Abstract
One approach to understanding common human diseases is to determine the genetic defects responsible for similar diseases in animal models and place those defective genes in their corresponding biochemical pathways. Our laboratory is working with an animal model for human rheumatoid arthritis called collagen-induced arthritis (CIA). We are particularly interested in determining the location of disease-predisposing loci. To that end, we performed experiments to localize susceptibility loci for CIA in an F2 cross between the highly susceptible mouse strain DBA/1j and the highly resistant mouse strain SWR/j. Specifically, a quantitative trait locus analysis was performed to localize regions of the mouse genome responsible for susceptibility/severity to CIA. One susceptibility locus, Cia1 in the major histocompatibility locus, had been identified previously. Two additional loci were detected in our analysis that contribute to CIA severity (Cia2, Cia3) on chromosomes 2 and 6. A third locus was detected that contributes to the age of onset of the disease. This locus (Cia4) was located on chromosome 2 and was linked to the same region as Cia2. Determining the identity of these loci may provide insights into the etiology of human rheumatoid arthritis.
Keywords: rheumatoid, genome scan, collagen-induced arthritis
One approach to solving the complex interactions of susceptibility genes for a particular human disease is to identify genes in an animal model for a similar disorder and then assess the relevance of those genes by looking at homologous genes or their pathways in humans. A number of rodent animal models exist for a variety of human diseases, such as diabetes, hypertension, systemic lupus erythrematosus, and rheumatoid arthritis. The genes predisposing to disease in the animal model may not be orthologous to the loci involved in the human pathologies. However, understanding the physiological mechanisms that lead to the disease in animals may provide insights into the etiology of their human counterparts. The NOD mouse has provided new insights for diabetes (1), the NZM2410 mouse has provided insights for systemic lupus erythrematosus (2, 3), and the stroke-prone spontaneously hypertensive rat (SHRSP) has provided insights for hypertension (4, 5). Our laboratory has been studying collagen-induced arthritis (CIA), a mouse animal model for human rheumatoid arthritis (6).
Autoimmune reactivity to type II collagen (CII) was first experimentally induced in the rat by Trentham et al. in 1977 (7). Intradermal injection of CII in complete Freund’s adjuvant (CFA) resulted in an inflammatory polyarthritis with characteristics similar to rheumatoid arthritis in man. A mouse model of CIA subsequently was established by Courtenay et al. (8). Most rodent studies immunize the animals with heterologous CII (e.g., bovine or chicken CII), which presents a pathology characterized as an arthritis that develops “explosively” 6–10 weeks after injection and never relapses after remission. Microscopic examination of the joints of diseased animals reveals many features that are similar to rheumatoid arthritis, including lymphocytic infiltration and synovial membrane hypertrophy.
The genetics of CIA has been under intense investigation for a number of years. Genetic crosses between susceptible and resistant inbred mouse strains have demonstrated that CIA is inherited in most inbred strains as a polygenic, dominant trait (9, 10). Several investigators have explored the role of the murine major histocompatibility complex (Mhc or H-2) in CIA development by immunization of congenic and inbred strains with heterologous and homologous CII (11–14). The results from these studies established that specific Mhc haplotypes are required for CIA susceptibility, namely H-2q and H-2r. Specifically, Wooley et al. (15) used recombinant inbred strains to map susceptibility to CIA within the I region of the Mhc. In this current study, the Mhc is given the CIA susceptibility locus name Cia1. Evidence localizing susceptibility to the β1 domain of the H2-Ab gene was described later (16).
Although susceptibility to CIA has been linked to H-2 inheritance, non-Mhc genes clearly influence the incidence and severity of CIA. This is demonstrated by the difference in the frequency of CIA in the H-2q strains DBA/1j (100%), B10.Q (84%), NFR/N (50%), B10.G (41%) and SWR/j (0%) (12, 14). Of interest, SWR/j mice are completely resistant to CIA induction with both heterologous and homologous CII, although they have the permissive H-2 haplotype. The underlying mechanisms responsible for their resistance have been under investigation for quite some time, with two principle loci being implicated in the resistance of SWR/j to CIA induction, complement C5 (Hc) and the T cell receptor beta locus (Tcrb) (17–21). The contribution of these loci to CIA susceptibility is still not clear and has been under debate (19, 20, 22). Further evidence of non-Mhc loci contributing to the severity of CIA was provided by Remmers et al. (23). They performed a genome scan to localize susceptibility loci for CIA in (DA × F344)F2 susceptible rats. In addition to the rat Mhc, five additional susceptibility loci were localized to five separate chromosomes (chromosomes 1, 4, 7, 8, and 10). Recently, Jirholt et al. (24) described two additional susceptibility loci for CIA in (B10.RII × RIIIS/j)F2 arthritic mice. These loci were located on chromosomes 3 and 13.
Given the disease heterogeneity and Mhc variability, we argue that mice of the H-2q and H-2r haplotypes represent different collections of polymorphism leading to CIA susceptibility. Our current study has focused on localizing the chromosomal positions of susceptibility loci for CIA for one of these groups by analyzing F2 progeny from the highly susceptible DBA/1j (H-2q) mouse strain and the completely resistant SWR/j (H-2q) mouse strain. We find that two loci, Cia2 and Cia3, on chromosomes 2 and 6, respectively, contribute to susceptibility and that a third locus (Cia4) on chromosome 2 contributes to the time of disease onset.
MATERIALS AND METHODS
Immunization and Monitoring.
All animals used in this study were bred at The Jackson Laboratory and were housed in the Department of Comparative Medicine’s SPF facility at the University of Washington. All procedures and assays were preapproved by the University of Washington’s Animal Care Committee. CIA was induced in control and experimental animals according to established protocols (8) with slight modifications. In brief, male control DBA/1j, SWR/j, and (DBA/1j × SWR/j)F1 and experimental male (DBA/1j × SWR/j)F2 progeny were immunized at 8 weeks of age at the base of the tail with 100 μg of chicken CII (Sigma) dissolved in 50 μl of 0.1 M acetic acid and mixed with an equal volume (50 μl) of CFA (CFA with 1 mg/ml Mycobacterium tuberculi; Sigma). At 4 weeks postimmunization, the animals were boosted i.p. with 100 μg of chicken CII in incomplete Freund’s adjuvant (Sigma). Negative control DBA/1j animals were immunized with 0.1 M acetic acid in CFA and were boosted with 0.1 M acetic acid in incomplete Freund’s adjuvant. All mice were kept for 20 weeks postimmunization to ensure that the animals were negative.
Animals were monitored on a weekly basis for signs of CIA. An arthritic index was assigned to each mouse by using the following criteria: 0, no signs of arthritis; 1, swelling and redness in a single joint; 2, inflammation in multiple joints; and 3, severe swelling, joint erosion, and/or ankylosis. Each paw was scored from 0–3, and the arthritic paws then were multiplied by their score with the index being the sum of all of the paws. For example, an animal with scores of 0:1:2:3 for each paw would have an arthritic index of 0 + 1(1) + 2(2) + 3(3) = 14. This scoring method was used to reflect the differences in the severity of the arthritis in affected paws. Without this method, an animal with three grade 1 paws (swelling in a single joint) would have the same severity index as an animal with one grade 3 paw (joint erosion and severe swelling).
The animals were killed after the 20-week observation period, and their paws were harvested for histologic examination. In brief, the paws from each animal were collected, fixed, and decalcified for a minimum of 72 hours in 3.0 ml of Cal-Ex II (Fisher Scientific) and were embedded in paraffin. Two sections (10 μm thick) along the longitudinal axis from each paw were mounted onto glass slides and were stained with hematoxylin and eosin. The slides were blindly scored independently by two pathologists using the following criteria: 0, normal synovial membrane and smooth cartilage surfaces; 1, synovial membrane hypertrophy and cellular infiltration; 2, grade 1 plus pannus formation with superficial cartilage erosions; 3, grade 2 plus major erosion of the cartilage and subchondral bone; and 4, loss of joint integrity through erosion, massive cellular infiltration, or the presence of ankylosis. The histology index for each animal was calculated in the same manner as the arthritic index described above.
DNA Isolation and Genotyping.
The genomic DNAs used for genotyping the mice were isolated from either a 1-cm tail clip or liver by using standard isolation protocols described elsewhere (25). Genomic DNA from each animal was genotyped for 182 commercially available microsatellite loci (Research Genetics, Huntsville, AL) by using PCR amplification procedures described previously (25). In brief, each reaction contained genomic DNA (50 ng), primers (0.2 μM each), 50 mM KCl, 10 mM Tris, 1–2.5 mM MgCl2, 0.2 mM dNTP, and 0.0–0.5 μM dATP-IRD40. The samples were cycled 35 times at 94°C for 15 s, 50–58°C for 15 s, and 72°C for 15 s. Markers were optimized for both their annealing temperature (50–58°C) and MgCl2 concentration (1–2.5 mM). The products from the first set of experimental animals (Table 1, Experiment 1) were resolved on denaturing polyacrylamide gels and were stained by using Silver Sequence staining reagents (Promega). Alternatively, the products from the second set of experimental animals was labeled with an infrared dye (dATP-IRD40), were resolved on denaturing polyacrylamide gels, and were detected by using a Li-Cor (Lincoln, NE) Model 4000S automated DNA sequencer (26, 27). The genotypes were scored independently by at least two people.
Table 1.
Incidence and severity of CIA in control and experimental animals
| Experiment | Strain | CII | CIA (%)* | Age of Onset, weeks | Mean age of onset† | Arthritic index‡ | Histology index§ |
|---|---|---|---|---|---|---|---|
| 1 | SWR/j | + | 0/6 (0) | − | − | 0 | 0 |
| 1 | DBA/1j | − | 0/10 (0) | − | − | 0 | 0 |
| 2 | DBA/1j | − | 0/10 (0) | − | − | 0 | 0 |
| 1 | DBA/1j | + | 10/10 (100) | 5 | 8.0 ± 0.8 | 20.4 ± 4.4 | 20.5 ± 4.6 |
| 2 | DBA/1j | + | 10/10 (100) | 5 | 7.0 ± 0.5 | 25.6 ± 3.7 | 24.5 ± 5.7 |
| 1 | (DBA/1j × SWR/j)F1 | + | 14/19 (73.7) | 6 | 9.5 ± 0.7 | 15.8 ± 2.1 | N.D. |
| 1 | (DBA/1j × SWR/j)F2 | + | 55/87 (63.2) | 5 | 8.1 ± 0.4 | 17.9 ± 1.4 | 12.8 ± 1.8 |
| 2 | (DBA/1j × SWR/j)F2 | + | 51/80 (63.8) | 5 | 8.0 ± 0.4 | 11.4 ± 1.4 | 13.2 ± 2.3 |
| Totals | |||||||
| DBA/1j | 20/20 (100) | 5 | 7.5 ± 0.5 | 23.0 ± 2.9 | 22.5 ± 3.7 | ||
| F2 Exp. | (DBA/1j × SWR/j)F2 | 106/167 (63.5) | 5 | 8.1 ± 0.3 | 14.8 ± 1.0 | 13.0 ± 1.5 |
N.D., Not done.
Number of arthritic animals/total number immunized.
Weeks after immunization ± S.E.M.
Arthritic and histology index ± S.E.M.
Linkage Analysis.
After genotyping the F2 progeny, linkage maps were constructed by using mapmaker/exp 3.0 (28). The linkage data and phenotype data were analyzed by using mapmaker/qtl 1.1b (29). The quantitative trait locus (QTL) analysis was conducted on the most severely arthritic (upper 15%) and least severely arthritic (lower 5%) F2 animals by using the histology index. The histology index was used because mistyping of nonarthritic animals would be minimized as macroscopic inspection will not reveal low level lymphocytic infiltration. Additional marker loci were typed in genomic intervals at which logarithm of odds (lod) scores exceeded 1.5, bringing the total number loci typed in these animals to 212. All intervals containing significant evidence of linkage were confirmed by two-point analysis by using bymarker (http://www.cityofhope.org/users/jlongmate/bymarker.htm.), a computer program that tests associations between marker data and phenotype data given either a normal or non-normal distribution (F statistic, ANOVA analysis).
RESULTS
Table 1 presents the results of two experiments showing the incidence and severity of CIA in DBA/1j control and (DBA/1j × SWR/j)F2 experimental animals. As expected, negative control DBA/1j mice immunized with 0.1 M acetic acid in CFA had no signs of arthritis (0/20) whereas positive control DBA/1j mice had an incidence of 100% (20/20) with a mean arthritic index of 23.0 ± 2.9 and age of onset of 7.5 ± 0.5 weeks. Consistent with a dominant mode of inheritance, the (DBA/1j × SWR/j)F1 progeny had an incidence of 73.7% (14/19), a mean arthritic index of 15.8 ± 2.1, and a mean age of onset of 9.5 ± 0.7. In contrast, the (DBA/1j × SWR/j)F2 animals had an incidence of 63.5% (105/168) with a significantly lower mean arthritic index (14.8 ± 1.0) than the positive control DBA/1j animals (23.0 ± 2.9, P < 0.0001) and similar age of onset (8.1 ± 0.3). The histology index was very similar to the arthritic index. DBA/1j positive control animals had a mean histology index of 22.5 ± 3.7, and the F2 experimental progeny had a mean histology index of 13.0 ± 1.5. The lower incidence and decreased severity in the F2 population would be consistent with a polygenic model for CIA susceptibility.
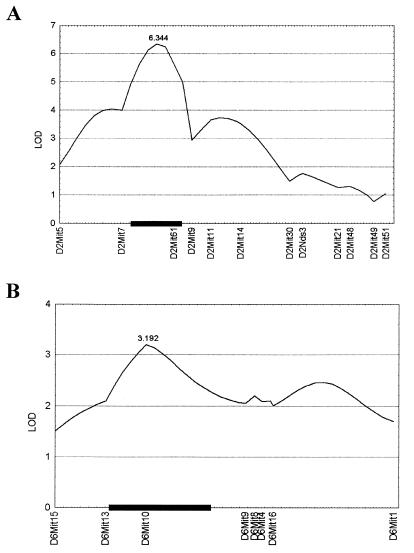
QTL analysis of the (DBA/1j × SWR/j)F2 progeny demonstrated the presence of two susceptibility loci in genomic intervals on chromosomes 2 and 6 (Fig. 1). The first locus (Cia2) on chromosome 2 had significant evidence of linkage to marker D2Mit61 (lod score of 6.34) whereas the second locus (Cia3) on chromosome 6 had suggestive evidence of linkage to marker D6Mit10 (lod score of 3.19). The support interval for Cia2 is ≈12 centimorgans whereas that of Cia3 is 23 centimorgans (Fig. 1). The fraction of the total variation in severity across the population of F2 animals (variance) explained by Cia2 and Cia3 was 29 and 9%, respectively. These values were calculated by mapmaker. To confirm the existence of these loci, a two-point analysis (F statistic, ANOVA) was accomplished by using bymarker. The data from this analysis yielded analogous results (Table 2). The loci identified in the analysis as significantly associated with CIA were D2Mit61 (F = 10.5, P < 0.0001) and D6Mit10 (F = 8.9, P < 0.00045).
Figure 1.
QTL plots for each chromosome containing non-Mhc linked loci using the histology index data. Log-likelihood values were determined by using mapmaker/qtl 1.1b. lod scores are presented on the y axis, and positions of the marker loci along the chromosome are given on the x axes. Support intervals are given by the black box. (A) Chromosome 2. Markers are presented centromere to telomere. (B) Chromosome 6. Markers are presented telomere to centromere.
Table 2.
Bymarker analysis of chromosomes with significant evidence of linkage for severity and age of onset
| Severity
|
Onset
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Locus | Centimorgans* | F† | P | Locus | Centimorgans* | F | P | |
| Chromosome 2 | ||||||||
| D2MIT5 | − | 2.1 | 0.13200 | D2MIT5 | − | 2.5 | 0.08 | |
| D2MIT7 | 14.3 | 6.5 | 0.00262 | D2MIT7 | 14.3 | 6.5 | 0.0023 | |
| D2MIT61 | 12.0 | 10.5 | 0.00010 | D2MIT61 | 12.0 | 15.8 | 0.0000011 | |
| D2MIT9 | 4.1 | 3.8 | 0.02802 | D2MIT9 | 4.1 | 8.0 | 0.00059 | |
| D2MIT11 | 4.4 | 7.2 | 0.00138 | D2MIT11 | 4.4 | 12.5 | 0.000014 | |
| D2MIT14 | 6.9 | 5.4 | 0.00641 | D2MIT14 | 6.9 | 7.6 | 0.00087 | |
| D2MIT30 | 11.3 | 2.7 | 0.07305 | D2MIT30 | 11.3 | 6.4 | 0.0024 | |
| D2NDS3 | 3.0 | 4.1 | 0.02041 | D2NDS3 | 3.0 | 8.7 | 0.00032 | |
| D2MIT21 | 8.1 | 2.8 | 0.06983 | D2MIT21 | 8.1 | 3.2 | 0.044 | |
| D2MIT48 | 2.9 | 2.5 | 0.09207 | D2MIT48 | 2.9 | 3.2 | 0.046 | |
| D2MIT49 | 5.4 | 2.4 | 0.09816 | D2MIT49 | 5.4 | 3.9 | 0.023 | |
| D2MIT51 | 2.7 | 2.6 | 0.08346 | D2MIT51 | 2.7 | 5.0 | 0.0083 | |
| Chromosome 6 | ||||||||
| D6MIT1 | − | 2.9 | 0.06182 | |||||
| D6MIT3 | 28.7 | 5.7 | 0.00572 | |||||
| D6MIT4 | 2.7 | 5.4 | 0.00716 | |||||
| D6MIT8 | 1.7 | 7.3 | 0.00153 | |||||
| D6MIT9 | 2.2 | 6.7 | 0.00254 | |||||
| D6MIT10 | 23.8 | 8.9 | 0.00045 | |||||
| D6MIT13 | 9.6 | 2.9 | 0.06503 | |||||
| D6MIT15 | 12.1 | 1.1 | 0.35456 | |||||
Genotype and phenotype data were analyzed by using bymarker under the assumption of a non-normal distribution. Data for both the severity and age of onset are presented.
Distance from the previous marker in centimorgans.
F test of ranks, 2 degrees of freedom.
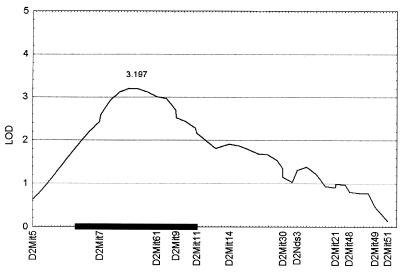
To determine whether specific genomic intervals were associated with the age of onset of CIA, we performed a QTL analysis of the affected F2 progeny by using animals exhibiting early (<6 weeks) and late (>12 weeks) CIA onset. In contrast to the above analysis, the results demonstrated that one genomic interval showed evidence of linkage to a locus that affects the age of onset of CIA on chromosomes 2 (Fig. 2). As with Cia2, the first locus also was linked to D2Mit61 (lod score of 3.2). The variance explained by this locus was 21%. Again, to confirm the presence of this locus, an association analysis was performed by using bymarker (Table 2). The locus associated with the age of onset from this analysis was D2Mit61 (F = 15.8, P < 0.000001).
Figure 2.
QTL plot for chromosome 2 containing a non-Mhc linked locus using the age of onset data. Log-likelihood values were determined by using mapmaker/qtl 1.1b. lod scores are presented on the y axis, and positions of the marker loci from centromere to telomere along the chromosome are given on the x axes. Support intervals are given by the black box.
DISCUSSION
The susceptibility and resistance of DBA/1j and SWR/j, respectively, to the induction of CIA is well documented (9, 10, 15). As with previously published data, the DBA/1j mice in these experiments had an incidence of 100% whereas the SWR/j mice were completely resistant (0% incidence). Consistent with a dominant mode of inheritance, our (DBA/1j × SWR/j)F1 progeny were susceptible to induction of CIA (Table 1). Of interest, there was incomplete penetrance of the susceptible phenotype (73% incidence). The further reduction in the number of affected F2 progeny (63.5%) is consistent with a polygenic model of CIA susceptibility. A comparison of the severity indexes (arthritic) between the F1 and F2 progeny did not show a significant difference. In contrast, the DBA/1j positive controls were significantly different in terms of severity when compared with either the F1 or F2 progeny (P < 0.0001). These data might indicate there are loci coming from SWR/j that have a protective affect for CIA. Alternatively, the decreased severity may be caused by random shuffling of loci during the breeding process. However, if this were the case, one would expect a difference between the F1 and F2 progeny. The localization of SWR/j derived protective loci could be determined by assessing CIA in (DBA/1j × SWR/j)F1 × SWR/j backcross progeny.
The two intervals associated with CIA severity (Fig. 1) contain a number of candidate loci (Table 3). The region of chromosome 2 containing Cia2 is syntenic to rat chromosome 3. Chromosome 3 did not show evidence of linkage to CIA in the (DA × F344)F2 rat QTL analysis (23), indicating that we have uncovered a unique locus for CIA susceptibility. There are two very interesting candidate loci in this region, prostagladin synthetase 1 (Pgs1) and complement C5 (Hc). Pgs1 is a key enzyme in prostaglandin biosynthesis and thus may impact modulation of the inflammatory responses. Complement C5 (Hc) is an important mediator of inflammation and has been implicated in the resistance of SWR/j mice to CIA induction (17, 21). The role complement C5 plays in the disease process of CIA has been debated (18, 30). However, recent genetic data using transgenic T cell receptor β chain mice have implicated C5 and this region in susceptibility to CIA (21).
Table 3.
List of candidate loci for each genomic interval
| QTL | Chromosome | Locus | Description |
|---|---|---|---|
| Cia2, Cia4 | 2 | Ptgs1 | Prostaglandin synthetase 1 |
| Dpp4 | Dipeptidylpeptidase 4 | ||
| Ssb | Sjogren syndrome antigen B | ||
| Hc | Complement C5 | ||
| Cia3 | 6 | Tnfr1 | Tumor necrosis factor receptor 1 |
| Il5ra | Interleukin 5 receptor α | ||
| Cd4 | CD4 | ||
| Cd27 | CD27 | ||
| Tgfa | Transforming growth factor α | ||
| Bphs | Bordetella pertussis-induced histamine sensitization |
Candidate loci were picked because of their immunologic relevance. This is not an exhaustive list.
The second locus (Cia3) is particularly interesting in that this region of chromosome 6 is syntenic to rat chromosome 4. This region of chromosome 4 has been shown to contain susceptibility loci for CIA (23) as well as IDDM in the rat (1). This observation would argue that there may be a major locus contributing to autoimmune inflammation in this interval. However, one cannot rule out that more than one locus is contained in this interval. There are a number of candidate genes found in this region (Table 3). Most notable are Il5ra, Tnfr1, Tgfa, and Cd27, one of which has been implicated in the modulation of CIA susceptibility. Mori et al. (31) used Tnfr1-deficient mice to demonstrate this receptor may play a role in the early inflammatory response that establishes CIA. Additionally, Le et al. (32) used a modified Tnfr1 gene to suppress CIA in experimental rats. The ligand of Tnfr1, Tnfa, affects the course of CIA induction (33, 34). Of interest, there is a (TC)9(TG)19 repeat polymorphism 67 bases 5′ of the first exon between DBA/1j and SWR/j at the Tnfr1 locus (35).
The QTL analysis comparing early- vs. late-age-of-onset arthritic mice produced one genomic interval with evidence of linkage. This locus (Cia4) was found in the same interval associated with CIA severity on chromosome 2. This could be the result of the same locus or a separate linked locus. Of interest, we did not detect Cia3 in this analysis. This may indicate Cia3 does not affect the age of onset or it is too weak to be detected in the current analysis. Candidate loci found in this region (chromosome 2) that would be predicted to affect the progression of CIA in the early stages of the disease would be Hc or Ptgs1.
One way to confirm that these genomic intervals contain susceptibility loci for CIA is to produce a congenic mouse in which the interval from the susceptible strain is moved onto the background of a resistant strain. These experiments are currently underway. Preliminarily data with an incipient congenic strain in which the DBA/1j derived genomic interval containing Cia2 (chromosome 2) was moved onto the resistant SWR/j strain (SWR.D1c2N3F1, 85% background) gave interesting results. Immunization of these animals with chicken CII resulted in ≈50% of the animals homozygous for the DBA/1j Cia2 region having severe CIA similar to control DBA/1j mice (G. Carlson, McClaughlin Institute, personal communication). Given the complete resistance of SWR/j to CIA induction, these results are extremely promising.
The loci we have described here contribute to ≈39% of the variance observed. This would indicate that there are more loci contributing to CIA severity; however, we were unable to detect them in the current data set. This is similar to the rat CIA QTL analysis in which 57% of the variance was explained by the loci detected in their screen (23). Our current analysis has identified one unique and possibly one common locus for CIA susceptibility when compared with the rat study as well as possibly one unique locus contributing to the age of onset of disease. Positional cloning efforts for these loci are also underway.
Acknowledgments
The authors thank Drs. Mark Hannibal, Leonid Kruglyak, and Elaine Ostrander for their helpful comments. This work was supported by the Stowers Institute (L.H.) and an National Research Service Award fellowship from National Institute of Arthritis and Musculoskeletal and Skin Diseases (R.A.M.).
ABBREVIATIONS
- CIA
collagen-induced arthritis
- CII
type II collagen
- CFA
complete Freund’s adjuvant
- QTL
quantitative trait locus
- lod
logarithm of odds
References
- 1.Jacob H J, Pettersson A, Wilson D, Mao Y, Lernmark A, Lander E S. Nat Genet. 1992;2:56–60. doi: 10.1038/ng0992-56. [DOI] [PubMed] [Google Scholar]
- 2.Morel L, Rudofsky U H, Longmate J A, Schiffenbauer J, Wakeland E K. Immunity. 1994;3:219–229. doi: 10.1016/1074-7613(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 3.Kono D H, Burlingame R W, Owens D G, Kuramochi A, Balderas R S, Balomenos D, Theofilopoulos A N. Proc Natl Acad Sci USA. 1994;91:10168–10172. doi: 10.1073/pnas.91.21.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacob H J, Lindpaintner K, Lincoln S E, Kusumi K, Bunker R K, Mao Y P, Ganten D, Dzau V J, Lander E S. Cell. 1991;67:213–224. doi: 10.1016/0092-8674(91)90584-l. [DOI] [PubMed] [Google Scholar]
- 5.Hilbert P, Lindpaintner K, Beckmann J S, Serikawa T, Soubrier F, Dubay C, Cartwright P, De Gouyon B, Julier C, Takahasi S, et al. Nature (London) 1991;353:521–529. doi: 10.1038/353521a0. [DOI] [PubMed] [Google Scholar]
- 6.Osman G E, Toda M, Kanagawa O, Hood L E. J Exp Med. 1993;177:387–395. doi: 10.1084/jem.177.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trentham D E, Dynesius-Trentham R A, Orav E J, Combitchi D, Lorenzo C, Sewell K L, Hafler D A, Weiner H L. Science. 1993;261:1727–1730. doi: 10.1126/science.8378772. [DOI] [PubMed] [Google Scholar]
- 8.Courtenay J S, Dallman M J, Dayan A D, Martin A, Mosedale B. Nature (London) 1980;283:666–668. doi: 10.1038/283666a0. [DOI] [PubMed] [Google Scholar]
- 9.Holmdahl R, Andersson M, Goldschmidt T J, Gustafsson K, Jansson L, Mo J A. Immunol Rev. 1990;118:193–232. doi: 10.1111/j.1600-065x.1990.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 10.Holmdahl R, Anders M E, Goldschmidt T J, Jansson L, Karlsson M, Malmstrom V, Mo J. APMIS. 1989;97:575–584. doi: 10.1111/j.1699-0463.1989.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 11.Holmdahl R, Vingsbo C, Hedrich H, Karls M, Kvick C, Goldschmidt T J, Gustafsson K. Eur J Immunol. 1992;22:419–424. doi: 10.1002/eji.1830220220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wooley P H, Chapedelaine J M. Crit Rev Immunol. 1987;8:1–22. [PubMed] [Google Scholar]
- 13.Wooley P H. Methods Enzymol. 1988;162:361–373. doi: 10.1016/0076-6879(88)62091-x. [DOI] [PubMed] [Google Scholar]
- 14.Holmdahl R, Klareskog L, Andersson M, Hansen C. Immunogenetics. 1986;24:84–89. doi: 10.1007/BF00373114. [DOI] [PubMed] [Google Scholar]
- 15.Wooley P H, Luthra H S, Stuart J M, David C S. J Exp Med. 1981;154:688–700. doi: 10.1084/jem.154.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmdahl R, Karls M, Andersson M E, Rask L, Andersson L. Proc Natl Acad Sci USA. 1989;86:9475–9479. doi: 10.1073/pnas.86.23.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spinella D G, Jeffers J R, Reife R A, Stuart J M. Immunogenetics. 1991;34:23–27. doi: 10.1007/BF00212308. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee S, Ander G D, Luthra H S, David C S. J Immunol. 1989;142:2237–2243. [PubMed] [Google Scholar]
- 19.Andersson M, Jansson L, Holmdahl R. Immunogenetics. 1992;35:71–72. doi: 10.1007/BF00216633. [DOI] [PubMed] [Google Scholar]
- 20.Spinella D G, Stuart J M. Immunogentics. 1992;35:73–74. [Google Scholar]
- 21.Mori L, De Libero G. Arthritis Rheum. 1998;41:256–262. doi: 10.1002/1529-0131(199802)41:2<256::AID-ART9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 22.David C S. Immunogenetics. 1992;35:69–70. doi: 10.1007/BF00216632. [DOI] [PubMed] [Google Scholar]
- 23.Remmers E F, Longman R E, Du Y, O’Hare A, Cannon G W, Griffiths M M, Wilder R L. Nat Genet. 1996;14:82–85. doi: 10.1038/ng0996-82. [DOI] [PubMed] [Google Scholar]
- 24.Jirholt J, Cook A, Emahazion T, Sundvall M, Jansson L, Nordquist N, Pettersson U, Holmdahl R. Eur J Immunol. 1998;28:3321–3328. doi: 10.1002/(SICI)1521-4141(199810)28:10<3321::AID-IMMU3321>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Dong P, Hood L, McIndoe R A. Genomics. 1996;31:266–269. doi: 10.1006/geno.1996.0047. [DOI] [PubMed] [Google Scholar]
- 26.McIndoe R A, Hood L, Bumgarner R. Electrophoresis. 1996;17:652–658. doi: 10.1002/elps.1150170405. [DOI] [PubMed] [Google Scholar]
- 27.McIndoe R A, Bumgarner R E, Welti R, Hood L. Proc Int Soc Opt Eng. 1996;2680:341–348. [Google Scholar]
- 28.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 29.Lander E S, Botstein D. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson M, Goldschmidt T J, Michaelsson E, Larsson A, Holmdahl R. Immunology. 1991;73:191–196. [PMC free article] [PubMed] [Google Scholar]
- 31.Mori L, Iselin S, De Libero G, Lesslauer W. J Immunol. 1996;157:3178–3182. [PubMed] [Google Scholar]
- 32.Le C H, Nicolson G, Morales A, Sewell K L. Arthritis Rheum. 1997;40:1662–1669. doi: 10.1002/art.1780400916. [DOI] [PubMed] [Google Scholar]
- 33.Thorbecke G J, Shah R, Leu C H, Kuruvilla A P, Hardi A M, Palladino M A. Proc Natl Acad Sci USA. 1992;89:7375–7379. doi: 10.1073/pnas.89.16.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brahn E, Peacock D J, Banquerigo M L, Liu D Y. Lymphokine Cytokine Res. 1992;11:253–256. [PubMed] [Google Scholar]
- 35.Takao S, Mykytyn K, Jacob C O. Immunogenetics. 1993;37:199–203. doi: 10.1007/BF00191885. [DOI] [PubMed] [Google Scholar]