Abstract
Protein prenylation is a widespread process that involves the transfer of either a farnesyl or a geranylgeranyl moiety to one or more C-terminal cysteines of the target protein. Rab geranylgeranyl transferase (RabGGTase) is responsible for the largest number of individual protein prenylation events in the cell. A decade-long effort to crystallize the catalytic ternary complex of RabGGTase has remained fruitless, prompting us to use a computational approach to predict the structure of this 200-kDa assembly. On the basis of high resolution structures of two sub-complexes, we have generated a composite model where the rigid parts of the protein are represented by precomputed grid potentials, whereas the mobile parts are described in atomic details using Internal Coordinate Mechanics. Selection of the best docking solution of the flexible parts on the grid is followed by explicit atomistic refinement of the lowest energy conformations enabling realistic modeling of complex structures. Using this approach we demonstrate that the flexible C terminus of Rab7 substrate forms a series of progressively weaker and less specific interactions that channel it into the active site of RabGGTase. We have validated the computational model through biochemical experiments and demonstrated that to be prenylated RabGTPase must possess at least nine amino acids between the prenylation motif and the hydrophobic sequence anchoring the beginning of the Rab C terminus on the enzyme. This sequence, known as the C-terminal interacting motif is shown to play a dual role in Rab prenylation by contributing a significant fraction of binding energy to the catalytic complex assembly and by orienting the C terminus of RabGTPase in the vicinity of the active site of RabGGTase. This mechanism is unique to RabGGTase when compared with other prenyltransferases, which encode the specificity for their cognate substrates directly at their active site.
Elucidation of structures of biomacromolecules is essential for understanding their functions. Structural biology provides us with submolecular level views of enzymatic reactions, protein-ligand and protein-protein interactions. Although the main methods of protein structure determination such as x-ray crystallography, NMR, and electron microscopy are biased toward well ordered structures, in reality conformational changes and transitions between ordered and disordered states are common and important features of protein function and regulation. It has become very clear over the last several years that sections of proteins and sometimes entire proteins do not display a defined structure in solution and become structured only in the presence of a ligand or a binding partner protein (1). Furthermore, the binding of an isolated disordered peptide fragment to a target protein without the support of the rest of the complex is frequently undetectable. If crystallographic studies can be performed on only a part of a large system, it may be exceedingly difficult to reproduce this ordered interaction in a crystal structure, and there are limitations to the application of NMR methods.
Recent advances in realistic computer simulations give us an opportunity to address this problem. However, the large number of degrees of freedom of large systems presents a distinct challenge. Here ICM3 global optimizer was applied as an efficient macromolecular docking procedure to predicting the behavior of functionally locally disordered protein fragments upon macromolecular assembly (2, 3). This has been successfully applied to peptide folding, protein docking and interface refinement (4), small molecule docking (5), and virtual screening (6). To deal with a large complex, a part of the system that can be assumed to stay relatively unchanged is replaced by precomputed grid potentials as is done routinely in small molecule docking (e.g. Ref. 5). In this case, a smaller molecule or a peptide can be docked to the grid potentials to generate a set of conformers for further refinement. This technique was demonstrated to correctly predict the docking of a series of peptides to PTB and SH2 domains in an unbiased simulation (7). Addition of explicit atomistic refinement to the best scored conformations was shown to reproduce the unusual binding geometries of the HLA peptides (8) to the major histocompatibility class I receptors.
A well documented case of a protein complex with a functionally important locally disordered region is Rab geranylgeranyl transferase (RabGGTase). RabGGTase is a member of the protein prenyltransferase family that catalyzes covalent attachment of either farnesyl or geranylgeranyl moieties onto the conserved C-terminal cysteines of intracellular proteins (9). RabGGTase attaches geranylgeranyl moieties to the C terminus of more than 60 members of the Rab GTPase family: central regulators of intracellular vesicular transport (10). The C terminus of Rab GTPases is naturally disordered, a feature that is important for their biological function (11). Unlike other protein prenyltransferases RabGGTase does not recognize a four-amino acid C-terminal sequence, known as a CAAX box, but requires an adaptor protein termed the Rab escort protein (REP) for substrate binding and selection. REP recruits newly synthesized Rab GTPases and then presents them to the RabGGTase. The proteins form a tight catalytic ternary complex in which two geranylgeranyl groups are transferred onto the C terminus of Rab GTPase (11-13). Recently, RabGGTase came into the spot-light due to the observation that its chemical inhibition induced apoptosis in cancer cells, promoting the search for new inhibitors of this enzyme (14, 15). Development of RabGGTase-specific inhibitors requires understanding of the mechanistic and thermodynamic basis of its function. Although we recently solved the structures of an isoprenoid stabilized REP·RabGGTase complex and of prenylated and unprenylated Rab7·REP complexes, the structure of the ternary Rab·REP·RabGGTase complex remains unknown (16, 17) (Fig. 1A). It transpired from the analysis of the available subcomplex structures that the REP molecule plays a central role in assembling the catalytic ternary complex by forming binding interfaces with both Rab GTPase and RabGGTase (17, 18). In the case of the REP-Rab interaction, one large interface is formed between the Rab-binding platform of REP and the GTPase domain of Rab, and a smaller one between the hydrophobic C-terminal binding region (CBR) of REP and the hydrophobic CBR interacting motif (CIM) of Rab GTPases (17, 18) (Fig. 1). Formation of the complex with Rab GTPases increases the affinity of REP for RabGGTase by nearly three orders of magnitude and leads to the formation of the catalytic ternary complex via interaction of domain 2 of REP with the α-subunit of RabGGTase (13, 16). Despite these advances, details of the Rab prenylation mechanism remain unknown, and the following important questions remain open. How does RabGGTase process all Rab family members despite the high variability in the sequence and the length of their C terminus? What structural mechanism, and which amino acids, ensures proper positioning of the Rab C terminus into the catalytic center of RabGGTase? To address these issues, we have generated a computational model of the entire Rab·REP·RabGGTase ternary complex and performed an extensive search for low energy conformations of the C terminus, which was invisible in structures of the individual proteins. Based on the resulting models we designed a series of biochemical experiments to validate the computational model and demonstrate that the CIM functions as an anchor concentrating the prenylatable Rab C terminus toward the active site of RabGGTase. The model provides an explanation for the lack of requirement for conservation of the C terminus of Rab GTPases and elucidates structural determinants of the assembly of the functional ternary complex.
FIGURE 1.
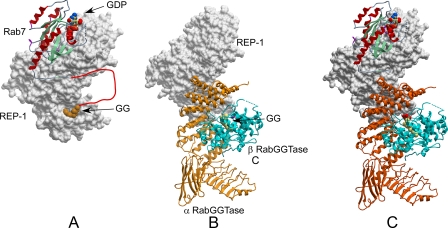
Structures used for model building. A, structure of the Rab7·REP-1 complex. REP-1 is displayed in surface representation and is shaded gray. Rab7 is displayed as ribbons and colored according to secondary structure. The disordered C terminus of Rab7 is drawn as a red line. B, REP-1·RabGTase complex. REP-1 is displayed as in A while RabGGTase is displayed in ribbon representation with the α subunit colored in orange and the β subunit in blue. C, model of the Rab7·REP-1·RabGGTase complex displayed as in A and B. The disordered C terminus of Rab7 is not displayed.
MATERIALS AND METHODS
Simulation—The stochastic simulation was performed with the Internal Coordinate Mechanics (ICM) program (Molsoft, La Jolla, CA). The stochastic optimization and sampling procedure was described previously (2, 3). The grid potentials included two van der Waals grids for hydrogen and non-hydrogen atoms, a hydrogen bonding grid that contained both donor and acceptor density, an electrostatic grid, and a hydrophobic grid, as described in Ref. 5. A surface-based implicit solvation model was used as described in Ref. 3, and the static part of the model was taken into account in the solvent accessibility calculations. The simulation was performed on a cluster of 200 Linux computers at the Scripps Research Institute.
Expression Constructs, Protein Expression, and Purification—Expression of rat REP-1 in SF21 cells and subsequent purification was performed as described previously (13, 19). Rab7 was expressed in Escherichia coli and purified as described before (13). The vectors for E. coli expression of Rab7 mutants were constructed by amplification of the open reading frame of Rab7 with the primers containing the respective mutations and subcloning the products into either previously described pGATEV vector (20) or pGATEV vector where the glutathione S-transferase gene was exchanged for maltose binding protein.4
Fluorescence Measurements—Fluorescence measurements were performed in 1-ml quartz cuvettes (Hellma) with continuous stirring and thermostatted at 25 °C on a Spex Fluoro-max-3 spectrofluorometer (Jobin Yvon Inc., Edison, NJ). Fluorescence titrations were carried out in titration buffer (50 mm Hepes, pH 7.2, 50 mm NaCl, and 5 mm dithioerythritol). Data analysis was performed with the program Grafit 5.0 (Erithacus Software) and Scientist (MicroMath Scientific Software) as described (21). To determine the affinities of Rab·REP binary complexes, REP-1 was titrated into 100 nm Rab7-NBD in the presence of 100-1000 nm Rab7 mutants or wild type. To determine the affinities of ternary complexes, RabGGTase was titrated into 100 nm Rab7-NBD·REP-1 complex in the presence of 500-1000 nm unlabeled Rab7·REP-1 complex. Rab7-NBD was excited at 479 nm, and the fluorescence was collected at 527 nm. The Kd value was obtained by fitting the data in a competitive model using program Scientist 2.0 as described elsewhere (22).
In Vitro Prenylation Assay—4 μm Rab wild type or mutants, 6 μm REP, 6 μm RabGGTase were incubated in prenylation buffer (50 mm HEPES, pH 7.2, 50 mm NaCl, 5 mm dithioerythritol, 2 mm MgCl2, 10 μm GDP) at 25 °C, and 50 μm NBD-FPP was added to initiate the reaction. At defined time intervals, 10-μl samples were withdrawn and quenched by addition of 10 μl of 2× SDS-PAGE sample buffer. For an end-point assay, 6 μm Rab7 wild type or mutants, 10 μm REP, and 6 μm were mixed with 40 μm NBD-FPP in prenylation buffer and incubated for 1.5 h at 25 °C, followed by quenching with 2× SDS-PAGE sample buffer. The samples were boiled at 95 °C for 3 min and were loaded onto 15% SDS-PAGE. The fluorescence bands corresponding to the NBD-farnesylated protein were visualized in the gel using a Fluorescent Image Reader FLA-5000 (Fuji, excitation laser: 473 nm, cut-off filter: 510 nm) followed by staining with Coomassie Blue and scanning. The fluorescence intensities of the bands were quantitatively analyzed using AIDA densitometry software. The traces were fitted to a single exponential equation using GraFit 5.0.
Isothermal Titration Calorimetry Measurements—Isothermal titration calorimetry measurements were carried out in isothermal titration calorimetry buffer (50 mm HEPES, pH 7.2, 50 mm NaCl, 0.2 mm β-mercaptoethanol) at 25 °C using a VP-isothermal titration calorimeter (MicroCal, Northampton, MA). A least-square fit of the titration data to a single-site binding model was performed using the program Origin, which includes routines designed to analyze isothermal titration calorimetry data (MicroCal), giving the binding constant (Ka), the standard enthalpy of binding (ΔH°), and the stoichiometry of binding (n).
RESULTS AND DISCUSSION
Simulation of the Rab·REP·RabGGTase Ternary Protein Complex—The structure of the ternary Rab·REP·RabGGTase complex has been sought for a long time by several groups due to its central role in the maintenance of membrane-bound organelle identity and the clinical relevance of Rab prenylation inhibitors (23). Although we previously reported crystallization of such a complex, it has remained refractory to structure solution due to its poor x-ray diffraction properties (24). This motivated us to seek an alternative approach for analysis of the structural details of RabGGTase-mediated protein prenylation reaction.
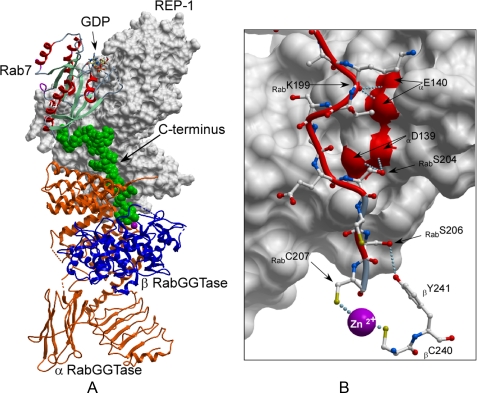
To this end we chose to perform a computational reconstruction of the ternary complex based on the available structures of the binary complexes (Fig. 1B). The model construction and stochastic simulation were performed with the ICM program (Molsoft). The ICM sampler operates in a space of arbitrary subset of internal coordinates of four types: bond (or virtual bond) length, bond angles, torsion angles, and phase angles that define an additional dihedral angle for the secondary branch. To create a full model of the catalytic ternary complex of RabGGTase, including the unprenylated substrate Rab7, we combined two high resolution crystal structures of RabGGTase·REP complex (PDB code 1LTX) and Rab7·REP complex (PDB code 1VG0). The two complexes were superimposed using overlapping atoms of the REP protein (Fig. 1). Fifteen C-terminal residues (DKNDRAKASAESCSC) that were invisible in the electron density map of the Rab7·REP complex were added to the substrate in an extended conformation. The resulting model of the biological ternary complex was then divided into two parts (ordered and disordered) and the ordered part was converted to a set of grid potentials (8). The C-terminal 22 residues of Rab7 were described in full atom representation, including hydrogens, and the first 4 residues were constrained to the crystallographic positions, whereas the last 18 residues were assumed to be fully flexible. For each C-terminal sequence, ten simulations from fully randomized structure were performed, and the solutions were clustered (25) to obtain a diverse set of the best energy representatives. Eight out of ten simulations converged in 70 h to virtually the same lowest energy conformation with energies within 1 kcal/mol. The energy difference between the preferred bound conformation and the next geometrically different solution was 4 kcal/mol. The simulation led to a lowest energy conformation as shown in Fig. 2. The simulated C terminus of Rab7 follows the surface of REP toward the bottom of domain II, where it bridges the solvent gap and contacts the β-subunit of RabGGTase at the loop connecting helices 11 and 12. The C-terminal cysteine protrudes into the active site, where the cysteine thiol coordinates the Zn2+ ion. Interestingly this interaction is not critical to drive the folding and docking procedure, because the omission of the cysteine residues did not significantly change the outcome of simulation (not shown). In the model obtained, the C terminus of Rab7 contacts the surface of the RabGGTase β subunit by forming a weak hydrogen bond between the side chain of Rab7S206 and side chain of βY241. Additional hydrogen bonds are formed between Rab7S204 and αD139 and Rab7K199 and αE140. It appears likely that, depending on the C-terminal sequence of individual Rabs, different hydrogen bonds can be formed between the Rab C terminus and the β subunit of RabGGTase.
FIGURE 2.
Model of the RabGGTase catalytic ternary complex. A, the lowest energy conformation of Rab7 C terminus docked onto the ternary complex. The model is displayed and colored as in Fig. 1, but the C terminus of Rab7 is displayed in space filling representation and colored in green. B, detailed view of interactions of the C terminus of Rab7 with the α and β subunits of RabGGTase. Rab 7 is displayed as a red worm, and the side chains are shown in ball-and-stick representation. The α subunit of RabGGTase is displayed as a gray molecular surface while the interacting residues of β subunit are displayed in ball-and-stick representation. The hydrogen bonds are displayed as in Fig. 1.
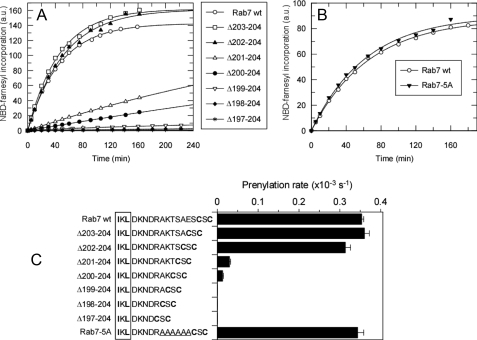
Effects of Rab C-terminal Sequence on the Prenylation and the Association of the Ternary Protein Complex—Several predictions can be made on the basis of the obtained model. First, the model predicts that to undergo prenylation the C terminus of Rab7 needs to be in an almost completely extended conformation. Shortening the C-terminal peptide by more than three amino acids would prevent prenylatable cysteines from reaching the active site of RabGGTase thereby reducing the prenylability of Rab, unless the RabGGTase can swing closer to the surface of REP. This conclusion is particularly interesting, because Rab proteins display no sequence conservation within last 40 C-terminal residues (26). To test this hypothesis experimentally, we generated a series of Rab7 mutants truncated by one to eight amino acids (Fig. 3). The mutants were designed in such a way that the prenylation (Cys-Ser-Cys) and CIM motifs remained unchanged in all cases while the number of spacing residues varied. To assess the kinetics and efficiency of the prenylation reaction we employed a recently developed Rab in vitro prenylation assay that makes use of NBD-FPP, a fluorescent analogue of GGPP (27). Fig. 3 (A and C) shows that truncation of the Rab7 C terminus by more than three residues leads to the dramatic reduction in Rab prenylation efficiency. Mutants truncated by four and five amino acids display residual prenylation activity, indicating that the ternary complex can probably undergo limited hinge movement. Further reduction in the sequence length completely removes the ability of Rab7 to act as the RabGGTase substrate. The inferred flexibility of the complex may explain why Rab7·REP-1·RabGGTase complex crystals diffract only to low resolution (24). The requirement for appropriate length of the linker peptide between CIM and the prenylation motif also finds its support in earlier in vivo data, where a truncation of 10 amino acid residues linking putative CIM (VDL) and prenylation motif (CC) of Rab5 rendered the protein cytosolic (28). The existence of the minimal length constraint in the Rab C terminus prompted us to analyze the hyper-variable C terminus of all mammalian Rab GTPases. In the majority of mammalian Rab GTPases putative CIM motif can be clearly identified, and supplemental Fig. S1 shows the list of aligned sequences. These data demonstrate that the minimal sequence between the CIM and the prenylatable cysteines in Rab proteins contains 9 amino acids, as featured by Rab9, Rab19, and Rab22. This is in agreement with the minimal artificial length of the Rab7 C terminus required for the efficient prenylation identified computationally and confirmed biochemically.
FIGURE 3.
Effect of Rab7 C terminus sequence composition and length on the prenylation efficiency. A, time course of prenylation of Rab7 wild-type and C-terminal truncation mutants with NBD-FPP. The solid lines show the fits to a single exponential function. B, as in A but comparing the prenylation rates of wt and Rab7 mutant with polyalanine C-terminal sequence. C, reaction rates of RabGGTase-mediated incorporation of NBD-farnesyl into the Rab7 mutants. The C-terminal sequences of the mutants are shown in the right-hand side. The CIM sequence is boxed while the prenylated cysteines are highlighted in bold. The polyalanine sequence is underlined.
Another prediction of our model is that the binding of the post CIM portion of the Rab C terminus to the RabGGTase should be rather weak due to the lack of extensive interactions with the RabGGTase. To test this we used a previously described fluorescent assay to measure the contribution of the C terminus to the affinities of the Rab·REP and the Rab·REP·RabGGTase complexes (29). As can be seen in Table 1, deletion of the prenylation motif has only a slight effect on the affinity of the ternary complex and no significant effect on the binary complex. Deletion of the last 14 residues does not affect the assembly of the Rab·REP complex but reduces the affinity of the ternary complex by a factor of 2, in accord with the computational model predicting the interaction of this part of C terminus with loop connecting helices 10 and 11 and the catalytic Zn2+ ion. It appears that interaction of the sequence down-stream of CIM with RabGGTase makes a small but measurable contribution to the affinity of the ternary protein complex. Such a weak association might be an advantage, because the overall reaction requires large rearrangements for the sequential double prenylation and translocation of the conjugated prenyl moieties to REP. Weak binding of the C terminus to the β-subunit may facilitate the dissociation of the prenylated product from the enzyme. This idea is indirectly supported by the observation that RabGGTase alone does not display detectable activity toward Rab GTPases (12). Moreover, a 10-mer and a 20-mer peptide mimicking Rab7 C terminus can neither interact with RabGGTase nor undergo prenylation by RabGGTase even when present in the reaction mixture at millimolar concentration.5
TABLE 1.
Summery of dissociation constants for interaction between Rab7wt/mutants and REP and RabGGTase
The sequence elements are marked as in Fig. 3.
| Rab wt/mutants | C-terminal sequence | Kd of binary complex, REP-1 | Kd of ternary complex, RabGGTase |
|---|---|---|---|
| nm | nm | ||
| Rab7wt | EFPEPIKLDKNDRAKTSAESCSC | 7.5 ± 2.7 | 130 ± 9.3 |
| Rab7Δ3 | EFPEPIKLDKNDRAKTSAES | 16.1 ± 1.0 | 191 ± 22 |
| Rab7Δ14 | EFPEPIKLD | 15.8 ± 2.1 | 321 ± 11 |
| Rab7Δ22 | E | 381 ± 37 | 491 ± 31 |
| Rab7-5A | EFPEPIKLDKNDRAAAAAACSC | 21.5 ± 1.1 | 188 ± 45 |
To further corroborate the model, we replaced the C-terminal sequence 199KTSAES204 between CIM and prenylation motif with a polyalanine stretch (Rab7-5A). The replacement does not impair the prenylation efficiency of the protein and does not significantly influence the affinity in both the binary and ternary complexes (Fig. 3, B and C). These observations suggest that the proper length rather than the sequence of the linker peptide between CIM and prenylation motif is critical for Rab prenylation, which explains the ability of RabGGTase to process Rab proteins containing C-terminal peptides of such diverse sequences and lengths. This finding demonstrates that the side chains of amino acid residues of the C-terminal linker peptide make only a limited contribution to the overall affinity of the ternary complex, which is consistent with the computational model.
Computational and Biophysical Analysis of Rab CIM Motif—In the x-ray structures of Rab7·REP-1 and the related YPT1·GDI complexes the C terminus of the GTPase can be traced only up to the CIM of Rab protein (17, 30, 31). We previously proposed that the interaction of the CBR with the CIM directs the C terminus of Rab proteins toward the active site of RabGGTase, because its mutation reduced prenylation efficiency (17). However, it remained unclear how the CIM performs this function and what role it plays in the assembly of the ternary complex and positioning of the C-terminal cysteines of Rab GTPases for catalysis. The developed computational model of RabGGTase ternary complex enabled us to test the role of CIM-CBR interactions in Rab prenylation. We repeated the stochastic sampling of the Rab7 C terminus when residues 190 and 192 were mutated to serines. Surprisingly, this dual mutation led to drastically different geometries of the lowest energy conformations. The RabGGTase-bound conformations of the C-terminal peptide were no longer at the previously identified positions. Instead, the peptide folded back on itself with its C terminus as far as 40 Å away from its active site location (not shown). Such a dramatic change highlights the importance of these two residues for the formation of the productive complex. This simulation suggests that the loss of C terminus coordination by CIM-CBR interactions would lead to reduction or inhibition of prenylation.
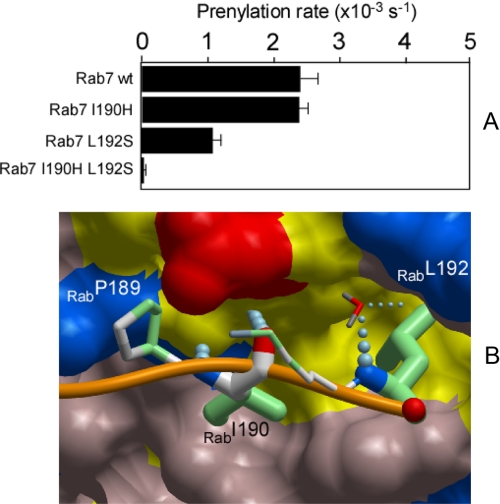
To test this experimentally we prepared mutants of Rab7 where either or both of the residues were mutated to polar amino acids. The resulting mutants were purified and subjected to in vitro prenylation analysis as described above. As can be seen in Fig. 4A, individual mutations led to a reduction in the prenylation reaction rates. The Rab7 L192S mutant displayed more significant impairment of prenylation than Rab7 I190H, suggesting that Leu-192 is a more critical residue in the CBR-CIM interaction. The mutations have an additive effect, because the double mutant Rab7 I190HL192S showed no detectable prenylation indicating that the CIM motif is strictly required for Rab prenylation (Fig. 4).
FIGURE 4.
Analysis of the CIM sequence contribution to the ability of Rab7 to undergo RabGGTase mediated prenylation. A, effect of the mutations in CIM sequence of Rab7 on the observed in vitro rates of RabGGTase mediated NBD-farnesyl incorporation. B, interaction of the C terminus of Rab7 with the C-terminal binding region of REP-1. The C terminus of Rab7 is displayed as an orange worm, and the residues involved in the interaction with REP-1 are displayed in ball-and-stick representation. The atoms involved in hydrophobic interactions are colored in green. The thickness of the side chains reflects the degree of interaction with REP-1. The order of contact area for these residues is: Leu-192>Ile-190>Pro-189>Lys-191. A water molecule is displayed as a dipole and colored according to atom type, and hydrogen bonds are shown as strings of small blue balls.
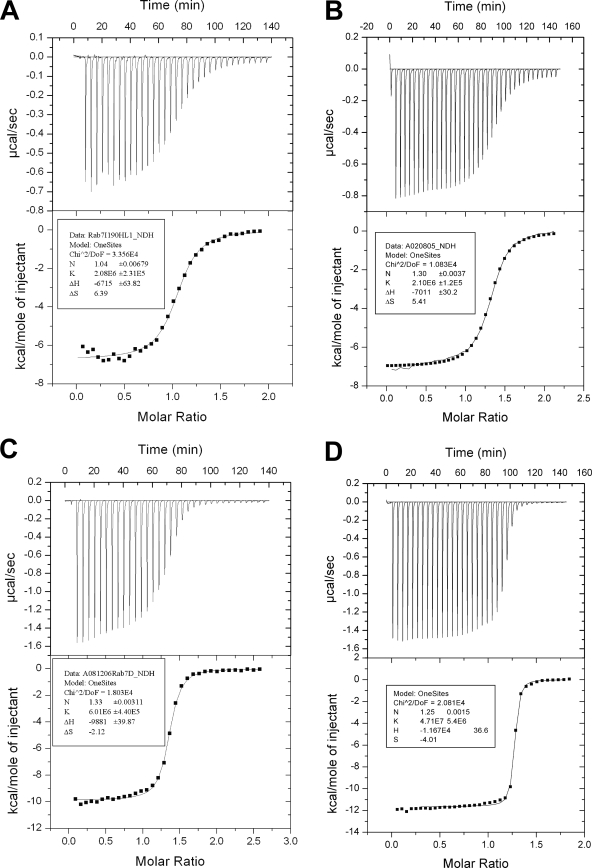
There are two most obvious explanations for the effect of CIM motif mutations on Rab prenylation. Because the CIM motif provides an additional contact between REP and Rab proteins, it may contribute to the overall affinity of the complex and thereby modulate the ability of RabGGTase to recruit and process the Rab protein. It is also possible that it is required for the coordination of the Rab C terminus, and the observed reduction in prenylation of CIM mutants reflects the failure of the C terminus to associate efficiently with the active site of RabGGTase. The latter model is more consistent with the results of our simulation. To discriminate between these possibilities, we quantified the contribution of the Rab7 CIM to the interaction energy of the Rab7·REP binary complex using isothermal titration calorimetry. The experiments give Kd values of 481 ± 53 nm, 476 ± 27 nm, 166 ± 12 nm, and 21 ± 2 nm for Rab7I190HL192S, Rab7L192S, Rab7Δ14, and Rab7 wild type, respectively (Fig. 5). This experiment clearly confirms that the CIM motif is an important quantitative contributor to the Rab·REP binary complex association, thereby ensuring the efficient assembly of the Rab·REP·RabGGTase ternary complex.
FIGURE 5.
Isothermal titration calorimetry titration of the Rab7I190H_L192S (A), Rab7L192S (B), Rab7Δ14 (C), and Rab7 wild type (D) with REP-1. The best fits for stoichiometry, binding constant, and enthalpy are shown in the inset box.
To test whether the affinity decrease alone is responsible for the failure of the mutants to undergo prenylation, we increased the concentration of the reaction components in in vitro prenylation reaction. This did not lead to an increase in product yield or the observed rate constant, suggesting that the observed rate reflects the single turnover reaction rate, and the failure of the double mutant to become prenylated is not a consequence of its lower affinity for REP (not shown). Therefore, we conclude that, in agreement with the simulation data, the CIM mutation changes the micro-environment within the ternary protein complex rather than the assembly of the whole complex. It appears that loss of coordination of the C terminus reduces the likelihood of its proper association with the catalytic site of RabGGTase, possibly resulting in its entrapment at other sites of the complex, where it is involved in non-productive stabilizing interactions.
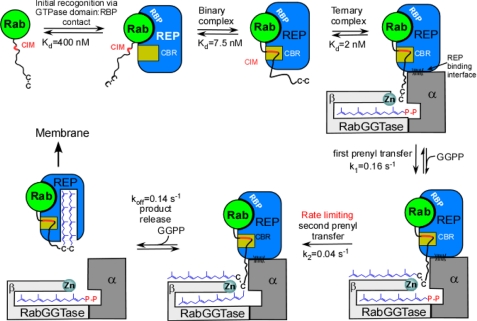
Functional Model of Rab Prenylation—Combination of the structural, simulation, and biochemical data help to generate a mechanistic model of RabGGTase function (Fig. 6). The assembly of the catalytic ternary Rab·REP·RabGGTase complex is triggered by the recognition of GTPase domain of Rab by the Rab-binding platform of REP. This results in a low to intermediate affinity complex, which is tightened by the interaction of the CIM with the CBR. The resulting binary complex forms a high affinity ternary complex with RabGGTase via the interactions between the α-subunit of RabGGTase and domain II of REP. The affinity of the complex is further increased by the weak and largely unspecific interactions of the C terminus with the active site of RabGGTase. This sequential assembly of the prenylation machinery implies progressive establishment of large and small binding interfaces and weak interactions. We have shown that the substrate specificity of RabGGTase is achieved via engagement of the REP molecule. REP, on one hand, selectively binds the GTPase core of the Rab proteins and, on the other hand, concentrates its C terminus in the vicinity of the active site of RabGGTase through the CIM·CBR anchor. Unlike the other structurally related protein prenyltransferases, RabGGTase does not engage its substrate in a tight interaction with the active site. Instead its activity is purely concentration driven, any cysteine-containing peptide concentrated in the active center undergoes modification (29). The cost of this feature of RabGGTase is a 5- to 100-fold reduction in reaction rate compared with that of FTase and GGTase-I (32-34). Despite this the arrangement allows RabGGTase to be active on >60 Rab GTPases with unrelated C-terminal sequences.
FIGURE 6.
Functional model of RabGGTase mediated Rab prenylation. CIM, CBR interacting motif; CBR, C terminus binding region; and RBP, Rab-binding platform.
CONCLUSIONS
We have built and tested a model of packing of the C-terminal sequence of Rab7 in the RabGGTase catalytic complex. A large transient complex of thee components was divided into a static precomputed part and an unstructured peptide and was subject to multiple simulations using stochastic optimization in internal coordinates with the ICM method. The simulation predicted the minimal functional length of the RabGTPase C terminus and a central role for the CIM sequence in funneling the prenylatable C-terminal cysteines into the active site of RabGGTase in the context of catalytic ternary complex. The model was confirmed experimentally and is in line with the notion that, in contrast to other prenyltransferases, RabGGTase has evolved an additional adaptor protein REP that enables it to accommodate GTPases with highly divergent C-terminal sequences.
The problem tackled in the presented study is of a general nature, because structural understanding of biochemical processes involving multiprotein assemblies becomes increasingly difficult as the number of components and protein-protein interactions goes up. In our case the biological complex was a transient assembly of three independently existing components: two protein monomers and an α/β heterodimer. However, there is little hope that the large number of weak transient complexes can be characterized by protein crystallography for a number of reasons. The formation of the transient complex leads to a specific rearrangement in which an unstructured element, typically an N and C terminus, a loop, or a short domain, undergoes a conformational change and acquires a specific productive conformation. Here we demonstrate that such biological events can be successfully approached with a combination of a structure prediction procedure, followed up by mutagenesis and detailed biophysical analysis. An extension of a peptide-docking technique, in which a system is divided into a large static part, represented by soft grid potentials, and a dynamic part, represented by explicit atomic models, leads to verifiable atomistic models of the transient complexes. The two-part approach makes it possible to consider large or multicomponent systems currently beyond the limits of de novo structure prediction. This approach can be applied to a variety of transient biological complexes containing disordered or alternatively packed ends, loops, or domains.
Acknowledgments
We gratefully acknowledge M. Terbeck, A. Sander, T. Rogowsky, S. Thuns, and N. Lupilova for excellent technical assistance. We are very grateful to T. Bergbrede and the Dortmund Protein Facility at the Max-Planck-Institute.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants AL 484/7-2 (to K. A.) and SFB 642 (to K. A., R. S. G., and H. W.).
Footnotes
The abbreviations used are: ICM, Internal Coordinate Mechanics; RabGGTase, Rab geranylgeranyl transferase; REP, Rab escort protein; CBR, C-terminal binding region; CIM, CBR interacting motif; NBD, nitrobenzoxadiazole; NBD-FPP, (3,7,11-trimethyl-12-(7-nitro-benzo[1,2,5]oxadiazo-4-ylamino)-dodeca-2,6,10-triem-1) pyrophosphate.
K. Alexandrov, unpublished data.
Y.-W. Wu and K. Alexandrov, unpublished.
References
- 1.Dyson, H. J., and Wright, P. E. (2005) Nat. Rev. Mol. Cell. Biol. 6 197-208 [DOI] [PubMed] [Google Scholar]
- 2.Abagyan, R., and Totrov, M. (1994) J. Mol. Biol. 235 983-1002 [DOI] [PubMed] [Google Scholar]
- 3.Abagyan, R. A., Totrov, M. M., and Kuznetsov, D. N. (1994) J. Comput. Chem. 15 488-506 [Google Scholar]
- 4.Totrov, M., and Abagyan, R. (1994) Nat. Struct. Biol. 1 259-263 [DOI] [PubMed] [Google Scholar]
- 5.Totrov, M., and Abagyan, R. (1997) Proteins Suppl. 1 215-220 [DOI] [PubMed]
- 6.Szewczuk, L. M., Saldanha, S. A., Ganguly, S., Bowers, E. M., Javoroncov, M., Karanam, B., Culhane, J. C., Holbert, M. A., Klein, D. C., Abagyan, R., and Cole, P. A. (2007) J. Med. Chem. 50 5330-5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou, Y., and Abagyan, R. (1998) Folding Des. 3 513-522 [DOI] [PubMed] [Google Scholar]
- 8.Bordner, A. J., and Abagyan, R. (2006) Proteins 63 512-526 [DOI] [PubMed] [Google Scholar]
- 9.Maurer-Stroh, S., Washietl, S., and Eisenhaber, F. (2003) Genome Biol. 4 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz, S. L., Cao, C., Pylypenko, O., Rak, A., and Wandinger-Ness, A. (2007) J. Cell Sci. 120 3905-3910 [DOI] [PubMed] [Google Scholar]
- 11.Goody, R. S., Rak, A., and Alexandrov, K. (2005) Cell Mol. Life Sci. 62 1657-1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andres, D. A., Seabra, M. C., Brown, M. S., Armstrong, S. A., Smeland, T. E., Cremers, F. P., and Goldstein, J. L. (1993) Cell 73 1091-1099 [DOI] [PubMed] [Google Scholar]
- 13.Alexandrov, K., Simon, I., Yurchenko, V., Iakovenko, A., Rostkova, E., Scheidig, A. J., and Goody, R. S. (1999) Eur. J. Biochem. 265 160-170 [DOI] [PubMed] [Google Scholar]
- 14.Lackner, M. R., Kindt, R. M., Carroll, P. M., Brown, K., Cancilla, M. R., Chen, C., de Silva, H., Franke, Y., Guan, B., Heuer, T., Hung, T., Keegan, K., Lee, J. M., Manne, V., O'Brien, C., Parry, D., Perez-Villar, J. J., Reddy, R. K., Xiao, H., Zhan, H., Cockett, M., Plowman, G., Fitzgerald, K., Costa, M., and Ross-Macdonald, P. (2005) Cancer Cell 7 325-336 [DOI] [PubMed] [Google Scholar]
- 15.Roelofs, A. J., Hulley, P. A., Meijer, A., Ebetino, F. H., Russell, R. G., and Shipman, C. M. (2006) Int. J. Cancer 119 1254-1261 [DOI] [PubMed] [Google Scholar]
- 16.Pylypenko, O., Rak, A., Reents, R., Niculae, A., Sidorovitch, V., Cioaca, M. D., Bessolitsyna, E., Thoma, N. H., Waldmann, H., Schlichting, I., Goody, R. S., and Alexandrov, K. (2003) Mol. Cell 11 483-494 [DOI] [PubMed] [Google Scholar]
- 17.Rak, A., Pylypenko, O., Niculae, A., Pyatkov, K., Goody, R. S., and Alexandrov, K. (2004) Cell 117 749-760 [DOI] [PubMed] [Google Scholar]
- 18.Alory, C., and Balch, W. E. (2003) Mol. Biol. Cell 14 3857-3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong, S. A., Brown, M. S., Goldstein, J. L., and Seabra, M. C. (1995) Methods Enzymol. 257 30-41 [DOI] [PubMed] [Google Scholar]
- 20.Kalinin, A., Thoma, N. H., Iakovenko, A., Heinemann, I., Rostkova, E., Constantinescu, A. T., and Alexandrov, K. (2001) Protein Expr. Purif. 22 84-91 [DOI] [PubMed] [Google Scholar]
- 21.Alexandrov, K., Scheidig, A. J., and Goody, R. S. (2001) Methods Enzymol. 329 14-31 [DOI] [PubMed] [Google Scholar]
- 22.Wu, Y. W., Tan, K. T., Waldmann, H., Goody, R. S., and Alexandrov, K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12294-12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson, M. A., Coulton, L., Ebetino, F. H., Vanderkerken, K., and Croucher, P. I. (2008) Biochem. Biophys. Res. Commun. 377 453-457 [DOI] [PubMed] [Google Scholar]
- 24.Rak, A., Niculae, A., Kalinin, A., Thoma, N. H., Sidorovitch, V., Goody, R. S., and Alexandrov, K. (2002) Protein Expr. Purif. 25 23-30 [DOI] [PubMed] [Google Scholar]
- 25.Abagyan, R., and Argos, P. (1992) J. Mol. Biol. 225 519-532 [DOI] [PubMed] [Google Scholar]
- 26.Colicelli, J. (2004) Sci. STKE 2004, RE13. [DOI] [PMC free article] [PubMed]
- 27.Wu, Y. W., Waldmann, H., Reents, R., Ebetino, F. H., Goody, R. S., and Alexandrov, K. (2006) Chembiochem 7 1859-1861 [DOI] [PubMed] [Google Scholar]
- 28.Chavrier, P., Gorvel, J. P., Stelzer, E., Simons, K., Gruenberg, J., and Zerial, M. (1991) Nature 353 769-772 [DOI] [PubMed] [Google Scholar]
- 29.Guo, Z., Wu, Y. W., Das, D., Delon, C., Cramer, J., Yu, S., Thuns, S., Lupilova, N., Waldmann, H., Brunsveld, L., Goody, R. S., Alexandrov, K., and Blankenfeldt, W. (2008) EMBO J. 27 2444-2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rak, A., Pylypenko, O., Durek, T., Watzke, A., Kushnir, S., Brunsveld, L., Waldmann, H., Goody, R. S., and Alexandrov, K. (2003) Science 302 646-650 [DOI] [PubMed] [Google Scholar]
- 31.Neu, M., Brachvogel, V., Oschkinat, H., Zerial, M., and Metcalf, P. (1997) Proteins 27 204-209 [DOI] [PubMed] [Google Scholar]
- 32.Thoma, N. H., Niculae, A., Goody, R. S., and Alexandrov, K. (2001) J. Biol. Chem. 276 48631-48636 [DOI] [PubMed] [Google Scholar]
- 33.Yokoyama, K., Zimmerman, K., Scholten, J., and Gelb, M. H. (1997) J. Biol. Chem. 272 3944-3952 [DOI] [PubMed] [Google Scholar]
- 34.Seabra, M. C., Reiss, Y., Casey, P. J., Brown, M. S., and Goldstein, J. L. (1991) Cell 65 429-434 [DOI] [PubMed] [Google Scholar]