobed nuclei and large refractile granules containing distinct, cationic secretory proteins.
Eosinophils differentiate from pluripotent stem cells in bone marrow, and they can be induced to develop from CD34 antigen-positive progenitor cells in vitro in response to cytokine stimulation (2). Whereas no unique events have been identified that define the eosinophil lineage specifically, transcription factors that contribute in various ways to eosinophil development include C/EBPα, C/EBPε, PU.1, and GATA-1 and -2, although the precise temporal and kinetic interplay between these factors remains to be elucidated (3, 4). The recent finding that ablation of a palindromic dblGATA enhancer-binding site in the promoter of the mouse Gata-1 gene results in eosinophil-lineage ablation in vivo (5) has resulted in a particular focus on the role of GATA transcription factors in promoting eosinophil development and differentiation. GATA-1 was first described as promoting transcription of erythroid genes (reviewed in Refs. 6-8), and Zon et al. (9) were the first to demonstrate expression of the hematopoietic GATA factors (GATA-1, GATA-2, and GATA-3) in isolated human eosinophils and eosinophilic cell lines. Kulessa et al. (10) found that overexpression of GATA-1 converts avian myelomonocytic cell lines into other lineages, including eosinophils, and Hirasawa et al. (11) discovered the same to be true for human primary myeloid progenitors, with GATA-2 compensating effectively for GATA-1 both in vitro and in vivo. Similarly, Iwasaki et al. (12) reported that GATA-2 overexpression in C/EBPα-expressing granulocyte/monocyte progenitor cultures likewise resulted in eosinophil commitment and differentiation. At the molecular level, functional GATA-1 enhancer sites direct the expression of the eosinophil granule major basic protein (13), and GATA-1 directs the eosinophil-specific expression of gp91phox, with GATA-2 functioning as a site-specific repressor (14).
Here we examine the role of GATA transcription factors in directing the expression of the human eosinophil secretory ribonuclease, the eosinophil-derived neurotoxin (EDN/RNase 2). EDN2 is a rapidly evolving RNase A ribonuclease with several intriguing biological activities (reviewed in Ref. 15). EDN was first identified as a neurotoxin (16, 17), as microgram quantities of this protein injected intrathecally into experimental animals resulted in Purkinje cell loss and concomitant ataxia. EDN also has antiviral properties (reviewed in Ref. 18), and recent studies suggest that EDN interacts with dendritic cells via signaling through the toll-like receptor TLR2 (19).
In an initial study utilizing minimal functional promoters, we determined that full transcriptional activity of EDN required the presence of one or more intron-based enhancer elements (20, 21). In subsequent studies, de Groot and co-workers (22, 23) demonstrated that both PU.1 and C/EBP consensus sites were crucial for EDN expression, and recently Wang et al. (24) identified regulatory elements in a 34-bp proximal promoter segment shared among primate EDNs. In this study, we focus on the extended 1000-bp 5′ promoter of EDN and utilize RNA silencing methods to explore the specific roles of GATA-1 and GATA-2 transcription factors in promoting its expression.
EXPERIMENTAL PROCEDURES
Preparation of Reporter Constructs—Human genomic DNA (Clontech) and DNA purified from clone BAC R-84C 10 (Bacpac Resources, Oakland, CA) were used as templates to amplify extended regions of genomic sequence 5′ to the known transcriptional start site of the EDN gene (20, 25). The 5′ regions were amplified by PCR in a 50-μl reaction containing 5 μl of 10× buffer, 1.5 μl of 10 mm dNTP, 1 μl of 50 mm MgSO4, 1.5 μl of 10 μm forward primer, 1.5 μl of 10 μm reverse primer, and 1.0 μl (1.25 units) of Pfx polymerase (Invitrogen). Reaction conditions were 94 °C for 2 min, followed by 35 cycles of 94 °C for 15 s, 55 °C for 30 s, and 68 °C for 3 min. Primers were as follows: EDN 1000 forward, 5′-CGG TAC CTT CCC TAA AGT CCC TGA AAA CTC C-3′; EDN 500 forward, 5′-CGG TAC CTA ATT ATC TAC AGA ATC TTG TGC CCC-3′; EDN 250 forward, 5′-CGG TAC CGC ATA TAG TTT TCA TCC AGA GTT T-3′; and EDN all reverse, 5′-GCT CGA GCT GTA AGA AAA GAA GAG AAG TAA C-3′. KpnI and XhoI restriction sites were added to the ends of both forward and reverse primers, respectively, to facilitate cloning. All the PCR products were gel-purified using GFX PCR DNA and Gel band purification kit (Amersham Biosciences Bioscience). A terminal nucleotide was added with the A-Addition kit (Qiagen), and the insert was ligated into the T/A cloning vector (Invitrogen), followed by transformation into TOPO 10 Escherichia coli (Invitrogen). Inserts were verified by sequencing, re-isolated by KpnI and XhoI digestion, and cloned into pGL3 Basic reporter vectors (Promega, Madison, WI).
Cell Culture—The HL-60 clone 15 human eosinophil promyelocytic leukemic cell line (26), purchased from American Type Culture Collection (Manassas, VA), was cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum, 25 mm HEPPSO (pH 7.6; Sigma), 2 mm l-glutamine (Invitrogen), 1% penicillin and streptomycin (Invitrogen) at 37 °C, and 5% CO2 in a humidified incubator. Unless otherwise indicated, cells at an initial concentration of 5 × 105 per ml were induced with 0.5 mm BA to differentiate them toward an eosinophilic phenotype for 2 days prior to transfection.
Transient Transfection Experiments—Culture medium as described was pre-warmed at 37 °C. Uninduced HL-60 clone 15 cells or cells that were BA-induced for 2 days as described above were centrifuged at 90 × g for 10 min at room temperature, and supernatant was discarded. Three million cells were suspended in 100 μl of room temperature Nucleofector solution V (Amaxa, Gaithersburg, MD), followed by addition of plasmid DNA (5 μg of pGL-3 constructs with 50 ng of pRL (Renilla luciferase) transfection control plasmid). The mixture was transferred to an Amaxa-certified cuvette, which was inserted into the cuvette holder of Nucleofector II machine (Amaxa). T-016 program was selected according to the manufacturer's recommendation, and cells were transfected, followed by suspension in 2 ml of pre-warmed culture medium and incubation for 5 h at 37 °C, 5% carbon dioxide prior to harvesting for luciferase assay.
Dual Luciferase Assays—The transiently transfected cells described above were harvested by centrifugation at 5000 rpm for 5 min. The cell pellets were washed with once with cold phosphate-buffered saline (PBS) and resuspended in 80 μl of 1× passive lysis buffer (PLB; Promega). The incubation was performed on a rocking platform at room temperature for 15 min, and then luciferase activity was measured using a TD20/20 luminometer (Turner Designs, Sunnyvale, CA). Briefly, 100 μl of Luciferase Assay Reagent II (LARII; Promega) was placed into luminometer tubes, and 20 μl of cell lysate was added and pipetted three times prior to reading firefly (FF) luciferase activity. Immediately, 100 μl of Stop & Glo Reagent (Promega) was added and vortexed, and then Renilla luciferase (RL) activity was read. All measurements were repeated three times for each data point. Data are reported as relative light units (FF/RL × 100).
Site-directed Mutagenesis of Consensus GATA-binding Sites—The primers for introducing mutations into the identified consensus GATA-binding sites in the extended 5′ promoter region of the EDN gene were designed based on sequence analysis results with the MatInspector program available on line. The primers for site-directed mutagenesis of GATA sites include the following: 5′-CTG AAA ACT CCC TGC GGC GAG TTT AGG GCC CTG CTG-3′ and 5′-CAG CAG GGC CCT AAA CTC GCC GCA GGG AGT TTT CAG-3′ for the GATA -1114 site; 5′-AGC AGA TTG TTT TAA GGC GAG ACA GAA TCT TGT GCC-3′ and 5′-GGC ACA AGA TTC TGT CTC GCC TTA AAA CAA TCT GCT-3′ for the GATA -535 site. Mutant strand synthesis reaction was performed in 50 μl of mixture containing 50 ng of DNA template, 125 ng of each primer, and 2.5 units of Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) at 95 °C for 30 s and 18 cycles of 95 °C for 30 s, 55 °C for 1 min, and 68 °C for 6 min. After the parental double-stranded DNA was digested with DpnI for 1 h at 37 °C, the synthesis products were transformed into XL1-Blue E. coli Supercompetent cells (Stratagene), and then the desired mutations were verified using DNA sequencing (3100 Sequencer, Applied Biosystems).
Western Blotting—HL-60 clone 15 cells differentiated for 2 days with 0.5 mm BA or uninduced cells transfected with GATA-2/pcDNA3.1 or pcDNA3.1 vector only (control) as described below were centrifuged at 8000 rpm for 5 min, and the pellet was washed with PBS and resuspended in lysis buffer (25 mm Tris, pH 8.0, 0.5% Triton X) plus 0.1% 4-(2-aminoethyl)benzenesulfonyl fluoride (ICN Biomedicals, Inc., Eschwege, Germany). The cells were lysed and heated for 5 min after adding the identical volume of 2× Tris-glycine SDS Sample Buffer (Invitrogen). The proteins were resolved in 14% Tris-glycine gel (Invitrogen) and transferred onto nitrocellulose membrane filter paper (Invitrogen). After blocking nonspecific protein binding with 5% nonfat dry milk, blots were incubated with 1:1000 dilutions of monoclonal antibodies to EDN (MBL, Naka-Ku, Japan), GATA-1 (Santa Cruz Biotechnology, Santa Cruz, CA), or GAPDH (US Biological, Swampscott, MA) or GATA-2-specific polyclonal antibody (Santa Cruz Biotechnology) followed by alkaline phosphatase-conjugated goat anti-mouse, -rat, and -rabbit IgG (Bio-Rad). Immunoconjugates were detected with alkaline phosphatase color-developing reagents (Bio-Rad) following the manufacturer's instructions.
Quantitative RT-PCR—The cells treated as described were homogenized in RNA-Bee RNA Isolation Solvent (at 1 ml per 106 cells, increased five times over the manufacturer's specifications to accommodate increased ribonuclease activity; Tel-Test, Inc., Friendswood, TX). The total RNA was prepared following the manufacturer's instruction, and its quality was monitored through gel electrophoresis analysis. Genomic DNA contamination was eliminated with DNA-free™ DNase treatment and removal reagent (Ambion, Austin, TX). RNA was reverse-transcribed in a mixture of 20 μl containing 5 mm MgCl2, 1 mm dNTP, 1.6 μg of oligo-p(dT)15, 50 μl of RNase inhibitor, and 20 units of avian myeloblastosis virus reverse transcriptase (Roche Applied Science) at 25 °C for 10 min, 42 °C for 1 h, and 99 °C for 5 min. After incubation at 50 °C for 2 min and 95 °C for 10 min, quantitative PCR was performed in 7500 real time PCR system (Applied Biosystems) for 40 cycles at 95 °C for 15 s and 60 °C for 1 min using Taqman® 2× universal PCR master mix (Applied Biosystems). The primers for EDN, GATA-1, and GATA-2 were purchased from Applied Biosystems. GATA-2 assay identification was Hs00927739_m1, and probe was 5′-CCC ACG GCT GCG TGT GGC CGT TGC C-3′; EDN assay identification was Hs00795553_s1, and probe was 5′-TCA CCA CAG TGG AAG CCA GGT GCC T-3′; GATA-1 assay identification was Hs00231112_m1, and probe was 5′-CAC CAG CCC AGG TTA ATC CCC AGA G-3′; the probes were labeled with FAM and NFQ. GAPDH expression assay part number was 4326317E used as the endogenous control (VIC/MGB probe).
Nuclear Extract Preparation—HL-60 clone 15 cells were differentiated with 0.5 mm BA for 2 days, harvested by centrifugation at 4 °C, and then washed with ice-cold PBS/phosphatase inhibitor (Active Motif, Carlsbad, CA). The pellet was gently suspended in 1× Hypotonic Buffer (Active Motif), incubated on ice for 15 min, and vortexed for 10 s after addition of detergent (Active Motif). The mixture was centrifuged for 30 s at 14,000 × g and the nuclear pellet was in Complete Lysis Buffer (Active Motif) prior to vortexing for 10 s. The suspension was incubated for 30 min at 4 °C on a rocking platform set at 150 rpm and then centrifuged for 10 min at 14,000 × g at 4 °C. The supernatant was aliquoted and stored at -80 °C for EMSA.
EMSA—The complementary oligonucleotides were labeled separately at their 3′ ends with Biotin-11-UTP using terminal deoxynucleotidyltransferase (Pierce), and then the terminal deoxynucleotidyltransferase was extracted by chloroform: isoamyl alcohol (24:1). The end-labeled or unlabeled complementary oligonucleotides were annealed to generate double-stranded DNA, which was stored at -20 °C until needed. The sequences of probes used in EMSA were as follows: GATA -1114, wild type, 5′-CTG AAA ACT CCC TGC TTA TCT TTT AGG GCC CTG CTG-3′, and mutant GATA -1114, 5′-CTG AAA ACT CCC TGC GGC GAG TTT AGG GCC CTG CTG-3′; GATA -535, wild type, 5′-AGC AGA TTG TTT TAA TTA TCT ACA GAA TCT TGT GCC-3′, and mutant GATA -535 5′-AGC AGA TTG TTT TAA GGC GAG ACA GAA TCT TGT GCC-3′. Four μg of nuclear extract was incubated with 4 μl of monoclonal anti-GATA-1 (Active Motif), polyclonal anti-GATA-2 (Santa Cruz Biotechnology) antibody, or no antibody on ice for 20 min prior to the addition of the biotin-labeled probe. The binding reactions contained 2.5% glycerol, 5 mm MgCl2, 1 μg of poly(dI·dT), 0.05% Nonidet P-40, labeled probe, nuclear extract, and antibody. For competition, 100-fold molar excess of unlabeled DNA fragment was added. The mixture was incubated for 20 min at room temperature and then electrophoresed at 100 V in 6% polyacrylamide DNA retardation gels (Invitrogen) in 0.5× TBE at 4 °C. The DNA was transferred to a positively charged nylon membrane (Pierce) and fixed to the membrane using UV cross-linking. The blots were probed with streptavidin-horseradish peroxidase conjugate (Pierce) through exposure to x-ray film (Eastman Kodak Co.) for 20-60 s.
Chromatin Immunoprecipitation—HL-60 Clone 15 cells were induced with 0.5 mm BA for 2 days, and 36% formaldehyde (Sigma) was added to a final concentration of 1% to cross-link proteins to DNA. Incubation was performed on a stir plate for 10 min at room temperature, and then the cross-linking reaction was stopped using Glycine Stop-Fix Solution as per manufacturer's instructions (Active Motif). The cells were pelleted by centrifugation for 10 min at 2500 rpm at 4 °C, washed with ice-cold PBS, and suspended in 1 ml of ice-cold Lysis Buffer supplemented with 5 μl of protease inhibitor mixture and 5 μl of 100 mm phenylmethylsulfonyl fluoride. Incubation was performed on ice for 30 min followed by homogenization in an ice-cold Dounce homogenizer. Pelleted nuclei were resuspended in 1.0 ml of Digestion Buffer supplemented with protease inhibitor mixture and phenylmethylsulfonyl fluoride as above; 50 μl of Enzymatic Shearing Mixture (200 units/ml; Active Motif) was added to shear the chromatin for 10 min. The sheared DNA was diluted 10-fold in Dilution buffer (Upstate; catalog number 20-153) and pre-cleared by adding 60 μl of protein G-agarose (Upstate) in 1 h of incubation at 4 °C with rotation, followed by removing agarose by a centrifugation of 4000 × g. Ten μl of pre-cleared DNA was saved as the input template (cross-linked DNA, and the other was used in the immunoprecipitation reaction. For the negative control, 5 μg of preimmune rabbit IgG or mouse IgG was added; the test antibodies included 10 μg of rabbit anti-human GATA-1 anti-peptide antibody (Active Motif) or mouse anti-human GATA-2 antibody (Santa Cruz Biotechnology). Incubations were performed overnight at 4 °C with rotation. 60 μl of protein G-agarose was added to the chromatin-protein with antibody mixture and incubated together for 1 h at 4 °C. The protein G-agarose-antibody/chromatin complex was pelleted, washed with Low Salt Immune Complex wash buffer, High Low Salt Immune Complex wash buffer, LiCl Immune Complex wash buffer, and TE Buffer (Upstate). Protein-DNA complex was eluted from protein G-agarose with 200 μl of elution buffer (10 μl 20% SDS, 20 μl of 1 m NaHCO3 and 170 μl of H2O). For all samples, including input, negative control and experimental reverse cross-linking of protein/DNA was performed to isolate free DNA. Specifically, for each sample, 8 μl of 5 m NaCl was added, and incubation was performed at 65 °C for 5 h, and then 1 μl of RNase A was added. After incubation at 37 °C for 30 min, 4 μl of 0.5 m EDTA, 8 μl of 1 m Tris-HCl, and 1 μl of proteinase K were added, and incubation was performed at 45 °C for 2 h. The resulting DNA was purified using spin columns (Upstate), and PCR was performed on a 50-μl sample containing 2 μl of DNA prepared as above, 5 μl of 10× buffer, 2 μl of 10 mm dNTP, and 0.5 μl of DNA polymerase. For the -1114 consensus GATA site in the EDN promoter, forward primer is 5′-CTGTGCCCAGAATGCTCATC-3′ and reverse primer is 5′-GCTTGAGGGACAAGGAAGACT-3′; for the -535 consensus GATA site, the forward primer is 5′-AGTGGATCCAATGCAAGAGG-3′ and reverse primer is 5′-TGAGCTATTAATTTCTTAGGGCACA-3′. PCR conditions were 3 min of 94 °C followed by 34 cycles of 20 s of 94 °C, 30 s of 58 °C, and 30 s of 72 °C.
RNA Silencing (siRNA)—The siRNA sequences commercially obtained from Ambion were 5′-GGU ACU CAG UGC ACC AAC UTT-3′ (GATA-1 siRNA), 5′-GGC UCG UUC CUG UUC AGA ATT-3′(GATA-2 siRNA 1), 5′-GGA GGA GGA UUG UGC UGA UTT-3′(GATA-2 siRNA 2). Negative control was Silencer Negative Control 1 siRNA (Ambion catalog number 4611). Three μg of siRNA was transfected into 2 × 106 clone 15 cells in 100 μl of Nucleofector Solution V (Amaxa) with T-016 program using Nucleofector II machine (Amaxa) as described above. The cells were then differentiated with BA and harvested 48 h later, and the total RNA was isolated with RNA-Bee RNA Isolation Solvent (1 ml per 106 cells) (Tel-Test, Inc). Quantitative PCR was used to analyze the extent to which levels of transcripts were reduced, and expression of protein encoding GATA-1, GATA-2, and EDN was evaluated by Western blotting.
GATA-2 Overexpression—cDNA encoding human GATA-2 (GenBank™ NM_032638) was isolated from BA-induced HL-60 clone 15 cells by RT-PCR using Pfx polymerase, forward primer 5′-CGG AAG CTT GCC GCC GGC CAT GGA GGT GGC-3′ and reverse primer 5′-GTT TCT AGA CTA CCC CAT GGC GGT CAC CA-3′, which includes restriction sites and a Kozak sequence at the start of translation. The insert was cloned into the HindIII/XbaI sites of pcDNA3.1/Zeo and confirmed by DNA sequencing. Three μg of GATA-2/pCDNA3.1 (pGATA-2) or pCDNA3.1 vector alone (pctrl) were transfected into 2 × 106 uninduced HL-60 clone 15 cells in 100 μl of Nucleofector solution V (Amaxa), T-016 program, Nucleofector II. Cells were resuspended in 2 ml of complete medium, and after 2 days, cells were harvested, and RNA was prepared using the qPCR-Grade RNA isolation kit (SuperArray Bioscience), and quantitative RT-PCR was performed as described.
Culture and Differentiation of Human Umbilical Cord and Peripheral Blood CD34+ Cells—Human umbilical cord CD34+ hematopoietic progenitors (Stemcell Technologies, catalog number CB008F) or CD34+ progenitors from peripheral blood from healthy human donors pre-stimulated with granulocyte colony stimulating factor (a generous gift from Dr. Arnold Kirshenbaum, MCBS, LAD (26)) were cultured at 0.3 × 106 cells per ml in Iscove's modified Dulbecco's medium (Invitrogen) with 10% fetal bovine serum, 50 μm β-mercaptoethanol, 2 mm glutamine, 10 units/ml penicillin, 10 units/ml streptomycin (basic medium), and cytokines, including stem cell factor (50 ng/ml), FLT-3L (50 ng/ml), granulocyte-macrophage colony-stimulating factor (5 ng/ml), IL-3 (5 ng/ml), and IL-5 (5 ng/ml, all cytokines from R & D Systems), as per Bedi et al. (27). After 3 days in culture, the medium was changed, and cells were maintained in the basic medium with cytokines IL-3 (5 ng/ml) and IL-5 (5 ng/ml) alone, at 0.5 × 106 cells/ml for the remainder of the experiment. Eosinophil differentiation over 21 days was monitored by modified Giemsa staining of cytocentrifuge cell preparations and EDN transcript levels.
Suppression of GATA-2 Transcription in Eosinophil Progenitors Derived from CD34+ Cells—Recombinant lentiviral particles delivering shRNA to suppress GATA-2 transcription together with puromycin resistance were assembled in HEK293 cells as per the manufacturer's instructions (Sigma Mission shRNA, with control plasmid pLK0.1 or with GATA-2 shRNA). 5 × 106 differentiating cells were transduced at day 14 of culture; 2 days later, fresh medium with cytokines IL-3 (5 ng/ml) and IL-5 (5 ng/ml) was supplemented with puromycin (2 μg/ml, the optimal concentration determined by serial dilution, as per manufacturer's instructions). Cells were harvested 5 days later for quantitative RT-PCR.
Statistical Considerations—All luciferase and quantitative PCR data represent compilations from three separate experiments with each data point from each experiment representing a trial performed in triplicate. Statistical significance determined by Student's t test and Mann-Whitney U test as appropriate.
RESULTS
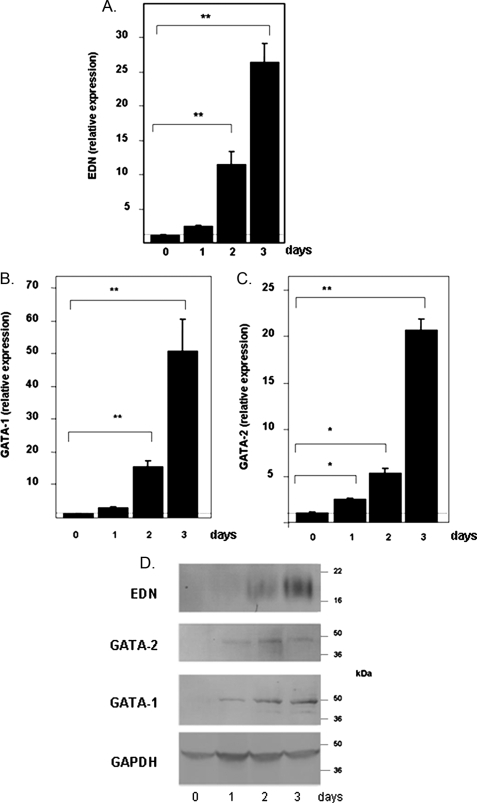
Expression of EDN, GATA-1, and GATA-2 in the HL-60 Clone 15 Eosinophil Promyelocyte Cell Line—The clone 15 subline of the HL-60 promyelocytic leukemia cell line has properties of an eosinophil progenitor when maintained under alkaline conditions. Addition of BA results in differentiation into cells with features characteristic of maturing human eosinophils (28, 29). We observe a 25-fold increase in EDN mRNA in response to BA at day 3 (Fig. 1A), consistent with previous results (9, 28). BA-induced differentiation of HL-60 clone 15 cells also results in augmented transcription of GATA-1 (Fig. 1B) and GATA-2 (Fig. 1C), reaching levels 50- and 20-fold over base line, respectively, at day 3. The GATA-2 expression pattern differs from that observed by Zon et al. (9) who performed Northern analysis and reported increased expression of GATA-1 but stable expression of GATA-2 in this cell line in response to 2 days of BA-induced differentiation. EDN, GATA-1, and GATA-2 proteins are not detectable at base line but are detected after 24 h of BA-induced differentiation (Fig. 1D). The indistinct banding pattern for EDN results from the heavy glycosylation that we described previously in this cell line (28).
FIGURE 1.
Expression of transcripts encoding EDN (A), GATA-1 (B), and GATA-2 (C) in the HL-60 clone 15 eosinophil promyelocyte cell line in response to BA-induced differentiation. Cells were induced to differentiate with 0.5 mm BA under alkaline conditions, and transcripts indicated were evaluated by quantitative RT-PCR; expression was normalized to GAPDH as described under “Experimental Procedures,” with relative expression of one sample from the t = 0 set at 1.0 (horizontal dotted line). Statistical significance is as follows: *, p < 0.05; **, p < 0.01. D, expression of immunoreactive EDN, GATA-1, and GATA-2. Western blots of extracts prepared from untreated cells (day 0) and from cells treated with BA for 1-3 days were probed with monoclonal anti-EDN, anti-GATA-1, or anti-GATA-2 antibodies. Relative protein loading was determined by probing with an anti-GAPDH antibody.
Identification of GATA Consensus Binding Sites in the 5′ Promoter Regions of the Gene Encoding EDN—We isolated a 1435-bp fragment of the EDN gene that extended the characterized minimal 5′ functional promoter (20) and also included exon 1, the single intron, and a fragment of exon 2 proximal to the ATG translational start site. There are two GATA consensus binding sites in the extended 5′ promoter region, one at a distal position (bp -1114) and another at a more proximal site (bp -535). Luciferase reporter constructs, including the entire 1435-bp fragment described above (EDN 1000) as well as serial 5′ truncations (EDN 500 and EDN 250), are shown in Fig. 2A. Promoter activities of the EDN reporter constructs were evaluated in HL-60 clone 15 cells induced for 48 h with BA prior to transfection (Fig. 2B). The activity of the EDN 1000 construct was nearly 50-fold over control (pGL3 basic reporter alone, no promoter included). The luciferase activities of the truncated EDN 500 and EDN 250 reporter constructs, with both GATA consensus sites eliminated, were reduced to ∼20-fold over control (p < 0.05 versus EDN 1000). Similarly, site-specific mutagenesis of the distal GATA consensus binding site (-1114) of the EDN 1000 construct reduced the promoter activity more than 3-fold; a similar reduction was observed upon introduction of mutations into the proximal GATA site (-535), but no further reduction was observed with the double mutation (Fig. 2C). Of note, the activity of the extended 5′ promoter (EDN 1000) was not elevated in uninduced HL-60 clone 15 cells and was indistinguishable from that of the truncated EDN 250 and from that of EDN 1000 with both GATA sites mutated (EDN 1000 mGATA -1114/-535; Fig. 2D).
FIGURE 2.
Contribution of GATA consensus binding sites to the transcription of the EDN gene. A, schematic of the gene encoding eosinophil-derived neurotoxin/RNase 2 indicating relative positions of consensus binding sites for GATA family transcription factors. The sequence of the GATA core and flanking sequences are as shown. Below each schematic are the segments of the EDN gene introduced 5′ to luciferase to create pGL3 reporter constructs. The EDN 1000 luciferase reporter construct includes 1138 bp of the 5′ promoter region, the 67-bp exon 1, and the 230-bp single intron (total 1435 bp); EDN 500 and EDN 250 are truncated versions of EDN 1000 luciferase that do not contain the GATA consensus binding sites. Numbering is based on original sequence data listed in GenBank™ (EDN, X16546). B-D, luciferase activity (relative light units = FF/RL × 100), where FF is firefly and RL is Renilla luciferase, detected in HL-60 clone 15 cells induced with 0.5 mm BA (B and C) and without BA (D). Cells were transfected with the full-length or truncated EDN promoter reporter constructs, with or without mutations in the GATA consensus binding sites. C, control pGL3 reporter alone; mutations in the core GATA sequences are as shown. Statistical significance is as follows: *, p < 0.05; **, p < 001 versus the unmutated full-length construct (EDN 1000).
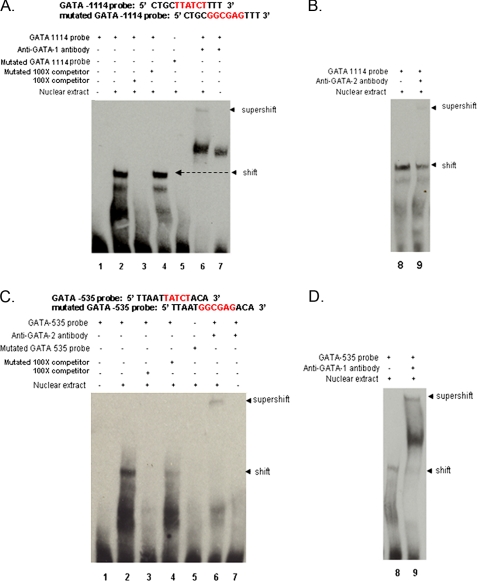
GATA-1 and GATA-2 Binding to Consensus Sites in the EDN Promoter—EMSA were performed with biotin-labeled probes that include the GATA sites within the EDN 5′ promoter (Fig. 3). We observed specific binding of proteins isolated from the BA-differentiated HL-60 clone 15 nuclear extract to the labeled probe that includes the GATA -1114 site (Fig. 3A, lane 2) but not to a probe with mutations within the core consensus GATA sequence (lane 5) nor in the presence of a 100-fold excess of unlabeled competing probe (lane 3). However, specific binding was detected in the presence of 100-fold excess unlabeled probe containing mutations (Fig. 3A, lane 4). Proteins specifically bound to the GATA -1114 probe were “super-shifted” by monoclonal antibody to GATA-1 (Fig. 3A, lane 6) and by polyclonal anti-GATA-2 (Fig. 3B, lane 8), indicating that both GATA-1 and GATA-2 proteins present in the nuclear extract can bind to this consensus site. The prominent band detected between the shift and supershift in Fig. 3A (lane 6) results from a nonspecific interaction between the antibody and the probe, as it can be detected even in the absence of nuclear extract (Fig. 3A, lane 7). We also observe specific binding of nuclear proteins isolated from BA-differentiated HL-60 clone 15 cells to a probe that includes the GATA -535 site, which also undergoes supershift with both polyclonal anti-GATA-2 (Fig. 3C) and monoclonal anti-GATA-1 (Fig. 3D).
FIGURE 3.
Electrophoretic mobility shift/supershift assays for evaluating interactions of consensus GATA sequences in the 5′ promoter of EDN with BA-treated HL-60 clone 15 nuclear extracts and anti-GATA-1 and anti-GATA-2 antibodies. A, lane 1, biotin-labeled GATA -1114 probe alone; lane 2, biotin-labeled GATA -1114 probe + nuclear extract; lane 3, biotin-labeled GATA -1114 probe + nuclear extract + 100× excess unlabeled GATA -1114 probe; lane 4, biotin-labeled GATA -1114 probe + nuclear extract + 100× excess mutated GATA -1114 probe; lane 5, biotin-labeled mutated GATA -1114 probe + nuclear extract; lane 6, biotin-labeled GATA -1114 probe + nuclear extract + anti-GATA-1 antibody; lane 7, biotin-labeled GATA -1114 probe + anti-GATA-1 antibody. B, lane 8, biotin-labeled GATA -1114 probe + nuclear extract + anti-GATA-2 antibody; lane 9, biotin-labeled GATA -1114 probe + anti-GATA-2 antibody. C, lanes 1-7 as in A, biotin-labeled GATA -535 probe, anti-GATA-2 antibody. D, lanes 8 and 9 as in B, biotin-labeled GATA -535 probe, anti-GATA-1 antibody. Arrows marked shift indicate primary DNA-nuclear protein interactions; arrows marked supershift indicate DNA-nuclear protein-antibody interactions. Probe sequences are as shown.
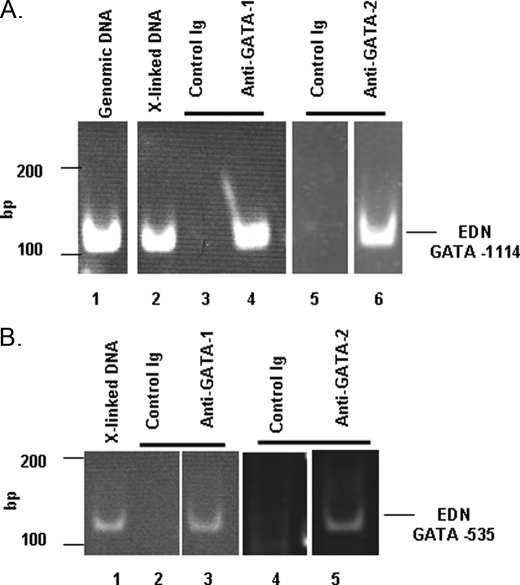
Chromatin Immunoprecipitation Analysis—Formaldehyde-cross-linked protein-DNA complexes were isolated from BA-differentiated HL-60 clone 15 cells, sheared, and subjected to immunoprecipitation with anti-GATA-1, anti-GATA-2, or control antibodies. Precipitated protein-DNA complexes were treated with proteases and used as a template to amplify consensus GATA-binding sites within the EDN promoter. Both GATA-1 and GATA-2 were detected in association with the distal -1114 GATA consensus binding site (Fig. 4A) and like-wise with the proximal -535 GATA site (Fig. 4B); no amplification was observed when precipitation was attempted with control antibody.
FIGURE 4.
Chromatin immunoprecipitation assay for evaluating interactions of consensus GATA sequences in the 5′ promoter of EDN. A, PCR amplification of the EDN GATA -1114 sequence from the following: lane 1, unmanipulated genomic DNA; lane 2, cross-linked DNA; lane 3, cross-linked DNA precipitated with rabbit IgG (control); lane 4, cross-linked DNA precipitated with rabbit anti-human GATA-1; lane 5, cross-linked DNA precipitated with mouse IgG (control); lane 6, cross-linked DNA precipitated with mouse anti-human GATA-2. Lanes 5 and 6 were examined simultaneously on the same gel; white bar indicates separation between nonadjacent lanes. B, PCR amplification of the EDN GATA -535 sequence from the following: lane 1, cross-linked DNA; lane 2, cross-linked DNA precipitated with rabbit IgG (control); lane 3, cross-linked DNA precipitated with rabbit anti-human GATA-1; lane 4, cross-linked DNA precipitated with mouse IgG (control); lane 4, cross-linked DNA precipitated with mouse anti-human GATA-2. Lanes 1-3 and lanes 4 and 5 were examined on two gels (lanes 1-3 on one, and lanes 4 and 5 on another); white bars indicate separations between nonadjacent lanes.
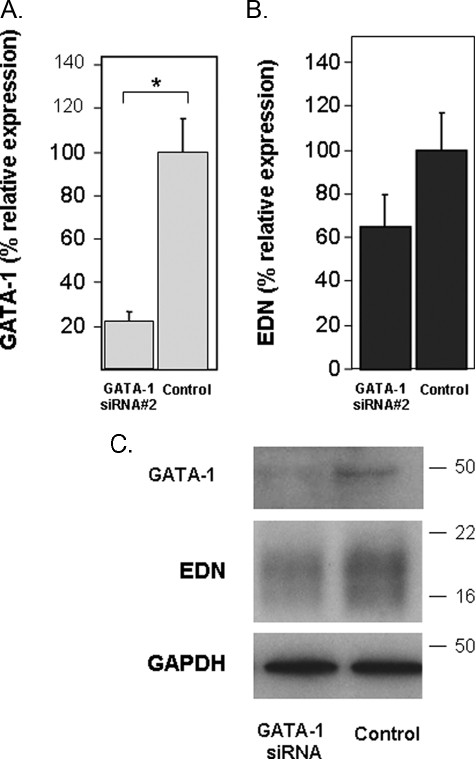
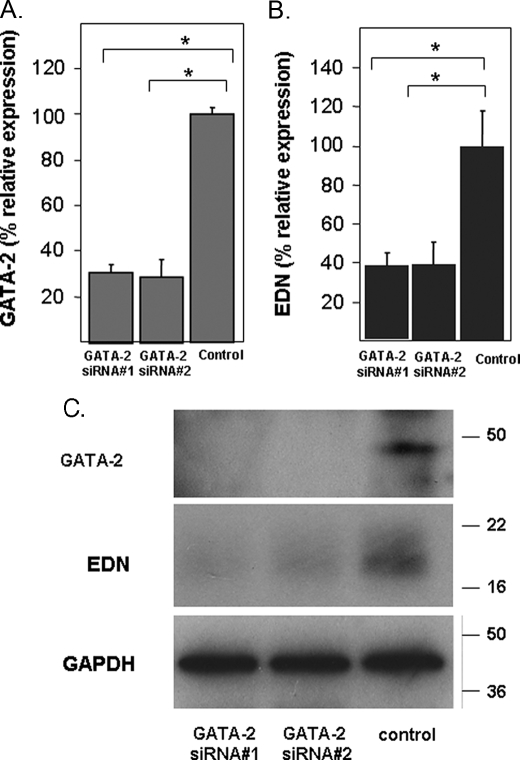
RNA Silencing of GATA-1 and GATA-2 Expression—HL-60 clone 15 cells were transfected with a GATA-1-specific siRNA (GATA-1 siRNA) or an irrelevant control sequence as described under “Experimental Procedures.” Transfected cells were then differentiated with BA and harvested 48 h later, and RNA and protein were isolated and evaluated. As shown, transcription of GATA-1 was suppressed by 80% when compared with control levels (Fig. 5A). Interestingly, suppression of GATA-1 transcription and concomitant reduction of GATA-1 protein synthesis had a small but not statistically significant effect on the expression of EDN transcript (Fig. 5B) and minimal impact on expression of EDN protein (Fig. 5C). In contrast, transfection with two independent GATA-2-specific siRNAs resulted in suppression of GATA-2 transcription by ∼70% when compared with control levels (Fig. 6A) and profound suppression of GATA-2 protein synthesis (Fig. 6C), accompanied by significant suppression of EDN (Fig. 6, B and C).
FIGURE 5.
Silencing of GATA-1 and its impact on EDN expression. A, transcription of GATA-1 in BA-differentiated HL-60 clone 15 cells transfected with irrelevant control sequence or a GATA-1-directed oligonucleotide was evaluated by quantitative RT-PCR. B, transcription of EDN was determined in cells described in A. C, extracts prepared from the cells described in A were subjected to Western blotting and probed with monoclonal anti-GATA-1, anti-EDN, or anti-GAPDH antibodies (loading control). Statistical significance is as follows: *, p < 0.05.
FIGURE 6.
Silencing of GATA-2 and its impact on EDN expression. A, transcription of GATA-2 in BA-differentiated HL-60 clone 15 cells transfected with control (irrelevant sequence) or one of two independent GATA-2-directed oligonucleotides was evaluated by quantitative RT-PCR. B, transcription of EDN was determined in the cells described in A. C, extracts prepared from the cells described in A were subjected to Western blotting and probed with anti-GATA-2, anti-EDN, or anti-GAPDH (loading control). Statistical significance is as follows: *, p < 0.05.
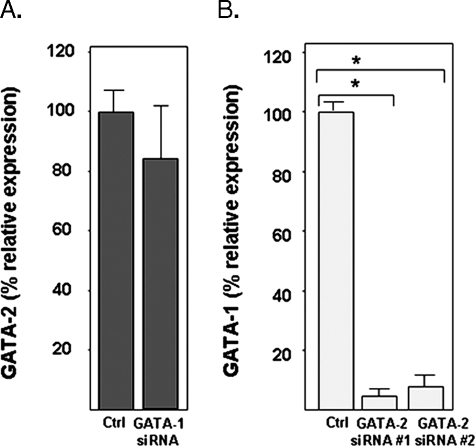
Given the critical role of GATA-2 in promoting expression of EDN, and findings suggesting that GATA-2 and GATA-1 have the potential to regulate one another's expression (30-32), we explored the relative expression of GATA-1 under conditions of GATA-2 suppression and vice versa. Here we find that suppression of GATA-1 had no impact on the expression of GATA-2 (Fig. 7A), but suppression of GATA-2 has a profound impact on the transcription of GATA-1, reducing transcription to less than 10% of levels observed at base line (Fig. 7B).
FIGURE 7.
A, silencing of GATA-1 and its impact on GATA-2 expression; B, silencing of GATA-2 and its impact on GATA-1 expression. Transcription of GATA-1 or GATA-2 in HL-60 clone 15 cells transduced with irrelevant control (Ctrl) or one of two independent GATA-1- or GATA-2-directed oligonucleotides was evaluated by quantitative RT-PCR. Statistical significance is as follows: *, p < 0.01.
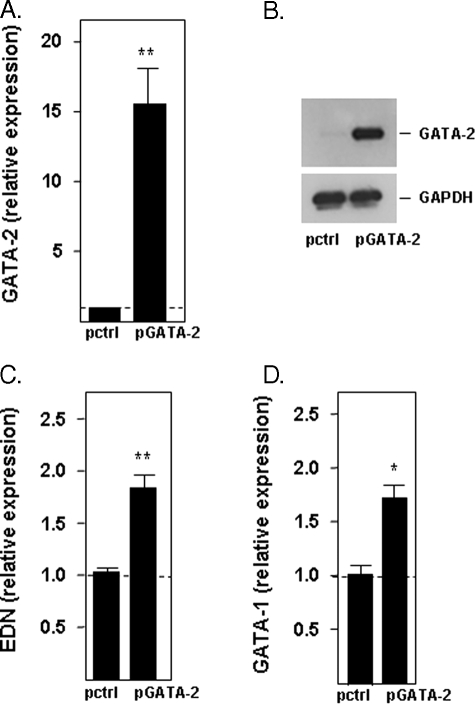
Overexpression of GATA-2—Transfection of uninduced HL-60 clone 15 cells with the expression vector pcDNA3.1/GATA-2 (pGATA-2) results in prominent expression of GATA-2 over background levels (Fig. 8, A and B). Consistent with the RNA silencing findings, expression of GATA-2 has a significant impact on the expression of both EDN and of GATA-1; elevated levels of transcript of both EDN (Fig. 8C) and GATA-1 (Fig. 8D) were detected in cells overexpressing GATA-2 versus cells transfected with the control vector (pctrl).
FIGURE 8.
Expression of EDN and GATA-1 in response to overexpression of GATA-2. Relative expression of GATA-2 in uninduced HL-60 clone 15 cells 2 days after transduction with vector only (pctrl) or with the GATA-2 expression vector (pGATA-2) was evaluated by quantitative RT-PCR (A) and by Western blotting (B). Relative expression of EDN (C) and GATA-1 (D) in uninduced HL-60 clone 15 cells transduced as described was determined by quantitative RT-PCR. Statistical significance is as follows: *, p < 0.05; **, p < 0.01.
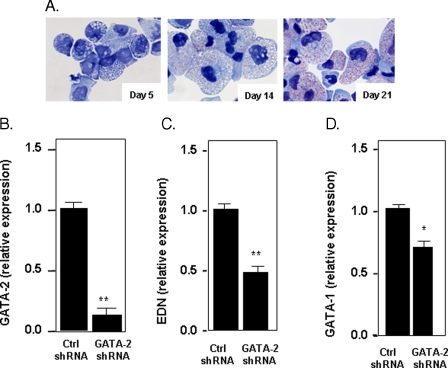
Suppression of GATA-2 in CD34+-derived Human Eosinophil Progenitors—CD34+ hematopoietic stem cells can be differentiated into eosinophilic progenitors using a specific cytokine regimen (“Experimental Procedures”) (27). The phenotypic progression of these cells is shown in Fig. 9A. By day 14, >70% of the cells have developed red-staining cytoplasmic granules, a hallmark of the eosinophilic promyelocyte; by day 21, the granulation has become more dense and intense, and many of the cells have developed rudimentary eccentric bilobed nuclei.
FIGURE 9.
Expression of EDN and GATA-1 in response to lentivirus-mediated suppression of GATA-2 in differentiating eosinophils from CD34+ hematopoietic progenitors. A, modified Giemsa-stained cytospin preparations of cells differentiating in eosinophilopoietic cytokines for 5, 14, and 21 days. B, expression of GATA-2 at day 21 in cells subjected to lentivirus shRNA (control or GATA-2 specific) on day 14 and puromycin selection beginning on day 16. C, expression of EDN; D, GATA-1 in response to GATA-2 suppression or control, as described in B. Statistical significance is as follows: *, p < 0.05; **, p < 0.01.
Lentivirus shRNAs were delivered to differentiating eosinophil cultures on day 14, and puromycin selection (2 μg/ml) was introduced on day 16, and cells were harvested for analysis on day 21. Under these conditions, expression of GATA-2 was reduced to ∼15-20% of control levels (Fig. 9B); of note, puromycin selection resulted in no differential impact on cell number or percentage eosinophils, determined phenotypically, in the control or GATA-2-suppressed cultures (data not shown). In these same cultures, EDN expression was reduced to 50% of control (Fig. 9C), which was similar to the extent to which EDN expression was diminished in response to suppression of GATA-2 in the BA-induced clone 15 cells. GATA-1 expression was also reduced in response to GATA-2 suppression (Fig. 9D), although interesting, not nearly as profoundly as in the HL-60 clone 15 cells, only by about 25% when compared with the control. The reason for this differential response remains unclear.
DISCUSSION
In this work, we evaluated the contribution of two consensus GATA-binding sites to the activity of the extended functional promoters of EDN and eosinophil cationic protein. Interestingly, both GATA consensus sites are crucial for full promoter activity of EDN in BA-induced cells of the eosinophil promyelocyte clone 15 HL-60 cell line. We detect binding of both GATA-1 and GATA-2 proteins to consensus sites in the EDN promoter, but silencing of GATA-1 alone had little to no impact on EDN transcription or translation, possibly due to compensation from GATA-2, which binds to the same consensus sites as GATA-1 both in tissue culture and in vivo. Interestingly, we found that silencing of GATA-2 reduced EDN transcription, but also reduced transcription of GATA-1 in both the HL-60 clone 15 eosinophil promyelocyte cell line and in differentiating eosinophils derived from human CD34+ hematopoietic progenitors. As such, our results are consistent with a more complex mechanism, one that requires coordinated actions of both GATA factors at one or more consensus binding sites.
GATA-1 and GATA-2 are members of a larger family of GATA transcription factors and co-factors (reviewed in Refs. 33-36), and although there is only limited amino acid sequence between the two proteins overall (∼52%), both bind to DNA sequences within the uniquely flanked internal GATA motif A/T (GATA) A/G (37). Initially explored vis à vis their role in promoting erythroid development, both GATA-1 and GATA-2 have been implicated in eosinophil hematopoiesis. Most of the recent focus has been on GATA-1 given the finding that ablation of a dblGATA enhancer in the mouse GATA-1 promoter, a site that has been implicated in autoregulation of the GATA-1 promoter (38), leads to selective eosinophil ablation in vivo (5), although this blockade can be circumvented in vitro (39). The human and mouse GATA-1 promoters have substantial sequence homology (40, 41), and the palindromic dblGATA consensus sequence is fully conserved. However, it is not at all clear how this consensus site functions within the human GATA-1 promoter, and whether it is similarly indispensable for eosinophil hematopoiesis in vivo.
Several earlier studies have addressed the transcriptional regulation of EDN. Our group has explored a minimal promoter-exon-intron reporter construct, which was 2-3-fold more active in BA-differentiated clone 15 cells than in undifferentiated counterparts, and featured active enhancer elements in the intron (20, 21). de Groot and co-workers (22, 23) have characterized the roles of PU.1 and C/EBP in promoting EDN transcription, and most recently Chang and co-workers (24) have shown that MAX and Sp1 interact with the 34-bp segment unique to the proximal promoter region of EDN. Interestingly, given issues of evolutionary divergence, it will be difficult to evaluate the role of any of these factors in promoting transcription of EDN via gene-deletion experiments in vivo. EDN and ECP are rapidly evolving genes that are unique to primate species (42, 43); the mouse eosinophil-associated RNases are a cluster of 10-15 related genes, but there is no one specific mouse ortholog of EDN (44). We have characterized a fragment of an active promoter of mEar2 (45), one of the two eosinophil ribonuclease orthologs that are expressed prominently in mouse eosinophils (46), and for the purposes of this work, we have identified an extended 5′ promoter region from the mouse genome data base. This extended 5′ promoter sequence has no substantial homology to the 5′ promoter of EDN (<50%), but it does include three consensus GATA sites that might be the subject of future exploration.
In our study, which features gene transcription in both a differentiating eosinophil cell line and in human CD34+ progenitor cells cultured in eosinophilopoietic cytokines, we find that GATA-2 regulates not only a crucial eosinophil granule protein gene but also regulates the transcription of GATA-1. To the best of our knowledge, this is the first study in which promoter expression is evaluated in the presence of silencing RNAs directed against GATA-1 and GATA-2. Among the previous studies on eosinophil hematopoiesis that have focused on GATA-2, Iwasaki et al. (12) found that GATA-2 overexpression in C/EBPα-expressing granulocyte/monocyte progenitors resulted in eosinophil differentiation, whereas expression of GATA-1 under these conditions resulted in commitment toward the megakaryocytic lineage. At the same time, GATA-1-dependent expression of the gp91phox gene in eosinophils is suppressed by GATA-2 (14), which is quite different from what is observed with respect to GATA-2 and EDN.
Consensus opinion is clear that eliminating eosinophils is a viable and meaningful approach toward the amelioration of symptoms of allergic disease (reviewed in Ref. 47). Given the disappointing results with agents directed at eliminating eosinophils via anti-IL-5 cytokine modulation (48, 49), attention may turn toward transcriptional control as a more direct means of eliciting temporary eosinophil lineage ablation. No one has successfully identified a single transcription factor or transcriptional event that uniquely defines the eosinophil lineage, although GATA factors are likely to play a pivotal role. Ongoing studies may identify unique, eosinophil-specific mediators that interact directly with GATA factors during hematopoietic differentiation.
Acknowledgments
We thank Dr. Arnold Kirshenbaum for the generous gift of peripheral blood CD34+ hematopoietic progenitor cells and Dr. Kirk Druey, Molecular Signal Transduction Section, Laboratory of Allergic Diseases, NIAID, National Institutes of Health, for reading the final draft and providing constructive suggestions.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FJ785402.
This work was supported, in whole or in part, by National Institutes of Health Grants AI00941 and AI00942 NIAID (Division of Intramural Research).
Footnotes
The abbreviations used are: EDN, eosinophil-derived neurotoxin; RNase, ribonuclease; siRNA, silencing RNA; HEPPSO, N-2-hydroxyethylpiperazine-N-2-hydroxypropanesulfonic acid; RT, reverse transcription; PBS, phosphate-buffered saline; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; BA, butyric acid; IL, interleukin; shRNA, short hairpin RNA; EMSA, electrophoretic mobility shift assay.
References
- 1.Rothenberg, M. E., and Hogan, S. P. (2006) Annu. Rev. Immunol. 24 147-174 [DOI] [PubMed] [Google Scholar]
- 2.Roboz, G. J., and Rafii, S. (1999) Curr. Opin. Hematol. 6 164-168 [DOI] [PubMed] [Google Scholar]
- 3.Escoubet-Lozach, L., Glass, C. K., and Wasserman, S. I. (2002) J. Allergy Clin. Immunol. 110 553-564 [DOI] [PubMed] [Google Scholar]
- 4.McNagny, K., and Graf, T. (2002) J. Exp. Med. 195 F43-F47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu, C., Cantor, A. B., Yang, H., Browne, C., Wells, R. A., Fujiwara, Y., and Orkin, S. H. (2002) J. Exp. Med. 195 1387-1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohneda, K., and Yamamoto, M. (2002) Acta Haematol. 108 237-245 [DOI] [PubMed] [Google Scholar]
- 7.Koury, M. J., Sawyer, S. T., and Brandt, S. J. (2002) Curr. Opin. Hematol. 9 93-100 [DOI] [PubMed] [Google Scholar]
- 8.Weiss, M. J., and Orkin, S. H. (1995) Exp. Hematol. 23 99-107 [PubMed] [Google Scholar]
- 9.Zon, L. I., Yamaguchi, Y., Yee, K., Albee, E. A., Kimura, A., Bennett, J. C., Orkin, S. H., and Ackerman, S. J. (1993) Blood 81 3234-3241 [PubMed] [Google Scholar]
- 10.Kulessa, H., Frampton, J., and Graf, T. (1995) Genes Dev. 9 1250-1262 [DOI] [PubMed] [Google Scholar]
- 11.Hirasawa, R., Shimizu, R., Takahashi, S., Osawa, M., Takayanagi, S., Kato, Y., Onodera, M., Minegishi, N., Yamamoto, M., Fukao, K., Taniguchi, H., Nakauchi, H., and Iwama, A. (2002) J. Exp. Med. 195 1379-1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki, H., Mizuno, S., Arinobu, Y., Ozawa, H., Mori, Y., Shigematsu, H., Takatsu, K., Tenen, D. G., and Akashi, K. (2006) Genes Dev. 20 3010-3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi, Y., Nishio, H., Kishi, K., Ackerman, S. J., and Suda, T. (1999) Blood 94 1429-1439 [PubMed] [Google Scholar]
- 14.Yang, D., Suzuki, S., Hao, L. J., Fujii, Y., Yamauchi, A., Yamamoto, M., Nakamura, M., and Kumatori, A. (2000) J. Biol. Chem. 275 9425-9432 [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg, H. F. (2008) Curr. Pharm. Biotechnol. 9 135-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durack, D. T., Ackerman, S. J., Loegering, D. A., and Gleich, G. J. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 5165-5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredens, K., Dahl, R., and Venge, P. (1982) J. Allergy Clin. Immunol. 70 361-366 [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg, H. F. (2008) J. Leukocyte Biol. 83 1079-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, D., Chen, Q., Su, S. B., Zhang, P., Kurosaka, K., Caspi, R. R., Michalek, S. M., Rosenberg, H. F., Zhang, N., and Oppenheim, J. J. (2008) J. Exp. Med. 205 79-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiffany, H. L., Handen, J. S., and Rosenberg, H. F. (1996) J. Biol. Chem. 271 12387-12393 [DOI] [PubMed] [Google Scholar]
- 21.Handen, J. S., and Rosenberg, H. F. (1997) J. Biol. Chem. 272 1665-1669 [DOI] [PubMed] [Google Scholar]
- 22.Baltus, B., Buitenhuis, M., van Dijk, T. B., Vinson, C., Raaijmakers, J. A., Lammers, J. W., Koenderman, L., and de Groot, R. P. (1999) J. Leukocyte Biol. 66 683-688 [DOI] [PubMed] [Google Scholar]
- 23.van Dijk, T. B., Caldenhoven, E., Raaijmakers, J. A., Lammers, J. W., Koenderman, L., and de Groot, R. P. (1998) Blood 91 2126-2132 [PubMed] [Google Scholar]
- 24.Wang, H. Y., Chang, H. T., Pai, T. W., Wu, C. I., Lee, Y. H., Chang, Y. H., Tai, H. L., Tang, C. Y., Chou, W. Y., and Chang, M. D. (2007) BMC Mol. Biol. 8 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg, H. F., Tenen, D. G., and Ackerman, S. J. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 4460-4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirshenbaum, A. S., Goff, J. P., Semere, T., Foster, B., Scott, L. M., and Metcalfe, D. D. (1999) Blood 94 2333-2342 [PubMed] [Google Scholar]
- 27.Bedi, R., Du, J., Sharma, A. K., Gomes, I., and Ackerman, S. J. (2009) Blood 113 317-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischkoff, S. A., Pollak, A., Gleich, G. J., Testa, J. R., Misawa, S., and Reber, T. J. (1984) J. Exp. Med. 160 179-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiffany, H. L., Li, F., and Rosenberg, H. F. (1995) J. Leukocyte Biol. 58 49-54 [DOI] [PubMed] [Google Scholar]
- 30.Tang, X. B., Liu, D. P., and Liang, C. C. (2001) Cell. Mol. Life Sci. 58 2008-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss, M. J., Keller, G., and Orkin, S. H. (1994) Cells 8 1184-1197 [DOI] [PubMed] [Google Scholar]
- 32.Grass, J. A., Boyer, M. E., Pal, S., Wu, J., Weiss, M. J., and Bresnick, E. H. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8811-8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantor, A. B. (2005) Int. J. Hematol. 81 378-384 [DOI] [PubMed] [Google Scholar]
- 34.Lowry, J. A., and Mackay, J. P. (2006) Int. J. Biochem. Cell Biol. 38 6-11 [DOI] [PubMed] [Google Scholar]
- 35.Migliaccio, A. R., Rana, R. A., Vannucchi, A. M., and Manzoli, F. A. (2005) Ann. N. Y. Acad. Sci. 1044 142-158 [DOI] [PubMed] [Google Scholar]
- 36.Shimizu, R., and Yamamoto, M. (2005) Semin. Cell Dev. Biol. 16 129-136 [DOI] [PubMed] [Google Scholar]
- 37.Ko, L. J., and Engel, J. D. (1993) DNA-binding Specificities of the GATA Transcription Factor Family 13 4011-4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai, S. F., Strauss, E., and Orkin, S. H. (1991) Genes Dev. 5 919-931 [DOI] [PubMed] [Google Scholar]
- 39.Dyer, K. D., Czapiga, M., Foster, B., Foster, P. S., Kang, E. M., Lappas, C. M., Moser, J. M., Naumann, N., Percopo, C. M., Siegel, S. J., Swartz, J. M., Ting-De Ravin, S., and Rosenberg, H. F. (2007) J. Immunol. 179 1693-1699 [DOI] [PubMed] [Google Scholar]
- 40.Nicolis, S., Bertini, C., Ronchi, A., Crotta, S., Lanfranco, L., Moroni, E., Giglioni, B., and Ottolenghi, S. (1991) Nucleic Acids Res. 19 5285-5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zon, L. I., and Orkin, S. H. (1992) Nucleic Acids Res. 20 1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenberg, H. F., Dyer, K. D., Tiffany, H. L., and Gonzalez, M. (1995) Nat. Genet. 10 219-223 [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg, H. F., and Dyer, K. D. (1995) J. Biol. Chem. 270 21539-21544 [DOI] [PubMed] [Google Scholar]
- 44.Zhang, J., Dyer, K. D., and Rosenberg, H. F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4701-4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyer, K. D., Nitto, T., Moreau, J. M., McDevitt, A. L., and Rosenberg, H. F. (2004) Mamm. Genome 15 126-134 [DOI] [PubMed] [Google Scholar]
- 46.Larson, K. A., Olson, E. V., Madden, B. J., Gleich, G. J., Lee, N. A., and Lee, J. J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12370-12375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster, P. S., Asquith, K. A., Rosenberg, H. F., and Kumar, R. K. (2008) Curr. Mol. Med. 8 585-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leckie, M. J. (2003) Am. J. Respir. Med. 2 245-259 [DOI] [PubMed] [Google Scholar]
- 49.Barnes, P. J. (2001) J. Allergy Clin. Immunol. 108 S72-S76 [DOI] [PubMed] [Google Scholar]