Abstract
Huntington disease (HD) is a fatal hereditary neurodegenerative disease caused by an expansion of the polyglutamine (polyQ) stretch in huntingtin (htt). Whereas the pathological significance of the expanded polyQ has been clearly established and a tremendous effort to develop therapeutic tools for HD has been exerted, there is yet no effective cure. Whereas many molecules able to reduce the polyQ accumulation and aggregation have been identified, including several Rho kinase (ROCK) inhibitors, it remains very important to determine the mechanism of action of the potential drugs. ROCK inhibitors, including Y-27632 were reported to decrease aggregation of htt and androgen receptor (AR) through ROCK1 and protein kinase C-related protein kinase-2 (PRK-2). A downstream effector of ROCK1, actin-binding factor profilin, was shown to inhibit the mutant htt aggregation but not AR by direct interaction. We found that the anti-aggregation effect of ROCK inhibitors was not limited to the mutant htt and AR and that Y-27632 was also able to reduce the aggregation of ataxin-3 and atrophin-1 with expanded polyQ. These results suggested that in addition to the mechanism reported for htt and AR, there might also be other common mediators involved in the reduced aggregation of different polyQ proteins. In this study, we show that Y-27632 not only reduced the mutant htt aggregation by enhancing its degradation, but surprisingly was able to activate the main cellular degradation pathways, proteasome, and macroautophagy. We also show that this unique effect was mediated by ROCK1 and ROCK2.
Huntington disease (HD)3 is a dominantly transmitted neurodegenerative disorder involving the basal ganglia and cerebral cortex that typically strikes in midlife, where survival from onset to death averages 17-20 years. Its prevalence is around 5-10 cases per 100,000 worldwide, which makes it one of the most common inherited neurodegenerative disorders. The characteristic symptoms of HD are involuntary choreiform movements, cognitive impairment, mood disorders, and behavioral changes that are chronic and progressive over the course of the illness. The underlying gene defect was proved to be a CAG repeat encoding polyglutamine (polyQ) in exon 1 of a 348-kDa protein named huntingtin (htt) (1, 2). In the unaffected population, the number of CAG repeats varies from 6 to 34 while repeats of 36 or more define an HD allele. The variability of the pathological alleles is quite wide, ranging from 36 to 121 repeats, displaying an inverse correlation with onset age (3).
Endogenous wild-type htt was shown to be essential for the normal development and health of the individual, but its mutated form confers a toxic gain-of-function (4). In 1997, polyQ aggregates were reported in the brains of transgenic mice and in the postmortem brains from patients with HD in the form of nuclear inclusions (5, 6).
Several models have been proposed to explain the mechanism by which the mutant htt causes neuronal degeneration (7), for example impairment of transcription and gene expression (8-11), impairment of axonal transport and synaptic transmission (12-14), suppression of energy metabolism (15, 16), and induction of apoptosis (17). Although controversial, mutant htt has also been proposed to impair the ubiquitin proteasome system (UPS) (18-21). Despite enormous progress in elucidating the molecular pathology of HD, the prognosis for patients has improved little since the first description of this disease, thus no effective treatments for HD patients have been developed.
Rho-associated kinases (ROCKs) are Ser/Thr protein kinases, which were found to be downstream targets of the small GTPase RhoA GTPase (22-24). In the mammalian system, ROCKs consist of two isoforms. ROCK1 (ROKβ, p160ROCK) is located on chromosome 18 and encodes a 1,354-amino acid protein (22). ROCK2 (ROKα), is located on chromosome 12 and contains 1,388 amino acids (25).
The ROCKs are important regulators of cell growth, migration, and apoptosis via control of actin cytoskeletal assembly. They regulate cell contraction through serine-threonine phosphorylation of adducin, ezrin-radixin-moesin (ERM) proteins, LIM kinase, myosin light chain phosphatase (MLCP), and Na/H exchanger 1 (NHE-1) (25, 26). RhoA/ROCK was shown to regulate the intracellular localization and phosphorylation of phosphatase and tensin homolog (PTEN), and RhoA/ROCK-mediated phosphorylation of PTEN is required for the phospholipid phosphatase activity of PTEN that antagonizes PI3K-mediated Akt signaling (27).
The RhoA/ROCK pathway was also reported to be implicated in the Aβ42 processing (28). Using APPswe-expressing Neuro2a mouse neuroblastoma cells, it was demonstrated that ROCK modulates shedding of sAPPα induced by statins. Constitutively active ROCK mutant attenuated sAPPα shedding from both untreated and statin-treated cells, whereas a ROCK mutant without kinase activity activated sAPPα shedding (29). Calorie restriction (CR)-induced SIRT1 expression promotes α-secretase activity, sAPPα generation, and diminishes Aβ generation by neurons from Tg2576 mice and CHO cells expressing APPswe. This effect of SIRT1 appears to be dependent on ROCK1 (30). The inhibition of ROCK activation was reported to be also an efficacious approach for the treatment of acute ischemic stroke (31).
Blocking the RhoA/ROCK pathway has been shown to markedly inhibit the polyQ protein aggregation and decrease its toxicity in cellular and Drosophila model of HD (32). ROCK1 and protein kinase C-related protein kinase-2 (PRK-2) have been identified to be the mediators of aggregation inhibition by Y-27632 (33). Moreover, a downstream effector of ROCK1, actin-binding factor profilin, was reported to inhibit the mutant htt aggregation by direct interaction via its polyproline-binding domain (34). Unlike htt, the inhibition of the mutant androgen receptor (AR) aggregation by profilin was not mediated by direct interaction (34).
We tested the effect of Y-27632 on several proteins with expanded polyQ and it was able to efficiently reduce the aggregation of these proteins. In addition to htt and AR, the tested proteins included mutant full-length and truncated ataxin-3 and full-length atrophin-1 with expanded polyQ (Fig. 1). Therefore in this study we investigated whether there might be a common mechanism by which the chemical ROCK inhibition leads to the reduced polyQ aggregation. We found that enhanced degradation of expanded polyQ protein largely contributes to this effect. Surprisingly, both major degradation systems, UPS and macroautophagy (hereafter referred to as autophagy), appeared to be activated by ROCK inhibition, and involved in reducing the polyQ aggregation.
FIGURE 1.
Y-27632 reduced aggregation of different proteins with expanded polyQ. Neuro2a cells were plated into 24-(for ArrayScan analysis) or 12-well plates (for Western blot analysis), transfected and 4-h later differentiated and treated with Y-27632. A, C, E, G, and I, ArrayScan analyses of the inclusion formation in Neuro2a cells transfected by plasmids coding different polyQ-containing proteins and treated with increasing concentrations of Y-27632 (10, 20, and 50 μm). Y-27632 reduced the inclusion formation of all tested proteins in a dose-dependent manner. Bars represent the relative mean values ± S.D. from three independent experiments. 0 μm values represent the control conditions (0 μm = 1). *, p < 0.05; **, p < 0.005; ***, p < 0.001. B, D, F, H, and J, Western blot analyses showed that Y-27632 reduced the levels of soluble and insoluble (gel top) forms of all tested proteins with expanded polyQ in a dose-dependent manner. A, and B, tNhtt-60Q, 24 h after transfection. C, and D, AR with 99Q, 24 h after transfection. E, and F, truncated ataxin-3 (trATX-3) with 130Q, 24 h after transfection. G and H, full-length ataxin-3 (FL ATX-3) with 130Q, 48 h after transfection. I, and J, DRPLA protein (atrophin-1) with 71Q, 48 h after transfection.
EXPERIMENTAL PROCEDURES
Materials—The ROCK inhibitors Y-27632, HA1077, and H89 were obtained from Sigma. H-1152, propidium iodide (PI), and the autophagy activator, rapamycin (35) were from Calbiochem. Fluorescent nucleic acid stain Hoechst 33258 was from Molecular Probes. MG-132 (Z-Leu-Leu-Leu-aldehyde) was from Wako Chemicals and 3-methyladenine (3MA) was from Sigma.
Mouse monoclonal antibody specific for N-terminal of huntingtin (EM48) and rat monoclonal anti-β-tubulin antibodies were from Chemicon. Anti-LC3, anti-GFP, and anti-RFP antibodies were from MBL and anti-ubiquitin antibody was from DAKO. Mouse monoclonal anti-ROCK1 and anti-ROCK2 antibodies were purchased from BD Transduction Laboratories and anti-ATG5 antibody was kindly provided by Dr. Mizushima. All other chemicals were from Sigma or Nacalai tesque unless otherwise specified.
Plasmids—Plasmids encoding the truncated N-terminal of human huntingtin (tNhtt) with 16, 60, and 150 glutamine repeats were introduced in pEGFP-N1 vector as previously described (36). The construction of plasmids encoding human truncated or full-length ataxin-3 containing 130Q (in pEGFP-N1 vector) was described previously (37). Human androgen receptor with 23Q (AR23Q) was amplified from human brain cDNA library by PCR using a set of primers BglII-AR-Fw (5′-AAAAGATCTATGGAAGTGCAGTTAGGGCT-3′) and SalI-AR-Rv (5′-AAAAAAGTCGACCTGGGTGTGGAAATAGATGG-3′), cleaved by BglII and SalI, and introduced into the BglII-SalI sites of the pEGFP-C1 vector (EGFP-AR23Q). The CAG repeat tract of EGFP-AR23Q was expanded via a method previously described (38). A primer set, BglII-AR-Fw and MmeI-AR-exp-Rv (CATCCTCACCCTGCTGCTGCTCCAACTGCCTGGGG) was used to amplify the 5′-coding sequence including the CAG repeat tract. Another primer set, MmeI-AR-exp-Fw (AGGCCGCGAGCGCAGCACCTTCCGACGCCAGTTTG) and AR-630Rv (TCTCCCGCTGCTGCTGCCTT) was used to amplify the CAG tract and its 3′-flanking region. These two fragments were digested by MmeI (New England Biolabs), gel-purified, and treated with T4 DNA ligase to connect them at their CAG repeat tracts. The ligated fragment was gel-purified and amplified by PCR using BglII-AR-Fw and AR-630Rv primers, and digested by BglII and AflII. The resulting fragment was ligated with EGFP-AR. By two cycles of expansion, EGFP-AR45Q and EGFP-AR99Q were obtained. The N terminus fragment of AR99Q, was amplified by PCR using primers BglII-AR-Fw and SalI-AR-396Rv (AAAAAAGTCGACGACGCAACCTCTCTCGGGGT), cleaved by BglII and SalI, and subcloned into the BglII-SalI sites of the pEGFP-C1. EGFP-DRPLA construct encoding atrophin-1 with Gln-71 was described previously (39) and was kindly provided by Dr. Masao Yamada. To prepare pcDNA3.1-tNhtt-60Q-EGFP for transient transfection, tNhtt-polyQ-EGFP fragment was cut from pIND-tNhtt-polyQ-EGFP (40) with HindIII-XbaI digestion, and the resulting fragment was inserted into pcDNA3.1-v5/His plasmid. The monomeric red fluorescence protein (mRFP) (41) and the ubiquitinated (Ub) Discosoma Red fluorescent protein (dsRed)2/N1 plasmids (42) were previously described.
Cell Culture and Treatment—Mouse neuroblastoma (Neuro2a; N2a) cells lines stably transfected with inducible expression of tNhtt-16Q-EGFP, tNhtt-60Q-EGFP, tNhtt-150Q-EGFP, and tNhtt-150Q-NLS-EGFP, which express a cDNA encoding htt exon 1 containing 16, 60 or 150 CAG repeats and fused with EGFP and eventually nuclear localization signal (NLS) (43), were previously established using the ecdysone-inducible mammalian expression system (Invitrogen) (36, 40). Neuro2a and mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37 °C in an atmosphere containing 5% CO2 and 95% air. Neuro2a cells were induced to express tNhtt-polyQ with 1 μm ponasterone A (ponA, Invitrogen) and differentiated to neuronal phenotype with 5 mm N6, 2′-O-dibutyryladenosine-3′, 5′-cyclic monophosphate sodium salt (dbcAMP) (Nacalai tesque). The differentiation status of the Neuro2 cells treated with dbcAMP is shown in supplemental Fig. S1. Except for the chase experiments, the cells were incubated with drugs at the time of differentiation and induction. MEFs were induced to ATG5(-/-) phenotype with 10 ng/ml doxycycline for 5 days as previously described (44).
Cells were transfected when they reached about 70-80% confluence. Transfection by Lipofectamine 2000 (Invitrogen) was done in accordance to the manufacturer's protocol in 24-well plates. Cells were used for experiments at indicated times after transfection.
Cell Death Assay—For quantification of cell death, 5 μg/ml each of Hoechst 33342 and PI were added to differentiated and induced Neuro2a cultures incubated with ROCK inhibitors. After 10 min at 37 °C, the PI-positive cells were quantified with ArrayScan (Cellomics).
RNA Interference—Each sense and antisense template short hairpin RNA (shRNA) for ROCK1 and ROCK2 was purchased from Operon, annealed and ligated into pSilencer1.0 vector with U6 promotor according to the manufacturer's instructions (Ambion). The target sequences were as follows: ROCK1, 5′-AAGTAGTGACATTGATACTAG-3′; ROCK2, 5′-AACAATAGAGATCTACAAGAT-3′. The plasmids containing shRNA were sequence-verified. Plasmids were transfected into Neuro2a cells using Lipofectamine 2000. Ater 2 days of silencing, cells were induced.
ArrayScan Quantification—For the inclusions (visible aggregates) quantification, cells were grown in 24-well plates for indicated periods, fixed in 4% paraformaldehyde, washed, and incubated with Hoechst 33258 at 1:1000 dilution in PBS. Cells were analyzed with ArrayScan®VTI High Content Screening (HCS) Reader using Target Activation BioApplication (TABA). TABA analyzes images acquired with an HCS Reader and provides measurements of the intracellular fluorescence intensity and localization on a cell-by-cell basis.
In each well, more than 10,000 cells were counted and quantified for the presence of the inclusions. Nuclei stained by Hoechst 33285 provided the autofocus target and a count gave the exact number of the quantified cells. The screening itself consisted of two scans using Hoechst and fluorescein isothiocyanate (for GFP) fluorescence. First, the number of inclusions was calculated when fluorescent spots of at least 5 pixels (magnification 20× for cytoplasmic and 40× for nuclear aggregates) and an average GFP intensity of more than 1500 were labeled as inclusions. Nuclei were then defined as the objects of interest and their number determined. The percentage of the cells with inclusions was then calculated. The reliability of the inclusion quantification by ArraScan was validated by test-counting of inclusions by eyes (supplemental Fig. S2). The number of mRFP- or Ub-dsRED-positive cells and fluorescent intensity was quantified by single scan by detecting the red fluorescence of each cell in the perinuclear region within a distance of 3 pixels from the nucleus. When the average intensity exceeded 50, the cell was considered mRFP/Ub-dsRED-positive. Scanning was performed with three or four times in each experimental condition. Data were generated from the quantification of more than 250,000 cells in each experimental set-up.
TaqMan Reverse Transcriptase-PCR (RT-PCR)—Total RNA and cDNAs were prepared from Neuro2a cells as described previously (45). The TaqMan primer and probe sets were designed and synthesized based on Primer Express Software (Applied Biosystems). The nucleotide sequences of the primers for EGFP were as follows: forward 5′-AGCAAAGACCCCAACGAGAA-3′, reverse 5′-GGCGGCGGTCACGAA-3′, TaqMan probe 5′-CGCGATCACATGGTCCTGCTGG-3′; TaqMan RT-PCR was performed as described previously (46). All values obtained were normalized against the levels of β-actin using the following primers: forward 5′-TCTTTGCAGCTCCTTCGTTG-3′, reverse 5′-ATCGTCATCCATGGCGAAC-3′, TaqMan probe 5′-CGGTCCACACCCGCCACC-3′.
Chase Experiments—To determine whether soluble tNhtt-polyQ degrades faster in the presence of ROCK inhibitors, chase experiments were performed. Neuro2a cells were first differentiated and induced to express tNhtt-polyQ for 24 h in case of 16Q and 60Q and 16 h in case of 150Q cells. Thereafter, ponA was removed, the cells were washed, and incubated in a medium containing dbcAMP (for maintaining differentiation status) with either water (control) or ROCK inhibitors at 20 μm concentration for 4 or 5 days. Medium was replaced every 2 days with the same concentration of the drugs, and cells were collected everyday. The cells were subsequently lysed, and the levels of tNhtt-polyQ analyzed using Western blotting.
Western Blot Analysis—Cells were washed twice with ice-cold PBS, scraped, and resuspended in lysis buffer (0.5% Triton X-100 in PBS, 0.5 mm phenylmethylsulfonyl fluoride, Complete protease inhibitor mixture (Roche Applied Sciences). After incubating on ice for 30 min lysates were briefly sonicated. Protein concentrations were determined according to the method of Bradford using Bio-Rad protein assay reagent (Bio-Rad). Equal amounts of protein were boiled for 5 min in 2× SDS-sample buffer and then separated by 5-12% gradient SDS-PAGE and electrophoretically transferred to a polyvinylidene difluoride (polyvinylidene difluoride) membrane. The membranes were blocked in 5% skim milk in 0.05% Tween 20/Tris-buffered saline (TBS-T) and then incubated with primary antibody (dilutions in accordance with manufacturer's recommendations) overnight at 4 °C. The membranes were washed three times in TBS-T and incubated for 1 h with horseradish peroxidase-conjugated secondary antibody (dilution 1:5000). Immunoreactive proteins were detected with enhanced chemiluminescence reagents (Amersham Biosciences).
Filter Trap Assay (FTA)—FTA was performed using a Hybri-Dot manifold (Bio-Rad) and cellulose acetate membrane filter with a pore size of 0.2 μm (Advantec). The cell lysates were prepared as for Western blotting. The same amount of protein from each experimental condition was diluted to 100 μl of PBS with 2% SDS and applied to the membrane. Soluble proteins were removed by vacuum suction while the SDS-resistant aggregates stayed trapped. Wells were washed three times with 2% SDS/PBS, and suction was maintained for 20 min to allow thorough and complete trapping of SDS insoluble material. Membranes were subsequently blocked with 5% skim milk, and immunostaining was performed.
In Vitro UPS Activity Assay—Neuro2a cells were transfected with ROCK1 and/or ROCK2 shRNA and 2 days later, 10 μm MG-132 was added. Eighteen hours later, cells were collected, and 5 μg of total protein from each lysate was pipetted to 96-well plate and 100 μl of fluorogenic UPS substrate I (trypsin-like activity) or II (peptidyl glutamyl peptide hydrolase (PGPH)-like activity) (Calbiochem). Plates were placed in the Arvo MX 1420 Multilabel Counter (Perkin Elmer), and absorbance was detected at 460 nm.
Statistical Analysis—We used the unpaired Student's t test for comparison between two samples. One-way ANOVA Fisher's test followed by Tukey's HSD test or two-way ANOVA test with pairwise contrast were performed using XLSTAT or Partek Genomic Solution Software. The statistical significance was confirmed by the non-parametric Mann-Whitney test where indicated. We considered the difference between comparisons to be significant when p < 0.05 for all the statistical analyses.
RESULTS
ROCK Inhibitors Inhibit PolyQ Aggregation—First, we investigated whether the effect of Y-27632 on polyQ aggregation is limited to htt and AR or if it is also able to inhibit the aggregation of other polyQ-containing proteins. We found that beside htt and AR, Y-27632 decreased the aggregation of truncated and full-length ataxin-3 and atrophin-1 in a dose-dependent manner (Fig. 1). To test whether the polyQ aggregation is decreased by more ROCK inhibitors or whether it is a specific effect of Y-27632, we examined four different drugs inhibiting ROCK. The ArrayScan analysis revealed that all of them were able to decrease the polyQ aggregation in 150Q and 150Q-NLS Neuro2a cells after 1 or 2 days of differentiation and induction in a dose-dependent manner (Fig. 2, A-D). The 150Q-NLS cell line was examined after 2 days because the nuclear inclusion formation sufficient for ArrayScan analysis did not appear earlier. Y-27632, H-1152, and HA1077 were very efficient in both cell lines, while H89 did not have so strong effect after 2 days of treatment. These results were confirmed by Western blot, as shown in a representative Western blot in Fig. 2E for Y-27632 and H-1152. Next, we examined the effect of Y-27632 on the polyQ-mediated cytotoxicity. After 3 days of 150Q and 150Q-NLS expression in Neuro2a cells, Y-27632 was able to decrease the percentage of propidium iodide-positive cells in both cell lines (Fig. 2F). These data confirmed the inhibitory effect of ROCK inhibitors on polyQ aggregation and polyQ cytotoxicity.
FIGURE 2.
ROCK inhibitors decrease the cytoplasmic and nuclear polyQ aggregation and toxicity in 150Q and 150Q-NLS Neuro2a cells. Cells were plated into 24-(for ArrayScan analysis) or 12-well plates (for Western blot analysis). On the following day, cells were differentiated with 5 mm dbcAMP and induced with 1 μm ponA for different time periods as indicated in the figure and below. Cells were fixed with 4% PFA and nuclei stained with Hoechst 33258. A, composite image generated by ArrayScan (20× magnification) shows a clear decrease in polyQ aggregation in 150Q Neuro2a cells after 24 and 48 h by 20 μm Y-27632. B-D, ROCK inhibitors Y-27632, H-1152, HA1077, and H89 significantly decreased the inclusion formation rate in 150Q N2a cells 24 h (B) or 48 h (C) after induction and treatment and in 150Q-NLS N2a cells 48 h (D) after induction and treatment in dose-dependent manner. E, 150Q Neuro2a cells were plated into 12-well tissue culture plates, and the next day the cells were differentiated with 5 mm dbcAMP, induced with 1 μm ponA, and treated with Y-27632 or H-1152. Cells were collected 24-h later and processed for immunoblot analysis using anti-GFP and anti-β-tubulin antibodies. Western blot analysis showed a reduction of the soluble and insoluble 150Q in Neuro2a cells by Y-27632 and H-1152 treatment in a dose-dependent manner. F, cell death in 150Q and 150Q-NLS Neuro2a cells. Cells were plated into 24-well tissue culture plates, and the following day, the cells were differentiated, induced, and treated with 20 μm Y-27632. After 3 days, PI and Hoechst 33258 were added to the medium in final concentrations of 5 μg/ml. Cells were incubated for 10 min at 37 °C before ArrayScan analysis was performed. Y-27632 reduced the PI-positive cells by 31% in 150Q and 36% in 150Q-NLS Neuro2a cells (n = 3) as compared with untreated cells. Bars in B-D represent the relative mean values ± S.D. from four different experiments. The control value of 1 represents the control conditions with no ROCK inhibitor added to the cells. *, p < 0.05; **, p < 0.005; ***, p < 0.001; p < 0.05 by non-parametric Mann-Whitney test for significant data by ANOVA.
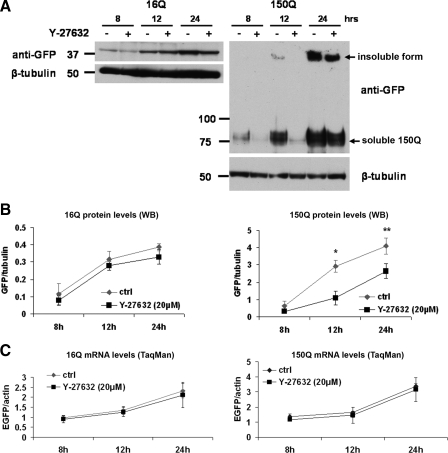
Y-27632 Decreases the Level of PolyQ Protein—Because the decrease in insoluble form of polyQ on gel top was not accompanied by an increase in the monomeric polyQ protein (Fig. 2E), we investigated the rate of htt expression. In 16Q Neuro2a cells, treatment with Y-27632 did not have significant effect on the protein level after 8, 12, and 24 h of induction (Fig. 3, A and B (left panel)). On the other hand, the level of 150Q protein decreased drastically when cells were incubated with the drug (Fig. 3, A and B (right panel)). The TaqMan real-time PCR showed no difference at the transcriptional level between treated and untreated cells (Fig. 3C).
FIGURE 3.
Y-27632 decreases the levels of htt with expanded polyQ without affecting the transcription. 16Q and 150Q Neuro2a cell were plated into 12-well plates. On the following day, cells were differentiated, induced, and treated with 20 μm Y-27632 for different time periods as indicated in the figure. Cells were collected, and total RNA for quantitative TaqMan RT-PCR or proteins for immunoblot analysis were prepared. A, representative Western blot showing the htt protein levels in 16Q and 150Q Neuro2a cells at different time points after induction and treatment using anti-GFP and anti-β-tubulin antibodies. B, quantification of band intensities of the anti-GFP immunoblots representing soluble forms of the proteins for 16Q (left panel) and 150Q Neuro2a cells (right panel) collected from three independent experiments. Levels of 16Q protein showed no significant difference between untreated and Y-27632-treated cells. Levels of soluble 150Q were significantly reduced upon Y-27632 treatment starting 12 h after induction. Data were normalized using β-tubulin. C, quantitative TaqMan RT-PCR analysis of the 16Q-(left panel) and 150Q-EGFP (right panel) mRNA levels showed no significant transcription alterations by 20 μm Y-27632 treatment at any time point in three independent experiments. Data were normalized using actin mRNA levels. Values in B and C are mean ± S.D. *, p < 0.05; **, p < 0.005.
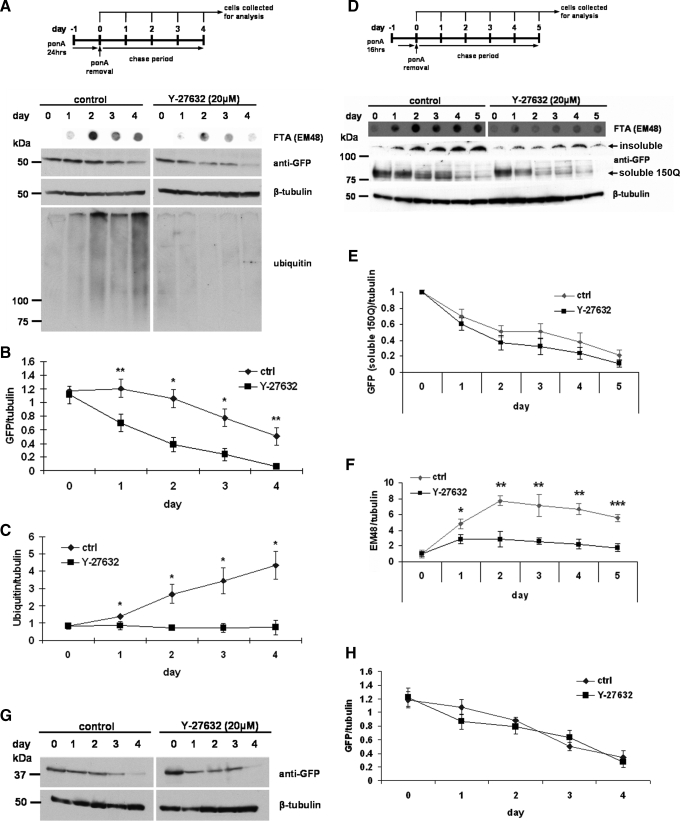
Y-27632 Enhances the Expanded PolyQ Degradation by UPS and Autophagy—We performed a set of chase experiments to investigate the mechanistic platform of the decreased expression of expanded htt. First, we induced the expression of htt in the 16Q and 60Q Neuro2a cells for 24 h, and after ponA removal, we collected the cells every 24 h. Y-27632 increased the turnover of expanded polyQ protein about 2.5 times compared with the control cells and markedly reduced the aggregation (Fig. 4, A and B). In case of 150Q, the effect on soluble form of the protein was not obvious, but the SDS-insoluble material was significantly decreased upon Y-27632 treatment (Fig. 4, D-F). This effect of Y-27632 was specific for expanded polyQ, because the levels of normal protein in 16Q Neuro2a cells were not affected (Fig. 4, G and H). Moreover, the drug was able to efficiently inhibit the accumulation of polyubiquitinated proteins after day 2 of the experiment (Fig. 4A (bottom panel) and C) suggesting the preservation of the UPS function. This data indicated that Y-27632 enhanced the degradation of mutant htt with expanded polyQ.
FIGURE 4.
Y-27632 enhances the degradation of htt with expanded polyQ. 16Q, 60Q, and 150Q Neuro2a cells were plated into 12-well plates and 1-day later differentiated with dbcAMP and induced with ponA. After 24 h in 16Q and 60Q cells and 16 h in 150Q Neuro2a cells, expression of htt polyQ was shut down by ponA removal, and cells were treated with 20 μm Y-27632 for the time periods indicated in the figure. Cells were collected and processed for immunoblot analysis using EM48, anti-GFP, anti-β-tubulin, or anti-ubiquitin antibodies. A, Western blot and FTA analysis of the chase experiment in 60Q Neuro2a cells showed enhanced degradation of soluble 60Q (anti-GFP) and reduced accumulation of the insoluble form of the protein (EM48; FTA) upon treatment with Y-27632. The top panel displays the experimental design of the chase experiments in 60Q and 16Q Neuro2a cells. B, densitometric quantification of a Western blot confirmed significant augmentation of soluble 60Q degradation in cells treated with Y-27632 comparing to untreated cells. C, quantification of ubiquitin immunoreactivity showed no accumulation of polyubiquitinated proteins in Y-27632-treated cells in contrast to untreated cells. D, Western blot and FTA analysis of the chase experiment in 150Q Neuro2a cell line revealed reduced accumulation of the insoluble form of the protein (anti-GFP, gel top in Western blot; EM48, FTA) by Y-27632 treatment. The top panel displays the experimental design of the chase experiment in 150Q Neuro2a cells. E, densitometric quantification of the soluble 150Q levels (anti-GFP, Western blot) revealed no significant difference between treated and untreated cells. F, quantification of the SDS-insoluble (aggregated) 150Q (EM48, FTA) exhibited a marked increase of the insoluble protein clearance in Y-27632-treated cells. G, Western blot analysis of the chase experiment in 16Q Neuro2a cell line. H, densitometric quantification revealed no difference of 16Q degradation between treated and untreated cells. All data were normalized using β-tubulin. All presented values are mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.005.
Next we examined which of the main degradation pathways is involved. We blocked autophagy and the UPS with 10 mm 3-methyladenine (3MA) and 10 μm MG-132, respectively (Fig. 5). The inhibition of the UPS, but not of autophagy, alleviated the effect of the drug on the soluble polyQ levels. Aggregation inhibition was partly abated when the cells were treated with 3MA or MG-132. When we blocked both pathways, the effect of Y-27632 on the soluble polyQ protein levels and aggregation was almost completely eliminated as observed using FTA (Fig. 5, A-C) and ArrayScan analysis (Fig. 5, D and E). To confirm this observation, we induced the ATG5(-/-) genotype in MEF 5-7 cells with doxycycline to block autophagy (Fig. 5F). After 5 days, the cells were transfected with 150Q-EGFP and treated. The effect of Y-27632 on polyQ aggregation was eliminated only upon the absence of autophagy (ATG5(-/-) cells) and treatment with 10 μm MG-132 (Fig. 5, F and G). These results confirmed that the effect of Y-27632 on polyQ turnover is mediated by both the UPS and autophagy.
FIGURE 5.
Inhibition of UPS and/or autophagy ameliorates the effects of Y-27632. A, 150Q Neuro2a cells were plated into 12-well plates. On the following day, cells were differentiated, induced, and treated with different combinations of 20 μm Y-27632, 10 μm MG-132, and 10 mm 3MA. Cells were collected 24 h later, and Western blot and FTA analysis were performed using anti-GFP, anti-β-tubulin, and EM48 antibodies. B, quantification of the 150Q band intensities in Western blot revealed that the inhibition of UPS but not that of autophagy ameliorated the effect of Y-27632 on soluble protein. C, densitometric quantification of FTA showed that the combination of the UPS and macroautophgy blockage was necessary to abolish the effect of Y-27632 on the accumulation of the insoluble form of 150Q. Inhibition of each of the degradation pathways separately ameliorated only partly the effect of Y-27632. Densitometric data were normalized using β-tubulin. D, 150Q Neuro2a cells were treated as described above and processed for the ArrayScan analysis. Compound images generated by ArrayScan show the effect of degradation pathways block on Y-27632-mediated polyQ aggregation. E, ArrayScan analysis quantification supported the data obtained by FTA analysis. F, MEF 5-7 cells were treated with 10 ng/ml doxycycline for 5 days to knockdown Atg5 and inhibit autophagy. To demonstrate the UPS inhibition by 10 μm MG-132 (MG), cells were transfected with ubiquitinated dsRED (Ub-dsRED) or mRFP and treated with MG for 1 day. Western blot analysis using anti-Atg5 antibody showed almost complete absence of Atg5 protein (in complex with Atg12) in doxycycline-treated cells. Anti-LC3 immunoblot revealed the consequent lack of autophagy activity in these cells. The levels of polyubiquitinated proteins and Ub-dsRED increased in cells treated with MG, while mRFP levels remained unchanged. G, untreated and doxycycline-treated MEF 5-7 cells were plated into 6-well plates and next day transfected with tNhtt-150Q-EGFP. Cells were then treated with 20 μm Y-27632 or 0.2 μm rapamycin and/or 10 μm MG-132 for 24 h. ArrayScan analysis confirmed that only the simultaneous inhibition of both UPS and autophagy negated the effect of Y-27632 (in dox +/Atg5-cells treated with MG-132 (MG)). Rapamycin, an mTOR inhibitor, activates autophagy and was used as a control. Bars represent the relative mean values ± S.D. from three independent experiments. The control value of 1 represents the control conditions without Y-27632 (B, C, and E) or without Y-27632 and rapamycin treatment (G). *, p < 0.05; **, p < 0.005; ***, p < 0.001.
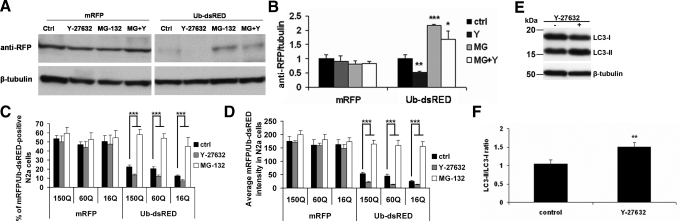
Y-27632 Increases the Activity of UPS and Autophagy—To investigate how the degradation systems are modulated by Y-27632, we first examined UPS activity using transient transfection of ubiquitin-dsRED (Ub-dsRED) and mRFP as a control. Y-27632 reduced the accumulation of Ub-dsRED in Neuro2a cells while it did not have an effect on mRFP levels (Fig. 6, A and B). When the UPS was blocked with MG-132, the levels of Ub-dsRED more than doubled over that of the control cells, and the effect of Y-27632 fell (Fig. 6, A and B). Quantification of the Ub-dsRED- or mRFP-positive 16Q, 60Q, and 150Q Neuro2a cells by ArrayScan revealed that Y-27832 treatment significantly reduced the number of Ub-dsRED-positive cells as well as average fluorescent intensity, while it had no effect on cells transfected with mRFP (Fig. 6, C and D). These results showed that Y-27632 was able to activate the UPS in Neuro2a cells lacking mutant protein and that it alleviated the UPS block caused by expanded polyQ in 60Q and 150Q Neuro2a cells.
FIGURE 6.
Y-27632 increases the UPS and autophagy activity. A, wild type Neuro2a cells were plated into 12-well plates. On the following day, cells were transfected with mRFP or Ub-dsRED and 4-h later differentiated and treated with 20 μm Y-27632 and/or 10 μm MG-132 for additional 24 h. Cells were then processed for immunoblot with anti-RFP and anti-β-tubulin. B, Western blot quantification confirmed that Y-27632 significantly decreased the levels of Ub-dsRED but not mRFP. C and D, 150Q, 60Q, and 16Q Neuro2a cells were plated into 24-well plates and transfected in a similar manner as described in A. Four hours later, the cells were differentiated, induced, and treated with drugs. ArrayScan analysis revealed that the percentage of Ub-dsRED-positive cells (C) and average red fluorescence in the cells (D) decreased in Y-27632-treated cells. Y-27632 had no effect on mRFP fluorescence. E, wild type Neuro2a cells were plated into 6-well plates, and 1-day later, cells were left untreated or treated with 20 μm Y-27632 for 24 h and processed for Western blot analysis of the LC3 conversion. F, densitometric quantification confirmed that Y-27632 significantly increased the conversion of LC3 representing increased autophagic activity in the treated cells. Bars in B and F represent the relative mean values ± S.D. from three independent experiments. The control value of 1 represents the control conditions without Y-27632 treatment. Bars in C and D represent mean values ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.001.
During autophagy, the cytosolic form of microtubule-associated protein 1 light chain 3 (LC3-I) is conjugated to phosphatidylethanolamine (PEA) to form LC3-PEA conjugate (LC3-II), which is recruited to autophagosomal membranes. Thus, lysosomal turnover of the autophagosomal marker LC3-II reflects autophagic activity (47). Y-27632 treatment of Neuro2a cells resulted in increased LC3-II/LC3-I ratio as observed by Western blot analysis (Fig. 6, E and F). The presented data suggested that Y-27632 is able to activate UPS and autophagy in Neuro2a cells.
ROCK1 and/or ROCK2 Knockdown Caused Activation of Degradation Systems—Knockdown of ROCKs in 150Q Neuro2a cells by shRNA resulted in reduction of polyQ aggregation (Fig. 7, A and B). ROCK1 silencing had stronger effect as compared with ROCK2 on visible aggregate formation (Fig. 7B). Although the knockdown of ROCK2 significantly decreased the amount of visible aggregates, we did not observe a marked decrease in insoluble fraction of 150Q on the gel top, probably because many of the large aggregates were removed by centrifugation during cell lysate preparation (Fig. 7A). Treatment with 20 μm Y-27632 produced additional effect at all knockdown conditions, suggesting that ROCKs are not the only targets of Y-27632 with antiaggregational activity.
FIGURE 7.
Knockdown of ROCK1 and/or ROCK2 reduces polyQ aggregation and activates the main degradation pathways in 150Q Neuro2a cells. 150Q Neuro2a cells were plated into 12-well plates and 1-day later transfected with different combinations of pSil vector, ROCK1 (R2) and ROCK2 (R2) shRNA (+mRFP as the transfection marker). After 4 h, the cells were differentiated for 48 h, then induced, and eventually treated with 20 μm Y-27632. Cells were processed 24 h later for Western blot or ArrayScan analysis. A, immunoblot using anti-GFP, anti-ROCK1, anti-ROCK2 antibodies showed decreased levels of 150Q (especially the soluble form) upon ROCKs silencing. LC3 conversion was enhanced only in the absence of both ROCKs. B, ArrayScan analysis showed a significant reduction of 150Q inclusion formation by ROCKs silencing. Y-27632 treatment had an additional affect in all ROCK knockdown conditions. C, quantification of the band intensities of LC3 blots revealed enhanced conversion of LC3 when both ROCK isoforms were knocked down. D and E, ROCKs knockdown was implemented in a similar manner as described above. After 2 days, cells were left untreated or treated with 10 μm MG-132 (for assay control) and incubated for 18 h. Cell lysates were then prepared for in vitro UPS activity assays. Knockdown of ROCK1 caused activation of trypsin-like activity, while ROCK2 appeared to be responsible for the PGPH activity of the UPS. The y axis represents the absorbance at 460 nm. Bars in B, D, and E represent mean values ± S.D., and bars in C represent the relative mean values ± S.D. from three independent experiments. The control value of 1 in C represents the control cells transfected with empty pSil vector. *, p < 0.05; **, p < 0.005; ***, p < 0.001.
Next, we investigated whether ROCKs influence the activity of the UPS and autophagy. The autophagy activity increased only when both isoforms of ROCK were silenced. Knockdown of a single ROCK had no effect on LC3-I to LC3-II conversion (Fig. 7, A and C). In vitro UPS activity analysis revealed that the knockdown of ROCKs activated the UPS (Fig. 7, D and E). The knockdown of ROCK1 significantly increased the trypsin-like activity, and that of ROCK2 increased the PGPH-like activity of the UPS. These results confirmed the involvement of ROCKs in the regulation of the cellular degradation pathways.
DISCUSSION
While the effect of Y-27632 on polyQ aggregation was previously identified in a screening study (32), the involvement of ROCKs was not confirmed until more recently (33, 34). In one of these reports, ROCK1 and PRK-2 were reported to be the effectors of Y-27632 in polyQ aggregation inhibition (33). The second study showed that the inhibition of profilin phosphorylation at Ser-137 by ROCK1 enhanced the direct binding of profilin to the polyproline region of htt. This interaction may sequester htt or stabilize it in a less aggregation-prone conformation. ROCK inhibitors were able to reduce the aggregation of AR as well, although the G-actin binding ability of profilin rather than direct interaction with AR appeared to be important (34).
In this report, we report on a novel effect of the ROCK inhibitor Y-27632 enhancing the clearance of mutant htt. We investigated the impact of Y-27632 on htt, AR, ataxin-3, and atrophin-1 with expanded polyQ and found a profound reduction of polyQ aggregation following Y-27632 treatment in all tested proteins. We hypothesize that in addition to previously published facts there might be yet unknown common mechanism responsible for this effect.
Therefore we investigated how mutant htt is processed after treatment with ROCK inhibitors, and found that the enhanced clearance of the expanded polyQ protein significantly contributed to the reduced polyQ aggregation. The levels of mutant htt fell markedly especially during the early phases after the induction of 150Q Neuro2a cells (Fig. 3). This change was not a consequence of modified transcription, since the TaqMan analysis showed no difference between the control and Y-27632-treated cells.
To investigate whether the decreased levels were caused by a higher turnover of the polyQ protein, we performed a set of chase experiments, which confirmed the enhanced degradation of htt (Fig. 4). Blocking of either the UPS or autophagy by MG-132 and 3MA, respectively, did not reverse the effect of Y-27632 completely. UPS inhibition suppressed the effect of the drug on the soluble form of 150Q and partially on the insoluble form. Autophagy inhibition had similar effect on the insoluble form of 150Q while the soluble form remained almost unchanged. Only simultaneous blockage of both main degradation systems reversed the effect of Y-27632 on both forms of 150Q (Fig. 5). When we overexpressed Ub-dsRED in wild type, 150Q, 60Q, or 16Q Neuro2a cell lines and treated them with Y-27632, the accumulation of Ub-dsRED markedly decreased as compared with untreated cells, suggesting the increased activity of the UPS (Fig. 6). Treatment of Neuro2a cells with Y-27832 caused enhanced conversion of LC3-I to LC3-II which suggested increased autophagy activity (Fig. 6). Our data show that the promotion of mutant htt degradation contributed in great extent to aggregation inhibition by Y-27632.
Reducing the intracellular levels of the mutant protein is the goal of many therapeutic approaches for polyQ diseases. This can be achieved by enhancing its degradation. The degradation of misfolded and damaged proteins by the UPS is essential to maintaining protein quality control, and the malfunctioning of the UPS has been linked to several neurodegenerative disorders (48). Mutant htt is a substrate of the UPS, but the presence of expanded polyQ inhibits the UPS, which results in further accumulation of htt (19-21, 41). We recently reported two drugs, amiloride and benzamil, as able to activate the UPS and enhance the clearance of mutant htt to reduce polyQ aggregation and toxicity in both the cellular and mouse models of HD (49). As well, attempts to increase the autophagic clearance of mutant htt resulted in reduced htt toxicity. Reports on several small molecules with therapeutic potential for HD activating autophagy, including rapamycin and lithium have been published (35, 50, 51). To the best of our knowledge, Y-27632 is the first chemical compound that is capable of enhancing both UPS and autophagy activity, though the effect on autophagy appeared weaker.
To investigate the direct involvement of ROCKs in the Y-27632 effect, we silenced ROCK1 and/or ROCK2. Surprisingly, both isoforms were able to reduce polyQ aggregation though the effect of ROCK1 appeared stronger (Fig. 7). Y-27632 had an additional effect in all knockdown conditions which is in agreement with the recent study identifying PRK-2 as another Y-27632 target (33). On the other hand, the effect of ROCK2 shRNA contradicts the study, which showed ROCK2 ineffective in inhibiting aggregation (33). This can be a consequence of using different methods for the aggregation detection. While the authors of the previous report utilized fluorescence resonance energy transfer (FRET), we counted visible aggregates using ArrayScan, or used immunoblotting (WB, FTA) to detect insoluble material. Further work will be necessary to clarify this discrepancy.
As well, identification of downstream effectors of ROCKs influencing degradation pathways will be necessary for the development of more effective therapies. ROCK regulates the activities of many target proteins by its kinase activity. Through some proteins, such as the myosin light chain (MLC), it induces actomyosin contraction which is an important step in cytoskeletal rearrangements (52). ROCK phosphorylates LIM kinases enhancing their activity, and the subsequent phosphorylation of cofilin proteins blocks their F-actin-severing activity (53-55). Another ROCK substrate, collapsin response mediator protein-2 (CRMP-2), is also involved in the regulation of cytoskeletal reorganization (56, 57). Taken together, ROCK activation leads directly to a number of actin-myosin-mediated processes including cell motility, adhesion, phagocytosis, neurite retraction, and smooth muscle contraction. These questions are thus raised: how does the cytoskeleton affect the activity of UPS and/or autophagy? Which of the ROCK targets are involved in this process?
We have observed that chemical ROCK inhibition efficiently disrupted aggresome formation in Neuro2a cells both in the presence and absence of htt with expanded polyQ (data not shown). Aggresome formation is a cellular response when UPS is overloaded by the production of aggregation-prone misfolded proteins (58) or UPS is chemically inhibited (59). ROCK has also been reported as a regulator of intracellular redistribution of lysosomes in invasive tumor cells (60), as we observed in Neuro2a cells (data not shown). The treatment of Neuro2a cells with Y-27632 hindered subcellular spreading of the lysosomes and promoted recruitment of the lysosomes in the perinuclear region. It is not clear however, if this redistribution contributes to the activation of autophagy, a matter still under investigation.
Another possible link between ROCKs and autophagy is the involvement of ROCKs in Akt signaling (27). Akt activation was shown to inhibit the mTOR-dependent autophagy activity in some cell lines including Neuro2a (61-63). Moreover, one recent study identified an actin-related protein (Arp2) to be a link between the actin cytoskeleton and autophagic machinery (64).
The downstream substrates responsible for degradation machinery activation are unknown and are the subjects of further investigation. This may potentially lead to identification or development of more potent and specific inhibitors providing effective treatment for polyQ-related diseases.
Supplementary Material
Acknowledgments
We thank Dr. Noboru Mizushima for the MEF 5-7 cell line and anti-ATG5 antibody. Also, we thank Dr. Masao Yamada for DRPLA (atrophin-1) plasmids. We would like to thank David W. Chapmon for the stylistic corrections in the manuscript.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas (Research on Pathomechanisms of Brain Disorders) from the Ministry of Education, Culture, Sport, Science, and Technology of Japan (17025044) and by a Grant-in-Aid for the Research on Measures for Intractable Diseases from the Ministry of Health, Welfare and Labor of Japan. This work was also supported in part by a grant from the JSPS (Japan Society for the Promotion of Science).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: HD, Huntington disease; polyQ, polyglutamine; htt, huntingtin; AR, androgen receptor; DRPLA, dentatorubropallidoluysian atrophy; tNhtt, truncated N-terminal huntingtin; ROCK, Rho kinase; EGFP, enhanced green fluorescent protein; NLS, nuclear localization signal; mRFP, monomeric red fluorescence protein; Ub-dsRED, ubiquitinated discosoma red fluorescent protein; PI, propidium iodide; 3MA, 3-methyladenine; MEF, mouse embryonic fibroblasts; dbcAMP, N6, 2′-O-dibutyryladenosine-3′, 5′-cyclic monophosphate sodium salt; RT-PCR, reverse transcriptase PCR; ponA, ponasterone A; FTA, filter trap assay; PBS, phosphate-buffered saline; shRNA, short hairpin RNA; PGPH, peptidyl glutamyl peptide hydrolase; UPS, ubiquitin proteasome system; PEA, phosphatidylethanolamine; PRK-2, protein kinase C-related protein kinase-2; ANOVA, analysis of variance.
References
- 1.Myers, R. H. (2004) NeuroRX, 1 255-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Huntington's Disease Collaborative Research Group (1993) Cell 72 971-983 [DOI] [PubMed] [Google Scholar]
- 3.Orr, H. T., and Zoghbi, H. Y. (2007) Annu. Rev. Neurosci. 30 575-621 [DOI] [PubMed] [Google Scholar]
- 4.Leavitt, B. R., van Raamsdonk, J. M., Shehadeh, J., Fernandes, H., Murphy, Z., Graham, R. K., Wellington, C. L., Raymond, L. A., and Hayden, M. R. (2006) J. Neurochem. 96 1121-1129 [DOI] [PubMed] [Google Scholar]
- 5.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L., and Bates, G. P. (1997) Cell 90 537-548 [DOI] [PubMed] [Google Scholar]
- 6.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P., and Aronin, N. (1997) Science 277 1990-1993 [DOI] [PubMed] [Google Scholar]
- 7.Landles, C., and Bates, G. P. (2004) EMBO Rep. 5 958-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka, T., Miyazaki, H., Oyama, F., Kurosawa, M., Washizu, C., Doi, H., and Nukina, N. (2008) EMBO J. 27 827-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kegel, K. B., Meloni, A. R., Yi, Y., Kim, Y. J., Doyle, E., Cuiffo, B. G., Sapp, E., Wang, Y., Qin, Z. H., Chen, J. D., Nevins, J. R., Aronin, N., and DiFiglia, M. (2002) J. Biol. Chem. 277 7466-7476 [DOI] [PubMed] [Google Scholar]
- 10.Nucifora, F. C., Jr., Sasaki, M., Peters, M. F., Huang, H., Cooper, J. K., Yamada, M., Takahashi, H., Tsuji, S., Troncoso, J., Dawson, V. L., Dawson, T. M., and Ross, C. A. (2001) Science 291 2423-2428 [DOI] [PubMed] [Google Scholar]
- 11.Steffan, J. S., Kazantsev, A., Spasic-Boskovic, O., Greenwald, M., Zhu, Y. Z., Gohler, H., Wanker, E. E., Bates, G. P., Housman, D. E., and Thompson, L. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6763-6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, W. C., Yoshihara, M., and Littleton, J. T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 224-3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, H., Li, S. H., Johnston, H., Shelbourne, P. F., and Li, X. J. (2000) Nat. Genet. 25 385-389 [DOI] [PubMed] [Google Scholar]
- 14.Usdin, M. T., Shelbourne, P. F., Myers, R. M., and Madison, D. V. (1999) Hum. Mol. Genet. 8 839-846 [DOI] [PubMed] [Google Scholar]
- 15.Beal, M. F. (2005) Ann. Neurol. 58 495-505 [DOI] [PubMed] [Google Scholar]
- 16.Ross, C. A. (2002) Neuron 35 819-822 [DOI] [PubMed] [Google Scholar]
- 17.Evert, B. O., Wullner, U., and Klockgether, T. (2000) Cell Tissue Res. 301 189-204 [DOI] [PubMed] [Google Scholar]
- 18.Davies, J. E., Sarkar, S., and Rubinsztein, D. C. (2007) BMC Biochem. 8 S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001) Science 292 1552-1555 [DOI] [PubMed] [Google Scholar]
- 20.Jana, N. R., Zemskov, E. A., Wang, G., and Nukina, N. (2001) Hum. Mol. Genet. 10 1049-1059 [DOI] [PubMed] [Google Scholar]
- 21.Bennett, E. J., Bence, N. F., Jayakumar, R., and Kopito, R. R. (2005) Mol. Cell 17 351-365 [DOI] [PubMed] [Google Scholar]
- 22.Ishizaki, T., Maekawa, M., Fujisawa, K., Okawa, K., Iwamatsu, A., Fujita, A., Watanabe, N., Saito, Y., Kakizuka, A., Morii, N., and Narumiya, S. (1996) EMBO J. 15 1885-1893 [PMC free article] [PubMed] [Google Scholar]
- 23.Leung, T., Manser, E., Tan, L., and Lim, L. (1995) J. Biol. Chem. 270 29051-29054 [DOI] [PubMed] [Google Scholar]
- 24.Matsui, T., Amano, M., Yamamoto, T., Chihara, K., Nakafuku, M., Ito, M., Nakano, T., Okawa, K., Iwamatsu, A., and Kaibuchi, K. (1996) EMBO J. 15 2208-2216 [PMC free article] [PubMed] [Google Scholar]
- 25.Riento, K., and Ridley, A. J. (2003) Nat. Rev. Mol. Cell Biol. 4 446-456 [DOI] [PubMed] [Google Scholar]
- 26.Denker, S. P., Huang, D. C., Orlowski, J., Furthmayr, H., and Barber, D. L. (2000) Mol. Cell 6 1425-1436 [DOI] [PubMed] [Google Scholar]
- 27.Li, Z., Dong, X., Wang, Z., Liu, W., Deng, N., Ding, Y., Tang, L., Hla, T., Zeng, R., Li, L., and Wu, D. (2005) Nat. Cell Biol. 7 399-404 [DOI] [PubMed] [Google Scholar]
- 28.Zhou, Y., Su, Y., Li, B., Liu, F., Ryder, J. W., Wu, X., Gonzalez-DeWhitt, P. A., Gelfanova, V., Hale, J. E., May, P. C., Paul, S. M., and Ni, B. (2003) Science 302 1215-1217 [DOI] [PubMed] [Google Scholar]
- 29.Pedrini, S., Carter, T. L., Prendergast, G., Petanceska, S., Ehrlich, M. E., and Gandy, S. (2005) PLoS Med. 2 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin, W., Yang, T., Ho, L. Zhao, Z., Wang, J., Chen, L., Zhao, W., Thiyagarajan, M., MacGrogan, D., Rodgers, J. T., Puigserver, P., Sadoshima, J., Deng, H., Pedrini, S., Gandy, S., Sauve, A. A., and Pasinetti, G. M. (2006) J. Biol. Chem. 281 21745-21754 [DOI] [PubMed] [Google Scholar]
- 31.Satoh, S., Toshima, Y., Hitomi, A., Ikegaki, I., Seto, M., and Asano, T. (2008) Brain Res. 1193 102-108 [DOI] [PubMed] [Google Scholar]
- 32.Pollitt, S. K., Pallos, J., Shao, J., Desai, U. A., Ma, A. A. K., Thompson, L. M., Marsh, J. L., and Diamond, M. I. (2003) Neuron 40 685-694 [DOI] [PubMed] [Google Scholar]
- 33.Shao, J., Welch, W. J., and Diamond, M. I. (2008) FEBS Lett. 582 1637-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao, J., Welch, W. J., Dispropero, N. A., and Diamond, M. I. (2008) Mol. Cell Biol. 28 5196-5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravikumar, B., Vacher, C., Berger, Z., Davies, J. E., Luo, S., Oroz, L. G., Scaravilli, F., Easton, D. F., Duden, R., O'Kane, C. J., and Rubinsztein, D. C. (2004) Nat. Genet. 36 585-595 [DOI] [PubMed] [Google Scholar]
- 36.Wang, G. H., Mitsui, K., Kotliarova, S., Yamashita, A., Nagao, Y., Tokuhiro, S., Iwatsubo, T., Kanazawa, I., and Nukina, N. (1999) Neuroreport 10 2435-2438 [DOI] [PubMed] [Google Scholar]
- 37.Wang, G. H., Sawai, N., Kotliarova, S., Kanazawa, I., and Nukina, N. (2000) Hum. Mol. Genet. 9 1795-1803 [DOI] [PubMed] [Google Scholar]
- 38.Peters, M. F., and Ross, C. A. (1999) Neurosci. Lett. 275 129-132 [DOI] [PubMed] [Google Scholar]
- 39.Tadokoro, K., Yamazaki-Inoue, M., Tachibana, M., Fujishiro, M., Nagao, K., Toyoda, M., Ozaki, M., Ono, M., Miki, N., Miyashita, T., and Yamada, M. (2005) J. Hum. Genet. 50 382-394 [DOI] [PubMed] [Google Scholar]
- 40.Zemskov, E. A., Jana, N. R., Kurosawa, M., Miyazaki, H., Sakamoto, N., Nekooki, M., and Nukina, N. (2003) J. Neurochem. 87 395-406 [DOI] [PubMed] [Google Scholar]
- 41.Machida, Y., Okada, T., Kurosawa, M., Oyama, F., Ozawa, K., and Nukina, N. (2006) Biochem. Biophys. Res. Commun. 343 190-197 [DOI] [PubMed] [Google Scholar]
- 42.Khan, L. A., Bauer, P. O., Miyazaki, H., Lindenberg, K. S., Landwehrmeyer, B. G., and Nukina, N. (2006) J. Neurochem. 98 576-587 [DOI] [PubMed] [Google Scholar]
- 43.Doi, H., Mitsui, K., Kurosawa, M., Machida, Y., Kuroiwa, Y., and Nukina, N. (2004) FEBS Lett. 571 171-176 [DOI] [PubMed] [Google Scholar]
- 44.Hosokawa, N., Hara, Y., and Mizushima, N. (2006) FEBS Lett. 580 2623-2629 [DOI] [PubMed] [Google Scholar]
- 45.Oyama, F., Kotliarova, S., Harada, A., Ito, M., Miyazaki, H., Ueyama, Y., Hirokawa, N., Nukina, N., and Ihara, Y. (2004) J. Biol. Chem. 279 27272-27277 [DOI] [PubMed] [Google Scholar]
- 46.Kotliarova, S., Jana, N. R., Sakamoto, N., Kurosawa, M., Miyazaki, H., Nekooki, M., Doi, H., Machida, Y., Wong, H. K., Suzuki, T., Uchikawa, C., Kotliarov, Y., Uchida, K., Nagao, Y., Nagaoka, U., Tamaoka, A., Oyanagi, K., Oyama, F., and Nukina, N. (2005) J. Neurochem. 93 641-653 [DOI] [PubMed] [Google Scholar]
- 47.Asanuma, K., Tanida, I., Shirato, I., Ueno, T., Takahara, H., Nishitani, T., Kominami, E., and Tomino, Y. (2003) FASEB J. 17 1165-1167 [DOI] [PubMed] [Google Scholar]
- 48.Ross, C. A., and Pickart, C. M. (2004) Trends Cell Biol. 14 703-711 [DOI] [PubMed] [Google Scholar]
- 49.Wong, H. K., Bauer, P. O., Kurosawa, M., Goswami, A., Washizu, C., Machida, Y., Tosaki, A., Yamada, M., Knopfel, T., Nakamura, T., and Nukina, N. (2008) Hum. Mol. Genet. 17 3223-3235 [DOI] [PubMed] [Google Scholar]
- 50.Sarkar, S., Krishna, G., Imarisio, S., Saiki, S., O'Kane, C. J., and Rubinsztein, D. C. (2004) Hum. Mol. Genet. 17 170-178 [DOI] [PubMed] [Google Scholar]
- 51.Zhang, L., Yu, J., Pan, H., Hu, P., Hao, Y., Cai, W., Zhu, H., Yu, A. D., Xie, X., Ma, D., and Yuan, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19023-19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown, M. E., and Bridgman, P. C. (2004) J. Neurobiol. 58 118-130 [DOI] [PubMed] [Google Scholar]
- 53.Ohashi, K., Nagata, K., Maekawa, M., Ishizaki, T., Narumiya, S., and Mizuno, K. (2000) J. Biol. Chem. 275 3577-3582 [DOI] [PubMed] [Google Scholar]
- 54.Sumi, T., Matsumoto, K., and Nakamura, T. (2001) J. Biol. Chem. 276 670-676 [DOI] [PubMed] [Google Scholar]
- 55.Scott, R. W., and Olson, M. F. (2007) J. Mol. Med. 85 555-568 [DOI] [PubMed] [Google Scholar]
- 56.Arimura, N., Inagaki, N., Chihara, K., Ménager, C., Nakamura, N., Amano, M., Iwamatsu, A., Goshima, Y., and Kaibuchi, K. (2000) J. Biol. Chem. 275 23973-23980 [DOI] [PubMed] [Google Scholar]
- 57.Arimura, N., Menager, C., Fukata, Y., and Kaibuchi, K. (2004) J. Neurobiol. 58 34-47 [DOI] [PubMed] [Google Scholar]
- 58.Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998) J. Cell Biol. 143 1883-1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagaoka, U., Kim, K., Jana, N. R., Doi, H., Maruyama, M., Mitsui, K., Oyama, F., and Nukina, N. (2004) J. Neurochem. 91 57-68 [DOI] [PubMed] [Google Scholar]
- 60.Nishimura, Y., Itoh, K., Yoshioka, K., Uehata, M., and Himeno, M. (2000) Cell Tissue Res. 301 341-351 [DOI] [PubMed] [Google Scholar]
- 61.Kuo, P. L., Hsu, Y. L., and Cho, C. Y. (2006) Mol. Cancer Ther. 5 3209-3221 [DOI] [PubMed] [Google Scholar]
- 62.Degtyarev, M., De Maziere, A., Orr, C., Lin, J., Lee, B. B., Tien, J. Y., Prior, W. W., van Dijk, S., Wu, H., Gray, D. C., Davis, D. P., Stern, H. M., Murray, L. J., Hoeflich, K. P., Klumperman, J., Friedman, L. S., and Lin, K. (2008) J. Cell Biol. 183 101-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng, M., and Zhou, J. N. (2008) Cell Signal. 20 659-665 [DOI] [PubMed] [Google Scholar]
- 64.Monastyrska, I., He, C., Geng, J., Hoppe, A. D., Li, Z., and Klionsky, D. J. (2008) Mol. Biol. Cell 19 1962-1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.