Abstract
Cyclic di-GMP (c-di-GMP) is a ubiquitous bacterial second messenger that is involved in the regulation of cell surface-associated traits and the persistence of infections. Omnipresent GGDEF and EAL domains, which occur in various combinations with regulatory domains, catalyze c-di-GMP synthesis and degradation, respectively. The crystal structure of full-length YkuI from Bacillus subtilis, composed of an EAL domain and a C-terminal PAS-like domain, has been determined in its native form and in complex with c-di-GMP and Ca2+. The EAL domain exhibits a triose-phosphate isomerase-barrel fold with one antiparallel β-strand. The complex with c-di-GMP-Ca2+ defines the active site of the putative phosphodiesterase located at the C-terminal end of the β-barrel. The EAL motif is part of the active site with Glu-33 of the motif being involved in cation coordination. The structure of the complex allows the proposal of a phosphodiesterase mechanism, in which the divalent cation and the general base Glu-209 activate a catalytic water molecule for nucleophilic in-line attack on the phosphorus. The C-terminal domain closely resembles the PAS-fold. Its pocket-like structure could accommodate a yet unknown ligand. YkuI forms a tight dimer via EAL-EAL and trans EAL-PAS-like domain association. The possible regulatory significance of the EAL-EAL interface and a mechanism for signal transduction between sensory and catalytic domains of c-di-GMP-specific phosphodiesterases are discussed.
The dinucleotide cyclic di-GMP (c-di-GMP) was discovered about 20 years ago when it was found to regulate the activity of cellulase synthase in Acetobacter xylinum (1). However, its prominent role as a global second messenger has been realized only upon the recent recognition of the omnipresence of genes coding for domains that catalyze c-di-GMP biosynthesis and degradation in eubacteria (2). GGDEF domains catalyze the condensation of two GTP molecules to the cyclic 2-fold symmetric dinucleotide (diguanylate cyclase activity (3-6)), whereas EAL domains are involved in its degradation to yield the linear dinucleotide pGpG (phosphodiesterase (PDE)4 A activity) (3, 7-9). Recently, also HD-GYP domains have been implicated in c-di-GMP-specific PDE activity (10). All the domains have been named according to their sequence signature motifs. They are typically found in combinations with various other, mostly sensory or regulatory, domains. It is thought that the balance between antagonistic diguanylate cyclase and PDE-A activities determines the cellular level of c-di-GMP and, thus, affects a variety of physiological processes in bacteria.
It has been shown that, in general, c-di-GMP regulates cell surface-associated traits and community behavior such as biofilm formation (for reviews see Refs. 11-12), and its relevance to the virulence of pathogenic bacteria has been demonstrated (11, 13, 14). In particular, the dinucleotide has been proposed to orchestrate the switch between acute and persistent phase of infection.
The best characterized diguanylate cyclase is PleD from Caulobacter
crescentus with a Rec-Rec-GGDEF domain architecture (Rec indicates
response regulator receiver domain). The structure of its GGDEF domain
revealed a single GTP-binding site and suggested that dimerization is the
prerequisite for enzymatic activity
(4). This has been corroborated
recently by crystallography showing directly that
 modification of the first Rec
domain, mimicking phosphorylation by the cognate kinase, induces formation of
a tightly packed dimer (15).
Additionally, an upper limit of c-di-GMP levels in the cell seems to be
ensured by potent allosteric product inhibition of the PleD cyclase
(4,
15,
16). Recently, the crystal
structure of another diguanylate cyclase, WspR from Pseudomonas
aeruginosa with a Rec-GGDEF domain architecture, has been determined
(17), which showed a
tetrameric quaternary structure and active and feedback inhibition sites that
are very similar to those in PleD.
modification of the first Rec
domain, mimicking phosphorylation by the cognate kinase, induces formation of
a tightly packed dimer (15).
Additionally, an upper limit of c-di-GMP levels in the cell seems to be
ensured by potent allosteric product inhibition of the PleD cyclase
(4,
15,
16). Recently, the crystal
structure of another diguanylate cyclase, WspR from Pseudomonas
aeruginosa with a Rec-GGDEF domain architecture, has been determined
(17), which showed a
tetrameric quaternary structure and active and feedback inhibition sites that
are very similar to those in PleD.
For EAL domains, it has been demonstrated that genetic knock-out results in phenotypes that are in line with the paradigm that an elevated cellular c-di-GMP concentration corresponds to a sessile and a low concentration to a motile bacterial life style (13, 18, 19). Only recently, EAL-mediated PDE-A activity has been measured in vitro (7-9, 20-22).
The Bacillus subtilis YkuI protein was targeted for structure determination by the Midwest Center for Structural Genomics as a member of the large sequence family that contains EAL (Pfam number PF00563) domains. Here we report the crystal structure of YkuI showing the fold of the N-terminal EAL domain and the C-terminal PAS-like domain. Co-crystallization with c-di-GMP revealed the substrate binding mode and allows the proposal of a catalytic mechanism. The PAS-like domain most probably has regulatory function, which is discussed. Recently, another EAL structure has been deposited in the Protein Data Bank by the Midwest Center for Structural Genomics, the EAL domain of a GGDEF-EAL protein from Thiobacillus denitrificans (tdEAL; PDB code 2r6o). Comparison of the two structures suggests a possible regulatory mechanism.
MATERIALS AND METHODS
Expression and Purification of Selenomethionine and Wild-type YkuI and Its EAL Domain—For expression and purification of selenomethionine (Se-Met) YkuI protein high through-put protocols developed at the Midwest Center for Structural Genomics were used. Overexpression of full-length B. subtilis YkuI protein with N-terminal His6 tag using the pMCSG7 vector transformed into the Escherichia coli host strain BL21-DE3 was performed as described (23, 24). The EALYkuI construct encompassed amino acids 1-259 and the same His6 tag and tobacco etch virus protease cleavage site as the full-length construct and carried the point mutation I258T (chosen to improve solubility).
The standard protocol was modified for the wild-type YkuI protein used in co-crystallization experiments and for EALYkuI. These were overexpressed in E. coli BL21(DE3) cells. Induction was performed at A600 of 0.6 with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and cells were harvested 4 h after induction by centrifugation at 6000 × g(max) (Sorval SLA 3000) for 5 min. The pellets were resuspended in the lysis buffer (50 mm NaH2PO4 (pH 8.0), 500 mm NaCl) containing EDTA-free protease inhibitor (Roche Diagnostics), and cells were then lysed by French press (Thermo Electron Corp.). The cell lysate was centrifuged at 30,000 × g(max) (Sorval SLA 1500) at 4 °C for 30 min, and the supernatant was collected. The charged nickel-nitrilotriacetic acid Superflow™ resin (Qiagen AG) pre-equilibrated with the lysis buffer was added to the cell lysates and incubated at 4 °C for 2 h. Then beads were loaded on a poly-prep chromatography column (Bio-Rad) and washed with lysis buffer. The eluted fractions from 50 and 100 mm imidazole were pooled and concentrated to around 10 mg/ml using Amicon Ultra device with a cutoff of 30 kDa (Millipore AG). The concentrated protein was centrifuged at 16,000 × g (max) (Vaudaux-Eppendorf, Centrifuge 5804 R) at 4 °C for 10 min and loaded onto a Superdex 75 gel filtration column (Amersham Biosciences Europe) equilibrated with 20 mm Tris-HCl (pH 7.6) and 500 mm NaCl. Fractions were pooled and concentrated to 10 mg/ml (assuming an ε280 of 65,335 m-1 cm-1
Crystallization and Data Collection—Crystallization conditions for Se-Met YkuI were found using the Hampton Research (Riverside, CA) Crystal Screen I. A hanging drop consisting of 2 μl of a protein solution (11 mg/ml) containing 10 mm Tris-HCl (pH 8.3), 250 mm NaCl, and 5 mm 2-mercaptoethanol mixed with 2 μl of a reservoir solution containing 0.2 m sodium acetate trihydrate, 0.1 m Tris-HCl (pH 8.5), and 30% w/v polyethylene glycol 4000 was equilibrated with 1 ml of the reservoir. Se-Met protein crystals suitable for data collection were obtained within a week. Single wavelength anomalous diffraction data were collected at the selenium absorption peak on a single Se-Met crystal to a resolution of 2.6 Å (Table 1). Diffraction data were collected at 100 K on the 5-ID-B beam line of the DuPont-Northwestern-Dow Collaborative Access Team at the Advanced Photon Source, Argonne, IL. Images were integrated and intensities scaled in the XDS suite (25).
TABLE 1.
Crystallographic data
| Se-Met | C-di-GMP complex | |
|---|---|---|
| Data collection | ||
| X-ray source | APS, 51D-B | SLS |
| Detector type | MAR CCD | MAR CCD |
| Data collection temperature | 100 K | 100 K |
| Space group | P212121 | P212121 |
| Unit cell dimensions | 46.3, 125.3, 168.0 Å | 46.1, 124.5, 168.7 Å |
| Wavelength | 0.97931 Å | 0.97800 Å |
| Data processing | ||
| Resolution range | 25.0-2.6 Å (2.76-2.61 Å)a | 70.0-2.8 Å (2.95-2.80 Å)a |
| No. of unique reflections | 30,096 (2,692)a | 21,459 (2,581)a |
| Completeness | 94.7% (84.7%)a | 87.2% (73.0%)a |
| Redundancy | 3.0 (3.0)a | 3.4 (2.8)a |
| I/σ(I) | 17.5 (3.7)a | 9.2 (1.6)a |
| Rmergeb | 4.4% (34.0%)a | 9.6% (42.4%)a |
| Phasing | ||
| No. of refined sites | 17 out of possible 20 | |
| Figure of merit initialc | 0.338 | |
| Solvent content | 48.2% | |
| Figure of merit density modifiedd | 0.528 | |
| Refinement | ||
| Rcryst/Rfreee | 20.7% (28.2%)/28.7% (37.2%) | 22.6% (36.7%)/27.5% (42.9%) |
| Mean B factor | 65.7 Å2 | 38.9 Å2 |
| Protein atoms/residues | 8265/799 | 8179/796 |
| Solvent molecules | 255 | 7 |
| Heteroatoms | 1 BME | 4 GMP, 2 Ca2+ |
| Stereogeometry | ||
| r.m.s.d. from ideal geometry | ||
| Bond length | 0.012 Å | 0.010 Å |
| Bond angle | 1.3° | 1.2° |
| Ramachandran plot | ||
| Most favored region | 89.8% | 90.5% |
| Additionally favored region | 10.2% | 8.9% |
| Generally allowed region | 0.0% | 0.4% |
| Disallowed region | 0.0% | 0.1% |
Highest resolution shell is given.
Rmerge = ∑|I(k) - (I)|/∑I(k), where I(k) is the value of the kth measurement of the intensity of a reflection; I is the mean of the intensity of that reflection, and the sum runs over all the measurements of that reflection.
Initial phases were calculated to 3.2 Å resolution.
Modified phases were extended to 2.61 Å resolution.
Rfactor = ∑||Fobs| - |Fcalc||/∑|Fobs|.
Structure Determination and Refinement of Se-Met YkuI—17 positions out of 20 possible selenium sites were determined with the program SOLVE (26) based on 2.6 Å resolution single wavelength anomalous data (Table 1). The asymmetric unit consists of two polypeptide chains, each containing 10 selenium atoms. The relation between the selenium sites helped to locate the noncrystallographic (NCS) 2-fold axis, which was present in the calculated self-rotation function (27) as a strong peak. Selenium sites were refined with the program SHARP (28), and initial phases were modified by multidomain NCS averaging using the program DM in the CCP4 suite (29). An initial model was built using RESOLVE (26). The model consisted of short 5-10-residue peptide chains, although the quality of modified map was relatively good. The main parts of polypeptide chain were built using poly(Ala) modules of α-helixes and β-strands, 15-20 residues long, fitted in the electron density maps in TURBO-FRODO (30). NCS information was used during model building and averaging to improve the electron density maps. Cycles of manual rebuilding were followed by positional simulated annealing and temperature factor refinement with the program CNS (31), which gradually improved the model. The free R factor was monitored by setting aside 5% of the reflections as a test data set (32). Final steps of the model refinement were performed in REFMAC5 (29) using bulk solvent and translation liberation spin correction (33). The final refinement statistics is given in Table 1.
Determination of the YkuI c-di-GMP Complex Structure—The YkuI c-di-GMP complex was formed by mixing 8.3 mg/ml YkuI with 2 mm chemically synthesized c-di-GMP (55) and 2 mm CaCl2 (all final concentrations) in 20 mm Tris-HCl (pH 7.6), 500 mm NaCl. Rod-like crystals were grown using the hanging-drop vapor diffusion method at 20 °C under the following condition. YkuI ligand complex was mixed with reservoir solution (28% (w/v) polyethylene glycol 4000, 3% (v/v) glycerol, 200 mm sodium acetate, 100 mm imidazole (pH 8.0)) at a ratio of 1:1. Diffraction data were collected to 2.8 Å from a single crystal at the Swiss Light Source, Villigen, Switzerland. The images were indexed and integrated using the program MOSFLM (34). Because both the native and the c-di-GMP complex of YkuI crystallized in the same space group with similar unit cell dimensions, the same set of test reflections as for the native data was used for subsequent Rfree calculations. The structure was solved by rigid body refinement starting with the Se-Met structure, followed by translation liberation spin and full atom refinement with strong restraints on the B factors and NCS restraints using the program REFMAC5 (29). The crystallographic statistic is given in Table 1.
Activity Measurements—Pure samples of c-di-GMP were obtained from N. Amiot, Department Chemistry, University of Basel. C-di-GMP-specific PDE activity was assayed indirectly by monitoring the production of phosphate using an enzyme-coupled spectrophotometric assay (35). The reaction mixture contained 5 μm YkuI in 50 mm Tris-HCl (pH 9.0), 250 mm NaCl, 100 mm MgCl2, 100 μm c-di-GMP, and alkaline phosphatase (900 milliunits/ml). The reaction was stopped by transferring 100 μl of reaction mixture into the phosphate assay reagent (pH ≈0; 1 ml final volume) containing molybdate and malachite green. As positive control, the activity of YahA from E. coli was verified. EALYkuI was assayed as described above. The measurements were performed at different protein concentrations (1, 10, 20, and 50 μm) using concentrations of c-di-GMP ranging from 100 μm to 1 mm. The reactions were kept at room temperature for different incubation times (10, 30, and 60 min and overnight) and then stopped as reported above.
Bioinformatics—Molecular interface areas were calculated with the PISA server (Protein Interfaces, Surfaces and Assemblies service (PISA) at EBI (36)). For domain and fold classifications, the CATH (version 3.2.0) and Pfam (version 23.0) data bases were interrogated (37, 38). Structural homologs to the YkuI_C domain were identified by the NCBI VAST server. Structure comparisons were performed with the SSM server (Protein Structure Comparison service SSM at EBI (39)) and topp (29).
RESULTS AND DISCUSSION
Overall Structure and Topology—The crystal structure of full-length YkuI from B. subtilis has been determined by single wavelength-anomalous-diffraction phasing using Se-Met substituted protein. Crystallographic data are given in Table 1. The entire main chain is defined by electron density with the exception of the purification tag and loop region 181-184 in both chains, as well as the N-terminal Met-1 of chain A and the C-terminal residues 401-407 of chain B. Additionally, there are one β-mercaptoethanol and 255 water molecules. Both chain structures are similar, but deviations are found for some loops (r.m.s.d. = 0.83 Å for all C-α positions after superpositions). In the following, the structure description refers to chain A.
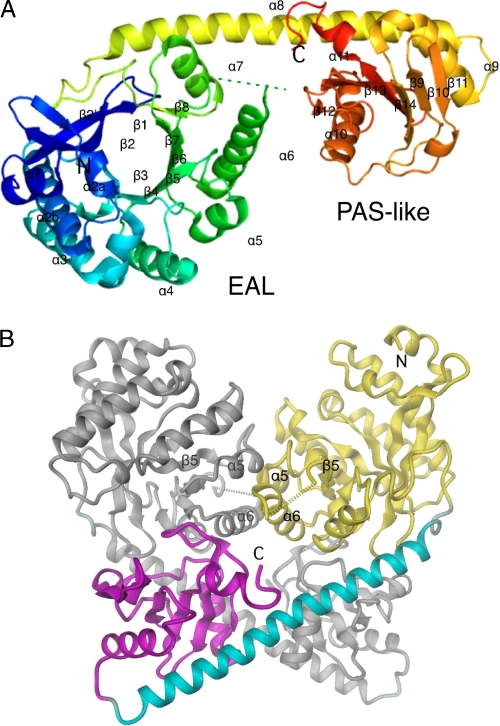
Fig. 1 shows the fold of YkuI. The 407-residue polypeptide is composed of an N-terminal EAL domain and a PAS-like C-terminal domain connected by a long α-helix (αlink). Secondary structure assignments are given in Fig. 2. The asymmetric unit contains a tight homodimer, which is formed by 2-fold association of the EAL domains and trans association of the PAS and EAL* domains (the asterisk indicates an element of the adjacent subunit in the YkuI dimer). The two EAL domains in the asymmetric unit of the tdEAL structure (PDB code 2r6o)5 show the same dimeric arrangement with an r.m.s.d. of 2.3 Å for 344 of the 470 C-α positions after superposition (the A-chains super-impose with 1.7 Å/162). This is surprising because the sequence similarity is modest, 21% identity, and the fragment of tdEAL protein that was crystallized was an isolated EAL domain.
FIGURE 1.
Crystal structure of YkuI. A, the monomer consists of two domains connected by a long helix (residues 246-289). The ribbon is colored from blue at the N terminus to red at the C terminus. The N-terminal EAL domain is folded to a TIM-barrel, and the C-terminal domain shows a PAS-like fold. Chain termini and secondary structure elements are labeled (see also Fig. 2). B, two monomers of YkuI form a noncrystallographic tight dimer. The 2-fold axis is vertical. The A-chain is colored in yellow (EAL domain), green (linking helix), and magenta (C-terminal PAS-like domain). The B-chain is shown in gray. The two EAL domains interact via isologous interactions across the molecular 2-fold axis involving the β5-α5 loop and the α6-helix.
FIGURE 2.
Sequence alignments of YkuI with structural homologs. Residue numbers correspond to the YkuI sequence (Swiss-Prot accession code O35014). The secondary structure elements of YkuI are displayed at the top. A, alignment with the EAL domains of an EAL-GGDEF protein from T. denitrificans (Q3SJE6), the GerE-EAL protein YahA from E. coli (P21514), the Rec-EAL protein RocR from P. aeruginosa (Q9HX69), and the EAL-GGDEF protein PdeA from C. crescentus (Q9A310). The alignment taken is that of EAL Pfam family (PF00563), with the exception of tdEAL (PDB code 2r6o) for which a structure-based alignment has been carried out. Partly and strictly conserved residues are highlighted by gray and black background, respectively. Residues that coordinate the divalent cation in YkuI and tdEAL have a magenta background; the putative general base Glu-209 is shown in red; the active site Lys-173 is in blue; the substrate-binding residues Gln-19 and Arg-37 are in green. B, structure-based alignment with PYP (PDB code 1otd) from Halorhodospira halophila (P16113) and the GAF B domain of phosphodiesterase 2a (1mc0 (49)) from Mus musculus (Q922S4). The secondary structure elements of PYP and GAF B are given below the alignment. The figure was produced with ESPRIPT (54).
In both proteins, the interface is mainly formed by the following: (a) an antiparallel β-strand arrangement of the β5-α5 loop with its symmetry mate, and (b) antiparallel packing of helix α6 with its symmetry mate (Fig. 1B). Between the two domains of the same subunit and between the two PAS-like domains, there is no significant contact. Intriguingly, the β5-α5 contact is not symmetric. Some inter-subunit H-bonds are not formed, and the rotation angle for superposition of the EAL subunits is 175°. The break-in symmetry probably reflects asymmetry in the crystal contacts, which are, however, remote from the EAL-EAL interface. This demonstrates that the β5-α5 interface is sensitive to large distance perturbations. The repercussions this may have for regulation is shown further below (see under “Regulation of PDE Activity”).
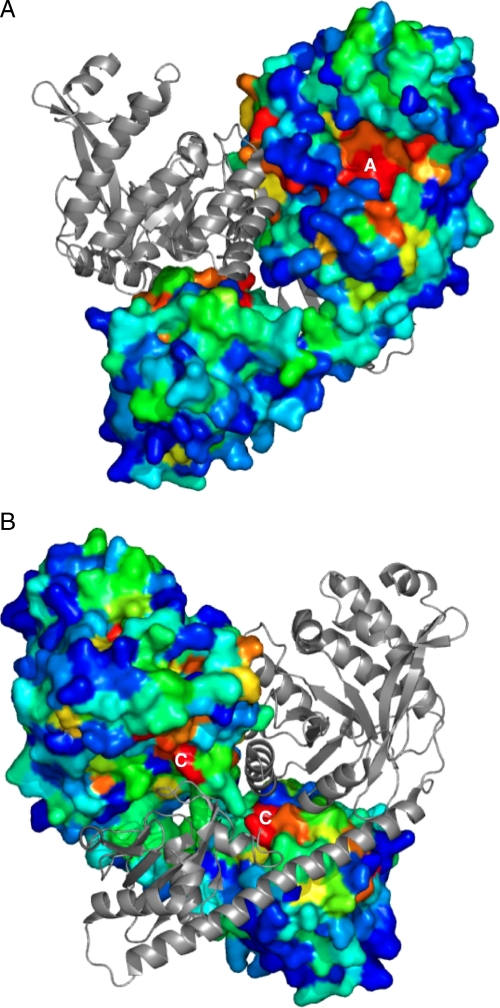
The crystal dimer exhibits an extensive interface area of about 3000 Å2 with a low ΔG of -30 kcal/mol and a good complementary surface score of 0.919 as measured by PISA (36). Other interfaces in the crystal lattice (not shown) are at least seven times smaller and show no significant complementary surface score. This suggests that the crystal dimer also represents the quaternary structure of YkuI in solution consistent with light scattering experiments (data not shown). Another indication that the crystal dimer likely represents a functional dimer is that conserved surface-exposed amino acids occur in patches at the points of contact (Fig. 3).
FIGURE 3.
Surface of YkuI colored by the conservation of amino acids. Forty sequences of YkuI homologs containing both EAL and PAS-like domains were aligned, and the relative conservation at each position was calculated. In one monomer, the surface is colored from blue (variable) to red (100% conserved); the other monomer is shown as a gray ribbon. A, most prominent conserved patch is seen above the C-terminal end of the central β-barrel of the EAL domain (top) forming the substrate-binding groove (labeled with A). B, view rotated 180° relative to A showing the conservation at the contacts between the two subunits of the dimer (labeled with C).
EAL Domain Structure—The crystal structure of YkuI provides the first view of an EAL domain. This domain, also known as DUF2 (Domain of Unknown Function 2) was originally detected through amino acid sequence comparisons (2), where it was found to be very common in bacterial species, often occurring in multiple genes and in the context of a variety of other domains.
The EAL domain exhibits the TIM-barrel fold (Fig. 1A) with, however, the first helix after strand β1 missing and β1 running antiparallel to the remaining strands. Thus, the succession of the major secondary structure elements is αβ(βα)6β with an N-terminal extra helix α1 and an extended loop instead of the canonical C-terminal helix. There is a small bifurcation of the central β-sheet formed by strands β1, β2, and β2′ (Fig. 1A). The β-barrel of canonical TIM-barrels, one of the most common protein superfamilies (40), is composed of entirely parallel β-strands. Among the exceptions, in the enolase family the second strand runs in the opposite direction (PDB code 3ENL (41)), whereas in glycosyl hydrolase family 25 (PDB code 1YFX (42)) and an uncharacterized bacterial protein (PDB code 1SFS), the eighth strand is anti-parallel to the other seven strands. To our knowledge, no other case with an antiparallel β1-strand has been reported so far. Indeed, in the CATH data base (37), the YkuI EAL domain has been classified as a new superfamily (entry 3.20.20.450).
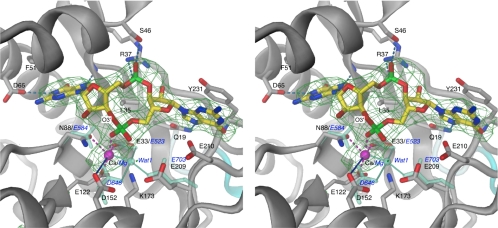
The EAL signature motif, which in YkuI as in many other EAL domains has the sequence EVL, locates to strand β2 with Glu-33 and Leu-35 forming part of the bottom of a shallow groove at the C-terminal end of the β barrel, the usual place of the active site in enzymes with the TIM-fold (40). Valine 34 is buried in the hydrophobic core of the domain. The groove is lined by a number of additional, highly conserved surface-exposed residues (Fig. 3A) making identification of the active site straightforward. c-di-GMP Bound to EAL Domain—To reveal the substrate binding details and to obtain insights to the catalytic mechanism, YkuI was crystallized in complex with c-di-GMP. To prevent substrate turnover, calcium instead of magnesium was added. It has been shown for other EAL proteins that Mg2+ is an essential cofactor for c-di-GMP-specific PDEs, whereas Ca2+ inhibits the enzyme activity (1, 7-8). The protein structure was found virtually unchanged upon complex formation (r.m.s.d. = 0.60 Å for all Cα) and displayed a well ordered c-di-GMP molecule bound to the anticipated active site at the C-terminal end of the β-barrel in each chain (Fig. 4). Sandwiched between substrate and protein, one calcium ion is found that has octahedral coordination with the carboxylates of Glu-33 and Glu-122, and the side chain carbonyl of Asn-88 (Table 2). Another coordination position is taken up by a substrate phosphate oxygen.
FIGURE 4.
Stereo view of the active site of the YkuI EAL domain with bound substrate c-di-GMP and Ca2+. The omit map for substrate and the divalent cation has been contoured at 3σ. Coordinating atoms are joint with the Ca2+ by dashed lines in magenta. Asp-152 has a distance of 3.7 Å to the metal, indicated by the blue line. Superimposed on the YkuI structure, the water molecule Wat1 of the tdEAL structure (PDB code 2r6o) is shown with its ligands Mg2+ (largely concealed by the YkuI calcium ion) and Glu-535, Asp-646, and Glu-703. These tdEAL residues are shown in aquamarine and labeled in blue. For clarity, the residues that are homologous to Asn-88, Glu-122, and Lys-173 of YkuI (Glu-616 and Lys-667 of tdEAL), are not shown, but would also superimpose closely. H-bonds and the Wat-Mg2+ coordination in tdEAL are drawn in light blue.
TABLE 2.
Ykul-ligand interactions
Only the values for the Ykul A-chain are given, those for the B-chain are similar. Cyclic di-GMP is composed of two GMP (5gp) molecules, called A501 and A502, that are linked by two P-O-3′ ester bonds.
| Atom 1 | Atom 2 | Distance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Å | |||||||||||
| Calcium coordination | |||||||||||
| Ca2 | A503A | CA | Glu | A33 | O-ε1 | 2.3 | |||||
| Asn | A88 | N-δ2 | 2.5 | ||||||||
| Glu | A122 | O-ε1 | 2.0 | ||||||||
| 5gp | A501 | O-1P | 2.2 | ||||||||
| Polar cyclic-di-GMP interactions (distance < 3.3 Å) | |||||||||||
| 5gp | A501A | O-1P | Asn | A88 | N-δ2 | 2.8 | |||||
| O-6 | Tyr | A231 | N | 3.2 | |||||||
| 5gp | A502A | N-1 | Asp | A65 | O-δ1 | 3.2 | |||||
| N-2 | Asn | A88 | O | 3.2 | |||||||
| N-7 | Gly | A36 | N | 2.8 | |||||||
| O-1P | Arg | A37 | N-H2 | 2.9 | |||||||
| O-2P | Ser | A46 | O-γ | 3.3 | |||||||
| Apolar c-di-GMP interactions (distance < 3.7 Å) | |||||||||||
| 5gp | A501 | C-2 | Tyr | A231 | C-ε2 | 3.4 | |||||
| C-3′ | Leu | A35 | C-δ2 | 3.8 | |||||||
| C-4 | Tyr | A231 | C-ζ | 3.4 | |||||||
| C-4 | Tyr | A231 | C-ε2 | 3.7 | |||||||
| C-5 | Tyr | A231 | C-ε1 | 3.7 | |||||||
| C-5 | Tyr | A231 | C-ε2 | 3.5 | |||||||
| C-5 | Tyr | A231 | C-ζ | 3.5 | |||||||
| C-6 | Tyr | A231 | C-ε2 | 3.5 | |||||||
| C-6 | Tyr | A231 | C-δ2 | 3.4 | |||||||
| C-6 | Tyr | A231 | C-γ | 3.8 | |||||||
| C-6 | Asp | A210 | C-α | 3.6 | |||||||
| C-8 | Tyr | A231 | C-ε1 | 3.6 | |||||||
| 5gp | A502 | C-4 | Phe | A51 | C-δ1 | 3.4 | |||||
| C-5 | Phe | A51 | C-δ1 | 3.5 | |||||||
| C-5 | Phe | A51 | C-ε1 | 3.9 | |||||||
| C-6 | Phe | A51 | C-ε1 | 3.7 | |||||||
The conformation of the 12-membered macrocycle of c-di-GMP, which covers Leu-35 of the EAL signature motif, is the same as found in PleD (4) or in small molecule crystals (43-44). The substrate interacts with Arg-37 through formation of a salt bridge with the second substrate phosphate moiety. Both guanine bases of the dinucleotide are in stacking interactions with aromatic residues (Phe-51 and Tyr-231) and are involved in base-specific H-bonds with main chain atoms (Table 2). All the aforementioned residues are highly conserved (see also Fig. 2A).
No detectable c-di-GMP-specific PDE activity was observed neither for full-length YkuI nor its EAL domain (EALYkuI) in vitro (data not shown). Possible reasons are discussed below. For other EAL proteins, the Km value for c-di-GMP hydrolysis has been measured to be in the micromolar to sub-micromolar range (7-9, 20), consistent with the low cellular c-di-GMP concentrations. The extended binding site involving hydrophobic, hydrophilic, and electrostatic interactions is consistent with the observed high substrate affinity. Furthermore, the entropic cost of substrate binding is low, because c-di-GMP is a molecule with low conformational flexibility. Previously, for the 12-membered macrocycle the same conformation has always been observed that probably represents a deep energetic minimum. This leaves only one torsion angle (angle χ around the C-1′-N-9 bond) per guanine base that can freely rotate.
Catalytic Mechanism—EAL domains catalyze the opening of the c-di-GMP macrocycle by hydrolysis of one of the O-3′-P ester bonds to yield the linear dinucleotide 5′-pGpG (7). Inspection of the active site shows that in line with the scissile ester bond there is a cavity that could well accommodate the water molecule needed for hydrolysis. Because of the modest resolution (2.8 Å), no water molecule could be assigned in the YkuI substrate complex structure. However, in the 1.8 Å resolution tdEAL/Mg2+ structure a well ordered water molecule that is coordinated to the magnesium ion is found at the right place (Wat1, see Fig. 5, bottom). As expected from the conservation of canonical EAL residues (Fig. 2A), the tdEAL/Mg2+ structure shows virtually the same active site geometry (except for Asp-152 (Asp-646), see below). This makes analysis of the relative position of the water molecule with respect to the bound c-di-GMP of the YkuI structure meaningful. For this, we translocated Wat1 and the Mg2+ ion into the YkuI active site (see Fig. 4). The Mg2+ position agrees very well (distance 0.3 Å) with the Ca2+ position in YkuI; and the 175° angle for the O-3′-P···Wat arrangement is close to the ideal geometry for an in-line attack of the water on the scissile bond. However, the 3.3 ÅP···Wat distance is rather long. Catalysis would proceed through a penta-coordinated transition state and eventual O-3′-P bond cleavage. In the tdEAL/Mg2+ structure, the water molecule is held in place by the aforementioned coordination with the metal and by H-bonds to the side chains of Glu-523, Asp-646, and Glu-703 (Fig. 4). These residues are strictly conserved within the family (see also Fig. 2A), with the former two additionally coordinating the metal ion. Note that Glu-523 and Glu-703 superimpose closely with their YkuI counterparts, Glu-33 and Glu-209, respectively. The side chain of YkuI Asp-152, however, appears to be pulled out of the active site by about 1.5 Å (with respect to tdEAL Asp-646) and thus can no longer coordinate the Ca2+ ion nor stabilize a water molecule at the tdEAL Wat1 position. The putative role of Asp-152 is discussed further below.
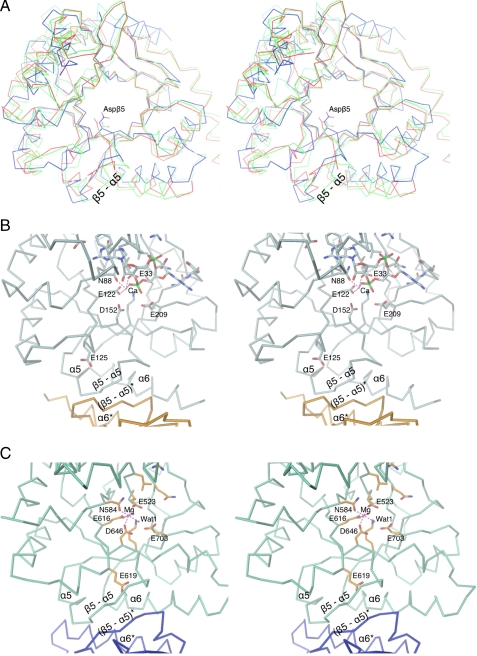
FIGURE 5.
Comparison of active site geometry and EAL-EAL dimer interface of YkuI and tdEAL (PDB code 2r6o). A, superposition of the two chains of YkuI (red and green) and the two chains of tdEAL (blue and light blue). The side chain of Asp-β5 (Asp-152 in YkuI and Asp-646 in tdEAL) is shown in full. The two Asp-β5 side chains of tdEAL superimpose closely, whereas the YkuI Asp-β5 side chain appears to be pulled out of the active site and shows variation in its position. B, view of a cut-out of the YkuI dimer. C, view of a cut-out of the tdEAL dimer. Note again the difference in the position of Asp-β5 (Asp-152 in YkuI and Asp-646 in tdEAL), which is correlated with a distinct β5-α5 loop conformation. In tdEAL this extended loop forms regular antiparallel β-strand interaction with its symmetry mate across the molecular 2-fold axis.
Generally, hydrolysis starts by water activation, i.e. deprotonation to generate a hydroxide ion ready for nucleophilic addition. Here, of the three carboxylates that are bonded to the water molecule, the carboxylate of Glu-209 is best suited to act as a general base. The other two side chains that also are H-bonded to the water molecule, i.e. Glu-33 and Glu-122, are simultaneously coordinating the divalent ion and should therefore have considerably lowered pK values. The role of Lys-173, another strictly conserved residue in the active site, is not clear. Its side chain amino group is equidistant (about 3 Å) to Glu-33, Glu-122, and Asn-88. Thus, its role may be fine-tuning the electrostatic potential within the active site.
The catalytic mechanism proposed here agrees with the one proposed recently by Rao et al. (9). In that study, the c-di-GMP macrocycle was docked to the tdEAL crystal structure to yield a model of the complex that agrees very well with the YkuI complex structure presented here. From that model, the catalytic mechanism was deduced and corroborated by mutagenesis and enzymatic characterization of the Rec-EAL protein RocR from P. aeruginosa (for an alignment with YkuI, see Fig. 2A). Reassuringly, mutagenesis of any of the residues that coordinate the metal or of the active site lysine rendered the enzyme inactive, but activity could partly be recovered at high magnesium concentration. Mutagenesis of the proposed general base Glu-352 in RocR (equivalent to the Glu-209 of YkuI), however, unrecoverably abolished enzyme activity, corroborating nicely the pivotal role of this residue.
A large number of monocyclic nucleotide-specific PDEs have been studied structurally (for a review, see Ref. 45). They contain a conserved catalytic core composed of three α-helical sub-domains and thus have no structural resemblance to the EAL domain.
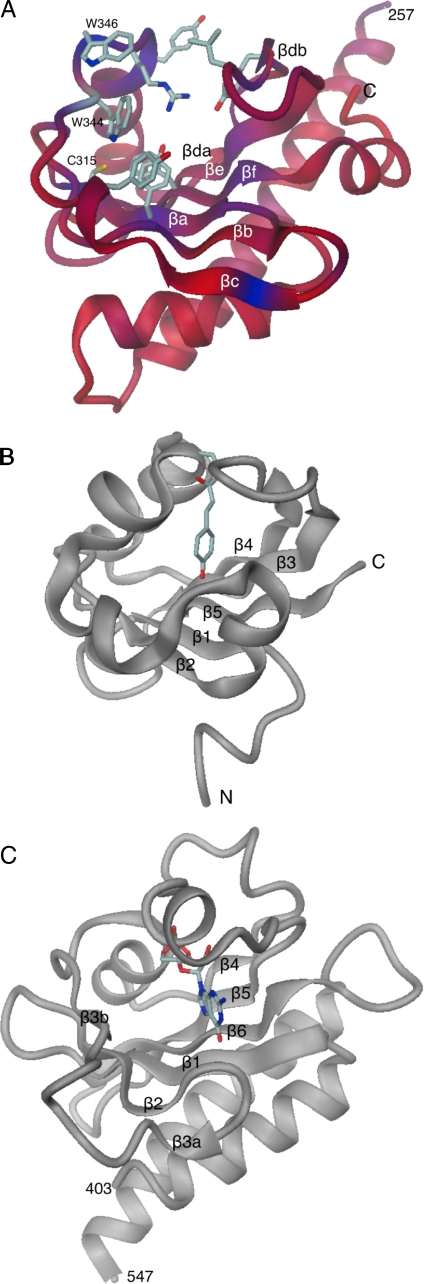
Structure of the PAS-like Domain—The C-terminal domain is folded into a 6-stranded antiparallel curved β-sheet (with strand order 3-2-1-6-5-4; Fig. 1A and Fig. 2B). Fig. 6 shows that the convex face of the sheet is packed against part of helix αlink and covered by helix αa. The concave face of the β-sheet together with loop βc-αb, helix αb, and loop βd-βe forms a pocket-like scaffold that appears not to be filled with side chains. The residue conservation score obtained from an alignment of 16 sequences, mostly from Bacillus sp. is mapped to the structure in Fig. 6. Clearly, the pocket lining and, in particular, the βc-αb loop with the sequence 343NWSWRPY349 are conserved. The opening of the pocket with its loop (top in Fig. 6) is intimately involved in contacts with the EAL* domain (helix α6 and α7-β8 loop located antipodal to the active site of the domain).
FIGURE 6.
Comparison of the folds of C-terminal PAS-like domain of YkuI (A), PYP (PDB 1otd (47)) (B), and second GAF domain of phosphodiesterase 2A (PDB 1mc0 (49)) (C). Chain/fragment termini and β-strands are labeled (see also Fig. 2B). For YkuI, the backbone is colored according to sequence conservation (red, variable, to blue, strictly conserved). Selected well conserved residues are shown in full. Note that the surface-exposed Trp-344 mediates, as other residues from the βc-αb loop, contact to the EAL* domain (not shown). For PYP and phosphodiesterase 2A, the chromophore and bound cGMP, respectively, are shown in full. Figs. 1B and 4, 5, 6 were produced with DINO (A. Philippsen, unpublished data.).
The closest structural homolog of the C-terminal domain is the N-terminal putative sensory box of a GGDEF protein from Vibrio parahaemolyticus (PDB code 2p7j6; 2.6 Å for 116 C-α positions). With 26% sequence identity for the structurally equivalent residues and the presence of a long N-terminal helix in the same position as αlink in YkuI, this domain is surely closely related in evolution. Fig. 6 shows two other structural homologs that have no significant sequence conservation. Photo yellow protein (PYP) (PDB code 1otd (47), see Fig. 6B) that is classified to include the PAS-fold ((48) Pfam PF00989) superimposes with an r.m.s.d. of 2.3 Å for 79 C-α of the 117 positions. The equivalent of the aforementioned pocket is harboring the chromophore that is covalently linked to Cys-69 of the α2-α3 loop (equivalent to the βc-αb loop of YkuI). The GAF B domain (Pfam PF01590) of phosphodiesterase 2a (PDB 1mc0 (49)), with cGMP bound to its pocket, superimposes equally well (r.m.s.d. of 2.3 Å/77).
Despite the structural homology to PAS-fold and GAF domains, the C-terminal domain is classified as a new motif in the Pfam data base (YkuI_C, PF10388). Fig. 6 shows that the topology of the β-strands is the same in all three domains (see also Fig. 2B), with the equivalent of the YkuI strand βc missing in the PAS-fold and split in two parts in GAF (β3a and β3b). Similarly, at the other edge of the sheet, strand βd of YkuI is split into βda and βdb. Noteworthy, loop βc-αb and helix αb of YkuI that form the “left” side of the pocket (Fig. 6A) have structural counterparts in the PAS-fold and GAF structures (Fig. 6, B and C) as has loop βd-βe on the “right” side.
PAS- and GAF-like domains frequently constitute sensors involved in signaling pathways (2). They can harbor chromophores or heme cofactors for light and oxygen sensing, respectively, or can accommodate various other small molecules, such a cGMP as in PDB code 1mc0. For YkuI, the natural ligand for the PAS-like domain is unknown, but by analogy a sensing function is likely. It remains to be investigated whether the YkuI_C domain expressed in the genuine Bacillus sp. background carries a chromophore (possibly attached to Cys-315) or, alternatively, shows affinity for specific small ligands. It is easy to envisage that, in latter case, ligand binding could induce βc-αb loop closure with concomitant modulation or disruption of the PAS-EAL* interaction. Thus, YkuI is probably a “one-component system” (50) whose putative PDE activity is regulated by the PAS-like sensor, similar to E. coli YcgF with its BLUF-EAL domain organization (51).
Regulation of PDE Activity—Can the lack of PDE activity of full-length YkuI and EALYkuI be reconciled with the structural data? A number of EAL proteins have been reported to be inactive (Ref. 9, see references therein), which could in most cases be traced back to a clearly corrupted active site lacking otherwise conserved residues. Such EAL proteins may represent a class of c-di-GMP-sensitive receptors that are involved in signaling, but not in degradation of the ligand. YkuI, however, appears to be a canonical EAL PDE with all residues that constitute the family present (Fig. 2A). Other well characterized members are YahA from E. coli, PdeA (CC3396) from C. crescentus, and RocR from P. aeruginosa (Fig. 2A) for which PDE-A activity has been demonstrated (7-9). Moreover, the structure analysis directly demonstrates substrate binding, i.e. one can infer that the Kd of binding is below 2 mm, the substrate concentration used for complex formation. It is also unlikely that the N-terminal His tag is detrimental for activity, because the N terminus is a large distance to the active site (Fig. 1).
A nonproductive arrangement of active site residues remains as a possible cause for the inactivity of YkuI. Indeed, Asp-152 of YkuI (homologous to Asp-295 of RocR and Asp-646 of tdEAL; called generally Asp-β5 in the following) appears to be in a nonproductive position compared with its counterpart in tdEAL/Mg2+ (see above; Figs. 4 and 5). Because mutagenesis of Asp-β5 in RocR renders the enzyme inactive (9), this may indeed be a severe disturbance in YkuI. At this stage, it cannot be ruled out that YkuI belongs to the class of catalytically noncompetent PDEs, but the other possibility that it can be activated appears attractive as outlined in the following.
What is the reason for the different position of Asp-β5 in the two crystal structures? Asp-β5 is adjacent to the β5-α5 loop that, in turn, is part of the dimer interface. Therefore, a coupling between interface conformation and Asp-β5 position is conceivable. Indeed, such coupling is evident when comparing the YkuI with the tdEAL/Mg2+ structure. Whereas tdEAL shows a regular antiparallel association of the β5-α5 loop with its symmetry mate, in YkuI the symmetry and several H-bonds are broken in this region (Fig. 5B). Note also that in tdEAL the loop is stabilized by Glu-619 (Fig. 5C), whereas the homologous residue in YkuI (Glu-125, Fig. 5B) is considerably shifted. Mutagenesis of the equivalent in RocR (E268A) rendered the enzyme inactive, and its putative effect on the β5-α5 loop conformation has been discussed (9).
If change in position of Asp-β5 is the decisive factor for the regulation of catalytic activity, how could this be coupled to the state of the PAS-like domain that putatively is the sensor for a yet unknown ligand? A direct inhibitory effect of the uncomplexed PAS-like domain appears unlikely, because it is rather remote from the β5-α5 loop. For the same reason, direct transmission of the information by the domain linking helix seems unlikely, although light-induced structural changes in the joining helix have been seen for a fragment of the BLUF-EAL protein YcgF by NMR (52). However, impact of the state of the PAS domain on the quaternary arrangement of YkuI (possibly transmitted by the linking helix) appears possible. A change in the quaternary structure, in turn, would affect the EAL-EAL interface and thus could change the β5-α5 loop conformation resulting finally in a shift of Asp-β5.
EALYkuI, which is monomeric under purification conditions as expected from the small (about 900 Å2) interface, was found to be inactive. Whether EAL-EAL dimerization per se could activate the domain may be tested by constructing a hybrid protein composed of a dimerization domain and EALYkuI. This would also test the feasibility of a general model in which EAL activation would proceed by signal-dependent homodimerization of a sensory domain that would synergistically promote EAL dimerization. Interestingly, light-induced dimerization of YcgF has been shown very recently by a spectroscopic technique (53).
Such a mechanism could apply to a number of other proteins, including YahA from E. coli where its GerE domain may dimerize only upon DNA binding; to RocR with its Rec domain that may dimerize upon phosphorylation, or to the large class of proteins with GGDEF-EAL domain composition. It has been shown for PdeA (CC3396) from C. crescentus (8) and FimX from P. aeruginosa (21) that GTP binding to the GGDEF domain activates the protein. It remains to be shown, but appears likely, that this would promote GGDEF dimer formation, the same way as substrate-loaded diguanylate cyclase domains have to approach each other to catalyze c-di-GMP formation (4).
Acknowledgments
Portions of this work were performed at the DuPont-Northwestern-Dow Collaborative Access Team located at Sector 5 of the Advanced Photon Source. DuPont-Northwestern-Dow Collaborative Access Team is supported by E.I. DuPont de Nemours & Co., The Dow Chemical Co., and the State of Illinois. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. We thank the staff of beamline X06SA of the Swiss Light Source (Villigen, Switzerland) for assistance with data acquisition; N. Amiot, Department of Chemistry, University of Basel, for the gift of chemically synthesized c-di-GMP; and I. Dubrovska for carrying out the dynamic light scattering experiments. We thank Z. Housley, C. Peneff, and P. Wassmann for critical reading of the manuscript and comments.
The atomic coordinates and structure factors (codes 2bas and 2w27) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant U54 GM074942 (Midwest Center for Structural Genomics). This work was also supported by Swiss National Science Foundation Grant 3100A0-10587.
Footnotes
The abbreviations used are: PDE, phosphodiesterase; NCS, noncrystallographic symmetry; EBI, European Bioinformatics Institute; r.m.s.d., rootmean-square deviation; Se-Met, selenomethionine; PDB, Protein Data Bank; PYP, photoactive yellow protein; TIM, triose-phosphate isomerase.
C. Chang, X. Xu, H. Zheng, A. Savchenko, A. M. Edwards, and A. Joachimiak, unpublished data.
R. Wu, A. James, and A. Joachimiak, unpublished data.
References
- 1.Ross, P., Weinhouse, H., Aloni, Y., Michaeli, D., Weinberger-Ohana, P., Mayer, R., Braun, S., de Vroom, E., van der Marel, G., Van Boom, J., and Benziman, M. (1987) Nature 325 279-381 [DOI] [PubMed] [Google Scholar]
- 2.Galperin, M. Y., Nikolskaya, A. N., and Koonin, E. V. (2001) FEMS Microbiol. Lett. 203 11-21 [DOI] [PubMed] [Google Scholar]
- 3.Tal, R., Wong, H. C., Calhoon, R., Gelfand, D., Fear, A. L., Volman, G., Mayer, R., Ross, P., Amikam, D., Weinhouse, H., Cohen, A., Sapir, S., Ohana, P., and Benziman, M. (1998) J. Bacteriol. 180 4416-4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, C., Paul, R., Samoray, D., Amiot, N. C., Giese, B., Jenal, U., and Schirmer, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17084-17089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul, R., Weiser, S., Amiot, N. C., Chan, C., Schirmer, T., Giese, B., and Jenal, U. (2004) Genes Dev. 18 715-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryjenkov, D. A., Tarutina, M., Moskvin, O. V., and Gomelsky, M. (2005) J. Bacteriol. 187 1792-1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt, A. J., Ryjenkov, D. A., and Gomelsky, M. (2005) J. Bacteriol. 187 4774-4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christen, M., Christen, B., Folcher, M., Schauerte, A., and Jenal, U. (2005) J. Biol. Chem. 280 30829-30837 [DOI] [PubMed] [Google Scholar]
- 9.Rao, F., Yang, Y., Qi, Y., and Liang, Z. X. (2008) J. Bacteriol. 190 3622-3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galperin, M. Y. (2006) J. Bacteriol. 188 4169-4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenal, U., and Malone, J. (2006) Annu. Rev. Genet. 40 385-407 [DOI] [PubMed] [Google Scholar]
- 12.Tamayo, R., Pratt, J. T., and Camilli, A. (2007) Annu. Rev. Microbiol. 61 131-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tischler, A. D., and Camilli, A. (2004) Mol. Microbiol. 53 857-869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamayo, R., Schild, S., Pratt, J. T., and Camilli, A. (2008) Infect. Immun. 76 1617-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wassmann, P., Chan, C., Paul, R., Beck, A., Heerklotz, H., Jenal, U., and Schirmer, T. (2007) Structure (Lond.) 15 915-927 [DOI] [PubMed] [Google Scholar]
- 16.Christen, B., Christen, M., Paul, R., Schmid, F., Folcher, M., Jenoe, P., Meuwly, M., and Jenal, U. (2006) J. Biol. Chem. 281 32015-32024 [DOI] [PubMed] [Google Scholar]
- 17.De, N., Pirruccello, M., Krasteva, P. V., Bae, N., Raghavan, R. V., and Sondermann, H. (2008) PLoS Biol. 6 e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simm, R., Morr, M., Kader, A., Nimtz, M., and Römling, U. (2004) Mol. Microbiol. 53 1123-1134 [DOI] [PubMed] [Google Scholar]
- 19.Bobrov, A. G., Kirillina, O., and Perry, R. D. (2005) FEMS Microbiol. Lett. 247 123-130 [DOI] [PubMed] [Google Scholar]
- 20.Tamayo, R., Tischler, A. D., and Camilli, A. (2005) J. Biol. Chem. 280 33324-33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazmierczak, B. I., Lebron, M. B., and Murray, T. S. (2006) Mol. Microbiol. 60 1026-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber, H., Pesavento, C., Possling, A., Tischendorf, G., and Hengge, R. (2006) Mol. Microbiol. 62 1014-1034 [DOI] [PubMed] [Google Scholar]
- 23.Korolev, S., Ikeguchi, Y., Skarina, T., Beasley, S., Arrowsmith, C., Edwards, A., Joachimiak, A., Pegg, A. E., and Savchenko, A. (2002) Nat. Struct. Biol. 9 27-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F. R., and Donnelly, M. I. (2002) Protein Expression Purif. 25 8-15 [DOI] [PubMed] [Google Scholar]
- 25.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795-800 [Google Scholar]
- 26.Terwilliger, T. C. (2003) Methods Enzymol. 374 22-37 [DOI] [PubMed] [Google Scholar]
- 27.Rossmann, M. G., and Blow, D. M. (1962) Acta Crystallogr. 15 24 [Google Scholar]
- 28.De La Fortelle, E., and Bricogne, G. (1997) Methods Enzymol. 276 472-493 [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 30.Roussel, A., and Cambillau, C. (1989) Silicon Graphics Partner Directory
- 31.Brunger, A. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 32.Brunger, A. T. (1992) Nature 355 472-475 [DOI] [PubMed] [Google Scholar]
- 33.Winn, M. D., Murshudov, G. N., and Papiz, M. Z. (2003) Methods Enzymol. 374 300-321 [DOI] [PubMed] [Google Scholar]
- 34.Leslie, A. (1992) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, No. 27
- 35.Baykov, A. A., Evtushenko, O. A., and Avaeva, S. M. (1988) Anal. Biochem. 171 266-270 [DOI] [PubMed] [Google Scholar]
- 36.Krissinel, E., and Henrick, K. (2007) J. Mol. Biol. 372 774-797 [DOI] [PubMed] [Google Scholar]
- 37.Greene, L. H., Lewis, T. E., Addou, S., Cuff, A., Dallman, T., Dibley, M., Redfern, O., Pearl, F., Nambudiry, R., Reid, A., Sillitoe, I., Yeats, C., Thornton, J. M., and Orengo, C. A. (2007) Nucleic Acids Res. 35 D291-D297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H. R., Ceric, G., Forslund, K., Eddy, S. R., Sonnhammer, E. L., and Bateman, A. (2008) Nucleic Acids Res. 36 D281-D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krissinel, E., and Henrick, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2256-2268 [DOI] [PubMed] [Google Scholar]
- 40.Nagano, N., Orengo, C. A., and Thornton, J. M. (2002) J. Mol. Biol. 321 741-765 [DOI] [PubMed] [Google Scholar]
- 41.Lebioda, L., Stec, B., and Brewer, J. M. (1989) J. Biol. Chem. 264 3685-3693 [DOI] [PubMed] [Google Scholar]
- 42.Rau, A., Hogg, T., Marquardt, R., and Hilgenfeld, R. (2001) J. Biol. Chem. 276 31994-31999 [DOI] [PubMed] [Google Scholar]
- 43.Egli, M., Gessner, R. V., Williams, L. D., Quigley, G. J., van der Marel, G. A., van Boom, J. H., Rich, A., and Frederick, C. A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3235-3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liaw, Y. C., Gao, Y. G., Robinson, H., Sheldrick, G. M., Sliedregt, L. A., van der Marel, G. A., van Boom, J. H., and Wang, A. H. (1990) FEBS Lett. 264 223-227 [DOI] [PubMed] [Google Scholar]
- 45.Jeon, Y. H., Heo, Y. S., Kim, C. M., Hyun, Y. L., Lee, T. G., Ro, S., and Cho, J. M. (2005) Cell. Mol. Life Sci. 62 1198-1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deleted in proof
- 47.Anderson, S., Crosson, S., and Moffat, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1008-1016 [DOI] [PubMed] [Google Scholar]
- 48.Hefti, M. H., Françoijs, K. J., de Vries, S. C., Dixon, R., and Vervoort, J. (2004) Eur. J. Biochem. 271 1198-1208 [DOI] [PubMed] [Google Scholar]
- 49.Martinez, S. E., Wu, A. Y., Glavas, N. A., Tang, X. B., Turley, S., Hol, W. G., and Beavo, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13260-13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulrich, L. E., Koonin, E. V., and Zhulin, I. B. (2005) Trends Microbiol. 13 52-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasegawa, K., Masuda, S., and Ono, T. A. (2006) Biochemistry 45 3785-3793 [DOI] [PubMed] [Google Scholar]
- 52.Schroeder, C., Werner, K., Otten, H., Krätzig, S., Schwalbe, H., and Essen, L. O. (2008) ChemBioChem 9 2463-2473 [DOI] [PubMed] [Google Scholar]
- 53.Nakasone, Y., Ono, T. A., Ishii, A., Masuda, S., and Terazima, M. (2007) J. Am. Chem. Soc. 129 7028-7035 [DOI] [PubMed] [Google Scholar]
- 54.Gouet, P., Courcelle, E., Stuart, D. I., and Métoz, F. (1999) Bioinformatics (Oxf.) 15 305-308 [DOI] [PubMed] [Google Scholar]
- 55.Amiot, N. C., Heintz, K., and Giese, B. (2006) Synthesis 24 4230-4236 [Google Scholar]