Abstract
Protein kinases often recognize their substrates and regulators through docking interactions that occur outside of the active site; these interactions can help us to understand kinase networks, and to target kinases with drugs. During mitogen-activated protein kinase (MAPK) signaling, the ability of MAPK kinases (MKKs, or MEKs) to recognize their cognate MAPKs is facilitated by a short docking motif (the D-site) in the MKK N terminus, which binds to a complementary region on the MAPK. MAPKs then recognize many of their targets using the same strategy, because many MAPK substrates also contain D-sites. The extent to which docking contributes to the specificity of MAPK transactions is incompletely understood. Here we characterize the selectivity of the interaction between MKK-derived D-sites and MAPKs by measuring the ability of D-site peptides to inhibit MAPK-mediated phosphorylation of D-site-containing substrates. We find that all MKK D-sites bind better to their cognate MAPKs than they do to non-cognate MAPKs. For instance, the MKK3 D-site peptide, which is a remarkably potent inhibitor of p38α (IC50 < 10 nm), does not inhibit JNK1 or JNK2. Likewise, MAPKs generally bind as well or better to cognate D-sites than to non-cognate D-sites. For instance, JNK1 and JNK2 do not appreciably bind to any D-sites other than their cognate D-sites from MKK4 and MKK7. In general, cognate, within-pathway interactions are preferred about an order of magnitude over non-cognate interactions. However, the selectivity of MAPKs and their cognate MKK-derived D-sites for each other is limited in some cases; in particular, ERK2 is not very selective. We conclude that MAPK-docking sites in MAPK kinases bind selectively to their cognate MAPKs.
Changes in protein kinase activity are associated with many human diseases, often contributing to, or even driving, disease pathology (1). Consequently, there is a considerable interest in targeting protein kinases with drugs (2-4), as well as in understanding the regulatory networks that kinases are a part of (5-8). For both these reasons, it is important to better understand how protein kinases select and recognize their substrates and regulators.
Naturally, a key aspect of kinase-substrate recognition is the interaction of the kinase catalytic cleft with the target phosphoacceptor residue (and with adjacent residues). Many kinases, however, augment the limited specificity of this catalytic cleft-target site interaction by also binding to one or more regions of the substrate that are distinct and separate from the target peptide (9, 10). Such “docking interactions,” mediated by “docking sites” on kinases and substrates, are thought to tether relatively low specificity catalytic domains to their proper substrates (11, 12). There is increasing interest in targeting these interactions as a drug development strategy (13-18).
Mitogen-activated protein kinase (MAPK)2 cascades (also sometimes called extracellular signal-regulated protein kinase (ERK) cascades), are crucial to the transmission of signals initiated by growth factors, developmental regulators, immune system mediators, cellular stresses, and many other stimuli. Central to such cascades is an MAPK kinase (MKK, also sometimes called MEK for MAPK/ERK kinase) that phosphorylates and activates a cognate MAPK. The activated MAPK then phosphorylates multiple downstream substrates, including transcriptional regulators. Major MAPK pathways in mammalian cells include the MEK½→ ERK½ pathway, which is typically involved in regulating growth and developmental signaling (19) (indeed, germ line mutations in components of this pathway are a cause of the hereditary disease cardiofaciocutaneous syndrome (20)), the MKK3/6→p38 pathway, which is typically involved in regulating inflammation (21), and the MKK4/7→ JNK pathway, which regulates apoptosis and many other disease-relevant processes (22, 23). There are active efforts to develop pharmacological inhibitors of MAPK signaling for use in treating cancer, chronic inflammatory conditions, neurological disorders, and many other diseases (24, 25).
Docking interactions are prominent at several steps of MAPK cascade-mediated signaling, including MKK activation of MAPKs and MAPK phosphorylation of substrates (26-28). During these transactions, short docking motifs on the MKK or substrate bind to binds to a complementary region on the cognate MAPK (9, 10, 13, 28).
The ability of an MEK/MKK to recognize its cognate MAPK is highly dependent on docking interactions involving a short motif (the “D-site”) found near the N terminus of MKKs (29-31). D-sites in MKKs reside in a small regulatory domain and are well separated from the much larger, C-terminal MKK catalytic domain. The D-site consensus consists of a cluster of about two or three basic residues, a short spacer, and a hydrophobic-X-hydrophobic submotif (K/R2-3-X1-6-ϕ-X-ϕ) (30, 31). The D-sites in several MKKs have been shown to be necessary and sufficient for relatively high affinity MAPK binding (i.e. dissociation constants that are typically in the low micromolar range) (31-33). Furthermore, preventing docking by mutation, or by blocking with competing D-site peptides, substantially compromises the ability of the MKK to phosphorylate and activate its cognate MAPK(s) (31-37). The human pathogen Bacillus anthracis has exploited the importance of MKK-MAPK docking: anthrax lethal factor protease severs MKK D-sites from the rest of the MKK polypeptide to disable the immune system during early infection (38, 39).
The above observations clearly demonstrate that MKK-MAPK docking is crucial for efficient signal transmission. However, less is known about the role of MKK-MAPK docking in specificity. MKKs are highly specific enzymes: they phosphorylate only their cognate (same pathway) MAPKs, and they do not target MAPKs in other pathways; for example MEK1/2 do not activate JNK or p38 MAPKs (40). MKKs phosphorylate a threonine and a tyrosine in a TXY motif found in the MAPK activation loop. In principle, the residues in and around this motif could provide a means for selective recognition. That is (for example), MEK1/2 might recognize the sequence FLTEY in ERK1/2, and MKK3/6 might recognize the sequence EMTGY in p38 MAPKs. However, loop-swap experiments have not supported this hypothesis (41-43). Thus, active site-target peptide recognition does not appear to play a major role in the specificity of MKK-MAPK transactions.
Another attractive possibility is that docking interactions could play a role in selective MKK-MAPK recognition. This mechanism would work best if docking sites were pathway specific. Such a clear-cut solution does exist in the human MEK5-ERK5 MAPK pathway, because these two proteins have been shown to dock using an acidic docking site on the MKK that bears no resemblance to the D-site (44). However, all other human MKK-MAPK docking interactions are mediated by D-sites in MKKs, and, as noted above, D-sites share a core consensus. Thus, the degree to which D-site-mediated docking interactions may provide specificity in mammalian MKK-MAPK transactions is unclear.
The fact that D-sites are found in both MKKs and substrates implies that MKKs and substrates may compete for MAPK binding, a prediction that we have verified by using MKK-derived D-site peptides to inhibit MAPK-mediated phosphorylation of D-site containing substrates (for example, the D-site from MEK2 inhibits ERK2-mediated phosphorylation of Elk-1) (36). Indeed, we have found that this competitive inhibition assay provides a sensitive method for quantifying the binding of a particular D-site peptide to a target MAPK. We have previously used this approach to examine the selectivity of MEK1/2 and MKK4/7 D-sites for ERK1/2 versus JNK1/2 (32, 33). Here we extend this analysis to include p38, allowing us to compare D-site selectivity between all three pathways and draw general conclusions about the selectivity of docking sites in MAPK kinases.
EXPERIMENTAL PROCEDURES
Proteins—Fusions of glutathione S-transferase (GST) to human c-Jun1-89, ATF219-96, and Elk-1307-428 were purchased from Cell Signaling Technology. GST-MEF2A261-315 and activated human JNK1α1 and JNK2α2 and were purchased from Upstate Cell Signaling Solutions/Millipore. Activated mouse ERK2 was purchased from New England Biolabs. Activated human p38α was purchased from BIOMOL International.
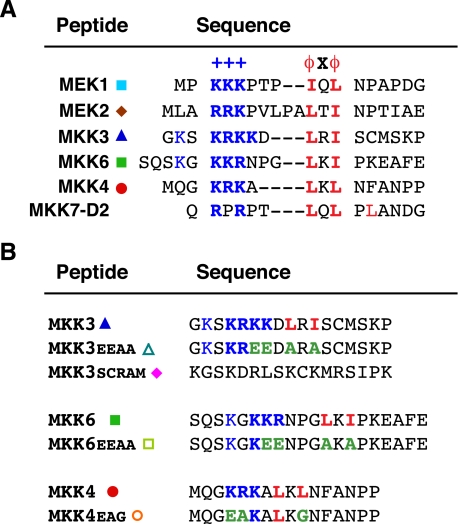
Peptides—The soluble peptides used in this study were synthesized by United Biochemical Research and by Mimotopes. Peptide sequences are shown in Fig. 2.
FIGURE 2.
Docking sites in MKKs. A, D-site peptides used in this study. Residues comprising the basic submotif (+++) are shown in bold and blue; residues comprising the hydrophobic-X-hydrophobic submotif (ϕXϕ) are shown in bold and red. Gaps have been introduced to maximize alignment of functionally similar residues; spaces are for visual clarity. The MEK1 peptide comprises residues 1-17 of the full-length protein; MEK2, 1-20; MKK3, 17-33; MKK6, 2-21; MKK4, 37-52; and MKK7-D2, 37-51. B, control peptides used in this study, with the wild-type version shown for comparison. Mutated residues are colored green.
Protein Kinase Assays—Protein kinase reactions (20 μl) contained kinase assay buffer (50 mm Tris-HCl (pH 7. 5), 10 mm MgCl2, 1 mm EGTA, and 2 mm dithiothreitol), 1 μm substrate (680 ng of GST-MEF2A, 740 ng of GST-c-Jun, 700 ng of GST-ATF2, or 820 ng of GST-Elk-1), active MAPK (0.9 milliunit (18 ng) of p38, 0.8 milliunit (2.6 ng) of JNK1, 0.8 milliunit (∼5 ng) of JNK2, or 10 units (∼1 ng) of ERK2), 50 μm ATP, 1 μCi of [γ-32P]ATP, and the indicated concentration of peptide. Reactions were for 20 min at 30 °C. Substrate phosphorylation was quantified by SDS-PAGE (12% gels), followed by analysis of relative incorporation using a PhosphorImager. All data points shown are averages from experiments repeated 3-7 times, with duplicate points in each experiment. We have found IC50 values measure by our protocol to be reproducible within ∼2-fold, even when re-measured several years later using different lots of all components.
RESULTS
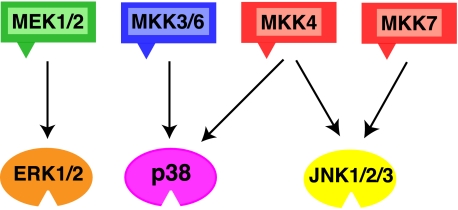
Specificity of MAPK Kinases—MKKs specifically phosphorylate their cognate MAPKs and do not appreciably phosphorylate MAPKs in other pathways (Fig. 1). For instance, MEK1 and MEK2 phosphorylate ERK (meaning ERK1/2), but not JNK or p38. Likewise, MKK3 and MKK6 phosphorylate p38 but not ERK or JNK (45, 46), and MKK7 phosphorylates JNK but not ERK or p38 (47, 48). MKK4 is notable in that it phosphorylates both JNK and p38 (45, 49).
FIGURE 1.
MAPK pathways. The schematic shows six of the seven human MAPK kinases (MKKs, or MEKs) with their cognate, within-pathway MAPKs indicated by the arrows. The triangles on the MKKs represent their D-sites. See text for details.
Competition Assay for Measuring D-site Selectivity—The ERK1/2, JNK1/2, and p38 pathways phosphorylate distinct but overlapping sets of substrates (50-52). In this study we use the MEF2A and ATF2 transcription factors as model p38α substrates, the c-Jun and ATF2 transcription factors as model JNK1/2 substrates, and the Elk-1 transcription factor as a model ERK2 substrate. All three of these substrates contain D-sites that have been shown to be important in MAPK targeting (26, 53, 54).
We have previously used a “peptide competition assay” to show that D-site peptides derived from MEK1 or MEK2 are able to inhibit ERK2-mediated phosphorylation Elk-1 (36). (In addition, these peptides can also inhibit MEK-mediated MAPK phosphorylation and MAPK phosphatase-mediated dephosphorylation (36)). We also showed that D-site peptides derived from MKK4 or MKK7 could inhibit JNK1/2-mediated phosphorylation of c-Jun and ATF2 (32, 33). These observations indicated that MKK-derived D-site peptides effectively bound to their cognate MAPKs and blocked docking interactions between the MAPK and its transcription factor substrate.
These conclusions have been supported by binding assays, by structural studies, and by mutagenesis (31-33, 36, 37, 55-59). However, we have found the peptide competition assay to be the most sensitive and reproducible approach to quantitatively compare the affinity of different D-sites for different MAPKs and have, therefore, used this approach exclusively in this study. The sequences of the peptides used in this study are shown in Fig. 2.
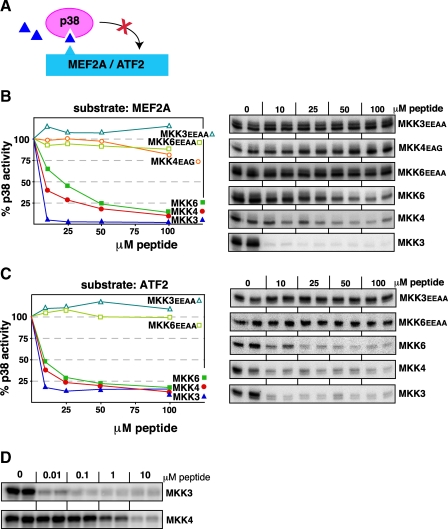
p38 Is Potently Inhibited by Cognate D-site peptides—To extend the above observations to the p38 pathway, we asked if D-site peptides derived from one of the three MKKs that can phosphorylate and thereby activate p38, MKK3, MKK4, and MKK6, could bind to p38 and thereby inhibit the ability of p38 to phosphorylate the D-site-containing substrates MEF2A and ATF2 (Fig. 3A). The p38α isoform, the most extensively studied isoform and the most prominent isoform in human cells and tissues, was used in all experiments.
FIGURE 3.
D-site peptides from MKK3, MKK4, or MKK6 inhibit p38 phosphorylation of the MEF2A and ATF2 transcription factors. A, D-site peptides (triangle) were used to inhibit p38α phosphorylation of MEF2A or ATF2. B, purified GST-MEF2A (1 μm) was incubated with purified active p38α (22.5 nm) and [γ-32P]ATP for 20 min in the absence or presence of the specific concentrations of the indicated peptides. In the graph, results are plotted as percent phosphorylation relative to that observed in the absence of any added peptide. Phosphate incorporation into MEF2A was analyzed by SDS-PAGE and quantified on a PhosphorImager. Data are the average of at least three experiments, with duplicate data points in each experiment. To the right is shown is an autoradiogram of a representative experiment. C, as in B, except that the substrate was purified GST-ATF2 (1 μm). D, comparison of MKK3 and MKK4 peptides at lower doses. Reaction conditions are as in B, above.
As shown in Fig. 3, all three cognate D-site peptides were effective inhibitors of p38α-mediated phosphorylation of both substrates. These results suggest that MKK3, MKK4, and MKK6 compete with transcription factors for binding to the same protein interaction site on p38; indeed, in the case of D-site peptides derived from MKK3 and MEF2A, there are crystallographic data that support this hypothesis (56).
Of the three D-site peptides, the MKK6 peptide was the least effective, with 50% inhibition (IC50) occurring at a concentration of 20 μm for inhibition of MEF2A phosphorylation (Fig. 3B) and 10 μm for ATF2 phosphorylation (Fig. 3C). The MKK4 peptide was next in potency, with an IC50 of ∼7 μm with respect to both substrates. These IC50 values in the low micromolar range are comparable to what we have previously observed when testing MEK1/2- and MKK4/7-derived peptides for inhibition of ERK2 and JNK1/2, respectively.
Remarkably, the MKK3-derived D-site peptide was a considerably more potent inhibitor of p38α-mediated phosphorylation of either substrate, with an IC50 of <10 nm (Fig. 3D). This degree of inhibition is over 100-fold more potent than we have observed with other D-site peptides.
Three peptides were employed as negative controls. These peptides, MKK3EEAA, MKK4EAG, and MKK6EEAA, have substitutions in the key basic and hydrophobic residues of the D-site consensus (see Fig, 2). As expected, none of these negative controls inhibited p38 (Fig. 3). We have previously shown that MKK4EAG also does not inhibit JNK1/2.
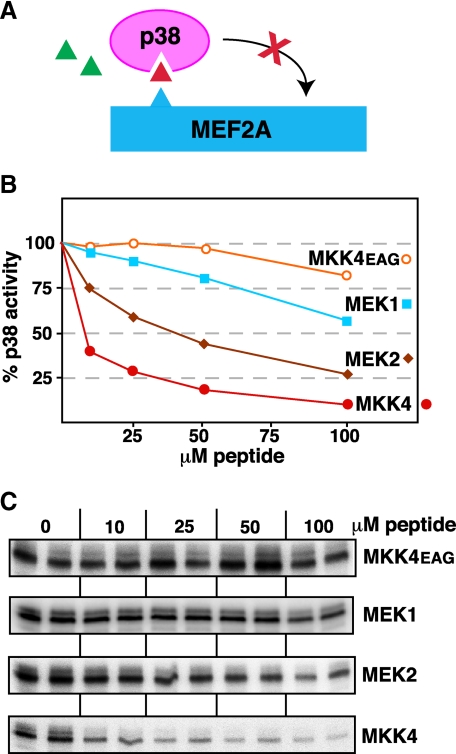
Specificity of D-site Binding to p38—To begin to explore the selectivity of D-sites for cognate versus non-cognate MAPKs, we have previously asked if the MEK1/2 D-site peptides could inhibit JNK1/2, and if MKK4/7 peptides could inhibit ERK1/2 (32, 33). To determine the specificity of p38α binding to MKK-derived D-site peptides, we compared ability of D-site peptides from MEK1 and MEK2 to inhibit p38α. As shown in Fig. 4, although both the MEK1 and (particularly) MEK2 D-sites were able to weakly inhibit p38α, they were considerable less effective than the cognate D-site from MKK4. Thus, p38α exhibits selectivity for cognate versus non-cognate D-sites.
FIGURE 4.
D-site peptides from MEK1 and MEK2 are weak inhibitors of p38. A, D-site peptides (triangle) were tested for their ability to inhibit p38α phosphorylation of MEF2A. B, graph representing an average of multiple experiments, experimental details as in Fig. 3. C, an autoradiogram of a representative experiment.
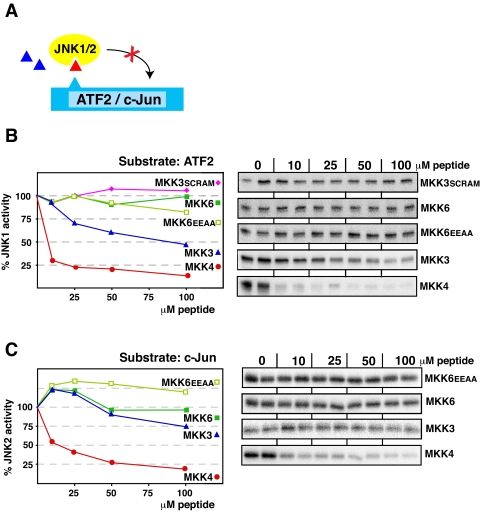
D-sites from MKK3 and MKK6 Do Not Inhibit JNK—We next asked if the MKK3- and MKK6-derived D-site peptides could inhibit the non-cognate MAPKs JNK1 and JNK2. As shown in Fig. 5, neither peptide was effective at inhibiting either JNK1 or JNK2. Indeed, the MKK6 peptide displayed absolutely no inhibitory activity toward JNK1/2, and the MKK3 peptide was only able to weakly inhibit JNK1. This was true whether ATF2 or c-Jun were used as the relevant JNK substrates (data for JNK1/c-Jun and JNK2/ATF2 are not shown but reflected the JNK1/ATF2 and JNK2/c-Jun data shown in Fig. 5).
FIGURE 5.
The MKK3 and MKK6 D-site peptides are poor inhibitors of JNK1/2. A, D-site peptides (triangle) were tested for their ability to inhibit JNK1 or JNK2-mediated phosphorylation of ATF2 or c-Jun. B, purified GST-ATF2 (1 μm) was incubated with purified active JNK1 (2.5 nm) and [γ-32P]ATP for 20 min in the absence or presence of the specific concentrations of the indicated peptides. Other details are as in Fig. 3. C, as in B, except that the substrate was purified GST-c-Jun (1 μm), and the kinase was JNK2 (5 nm).
D-site Peptide from MKK4 Inhibits Two Classes of MAPKs—MKK4 protein activates two families of MAPKs: JNK and p38 (45, 49, 60). Consistent with the dual function of the full-length kinase, the D-site from MKK4 is an effective inhibitor of both p38α (Fig. 3) and JNK1/2 (Fig. 5).
MKK7-D2 Peptide Is Selective for JNK—Although MKK4 activates JNK and p38, MKK7 protein activates only JNK (47, 48). MKK4, like MEK1/2 and MKK3/6, contains only a single high affinity D-site. However, MKK7 contains three low affinity D-sites its N-terminal domain, which interact to create a high affinity JNK-binding platform (33). We chose to focus on the MKK7-D2 D-site for this report, because we have previously found it to exhibit the highest binding affinity for JNK1/2 of the three MKK7 D-sites (33). Interestingly, the selectivities of the MKK4 and MKK7-D2 D-sites mirror the preferences of the full-length kinases: the MKK4 D-sites bind well to both JNK1/2 and p38 (IC50 < 10 μm), whereas MKK7-D2 binds to JNK1 and JNK2 (IC50 values of 8 and 30 μm) but does not bind to p38 (IC50 > 100 μm).
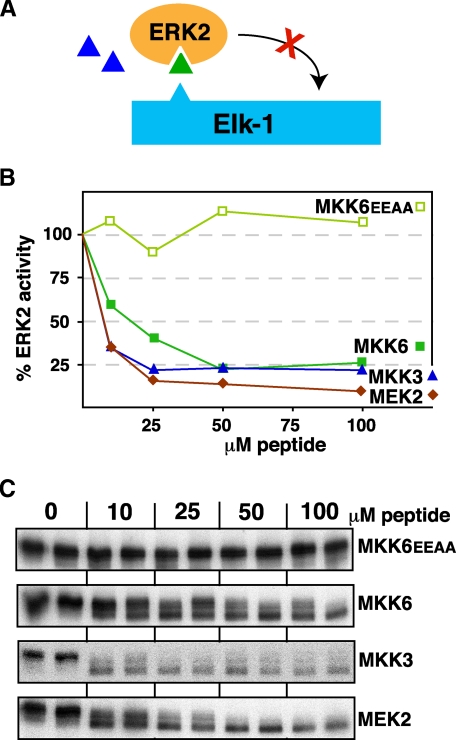
D-sites from MKK3 and MKK6 Can Inhibit ERK2—Having shown that the MKK3 and MKK6 peptides could not inhibit JNK1/2, we next ask if they could inhibit ERK2. Surprisingly, both the (non-cognate) MKK3 and MKK6 peptides were almost as effective at inhibiting ERK2 phosphorylation of Elk-1 as was the (cognate) MEK2 peptide (Fig. 6). Thus, ERK2 cannot selectively discriminate cognate D-sites from non-cognate, p38 pathway D-sites.
FIGURE 6.
The MKK3 and MKK6 D-site peptides are effective inhibitors of ERK2. A, D-site peptides (triangle) were tested for their ability to inhibit ERK2-mediated phosphorylation of Elk-1. B, purified GST-Elk-1 (1 μm) was incubated with purified active ERK2 (∼1 nm) and [γ-32P]ATP for 20 min in the absence or presence of the specific concentrations of the indicated peptides. Other details are as in Fig. 3. C, an autoradiogram of a representative experiment. Elk-1 is phosphorylated by ERK2 on multiple residues, resulting in a ladder of bands displaying retarded electrophoretic mobility (36). Peptide-dependent inhibition was revealed both by the progressive collapsing of this ladder and by reduced phosphate incorporation overall. The fastest migrating band, which was largely unaffected by peptide inhibition, was excluded from quantification.
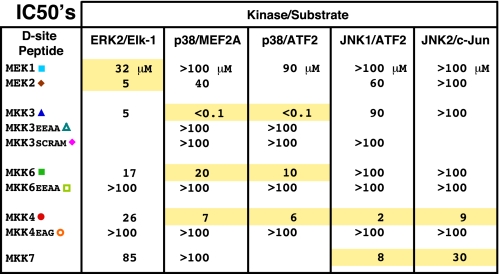
Comparison of D-site Binding Affinity—Fig. 7 shows the compiled IC50 values for the inhibition of MAPKs by MKK-derived D-sites. Assuming a simple competitive inhibition scheme, which seems entirely reasonable here (61-64), IC50 values are a measure of the dissociation constant (Kd) of the peptide-kinase binding interaction, with a lower IC50 indicating tighter binding. Indeed, IC50 values as measured in the competition assay used here have generally correlated well with Kd values measured in direct binding assays (32, 33). As can be seen (Fig. 7), IC50 values for cognate interactions range from 0.1 μm (MKK3 D-site to p38) to 32 μm (MEK1 D-site for ERK2), whereas IC50 values for non-cognate interactions range from 5 μm (MKK3 D-site for ERK2) to >100 μm (many examples, e.g. all non-cognate D-sites for JNK2).
FIGURE 7.
IC50 values for the inhibition of the given kinase/substrate pair by the indicated D-site peptide. The IC50 is the concentration of peptide required to inhibit kinase activity by 50%; a lower IC50 indicates better binding. Cognate interactions are highlighted in yellow, while non-cognate and control interactions are not highlighted.
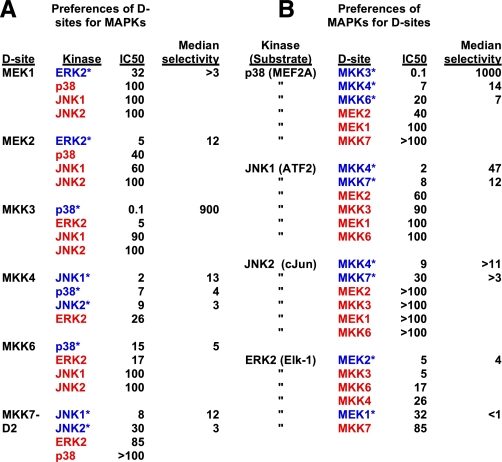
Notably, almost all of the MKK-derived D-sites bound better to their cognate MAPK(s) than to non-cognate MAPKs (Fig. 8A). The single exception was the MKK6 D-site, which inhibited non-cognate ERK2 about as well as it inhibited cognate p38. Similarly, with the exception of ERK2, the MAPKs strongly preferred cognate D-sites to non-cognate D-sites (Fig. 8B).
FIGURE 8.
Preferences of D-sites for MAPKs and of MAPKs for D-sites. A, all MAPKs that each D-site had been tested against are listed in order of preference, with highest to lowest binding affinity (i.e. lowest to highest IC50) running from top to bottom. The one or more cognate MAPK(s) for each D-site are indicated with blue text and an asterisk. Non-cognate MAPKs are indicated with red text. See Tables 1 and 2 for selectivity calculations. B, all D-sites that each MAPK had been tested against are listed in order of preference, as in A. Cognate D-sites for each MAPK are indicated with blue text and an asterisk. Non-cognate D-sites are indicated with red text.
Selectivity of Docking Sites in MKKs—To obtain additional quantitative insights into D-site specificity, selectivity ratios were calculated. One type of selectivity ratio evaluates how well a particular D-site peptide inhibits the kinase reaction of a cognate MAPK versus a non-cognate MAPK. This ratio consists of the IC50 with which a given peptide inhibits a non-cognate kinase divided by the IC50 with which the same peptide inhibits a cognate kinase. Thus, a higher ratio indicates that the D-site peptide is significantly less effective inhibiting the non-cognate kinase reaction versus the cognate reaction, and is therefore more selective. In essence, this comparison asks “How much better does a given D-site (e.g. MKK3) bind its cognate MAPK (e.g. p38) compared with its binding to non-cognate MAPKs (e.g. JNK1, JNK2, and ERK2)?”
The results from such calculations are summarized in Table 1 and Fig. 8. In general, all the D-sites were selective for their cognate MAPKs, with a median selectivity of 8 (we use the median rather than the mean because the mean is so strongly influenced by the extremely high selectivity of the MKK3 D-site). The MKK3 D-site was the most selective peptide, with a median selectivity of 900, driven by its highly potent inhibition of p38. Next, in order, were the D-sites of MEK2, MKK7-D2, MKK6, MKK4, and MEK1, with median selectivities of 12, 7, 5, 4, and 3, respectively. These comparisons indicate that, in general, MKK D-sites bind ∼8-fold better to their cognate MAPKs than they do to non-cognate MAPKs.
TABLE 1.
Selectivity ratios of D-sites for MAPKs
| D-site peptide | Cognate MAPK | Cognate IC50 | Non-cognate MAPK | Non-cognate IC50 | Selectivity ratioa |
|---|---|---|---|---|---|
| MEK1 | ERK2 | 32 | p38/MEF2Ab | >100 | >3 |
| MEK1 | ERK2 | 32 | p38/ATF2 | 90 | 3 |
| MEK1 | ERK2 | 32 | JNK1 | >100 | >3 |
| MEK1 | ERK2 | 32 | JNK2 | >100 | >3 |
| MEK1 mean | 3 | ||||
| MEK1 median | 3 | ||||
| MEK2 | ERK2 | 5 | p38/MEF2A | 40 | 8 |
| MEK2 | ERK2 | 5 | p38/ATF2 | NDc | ND |
| MEK2 | ERK2 | 5 | JNK1 | 60 | 12 |
| MEK2 | ERK2 | 5 | JNK2 | >100 | >20 |
| MEK2 mean | 13 | ||||
| MEK2 median | 12 | ||||
| MKK3 | p38b | <0.1 | JNK1 | 90 | >900 |
| MKK3 | p38 | <0.1 | JNK2 | >100 | >1000 |
| MKK3 | p38 | <0.1 | ERK2 | 5 | >50 |
| MKK3 mean | 650 | ||||
| MKK3 median | 900 | ||||
| MKK4 | JNK1 | 2 | ERK2 | 26 | 13 |
| MKK4 | JNK2 | 9 | ERK2 | 26 | 3 |
| MKK4 | p38/MEF2A | 7 | ERK2 | 26 | 4 |
| MKK4 | p38/ATF2 | 6 | ERK2 | 26 | 4 |
| MKK4 mean | 6 | ||||
| MKK4 median | 4 | ||||
| MKK6 | p38/MEF2A | 20 | JNK1 | >100 | >5 |
| MKK6 | p38/MEF2A | 20 | JNK2 | >100 | >5 |
| MKK6 | p38/MEF2A | 20 | ERK2 | 17 | 1 |
| MKK6 | p38/ATF2 | 10 | JNK1 | >100 | >10 |
| MKK6 | p38/ATF2 | 10 | JNK2 | >100 | >10 |
| MKK6 | p38/ATF2 | 10 | ERK2 | 17 | 2 |
| MKK6 mean | 5 | ||||
| MKK6 median | 5 | ||||
| MKK7 | JNK1 | 8 | p38/MEF2A | >100 | >13 |
| MKK7 | JNK1 | 8 | ERK2 | 85 | 11 |
| MKK7 | JNK2 | 30 | p38/MEF2A | >100 | >3 |
| MKK7 | JNK2 | 30 | ERK2 | 85 | 3 |
| MKK7 mean | 7 | ||||
| MKK7 median | 7 | ||||
| All mean | 150 | ||||
| All median | 8 |
Selectivity ratio = IC50 (D-site peptide with non-cognate MAPK)/IC50 (D-site peptide with cognate MAPK).
The p38 the substrate used is shown. For p38 inhibition by the MKK3 peptide, both substrates gave the same results.
ND, not determined.
A second type of selectivity ratio evaluates how a given kinase is inhibited by a cognate peptide as compared with another peptide. This selectivity ratio consists of the IC50 of the comparison peptide divided by the IC50 of the cognate peptide, assessed against the same kinase. Thus, a higher ratio signifies a more selective cognate peptide; that is, the peptide has a proportionally lower IC50 for the chosen reaction than the comparison peptide. In essence, this comparison asks “How much better does a given kinase (e.g. ERK2) bind to cognate D-sites (e.g. MEK1/2) compared with its binding to non-cognate D-sites (e.g. MKK3/4/6)?”
In general (see Table 2 and Fig. 8), all the MAPKs were selective for their cognate D-sites, with a median selectivity of 11. JNK1 was the most selective MAPK, with a median selectivity of 21 for its cognate D-sites MKK4 and MKK7. Next, in order, were p38 and JNK2, with median selectivities of 14 and 7, respectively. ERK2 was not highly selective: although it did prefer its cognate MEK2 D-site the most, it preferred the non-cognate D-sites from MKK3, MKK4, and MKK6 over its cognate D-site MEK1.
TABLE 2.
Selectivity ratios of MAPKs for D-sites
| Kinase | Cognate peptide | Cognate IC50 | Non-cognate peptide | Non-cognate IC50 | Selectivity ratioa |
|---|---|---|---|---|---|
| p38/MEF2Ab | MKK3 | 0.1 | MEK1 | >100 | >1000 |
| p38/MEF2A | MKK3 | 0.1 | MEK2 | 40 | 400 |
| p38/MEF2A | MKK3 | 0.1 | MKK7 | >100 | >1000 |
| p38/MEF2A | MKK6 | 20 | MEK1 | >100 | >5 |
| p38/MEF2A | MKK6 | 20 | MEK2 | 40 | 2 |
| p38/MEF2A | MKK6 | 20 | MKK7 | >100 | >5 |
| p38/MEF2A | MKK4 | 7 | MEK1 | >100 | >14 |
| p38/MEF2A | MKK4 | 7 | MEK2 | 40 | 6 |
| p38/MEF2A | MKK4 | 7 | MKK7 | >100 | >14 |
| P38/ATF2b | MKK3 | 0.1 | MEK1 | 90 | 900 |
| P38/ATF2 | MKK6 | 10 | MEK1 | 90 | 9 |
| P38/ATF2 | MKK4 | 6 | MEK1 | 90 | 15 |
| p38 mean | 281 | ||||
| p38 median | 14 | ||||
| JNK1 | MKK4 | 2 | MEK1 | >100 | >50 |
| JNK1 | MKK4 | 2 | MEK2 | 60 | 30 |
| JNK1 | MKK4 | 2 | MKK3 | 90 | 45 |
| JNK1 | MKK4 | 2 | MKK6 | >100 | >50 |
| JNK1 | MKK7-D2 | 8 | MEK1 | >100 | >13 |
| JNK1 | MKK7-D2 | 8 | MEK2 | 60 | 8 |
| JNK1 | MKK7-D2 | 8 | MKK3 | 90 | 11 |
| JNK1 | MKK7-D2 | 8 | MKK6 | >100 | >13 |
| JNK1 mean | 27 | ||||
| JNK1 median | 21 | ||||
| JNK2 | MKK4 | 9 | MEK1 | >100 | >11 |
| JNK2 | MKK4 | 9 | MEK2 | >100 | >11 |
| JNK2 | MKK4 | 9 | MKK3 | >100 | >11 |
| JNK2 | MKK4 | 9 | MKK6 | >100 | >11 |
| JNK2 | MKK7-D2 | 30 | MEK1 | >100 | >3 |
| JNK2 | MKK7-D2 | 30 | MEK2 | >100 | >3 |
| JNK2 | MKK7-D2 | 30 | MKK3 | >100 | >3 |
| JNK2 | MKK7-D2 | 30 | MKK6 | >100 | >3 |
| JNK2 mean | 7 | ||||
| JNK2 median | 7 | ||||
| ERK2 | MEK1 | 32 | MKK3 | 5 | 0.2 |
| ERK2 | MEK1 | 32 | MKK6 | 17 | 0.5 |
| ERK2 | MEK1 | 32 | MKK4 | 26 | 0.8 |
| ERK2 | MEK1 | 32 | MKK7 | 85 | 3 |
| ERK2 | MEK2 | 5 | MKK3 | 5 | 1 |
| ERK2 | MEK2 | 5 | MKK6 | 17 | 3 |
| ERK2 | MEK2 | 5 | MKK4 | 26 | 5 |
| ERK2 | MEK2 | 5 | MKK7 | 85 | 17 |
| ERK2 mean | 4 | ||||
| ERK2 median | 2 | ||||
| All mean | 102 | ||||
| All median | 11 |
Selectivity ratio = IC50 (MAPK with non-cognate D-site peptide)/IC50 (MAPK with cognate D-site peptide).
The p38 the substrate used is shown.
DISCUSSION
Apparently, it is crucial that MKKs do not make mistakes. In other words, there appears to have been a great deal of selective pressure for fidelity in MKK-MAPK transactions: MKKs are dedicated to phosphorylating their cognate MAPKs and do not phosphorylate other targets. MAPK-docking sites are found in the N termini of all seven human MKKs and have been thought to contribute to selective recognition of MAPKs. However, with the exception of MKK5, all the docking sites found in human MKKs are of a single class (the D-site class) and share a core consensus. Thus, it was reasonable to entertain the hypothesis that D-sites might contribute only to binding energy, and not to specificity.
The data presented herein, combined with our previous studies, allow us for the first time to systematically assess the selectivity of D-sites in human MKKs for their cognate MAPKs. From this assessment, we draw three main conclusions.
First, MKK-derived D-sites are selective for their cognate MAPKs. Each of the MKK-derived D-sites preferred its cognate MAPK(s) to non-cognate partners (Fig. 8A). This selectivity ranged from a high of >1000 (the preference of the MKK3 D-site for p38 over JNK2) to a low of just over 1 (the preference of the MKK6 D-site for p38 over ERK2) and had a median value of 8, meaning that MKK-derived D-sites bind ∼8-fold better to their cognate MAPKs than to non-cognate MAPKs.
Second, the MAPKs JNK1, JNK2, and p38α are selective for their cognate D-sites: they bound better to D-sites derived from their cognate, within-pathway MKKs than to non-cognate D-sites. This selectivity ranged from a high of >1000 (the preference of p38 for the MKK3 D-site over that of MEK1 or MKK7) to a low of 2 (the preference of p38 for the MKK6 D-site over that of MEK2), and had a median value of 11, meaning that these MAPKs bind ∼11-fold better to their cognate D-sites than to non-cognate D-sites.
Our third conclusion is that the ERK2 MAPK is not highly selective for cognate versus non-cognate D-sites. ERK2 bound just about as well to cognate MEK2 as it did to non-cognate MKK3 and bound better to non-cognate MKK3, MKK4, and MKK6 than it did to cognate MEK1.
Model for MKK-MAPK Specificity—Our results support a bipartite model for MKK/MAPK specificity, in which both the MKK D-site and the MKK catalytic domain play important roles. The specificity cannot be determined uniquely by the D-site-MAPK interaction, because some such interactions are not selective enough (in particular, those involving ERK2). However, it is also unlikely that docking interactions have no role in MKK/MAPK specificity, because in most cases these interactions are quite selective.
In the case of ERK2, deletion of the D-site impairs but does not completely eliminate MEK1/2 binding (31). Consistent with this, mutations in two distinct regions of the kinase domain diminish the physical interaction with MEK1/2, suggesting at least two additional regions of contact in addition to the docking interaction (65, 66). Indeed, ERK2 would seem to need these additional contacts for specificity, because the docking interaction is not very selective.
Flexibility of D-site Binding—The present results highlight the flexibility of D-site binding. Apparently, it has been possible to evolve D-sites that fit JNK but not p38 (i.e. MKK7-D2), p38 but not JNK (i.e. MKK3 and MKK6), and both JNK and p38 (MKK4). Likewise, some D-sites in transcription factors bind to more than one family of MAPK, e.g. the Elk-1 D-site, which binds to both ERK and JNK, and the ATF2 D-site, which binds to both JNK and p38 (26). The D-site-MAPK interaction can apparently be tuned to be highly selective for a single MAPK family or to involve multiple families. On the other hand, it seems possible that selective pressure to recognize multiple different D-sites on multiple different substrates might ultimately compromise the ability of a MAPK to bind in a highly selective manner to some D-sites and not others. Indeed, perhaps such a scenario may explain the promiscuity of ERK2-D-site interactions.
MKK3 D-site Is a Highly Potent Inhibitor—In the course of these studies, we found that the MKK3-derived D-site peptide was a particularly potent inhibitor of p38α, with a degree of inhibition over 100-fold greater than observed with other D-site peptides. p38α continues to generate a great deal of interest as a drug target (16, 21, 24). Our results suggest that using an MKK3-derirved peptide to inhibit p38 may be a therapeutic approach worth considering, as is currently being explored with the JIP-1 peptide that targets JNK (67). Even if this ultimately proves impractical, a better understanding of the high efficiency of this inhibition may provide insight useful for designing more traditional small molecule agents.
A co-crystal structure of unphosphorylated p38 bound to an MKK3-dervied D-site peptide has been solved (56), but this structure does not show any interactions involving the basic residues in the MKK3 D-site, and thus does not provide much insight as to why this interaction may be particularly high affinity. It should be noted that the lack of contacts involving the basic submotif in this structure may well be due to the particular crystallization conditions chosen, because such contacts have been observed in other MAPK/D-site peptide complexes (37, 57, 58). Further structure-function studies will be required to dissect the mechanism of MKK3 D-site binding/inhibition.
Implications for Drug Design—Although many kinase inhibitors bind to the ATP binding pocket of their target kinase, there is increasing interest in developing drug candidates that modulate kinase function by a different mechanism, such as by blocking kinase-substrate interactions (68, 69). Indeed, there are current efforts to target MAPK/D-site interactions for therapeutic purposes, using either peptides or more traditional small molecule compounds (13-18). Clearly, such efforts will be facilitated by information regarding D-site selectivity, such as we have provided here.
More particularly, as noted above, our results indicate that the MKK3 D-site peptide may be a particularly promising reagent along these lines. Another implication for drug design follows from our observation that JNK1 and particularly JNK2 do not appreciably bind to non-cognate D-sites. Thus, targeting the D-site-binding regions of JNK1 and JNK2 would seem to have a reasonable probability of a highly selective outcome.
Summary—We have systematically and quantitatively compared the binding of docking sites derived from MAPK kinases (MKKs) for their cognate (within-pathway) and non-cognate MAPKs. We find that cognate interactions are preferred ∼10-fold over non-cognate interactions. These results are consistent with a “double selection” model, in which both the docking-site interaction and interactions involving the catalytic domain of the MKK are important for specificity.
Acknowledgments
We thank W.-L. Blackwell for assistance with preliminary studies on p38.
This work was supported, in whole or in part, by National Institutes of Health Grant GM60366 from the NIGMS.
Footnotes
The abbreviations used are: MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated protein kinase; MKK, MAPK kinase; MEK, MAPK/ERK kinase; JNK, c-Jun N-terminal kinase; GST, glutathione S-transferase.
References
- 1.Blume-Jensen, P., and Hunter, T. (2001) Nature 411 355-365 [DOI] [PubMed] [Google Scholar]
- 2.Cohen, P. (2002) Nat. Rev. Drug Discov. 1 309-315 [DOI] [PubMed] [Google Scholar]
- 3.Force, T., Kuida, K., Namchuk, M., Parang, K., and Kyriakis, J. M. (2004) Circulation 109 1196-1205 [DOI] [PubMed] [Google Scholar]
- 4.Johnson, L. (2007) Biochem. Soc Trans 35 7-11 [DOI] [PubMed] [Google Scholar]
- 5.Pawson, T. (2004) Cell 116 191-203 [DOI] [PubMed] [Google Scholar]
- 6.Papin, J. A., Hunter, T., Palsson, B. O., and Subramaniam, S. (2005) Nat. Rev. Mol. Cell Biol. 6 99-111 [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya, R. P., Remenyi, A., Yeh, B. J., and Lim, W. A. (2006) Annu. Rev. Biochem. 75 655-680 [DOI] [PubMed] [Google Scholar]
- 8.Legewie, S., Schoeberl, B., Bluthgen, N., and Herzel, H. (2007) Biophys. J. 93 2279-2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remenyi, A., Good, M. C., and Lim, W. A. (2006) Curr. Opin. Struct. Biol. 16 676-685 [DOI] [PubMed] [Google Scholar]
- 10.Goldsmith, E. J., Akella, R., Min, X., Zhou, T., and Humphreys, J. M. (2007) Chem. Rev. 107 5065-5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grewal, S., Molina, D. M., and Bardwell, L. (2006) Cell. Signal. 18 123-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannan, N., Haste, N., Taylor, S. S., and Neuwald, A. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1272-1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akella, R., Moon, T. M., and Goldsmith, E. J. (2008) Biochim. Biophys. Acta 1784 48-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock, C. N., Macias, A. T., Mackerell, A. D., Jr., and Shapiro, P. (2006) Med. Chem. 2 213-222 [DOI] [PubMed] [Google Scholar]
- 15.Kaidanovich-Beilin, O., and Eldar-Finkelman, H. (2006) Physiology (Bethesda) 21 411-418 [DOI] [PubMed] [Google Scholar]
- 16.Mayor, F., Jr., Jurado-Pueyo, M., Campos, P. M., and Murga, C. (2007) Cell Cycle 6 528-533 [DOI] [PubMed] [Google Scholar]
- 17.Parang, K., and Sun, G. (2004) Curr. Opin. Drug Discov. Dev. 7 617-629 [PubMed] [Google Scholar]
- 18.Waetzig, V., and Herdegen, T. (2005) Trends Pharmacol. Sci. 26 455-461 [DOI] [PubMed] [Google Scholar]
- 19.Lawrence, M. C., Jivan, A., Shao, C., Duan, L., Goad, D., Zaganjor, E., Osborne, J., McGlynn, K., Stippec, S., Earnest, S., Chen, W., and Cobb, M. H. (2008) Cell Res. 18 436-442 [DOI] [PubMed] [Google Scholar]
- 20.Senawong, T., Phuchareon, J., Ohara, O., McCormick, F., Rauen, K. A., and Tetsu, O. (2008) Hum. Mol. Genet. 17 419-430 [DOI] [PubMed] [Google Scholar]
- 21.Cuenda, A., and Rousseau, S. (2007) Biochim. Biophys. Acta 1773 1358-1375 [DOI] [PubMed] [Google Scholar]
- 22.Johnson, G. L., and Nakamura, K. (2007) Biochim. Biophys. Acta 1773 1341-1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, X., Destrument, A., and Tournier, C. (2007) Biochim. Biophys. Acta 1773 1349-1357 [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., Boehm, J., and Lee, J. C. (2003) Nat. Rev. Drug Discov. 2 717-726 [DOI] [PubMed] [Google Scholar]
- 25.Roberts, P. J., and Der, C. J. (2007) Oncogene 26 3291-3310 [DOI] [PubMed] [Google Scholar]
- 26.Sharrocks, A. D., Yang, S. H., and Galanis, A. (2000) Trends Biochem. Sci. 25 448-453 [DOI] [PubMed] [Google Scholar]
- 27.Enslen, H., and Davis, R. J. (2001) Biol. Cell 93 5-14 [DOI] [PubMed] [Google Scholar]
- 28.Bardwell, L. (2006) Biochem. Soc. Trans. 34 837-841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bardwell, L., Cook, J. G., Chang, E. C., Cairns, B. R., and Thorner, J. (1996) Mol. Cell. Biol. 16 3637-3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bardwell, L., and Thorner, J. (1996) Trends Biochem. Sci. 21 373-374 [PubMed] [Google Scholar]
- 31.Bardwell, A. J., Flatauer, L. J., Matsukuma, K., Thorner, J., and Bardwell, L. (2001) J. Biol. Chem. 276 10374-10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho, D. T., Bardwell, A. J., Abdollahi, M., and Bardwell, L. (2003) J. Biol. Chem. 278 32662-32672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho, D. T., Bardwell, A. J., Grewal, S., Iverson, C., and Bardwell, L. (2006) J. Biol. Chem. 281 13169-13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, B., Wilsbacher, J. L., Collisson, T., and Cobb, M. H. (1999) J. Biol. Chem. 274 34029-34035 [DOI] [PubMed] [Google Scholar]
- 35.Enslen, H., Brancho, D. M., and Davis, R. J. (2000) EMBO J. 19 1301-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bardwell, A. J., Abdollahi, M., and Bardwell, L. (2003) Biochem. J. 370 1077-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remenyi, A., Good, M. C., Bhattacharyya, R. P., and Lim, W. A. (2005) Mol. Cell 20 951-962 [DOI] [PubMed] [Google Scholar]
- 38.Bardwell, A. J., Abdollahi, M., and Bardwell, L. (2004) Biochem. J. 378 569-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turk, B. E. (2007) Biochem. J. 402 405-417 [DOI] [PubMed] [Google Scholar]
- 40.Lewis, T. S., Shapiro, P. S., and Ahn, N. G. (1998) Adv. Cancer Res. 74 49-139 [DOI] [PubMed] [Google Scholar]
- 41.Brunet, A., and Pouyssegur, J. (1996) Science 272 1652-1655 [DOI] [PubMed] [Google Scholar]
- 42.Jiang, Y., Li, Z., Schwarz, E. M., Lin, A., Guan, K., Ulevitch, R. J., and Han, J. (1997) J. Biol. Chem. 272 11096-11102 [DOI] [PubMed] [Google Scholar]
- 43.Robinson, M. J., Cheng, M., Khokhlatchev, A., Ebert, D., Ahn, N., Guan, K. L., Stein, B., Goldsmith, E., and Cobb, M. H. (1996) J. Biol. Chem. 271 29734-29739 [DOI] [PubMed] [Google Scholar]
- 44.Seyfried, J., Wang, X., Kharebava, G., and Tournier, C. (2005) Mol. Cell. Biol. 25 9820-9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Derijard, B., Raingeaud, J., Barrett, T., Wu, I. H., Han, J., Ulevitch, R. J., and Davis, R. J. (1995) Science 267 682-685 [DOI] [PubMed] [Google Scholar]
- 46.Han, J., Lee, J. D., Jiang, Y., Li, Z., Feng, L., and Ulevitch, R. J. (1996) J. Biol. Chem. 271 2886-2891 [DOI] [PubMed] [Google Scholar]
- 47.Tournier, C., Whitmarsh, A. J., Cavanagh, J., Barrett, T., and Davis, R. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7337-7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, Z., Wu, J., Jacinto, E., and Karin, M. (1997) Mol. Cell. Biol. 17 7407-7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin, A., Minden, A., Martinetto, H., Claret, F. X., Lange-Carter, C., Mercurio, F., Johnson, G. L., and Karin, M. (1995) Science 268 286-290 [DOI] [PubMed] [Google Scholar]
- 50.Bogoyevitch, M. A., and Kobe, B. (2006) Microbiol. Mol. Biol. Rev. 70 1061-1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turjanski, A. G., Vaque, J. P., and Gutkind, J. S. (2007) Oncogene 26 3240-3253 [DOI] [PubMed] [Google Scholar]
- 52.Yoon, S., and Seger, R. (2006) Growth Factors 24 21-44 [DOI] [PubMed] [Google Scholar]
- 53.Kallunki, T., Deng, T., Hibi, M., and Karin, M. (1996) Cell 87 929-939 [DOI] [PubMed] [Google Scholar]
- 54.Yang, S. H., Yates, P. R., Whitmarsh, A. J., Davis, R. J., and Sharrocks, A. D. (1998) Mol. Cell. Biol. 18 710-720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kusari, A. B., Molina, D. M., Sabbagh, W., Jr., Lau, C. S., and Bardwell, L. (2004) J. Cell Biol. 164 267-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang, C. I., Xu, B., Akella, R., Cobb, M., and Goldsmith, E. J. (2002) Mol. Cell 9 1241-1249 [DOI] [PubMed] [Google Scholar]
- 57.Heo, Y. S., Kim, S. K., Seo, C. I., Kim, Y. K., Sung, B. J., Lee, H. S., Lee, J. I., Park, S. Y., Kim, J. H., Hwang, K. Y., Hyun, Y. L., Jeon, Y. H., Ro, S., Cho, J. M., Lee, T. G., and Yang, C. H. (2004) EMBO J. 23 2185-2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu, S., Sun, J. P., Zhou, B., and Zhang, Z. Y. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5326-5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanoue, T., Adachi, M., Moriguchi, T., and Nishida, E. (2000) Nat. Cell Biol. 2 110-116 [DOI] [PubMed] [Google Scholar]
- 60.Brancho, D., Tanaka, N., Jaeschke, A., Ventura, J. J., Kelkar, N., Tanaka, Y., Kyuuma, M., Takeshita, T., Flavell, R. A., and Davis, R. J. (2003) Genes Dev. 17 1969-1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callaway, K., Abramczyk, O., Martin, L., and Dalby, K. N. (2007) Biochemistry 46 9187-9198 [DOI] [PubMed] [Google Scholar]
- 62.Ember, B., Kamenecka, T., and LoGrasso, P. (2008) Biochemistry 47 3076-3084 [DOI] [PubMed] [Google Scholar]
- 63.Niu, L., Chang, K. C., Wilson, S., Tran, P., Zuo, F., and Swinney, D. C. (2007) Biochemistry 46 4775-4784 [DOI] [PubMed] [Google Scholar]
- 64.Rainey, M. A., Callaway, K., Barnes, R., Wilson, B., and Dalby, K. N. (2005) J. Am. Chem. Soc. 127 10494-10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson, F. L., Whitehurst, A. W., Raman, M., and Cobb, M. H. (2002) J. Biol. Chem. 277 14844-14852 [DOI] [PubMed] [Google Scholar]
- 66.Wilsbacher, J. L., Goldsmith, E. J., and Cobb, M. H. (1999) J. Biol. Chem. 274 16988-16994 [DOI] [PubMed] [Google Scholar]
- 67.Borsello, T., Clarke, P. G., Hirt, L., Vercelli, A., Repici, M., Schorderet, D. F., Bogousslavsky, J., and Bonny, C. (2003) Nat. Med. 9 1180-1186 [DOI] [PubMed] [Google Scholar]
- 68.Molina, D. M., Grewal, S., and Bardwell, L. (2005) J. Biol. Chem. 280 42051-42060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bogoyevitch, M. A., and Fairlie, D. P. (2007) Drug Discov. Today 12 622-633 [DOI] [PubMed] [Google Scholar]