Abstract
A new web portal for the CHARMM macromolecular modeling package, CHARMMing (CHARMM interface and graphics, http://www.charmming.org), is presented. This tool provides a user friendly interface for the preparation, submission, monitoring, and visualization of molecular simulations (i.e., energy minimization, solvation, and dynamics). The infrastructure used to implement the web application is described. Two additional programs have been developed and integrated with CHARMMing: GENRTF, which is employed to define structural features not supported by the standard CHARMM force field, and a job broker, which is used to provide a portable method for using grid and cluster computing with CHARMMing. The use of the program is described with three proteins: 1YJP, 1O1O and 1UFY. Source code is provided allowing CHARMMing to be downloaded, installed, and used by supercomputing centers and research groups that have a CHARMM license. Although no software can replace a scientist’s own judgment and experience, CHARMMing eases the introduction of newcomers to the molecular modeling discipline by providing a graphical method for running simulations.
Keywords: CHARMM, CHARMMing, Portal, Interface, Software, Visualization
1 Introduction
A common lament amongst novice and more experienced computational biologists is the lack of easy to use tools for preparing structures and setting up simulations. Although a wide variety of graphical and command line driven molecular modeling software exists,1–6 in most cases successful use requires years of training. The end result is that newcomers to the field, be they students or scientists from other disciplines, face a significant learning curve. While detailed knowledge of the software is ultimately necessary, a more user friendly interface would help less experienced users become independent. To provide such an interface, a new web-based tool named CHARMMing (CHARMM interface and graphics) is introduced. This new tool provides a friendly interface for the CHARMM (Chemistry at HARvard Macromolecular Mechanics)6 software package and is designed to facilitate the preparation, execution, and visualization of molecular simulations.
CHARMMing is not the first web-based tool developed to facilitate molecular modeling and simulation. For example, perhaps the first web-based tool designed to facilitate computational work is WebMO, a free interface to numerous computational chemistry packages.7 The online ProPKA8 web site (http://propka.ki.ku.dk/~drogers/) allows for quick calculation of protein pKa values. The ProBis web site (http://tyr.cmm.ki.si/probis/bin/probis.php) allows structural similarity searching in multiple related proteins to find conserved binding regions involved in protein-protein interactions.9, 10 The GLYCAM Biomolecule Builder11 (http://www.glycam.com/CCRC/biom-builder/biomb index.jsp) provides an interactive carbohydrate builder as well as tools to simplify the use of different classes of biomolecules. It facilitates building glycoprotein and glycolipid structure files for use with CHARMM or AMBER. Moreover it has the ability to generate Sander input files for running molecular dynamics simulations. CHARMM-GUI12, 13 (http://www.charmm-gui.org) provides a graphical interface for setting up protein and membrane simulations with CHARMM. Tools are supplied that assist with reading in structures, solvating them, setting up molecular dynamics input files, performing PBEQ calculations, and building protein-membrane complexes.
CHARMMing differs from the aforementioned interfaces in two respects: firstly, unlike ProPKA, it is focused on a wide variety of methods, and secondly unlike the GLYCAM or CHARMM-GUI tools it is designed to allow flexible combinations of different methods. CHARMMing contains a set of tools for uploading protein structures or retrieving them from the Protein Data Bank (PDB),14 performing energy minimizations and simulations, solvation, and viewing the results. Calculations can be performed in arbitrary order, thus giving the end user maximum flexibility; for example, results of one operation can be used as input to another. The interface also incorporates a number of freely available tools such as Jmol15 for visualization and an automatic residue topology file (RTF) generator (GENRTF),16 which creates topology and parameter information for residues not currently supported by the CHARMM force field.17 User supplied topology and parameter files are also supported.
Two additional important and unique features of the current work are; this package is open source software and the community as a whole is encouraged to contribute to its development, and the integration with popular queuing systems that allows CHARMMing to be installed as a front-end to computational resources in individual research groups and computational centers alike.
Given its flexibility, research groups will find this new tool useful for many tasks:
Job preparation. CHARMMing contains a number of tools that parse PDB format files and create CHARMM input scripts. The currently supported functionality includes energy minimization, solvation (implicit18 and explicit), molecular dynamics, Langevin dynamics, and Self-guided Langevin dynamics.19 Each of these functions allow extensive customization (i.e. adding patches, applying restraints, charge neutralization).
Job submission and monitoring. Currently the software supports job submission to Condor20 and PBS21 (Portable Batch Systems) based (including TORQUE22 and PBSPRO23) queuing systems. Additionally, jobs are actively monitored and the status is updated continuously. As with standard job submission there is no requirement that users maintain an active session; logging out of the website and returning later will not affect jobs that have been submitted.
Visualization of structures and trajectories. CHARMMing can illustrate structures that have been uploaded and can create movies from trajectory files generated from dynamics simulations. The primary visualization package used and distributed is Jmol,15 but similar packages with JavaScript application programming interfaces (APIs) can also be integrated. For example, an interface to ChemAxom’s MarvinSpace24 visualization software is also distributed.
Testing and deployment platform for new methods. A web-based interface allows new methods to be accessed without forcing users to learn complicated command line syntax. It is also straightforward for those with some software development experience to add functionality from other packages that have an existing interface with CHARMM.
Tutorial and instructional aid. A tutorial/introduction to the use of CHARMM has been integrated into the site. This tutorial also provides conceptual foundations to molecular simulation techniques such as energy minimization, solvation, molecular dynamics, and the use of periodic boundary conditions that are implemented in CHARMMing. Thus the web site may be used as an introduction to molecular simulations in general and CHARMM in particular. With a web interface, students can concentrate on learning the techniques as opposed to software commands. For example, the interface guides the student through a simulation and provides pop-up descriptions of the various areas of the site.
Section 2 describes the fundamental design goals and tools used to implement the current application. Section 3 shows examples of various proteins, highlighting CHARMMing’s unique features, and section 4 summarizes the existing functionality and describes planned enhancements to the software.
2 Software Design and Implementation
2.1 Application Framework
The primary design considerations of CHARMMing are flexibility, transparency, and adaptability. It is intended to be used with an up to date web browser.1 The CHARMMing software is built on top of the freely available Django25 web application framework. The application framework handles various low level details of web programming so that developers can concentrate on implementing functionality. Django was chosen for its technical capabilities and because it is built around the popular Python programming language.
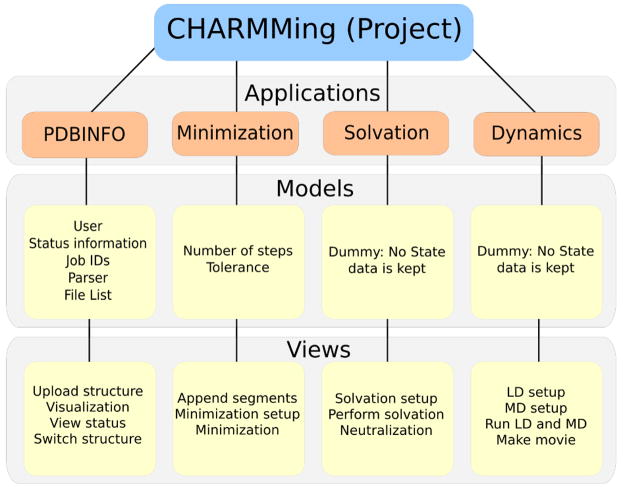
CHARMMing is divided into applications, each of which encapsulate some important part of the program (Figure 1). Each application has its own data model which stores information about the particular task being performed and a set of functions which process commands and are collectively called the application’s views. Data generated by one CHARMMing application can be used in another. For example, a protein can be solvated and the resulting structure minimized. The end result of this architecture is a highly flexible and easy to modify code.
Figure 1.
The structure of CHARMMing. The program is divided into several applications, the most important of which are shown here.
To give an example of the internal structure of the program, consider the pdbinfo application. The data model contains the location of data files such as the PDB, CRD (CHARMM card formatted file), and PSF (protein structure file) files, information about how many segments or chains are present, and what operations have been performed. The views contain functions that handle requests to visualize or download files for the structure, parse a newly uploaded PDB into its segments, and switch to a different PDB, among others.
One of the benefits of using Django is that it supports multiple independent accounts. Therefore users can run simulations without interfering with the work of others. It is necessary to register for an account, and Django provides customizable registration forms; currently, basic contact information and institutional affliation must be given. Administrators have special accounts that are given access to the Django administration interface. This allows them to modify or delete data stored in the system as well as approve new users.
CHARMMing, sitting on top of Django, must be run on a web server. Although, Django contains its own built in web server, that is suitable for development and testing, integration with the popular Apache26 web server is recommended. Since Django is designed for developing web applications, it contains a number of features that assist in software debugging. The information given, however, could allow malicious misuse of the web server and therefore these features should be disabled on production sites. In this case, errors encountered will appear to the user as non-specific internal web server errors. One important point with respect to deployment is that the user who initially installs the server automatically receives administrative privileges.
2.2 Installation Procedure
A Python script has been developed to facilitate the installation of CHARMMing on a web server running a Unix-like operating system. For Linux distributions that have native Django packages (currently Debian, Fedora, Gentoo, and Ubuntu), the installer will attempt to use the native package manager; therefore, it is assumed that the installer will be run with administrative privileges. The installer will perform the following tasks in order: (1) install Django (this step is skipped if it is already installed), (2) copy the CHARMMing source distribution into a user-specified directory, (3) download and install third-party software not distributed with CHARMMing that does not require additional license agreements, (4) configure limits to the use of CHARMMing (e.g. the number of dynamics steps allowed), and (5) optionally configure Apache. In cases where the user’s intervention is necessary, the install script gives clear directions on what must be done and proceeds from the same point once the required action is taken. This is accomplished by writing a configuration file that is read when restarting installations.
2.3 GENRTF
GENRTF is used to generate the necessary CHARMM input scripts for non-standard or small molecules not supported by the CHARMM all-atom protein force field.17 GENRTF reads and analyzes arbitrary Cartesian coordinate files in either PDB, XYZ, MOL, or CHARMM’s CRD format. Using the information obtained, input scripts are generated from the standard CHARMM topology file. In the case where an unknown residue is encountered a new topology is created and appended to the RTF information. In CHARMMing’s implementation, GENRTF is only passed the structural features that are non-native to the CHARMM force field.
For example, using a PDB file, GENRTF creates an input script to generate the native CHARMM coordinate and PSF files. If an unsupported substrate is identified, GENRTF builds the bonding structure for this substrate and creates new topology and parameter information for all such residues. This information can subsequently be read into CHARMM and used for calculations that include the previously undefined parts of the structure. Currently, the code assigns distinct parameters only for undefined atomic elements. While generating the PSF from PDB files, it is convenient that the residue numbering is preserved. Therefore, the original PDB file is separated, one file containing each chain. The generated input script then combines these segments into the complete system. In addition, GENRTF can add the necessary control file and commands and set up ab initio QM/MM calculations using CHARMM’s interface to either the Q-Chem or GAMESS quantum chemistry programs.27–30 In this case the accuracy of the parameters provided by GENRTF are not a concern as the undefined portion of the structure will likely be treated quantum mechanically. In other cases generic parameters are provided; a list of these can be found in the supporting information (Table 1). Although these parameters are not intended for use in meaningful simulations, they do provide a bridge for two important steps; they serve as a good starting point for parameter refinement and allow CHARMMing to create workable CHARMM compatible files for future use.
In CHARMMing, all of the unknown heteroatoms are separated into their own segment, and a PDB for that segment is created and run through GENRTF. The input file produced is not used directly, instead the CHARMM formatted topology and parameter information is parsed and used to create new RTF and parameter files. Incorporating the newly generated files provides functioning RTF and parameter files in a facile manner that can be subsequently improved.
2.4 Job Broker
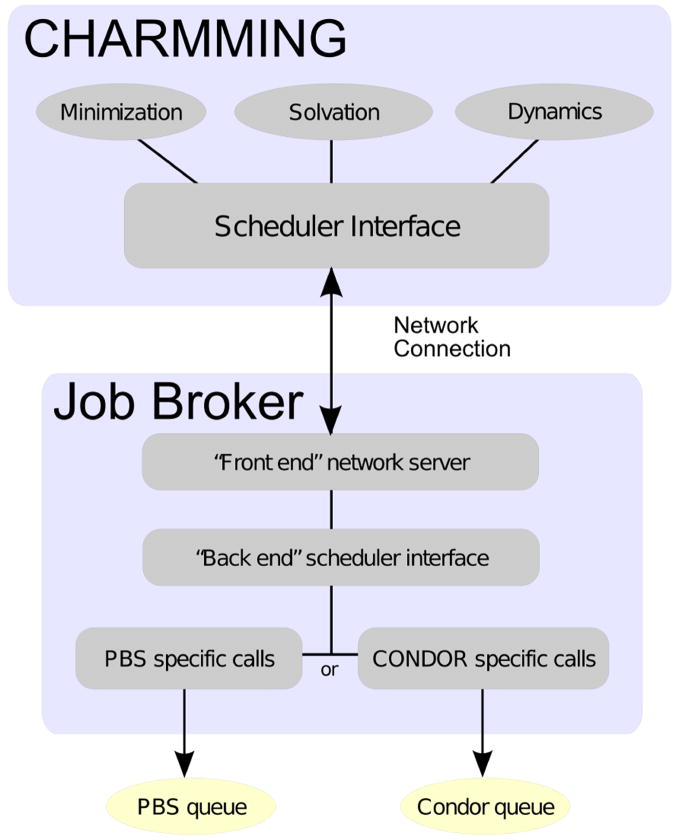
In addition to the aforementioned code, CHARMMing includes a standalone program called the “job broker”. The purpose of this component is to provide a mechanism to submit jobs to a local or remote computing cluster or grid. There are two parts to the job broker: a front-end which is called upon by the applications, and a back-end which interfaces with the grid or cluster queuing system. An illustration of this scheme is presented in Figure 2.
Figure 2.
The structure of the job broker. The part of the code that is specific to the queuing system is abstracted out, allowing support for different back ends to be easily added.
Applications interact with the job broker via a common network interface. The advantage of this design is that the job broker need not run on the web server itself, which will be useful at high trafic sites. Currently, submitting new jobs and checking the status of running jobs is supported; however, job termination is a planned feature. If a series of CHARMM scripts needs to be run sequentially, they can be submitted as a single batch and the job broker will guarantee that they are run in order. This is possible because of the directed acyclic graph functionality of CONDOR and job dependencies in PBS. If one of the preliminary jobs fails, the entire operation will be marked as having failed. This simplifies the submission of interdependent tasks.
The job broker back end is responsible for passing jobs from the front-end to a queuing system. Currently PBS and Condor are supported; however, the back end is decoupled from the job broker, allowing for easy integration with additional queuing systems. The benefit of integrating with PBS and Condor is their widespread use on traditional supercomputers (i.e. supercomputing centers), workstation clusters, and computational grids.
3 Results and Discussion
3.1 Preparing a Structure
3.1.1 Submitting the Structure
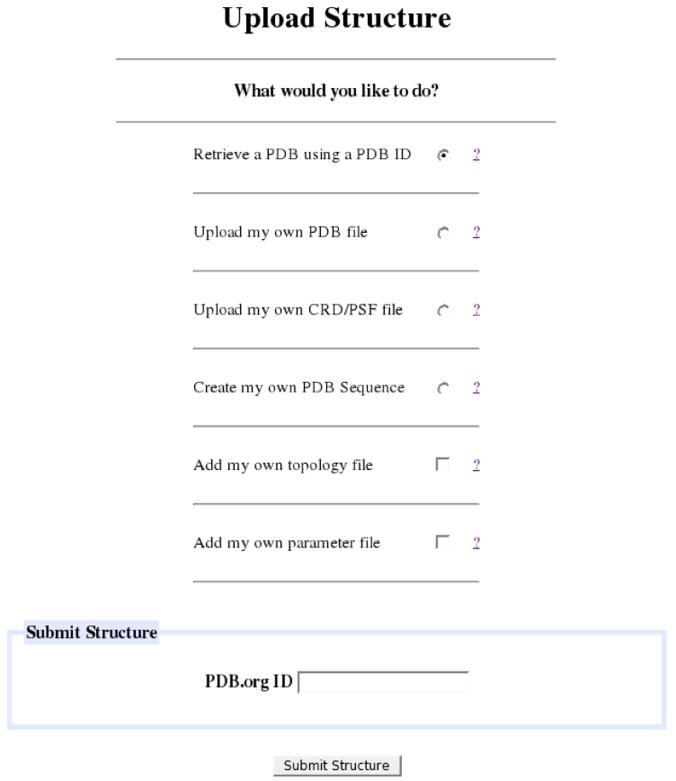
The first task in any project is to obtain a structure in usable format and resolve inconsistencies between PDB format and CHARMM card format. The CHARMMing coordinate parser provides access to a robust input tool. Structures are submitted to the web site using the Submit Structure form (Figure 3), which is located under the File Manager submenu. Users may upload their own PDB, CHARMM formatted protein structure (PSF) and coordinate (CRD) files, amino acid sequence (e.g. “ALA GLY PRO”), or provide a PDB ID and allow the file to be downloaded automatically from http://www.pdb.org.14 As an advanced feature, custom topology and parameter files can be uploaded to define parts of the structure that are non-standard.
Figure 3.
The structure submission page. There are four methods for uploading structures, by having CHARMMing: download them automatically by PDB code, upload a PDB file, upload CHARMM-formatted protein structure and coordinate files, and read a custom sequence.
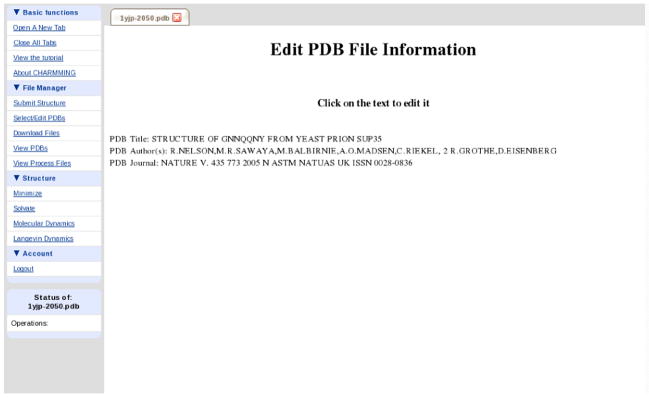
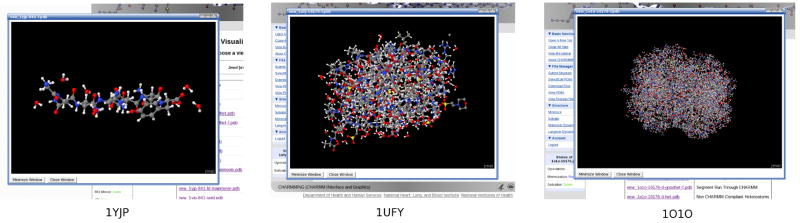
If the structure obtained is in PDB format, the header of the PDB file will be parsed and basic information such as the name of the structure, the author, and the journal in which it was published will be determined. The information gleaned is displayed with an opportunity to modify it. Figure 4 shows a structure from the yeast prion Sup35 (PDB code 1YJP).31 This is an excellent test case as it is small (7 residues) and all chains are CHARMM compliant. To illustrate the unique features of the software, we will also demonstrate calculations on human deoxy hemoglobin and chorismate mutase from Thermus Thermophilus (PDB codes 1O1O32 and 1UFY,33 respectively). These proteins are shown in CHARMMing’s Jmol visualizer in Figure 5.
Figure 4.
The screen that is displayed after a structure is submitted. It is possible to click on any of the lines to edit the information. For example, unique names may be given to custom generated structures.
Figure 5.
1YJP (left), a yeast prion, 1UFY (middle), a chorismate mutase, and 1O1O (right), a deoxy Hemoglobin as shown in CHARMMing’s built-in Jmol viewer.
3.1.2 Structure Parsing and Appending
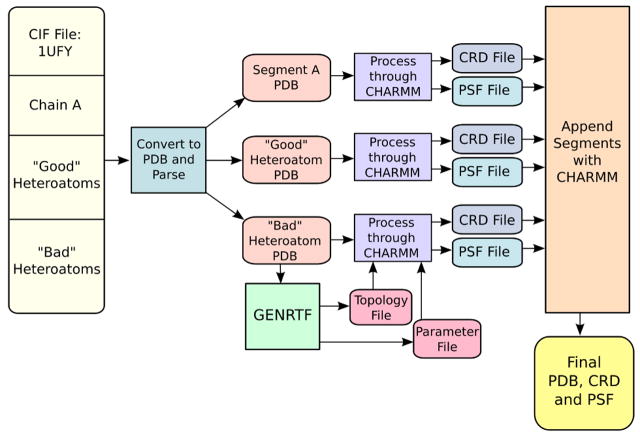
Once CHARMMing obtains a structure, a significant amount of work is performed automatically. If the file is to be retrieved from the PDB,14 the Crystallographic Information File (CIF)34 format file is fetched and the CIFtr program35 converts it to PDB format. The CIF format is used because it is more standardized than the PDB format, thus increasing the chances of successful parsing. The subsequent PDB is split up into its component segments, which consist of three types: protein segments, “good” heteroatoms, and “bad” heteroatoms. The “good” heteroatoms are defined as those for which the CHARMM protein all-atom force field has topology and parameter data. Examples of good heteroatom types include waters (TIP3P) and heavy metals such as zinc and iron. When unknown (“bad”) heteroatoms are present, CHARMMing invokes GENRTF to create topology and parameter files. This process is shown in Figure 6. While the parameters given by GENRTF are generic, they provide enough information to conduct basic operations. This is required for 1UFY and 1O1O as both contain substrates not present in the standard CHARMM topology files.
Figure 6.
The workflow for structure preparation. When a new PDB file is submitted, it is parsed, unknown residues are passed through GENRTF, PSF and CRD files are produced, and segments are appended together as needed.
During the initial processing, all residues with multiple protonation states (histidine, lysine, glutamic acid, and aspartic acid) are displayed and the user is given the opportunity to edit them individually. If changes are made to histidine residues, the PDB files are modified to reflect the new sequences. The CHARMM force field does not contain separate topologies for all states that are listed, therefore, a stream file is prepared containing patches that change the protonation states of the selected residues.
When the first task (e.g. minimization) is requested individual segments must be concatenated. A PSF and CRD file for each desired segment is prepared and the custom protonation stream files are processed on the appropriate segments. Following this process all hydrogen position are re-built using CHARMM’s HBUIld utility. This is an essential step as crystal structures typically do not have hydrogen positions defined and the positions of the hydrogens are dependent upon which segments are combined into the final structure. At this point, final PSF and CRD files for the appended structure are created.
3.1.3 Energy Minimization
Once a structure is prepared, the first step is usually to perform energy minimization to remove areas that may be subject to undesired stresses (i.e. bad van der Waals contacts). On the left hand menu, Minimize can be selected from the Structure/System submenu to bring up the minimization dialog (Figure 7). Here it is possible to select which chain(s) to minimize; in the case of 1YJP there are two options: the protein itself (segment a) and its CHARMM-compliant heteroatoms (a-goodhet). The latter are crystal waters. Similar segments exist for 1UFY and 1O1O, also representing embedded water.
Figure 7.
The minimization display.
CHARMMing supports two of the several minimization algorithms available in CHARMM: steepest descent (SD) and the adopted basis Newton-Raphson (ABNR) method, although support for other minimizers can easily be added. From the minimization dialog the number of steps for each type of minimization, the gradient tolerance, and additional parameters can be specified. Custom patches (e.g. disulfide bonds) and the SCPISM18 implicit solvent model may also be applied. To further control the minimization, various portions of the structure can be selected and restrained. An option is also provided to fix heavy atoms and thus only minimize the hydrogen positions. This is often necessary as hydrogens placed by HBUIld are often not in optimal positions.
3.1.4 Solvation
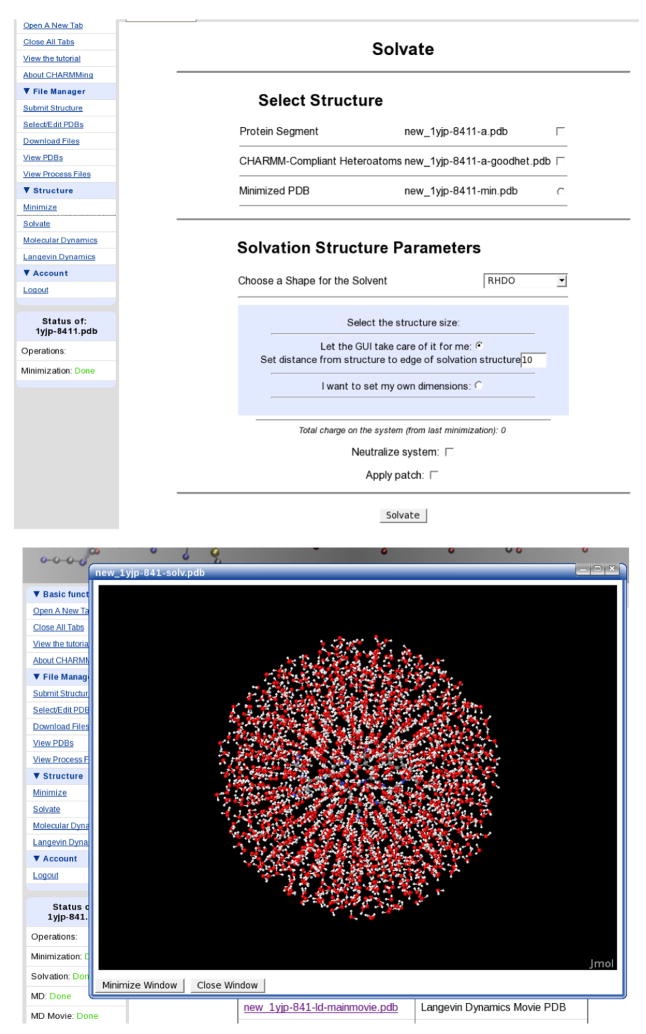
Although an implicit solvation model can be used, it is more typical that explicit waters are added to solvate the system before performing molecular dynamics. This can be a dificult process and much effort has been made to simplify this task. As with minimization, the Solvation page (Figure 8) is available under the Structure/System submenu and allows the selection of segments to be solvated. The shape of the water structure may be chosen from cubic, spherical, hexagonal, and rhombic dodecahedronal (RHDO). CHARMMing can also automatically select an appropriate solvation structure for the user. If this option is chosen a check is done on the dimensions of the protein. If the either the shortest or longest axes are more than 30% different from the median axis then the system is solvated in a hexagon, otherwise an RHDO is used. The software uses CHARMM’s COORdinate commands to determine an appropriate dimension for the box based on two possible criteria; how far the water is specified to extend beyond the structure or exact user specified dimensions. Once the form is submitted, an input script is generated that reads in water coordinates and creates the correct geometric shape, deleting waters that overlap with the solute. Note that this procedure can also introduce internal waters within cavities, but not in a measured manner. That is the number of water molecules is likely to be non-optimal.
Figure 8.
Top: The solvation page, which allows proteins to be solvated in various water structures. Bottom: A view of 1YJP solvated in a water ball.
An additional option given during the solvation procedure is the ability to automatically neutralize the system by adding counter ions. This is a useful procedure if Particle-Mesh Ewald is subsequently employed as charge neutrality is desired. Currently, six ions are supported; potassium, sodium, cesium, magnesium, calcium and chloride. By default, KCl is used and a concentration of approximately 0.15M is applied, however, these are user defined parameters and any combination of positive and negative ions can be used in addition to setting the concentration. For example, 0.15M KCl is indicative of cellular concentration whereas much lower KCL concentrations are found in the blood stream. Therefore, by making the concentration an adjustable parameter simulation conditions can easily be varied.
The neutralization procedure is carried out via the following steps; concentration and counter ions are defined, the number of ions required to reach that concentration is calculated using the difference of the protein volume and the total volume of the box, and the ions are added via a Monte-Carlo like procedure where random waters are replaced by ions and the energy (following brief minimization) is compared to the previous step with higher energy conformations being rejected. The number of Monte-Carlo steps is an adjustable parameter in the setup procedure. After solvation and neutralization, it may be necessary to re-minimize the structure to remove bad contacts (those with high energy). This is easily accomplished by revisiting the minimization dialog, selecting the newly solvated structure and applying periodic boundary conditions to a new minimization.
3.2 Running Dynamics
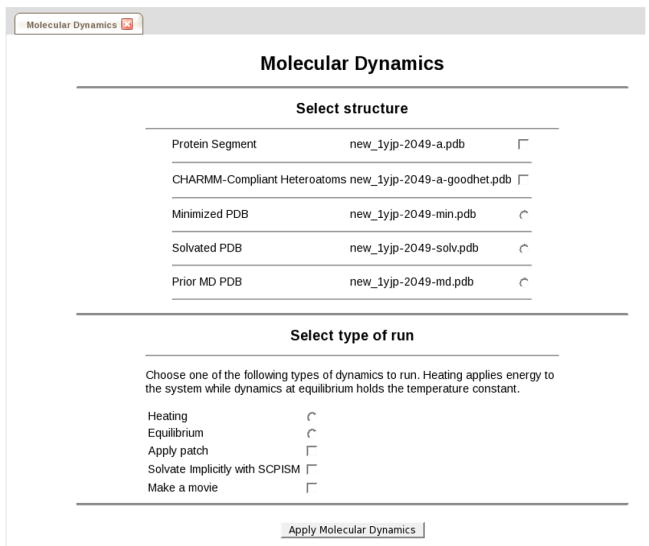
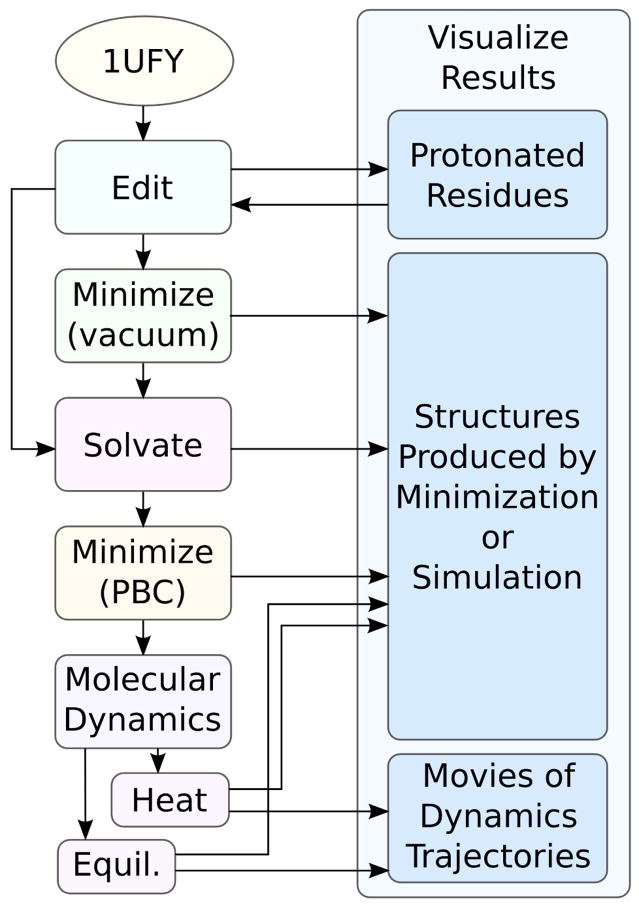
CHARMMing provides interfaces to both molecular dynamics and Langevin dynamics. Like minimization and solvation, both of the options are available from the Structure/System submenu (Figure 10). Periodic boundary conditions for molecular dynamics are employed via CHARMM’s CRYStal and IMAGe commands when available (i.e. for all non-vacuum systems except those solvated in a spherical water structure or that employ implicit solvation). This effectively replicates the simulated structure, forming a perfect lattice. This eliminates both vacuum/boundary effects and problems caused by atoms moving outside the unit cell.36 The suggested progression for running molecular dynamics simulations in explicit solvent is illustrated in Figure 9.
Figure 10.
The form used to set up molecular dynamics calculations.
Figure 9.
Suggested sequence of steps for running an explicitly solvated molecular dynamics simulations of choris-mate mutase from Thermus Thermophilus (PDB code: 1UFY).
Heating runs, in which energy is gradually added to the system (the frequency and scale of the temperature increases may be configured), or equilibration runs, in which temperature is kept constant at a user specified value, can be specified. Heating is useful for applying thermal energy to a minimized structure while equilibration is a useful precursor for production runs. CHARMM’s velocity rescaling functionality is employed to increase system energy in heating runs. Constant temperature and pressure (CPT) simulations with the Hoover thermostat37, 38 are used for equilibrium runs other than for spherically solvated, vacuum, and implicitly solvated systems. The Langevin piston method39 is used for all equilibrium runs that also employ periodic boundary conditions. The pressure piston mass (PMASs) is set to 2% of the molecular weight of the system while the mass of the thermal piston (TMASs) is set to 10 times that value. Constant pressure (PCONs) is also used for heating non-spherically solvated structures, but CPT and the Hoover thermostat are not used as these are inappropriate in this case. For structures that do not use periodic boundary conditions and all heating runs, molecular dynamics with the leapfrog Verlet integrator and no pressure or temperature constraints are used. Currently, molecular dynamics runs are limited to 10000 steps (with a 1 fs time step, a maximum of 10 ps). This value was chosen to prevent overloading the computational resources dedicated to this project; however, sites that want to use CHARMMing as a full scale front-end can easily increase the limit during the installation procedure.
For systems solvated in a cube, hexagon or RHDO, CHARMM’s Particle-Mesh Ewald (PME) capabilities are employed in molecular dynamics simulations to efficiently calculate electrostatic interactions. An external program is used to determines appropriate Fast Fourier Transform (FFT) dimensions for the Ewald summation and charge neutrality is tested. Finally, the SHAKe command is used to fix hydrogen bond lengths. Applying SHAKe constraints requires that the structure be well-minimized after all hydrogens (including ones in solvent) have been added. Failure to do so may lead to errors and/or failure of the simulation.
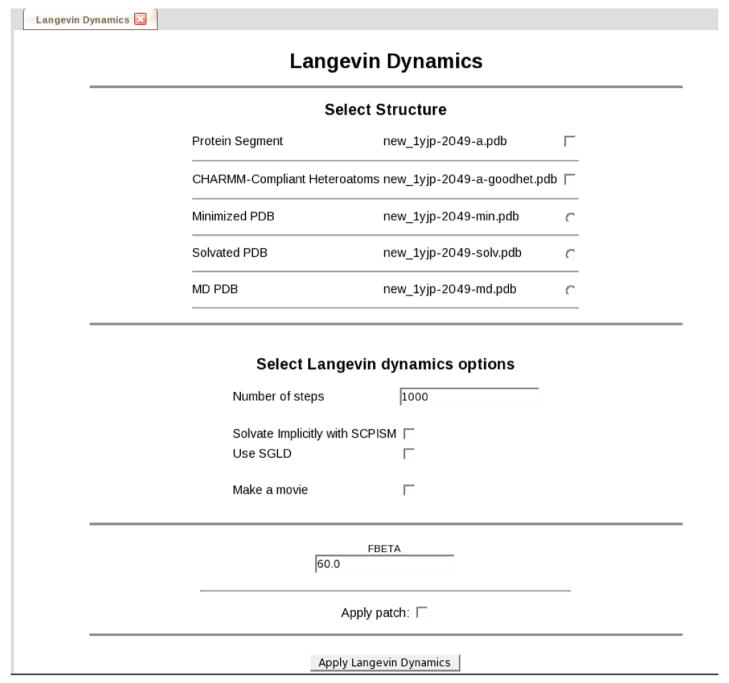
For Langevin dynamics, the leap-frog Verlet integrator is used with the LANGevin keyword.40 This adds random forces into the system as specified by the Langevin equation and simulates the presence of solvent drag on the system. The collision frequency is specified via the FBETa scalar value. Although periodic boundary conditions are not used, SHAKe is still applied as for molecular dynamics. Additionally, the self-guided Langevin dynamics method of Wu and Brooks is available via the SGLD keyword.19 This adds a guiding force to the Langevin equation based on the momentum of the system, causing more conformations to be sampled in a given time period. The form used to set up Langevin dynamics is shown if Figure 11.
Figure 11.
The form used to set up Langevin dynamics calculations.
Beyond the normal process of running jobs, there is an additional step that must be taken if a movie showing the motion of the system during dynamics is requested. A multi-frame PDB, is extracted from the dynamics trajectory using CHARMM’s trajectory I/O mechanism. Jmol is very particular about the format of PDBs used to create movies; therefore, a separate parser exists to combine the coordinates into a properly formatted file. Movies are currently limited to 10 frames, but can easily extended for longer simulations during the installation procedure.
3.3 Viewing and Analyzing Output
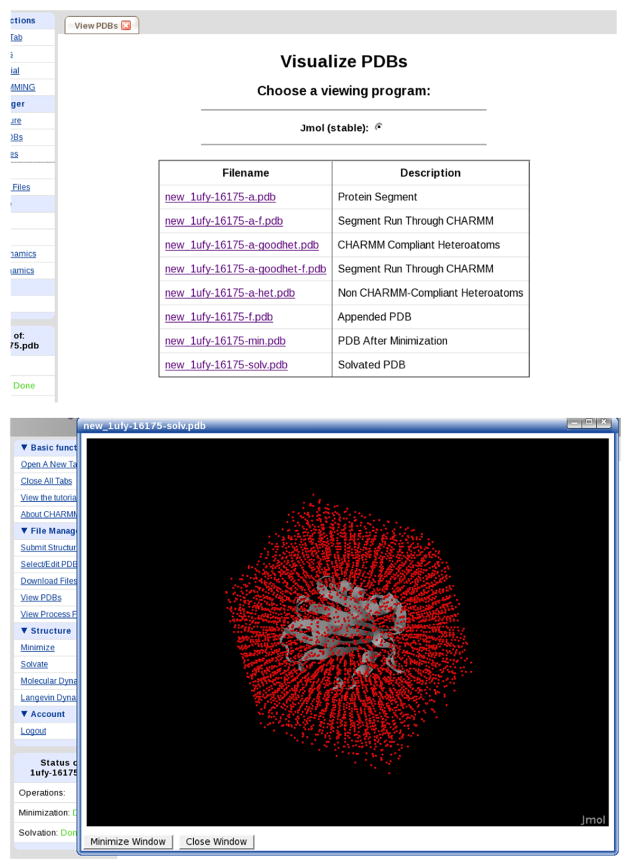
CHARMMing tracks submitted jobs in a database and provides a continuously updated status display. Once jobs have completed there are several methods available for accessing the results, all of which are available under the File Manager submenu. The input, output, structure, and coordinate files may be downloaded from the Download Files page. Files can be downloaded individually or concatenated as a tar archive. The input and output files can also be viewed directly in a web browser via the View Process Files page (Figure 12). Even if a job has failed, the input and output files are provided here to facilitate debugging.
Figure 12.
Top: CHARMMing’s visualization page. Jmol is available and an interface to ChemAxom has also been developed. Bottom: The result of visualizing 1UFY solvated in a rhombic dodecahedronal water structure. The red dots are the water oxygens.
Visualization options are available on the View PDBs page for runs that have succeeded. Multiple viewing programs, such as Jmol or MarvinSpace, can be selected if installed. The default viewer used in all of the figures is Jmol, which is distributed with CHARMMing. When a PDB is selected from the list, a new window containing the Jmol application appears. By right-clicking within this window Jmol’s standard functionality can be accessed. When a movie PDB is selected, Jmol is set to automatically play it in a loop, although this behavior can be changed.
4 Conclusion
CHARMMing, a new graphical portal to CHARMM, has been presented. It is anticipated that this new tool will assist both novice and expert computational biologists in efficiently and effectively setting up and carrying out complex simulations. No software, however, can replace a scientist’s in-depth knowledge of both the simulation methods and the tools used to carry them out. For this reason, results obtained from CHARMMing should not be accepted without detailed consideration. Fortunately, by providing easy access to raw input and output files, the software eases this task considerably. Additionally, a 50 page integrated tutorial is included as an introduction to CHARMM which further assists both novice and advanced users.
The software currently supports several different simulation techniques: energy minimization, solvation (implicit and explicit), molecular dynamics, Langevin dynamics, and self-guided Langevin dynamics. For each of these actions, several parameters related to the calculations can be customized. Based on this input, CHARMMing will produce and run CHARMM scripts. Back end computing resources such as Beowulf clusters, grids and supercomputers can be accessed via PBS or Condor based batch queuing systems. While other web interfaces exist, CHARMMing is well suited for setting up and managing protein simulations. CHARMMing’s source code is available for download from http://source.charmming.org and contributions from the community to expand its functionality or fix bugs will be gratefully accepted, as befitting the project’s open source nature. The web-based version of CHARMMing can be found at http://www.charmming.org. A default user name and password is listed on the website and user accounts can also be created.
There are already plans to increase CHARMMing’s functionality and improve the user experience. One key area is to add support for multi-model PDBs and allow the selection of model. Another area is to improve error checking and correction once the structure has been submitted. This is particularly important in the interface with GENRTF. Furthermore, improved auto typing and charge assignment is planned as this is an area of needed for throughput screening of potential drug targets. Although this project has already implemented a significant amount of functionality, it has only scratched the surface of CHARMM’s capabilities and, by its very nature, will never be truly finished.
Acknowledgments
We wish to acknowledge Richard M. Venable and Xiongwu Wu for their assistance with preparing the CHARMM input scripts. We also wish to thank the developers of Django (an open source Web application framework), Bubble tooltips (provides context-sensitive help), TableKit (For table sorting) and GreyBox (for various display functionality). This research was supported by the Intramural Research Program of the NIH, NHLBI.
Footnotes
Mozilla Firefox version 2 or later, Internet Explorer 7 or later, Safari
References
- 1.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 2.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theor Comp. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 4.Sayle RA, Milnerwhite EJ. RASMOL - Biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 5.Case DA, Cheatham TE, III, Darden T, Gohlke H, Lou R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods R. The Amber biomolecular simulation programs. J Comp Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaninathan S, Karplus M. CHARMM -A program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 7.Webmo, the free world wide web-based interface to computational chemistry packages. http://www.webmo.net, 2007.
- 8.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pk(a) values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 9.Konc J, Janezic D. Protein-protein binding-sites prediction by protein surface structure conservation. J Chem Inform Mod. 2007;47:940–944. doi: 10.1021/ci6005257. [DOI] [PubMed] [Google Scholar]
- 10.Carl N, Konc J, Janezic D. Protein surface conservation in binding sites. J Chem Inform Mod. 2008 doi: 10.1021/ci8000315. Submitted. [DOI] [PubMed] [Google Scholar]
- 11.Glycam biomolecule builder: The glycam biomolecule builder is a free, online builder for carbohydrates and related molecules (e.g., glycoproteins). http://www.glycam.com/CCRC/biombuilder/biombindex.jsp, 2008.
- 12.Jo S, Kim T, IM W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo S, Taehoon K, Iyer VG, Im W. Charmm-gui: A web-based graphical user interface for charmm. J Comp Chem. 2008 doi: 10.1002/jcc.20945. in press. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nuc Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinbeck C, Han YQ, Kuhn S, Horlacher O, Luttmann E, Willighagen E. The chemistry development kit (cdk): An open-source java library for chemo- and bioinformatics. J Chem Inform Comp Sci. 2003;43:493–500. doi: 10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodoscek M. Genrtf is a program to convert cartesian coordinates into charmm input files. 2008 http://a.cmm.ki.si/genrtf/
- 17.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem A. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 18.Hassan SA, Guarnieri F, Mehler EL. A general treatment of solvent effects based on screened Coulomb potentials. J Phys Chem B. 2000;104:6478–6489. [Google Scholar]
- 19.Wu XW, Brooks BR. Self-guided langevin dynamics simulation method. Chem Phys Lett. 2003;381:512–518. [Google Scholar]
- 20.Thain D, Tannenbaum T, Livny M. Distributed computing in practice: the condor experience. Concurrency: Pract Exper. 2005;17:323–356. [Google Scholar]
- 21.OpenPBS is the original version of the portable batch system, a queuing system developed for nasa in the early 1990s. OpenPBS: http://www.openpbs.org, 2008.
- 22.TORQUE is an open source resource manager providing control over batch jobs and distributed compute nodes. Cluster Resources, Inc.: http://www.clusterresources.com/pages/products/torque-resource-manager.php, 2001.
- 23.PBS professional is a service orientated architecture and field-proven grid infrastructure software. Altair Inc.: http://www.pbsgridworks.com, 2008.
- 24.Marvin was used for drawing, displaying and characterizing chemical structures, substructures and reactions, marvin 4.1.11, 2007, chemaxon (http://www.chemaxon.com) ChemAxom: http://www.chemaxon.com/, 2007.
- 25.Django is a framework that makes it easier to build better World Wide Web applications more quickly and with less code. Lawrence Journal-World: http://www.djangoproject.com, 2008.
- 26.The Apache HTTP server an open-source HTTP server for modern operating systems including UNIX and Windows NT. Apache Software Foundation: http://httpd.apache.org, 2008.
- 27.Shao Y, Fusti-Molnar L, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O’Neill DP, Distasio RA, Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Lin CY, Van Voorhis T, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EFC, Daschel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu CP, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill DC, Simmonett AC, Subotnik JE, Woodcock HL, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJ, Schaefer HF, Kong J, Krylov AI, Gill PMW, Head-Gordon M. Advances in methods and algorithms in a modern quantum chemistry program package. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, Dupuis M, Montgomery JA. General atomic and molecular electronic-structure system. J Comp Chem. 1993;14:1347–1363. [Google Scholar]
- 29.Woodcock HL, Hodoscek M, Gilbert AT, Gill PM, Schaefer HF, Brooks BR. Interfacing Q-Chem and CHARMM to perform QM/MM reaction path calculations. J Comp Chem. 2007;28:1485–1502. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]
- 30.Eurenius KP, Chatfield DC, Brooks BR, Hodoscek M. Enzyme mechanisms with hybrid quantum and molecular mechanical potentials .1. theoretical considerations. Int J Quant Chem. 1996;60:1189–1200. [Google Scholar]
- 31.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brucker EA. Genetically crosslinked hemoglobin: a structural study. Acta Cryst Sec D. 2000;56:812–816. doi: 10.1107/s0907444900006557. [DOI] [PubMed] [Google Scholar]
- 33.Crystal analysis of chorismate mutase from thermus thermophilus. 1UFY: http://www.pdb.org, 2003.
- 34.Hall SR, Allen FH, Brown ID. The crystallographic information file cif - a new standard archive file for crystallography. Acta Crystallographica Section A. 1991;47:655–685. [Google Scholar]
- 35.CIFTr is an application program for translating files in mmCIF format into files in PDB format. http://sw-tools.pdb.org/apps/CIFTr/index.html, 2007.
- 36.Cheatham TE, Brooks BR. Recent advances in molecular dynamics simulation towards the realistic representation of biomolecules in solution. Theor Chem Acc. 1998;99:279–288. [Google Scholar]
- 37.Nose S. A unified formulation of the constant temperature molecular-dynamics methods. J Chem Phys. 1984;81(1):511–519. [Google Scholar]
- 38.Hoover WG. Canonical dynamics - equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 39.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-pressure molecular-dynamics simulation - the langevin piston method. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 40.Allen MP, Tildesley DJ. Computer Simulations of Liquids. Oxford Science Publications; Oxford, N.Y: 1987. [Google Scholar]