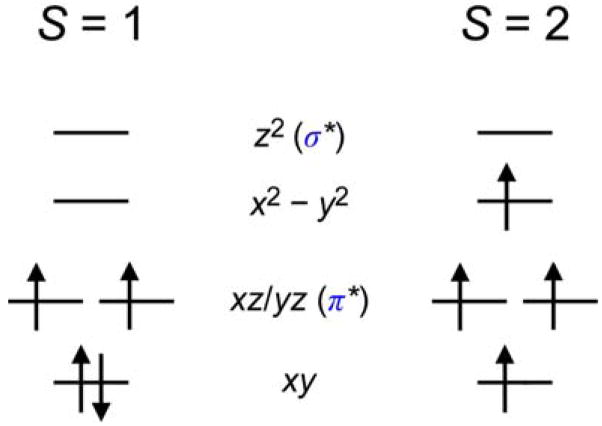Summary
FeIII–OOH and FeIV=O intermediates have now been documented in a number of non-heme iron active sites. In this Opinion we use spectroscopy combined with electronic structure calculations to define the frontier molecular orbitals (FMOs) of these species and their contributions to reactivity. For the low-spin FeIII–OOH species in activated bleomycin we show that the reactivity of this non-heme iron intermediate is very different from that of the analogous Compound 0 of cytochrome P450. For FeIV=O S = 1 model species we experimentally define the electronic structure and its contribution to reactivity, and computationally evaluate how this would change for the FeIV=O S = 2 intermediates found in non-heme iron enzymes.
Introduction
The presently-known classes of mononuclear non-heme iron enzymes that react with O2 are summarized in Table 1. Their reaction mechanisms divide into two broad types: either O2 activation by FeII active sites or substrate activation by FeIII active sites [1,2]. We focus on the former in this review. For the pterin- and α-ketoglutarate(αKG)-dependent enzymes, an FeII and a reduced cofactor react with O2 to form an FeIV=O (S = 2) intermediate that has been observed in a number of these enzymes [3–8]. This goes on to react with the substrate either by H-atom abstraction or electrophilic attack on a ring [9]. For bleomycin (BLM, Fig. 1A), a glycopeptide antibiotic that cleaves DNA by H-atom abstraction [10–15], used in the clinical treatment of a variety of cancers, the FeII form reacts with O2 and an exogenous electron to generate activated BLM (ABLM), a species shown to be kinetically competent in cleaving DNA [16]. ABLM has been determined to be a low-spin FeIII-hydroperoxide species similar to Compound 0 in P450 [17–24]. In this Current Opinion, we first consider the chemistry of ABLM and contrast it with that of hydroperoxy-heme enzymes. We then consider the nature of the FeIV=O bond in a series of S = 1 model complexes which have variable reactivity in H-atom abstraction, and computationally extend these results to FeIV=O (S = 2) enzyme systems. In both broad classes of enzyme intermediates, low-spin FeIII–OOH and FeIV=O, we elucidate the frontier molecular orbitals (FMOs) involved in reactivity. At this time, less is known about the reaction coordinates of the extradiol dioxygenases and Rieske dioxygenases. However, peroxy intermediates have been crystallographically defined in both [25,26], and these combined with spectroscopy and calculations should allow mechanistic insight into their reactivities. For the FeIII substrate-activating non-heme enzymes, the reader is referred to Reference 27.
Table 1.
Classes of mononuclear non-heme iron enzymes.
| A. Oxygen Activating (FeII) | |
| Extradiol Dioxygenases |
|
| Pterin-Dependent Hydroxylases |
|
| α-KG-Dependent Dioxygenases |
|
| Rieske Dioxygenases |
|
| Bleomycin |
|
| B. Substrate Activating (FeIII) | |
| Lipoxygenases |
|
| Intradiol Dioxygenase |
|
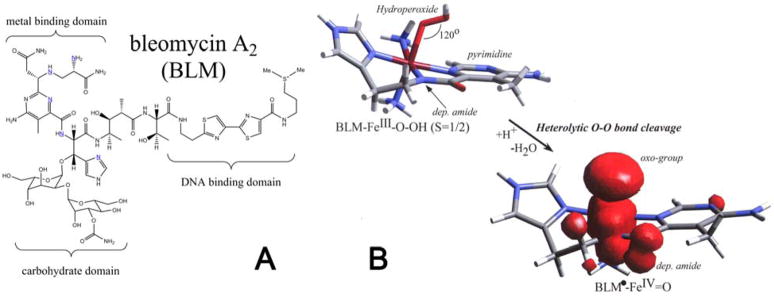
Figure 1.
(A) Structure of the BLM ligand and (B) geometric structures of ABLM and the hypothetical (BLM•)Fe(IV)=O (spin density overlaid.)
FeIII–OOH Intermediates
Spectroscopy combined with DFT calculations have led to the geometric and electronic structure description of ABLM. The bleomycin ligand is given in Figure 1A. In Figure 1B top, the hydroperoxide binds end-on to a low-spin FeIII center with an Fe–O–O angle of ~120°. It bisects the two chemically-interesting functional groups: the deprotonated amide nitrogen and a pyrimidine nitrogen [24]. In heme chemistry, P450 in particular, Compound 0 is also a low-spin FeIII–OOH, which is protonated and the O–O bond is cleaved heterolytically to give Compound I (FeIV=O, S = 1 with the porphyrin oxidized to the monoanionic radical state). If ABLM were protonated and the O–O bond cleaved, the Compound I equivalent of BLM would be obtained; this is given in Figure 1B bottom. As for P450, an FeIV=O (S = 1) would be produced and the ligand oxidized by one electron. The hole would localize on the deprotonated amide N ligand but with some delocalization over the tail.
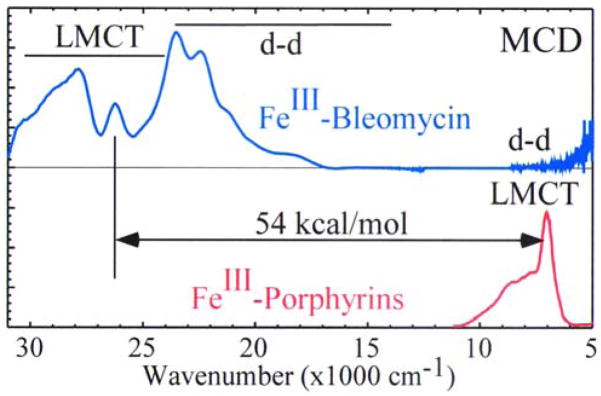
Importantly, there is a large difference in the energetics of the reactions of the heme and ABLM sites. For the low-spin hydroperoxy–FeIII site of P450, the reaction is highly exothermic and exergonic (Table 2). This is because protonation is favored due to the −1 charge (of the thiolate-bound low-spin heme FeIII–OOH complex) and the heterolytic cleavage produces a hole which is delocalized over the porphyrin ligand. In contrast, this protonation and heterolytic cleavage of ABLM is energetically disfavored by 99 kcal/mol for ABLM [28]. This is due to both the charge of +1 (of the deprotonated-BLM-ligated low-spin FeIII–OOH complex) disfavoring protonation and the fact that it is far more difficult to oxidize a non-heme ligand system. This can be seen experimentally in Figure 2, where low-temperature magnetic circular dichroism (LT MCD) correlated with absorption data (not shown) allows assignment of the lowest energy ligand-to-metal charge transfer (LMCT) bands for both the low-spin FeIII –heme and low-spin FeIII –BLM [24]. The deprotonated amide N-to-FeIII CT transition is almost 2.5 eV higher in energy than the heme HOMO CT, indicating that at least this much additional energy is required to oxidize the non-heme ligand system of BLM.
Table 2.
Comparison of heterolytic cleavage energetics of ABLM and P450.
B3LYP/TZV; all values in kcal/mol;
solvent: protein, ε = 4.0;
includes zero-point correction energy.
Figure 2.
Comparison of MCD spectra of Fe(III)–BLM and a prototypical FeIII–heme (adapted from Reference 29.)
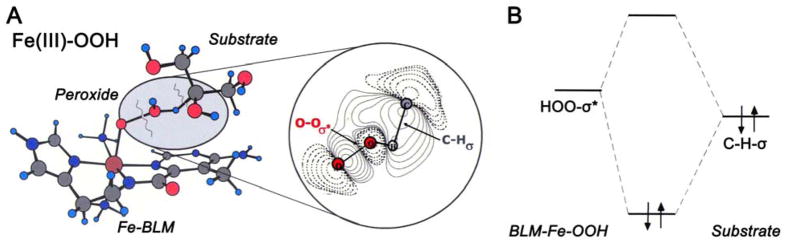
Therefore, we proposed the alternative reaction mechanism for the ABLM cleavage of DNA shown in Figure 3A, where the FeIII–OOH directly abstracts an H-atom from the H–C bond of the DNA sugar. The FMO for the reaction is shown in the expansion and in Figure 3B. Protonation of the peroxide lowers the energy of its σ* orbital, activating it for electrophilic attack on the H–C bond which would shift electron density out of a bonding and into an antibonding orbital and cleave both bonds. We then proceeded to evaluate the coordinate for the FeIII–OOH H-atom abstraction reaction by a non-heme iron site both experimentally and computationally.
Figure 3.
(A) Direct H-atom abstraction from sugar substrate by ABLM and (B) FMOs involved in the reaction.
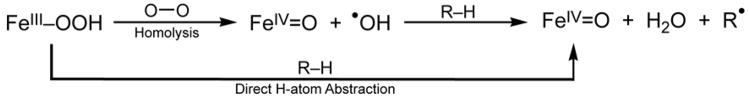
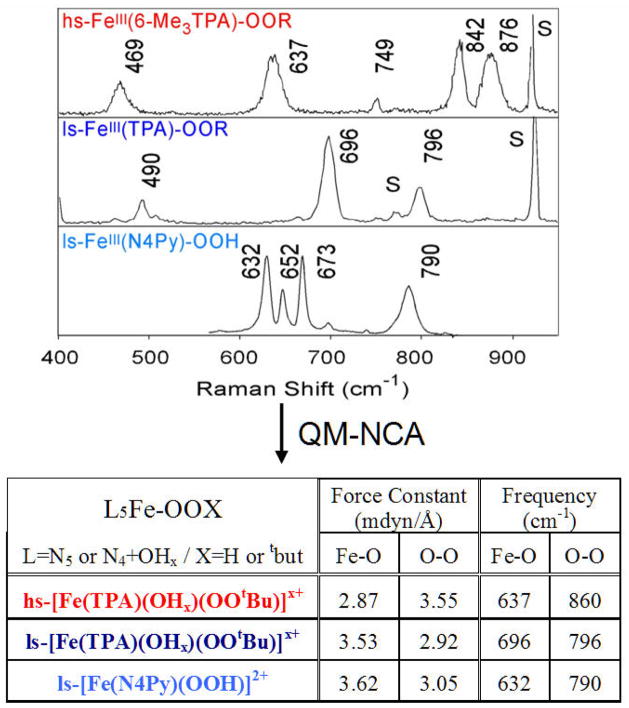
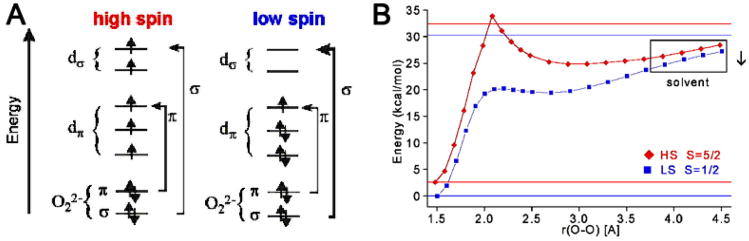
We consider this reaction in two steps: first, the O–O bond homolysis and then the formation of the O–H (and cleavage of the H–C) bond along this reaction coordinate (Scheme 1.) It is interesting to compare the hydroperoxide–FeIII bond in a low-spin complex to that of a high spin FeIII–OOH analog. From Figure 4 and the associated Table of force constants from a normal coordinate analysis of the data, for the low spin complex, the FeIII–(OOH) bond is very strong and the O–O bond is weak [30–32]. Thus the low-spin FeIII–OOH species is activated for homolytic O–O bond cleavage. The origin of this can be seen from the electronic structure description in Figure 5A, where in the low-spin complex the d σ orbitals are unoccupied and allow strong σ donation from σ- and π-bonding orbitals on the hydroperoxide, which strengthens the peroxide–FeIII bond and weakens the O–O bond. This activates the O–O bond for cleavage and from Figure 5B, in contrast to the high-spin case, the reaction coordinate of the low-spin FeIII–OOH complex has no additional barrier to the O–O bond homolysis. The energetics for this reaction are summarized in Table 3. The electronic energy for O–O bond homolysis is ~29 kcal/mol, much lower than the 99 kcal/mol for the heterolytic cleavage in Table 2, since this reaction gives an FeIV=O S = 1, with no oxidation of the ligand. As there is no change in charge in this reaction, there is little solvent effect and the increase in entropy due to the bond cleavage gives a reaction that is endergonic by only 13 kcal/mol. The bottom row of Table 3 includes the energetics of C–H cleavage and H–O bond formation for the H-atom abstraction reaction which is exergonic by 7 kcal/mol and based on calculated energetics, direct reaction of ABLM, a low spin FeIII–OOH species, with the H–C bond of the DNA sugar is most favored.
Scheme 1.
Homolysis and direct H-atom abstraction pathways of FeIII–OOR species.
Figure 4.
Resonance Raman spectra of three FeIII–OOH/R model complexes and calculated force constants from normal coordinate analyses of the Raman data.
Figure 5.
Comparison of (A) bonding character and (B) reaction coordinates of O–O bond homolysis for high- and low-spin FeIII–OOH/R.
Table 3.
Comparison of homolytic cleavage energetics of P450 and ABLM, and the energetics of direct H-atom abstraction by ABLM.
| Reactant | ΔE(gas) | ΔE(solv)a | −TΔSb | ΔG(solv) | |
|---|---|---|---|---|---|
| Homolysis: | FeIII(P450)OOH⌉− | +28 | +25 | −14 | +11 |
| Homolysis: | FeIII(BLM)OOH⌉+ | +29 | +27 | −14 | +13 |
| H-Atom Abstraction: | FeIII(BLM)OOH⌉+ | +13 | +9 | −16 | −7 |
B3LYP/TZV; all values in kcal/mol;
solvent: protein, ε = 4.0;
includes zero-point correction energies.
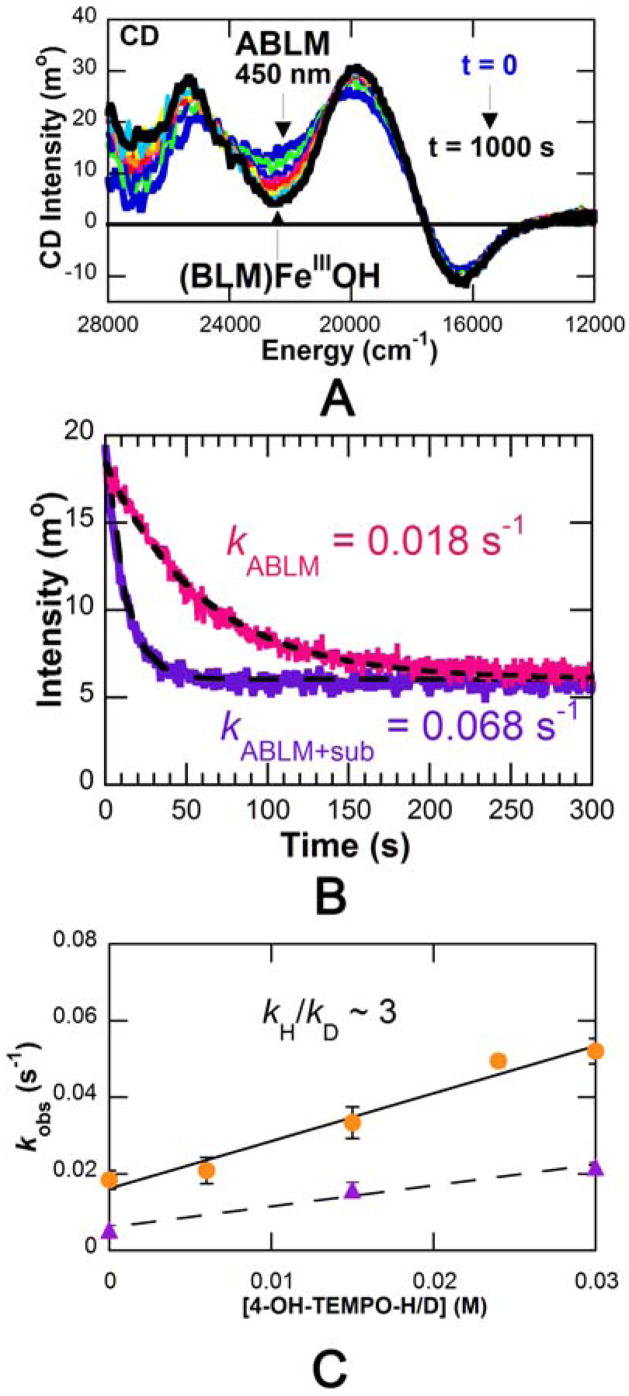
Thus we proceeded to evaluate this experimentally. While it had previously been difficult to monitor the reaction kinetics of ABLM in real time as ABLM has no characteristic absorption feature, it does have a characteristic feature in its CD spectrum at 450 nm [33]. We were able to use this to monitor the decay of ABLM with time and evaluate how it is affected by exogenous substrates. Importantly, from Figure 6, addition of H-atom donor substrates (4-OH-TEMPO-H, ascorbate) accelerates the rate of decay of ABLM proportional to the amount of added substrate. This substrate-dependent reaction of ABLM shows a kinetic isotope effect (KIE) of 3. This strongly indicates that the substrate participates in the transition state and that the decay of ABLM involves H-atom abstraction from these substrates.
Figure 6.
ABLM reaction kinetics studied by CD spectroscopy. (A) Time-dependent CD spectra of reaction. (B) Decay of ABLM (pink trace), and reaction of ABLM with substrate (purple trace.) (C) Rate constant versus 4-OH-TEMPO-H concentration (circles), and rate constants versus 4-OH-TEMPO-D concentration (triangles.)
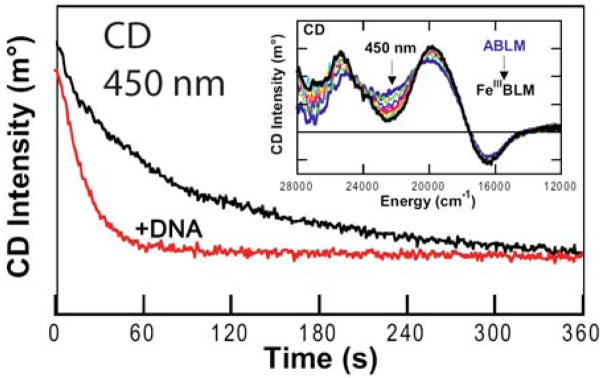
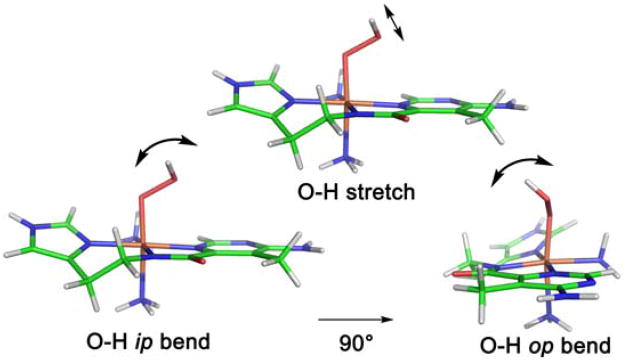
It is important to notice from the left of Figure 6C that the decay of ABLM even in the absence of substrate shows a KIE of 3.6. We ascribe this to an H-atom abstraction reaction of ABLM with the H–N bonds of its bithiazole tail (Fig. 1A) which are exchangeable with solvent [33]. However, the alternative mechanism of heterolytic cleavage of ABLM to produce a formally FeV=O species has continued to be considered in the reaction with DNA [34]. Thus we used our CD kinetic approach to evaluate whether binding of DNA to ABLM affects the reaction kinetics [35]. From Figure 7, the rate of the decay of ABLM is clearly accelerated in the presence of DNA and all the kinetic parameters determined for this reaction (Table 4) have changed, indicating that the DNA substrate also participates in the transition state of this reaction. The KIE is now reduced from 3.6 in the absence of DNA to 1.7 in the reaction with DNA. Since the DNA sugar H–C bond is not exchangeable, this must a secondary KIE involving the proton of the hydroperoxide. From the calculated frequencies of the transition state for this reaction (vide infra), the dominant contribution to this KIE is the in-plane (ip) OO–H bend (Figure 8 and Table 5) which decreases at the transition state due to elongation of O–O bond. The calculated secondary KIE for the DNA reaction is 1.47. For the reaction where the substrate also can have a KIE (as in the decay of ABLM in the absence of substrate), the combined primary and secondary KIEs are calculated to be 3.84 (Table 5), very similar to the 3.6 KIE experimentally observed for ABLM decay. Also from Table 4, the activation energy of the reaction with DNA decreases relative to that of the decay of ABLM in the absence of DNA, which can be correlated to the effects of the H–C versus H–N bond strength on the transition-state energy (bond-dissociation energies: H–C, 92 kcal/mol; H–N, 105 kcal/mol.)
Figure 7.
Time-dependent CD spectra (at 450 nm) of ABLM self-decay (black) and the reaction of ABLM with DNA (red.) Inset: interval scans showing CD spectral changes for ABLM decaying to FeIII–BLM.
Table 4.
Kinetic and energetic parameters of ABLM reaction with DNA and ABLM self-decay, calculated from CD kinetic data (see Figure 7.)
| Reaction | ABLM + DNA | ABLM decay |
|---|---|---|
| Rate (s−1) | 0.044 ± 0.02 | 0.018 ± 0.003 |
| kH/kD | 1.7 ± 0.2 | 3.6 ± 0.9 |
| Ea (kcal/mol) | 4.7 ± 0.9 | 9.3 ± 0.9 |
| ΔG‡ (kcal/mol) | 19.0 ± 0.02 | 19.5 ± 0.03 |
| ΔH‡ (kcal/mol) | 4.4 ± 1.6 | 9.0 ± 3 |
| ΔS‡ (cal/mol−1K−1) | −48 ± 6 | −36 ± 10 |
Figure 8.
Transition state O–H vibrational modes in ABLM sensitive to H/D substitution, contributing to the 2H-KIE of H-atom abstraction.
Table 5.
Calculated contributions of vibrational modes to the 2H-KIEs of H-atom abstraction of ABLM (modes described in Figure 8).
| Vib. mode | kH/kD |
|---|---|
| O-H stretch | 1.02 |
| OO-H ip bend | 1.53 |
| OO-H op bend | 0.93 |
| Secondary KIE | 1.47 |
| Primary KIE | 2.61 |
| Total KIE | 3.84 |
Thus, spectroscopy and calculations on ABLM support a model where ABLM reacts differently from Compound 0 of P450 in heme chemistry. Rather than heterolytic O–O bond cleavage, this non-heme FeIII–OOH directly abstracts an H-atom from the substrate. These different reaction pathways are due to differences in the energy of protonation of the hydroperoxide and the difficulty of oxidizing a non-heme ligand environment to generate Compound I. Experiments with H-atom donor substrates including DNA now demonstrate that the H–X bond participates in the transition state. These experiments then lead to the computational definition of the coordinate for this reaction and elucidation of the transition state.
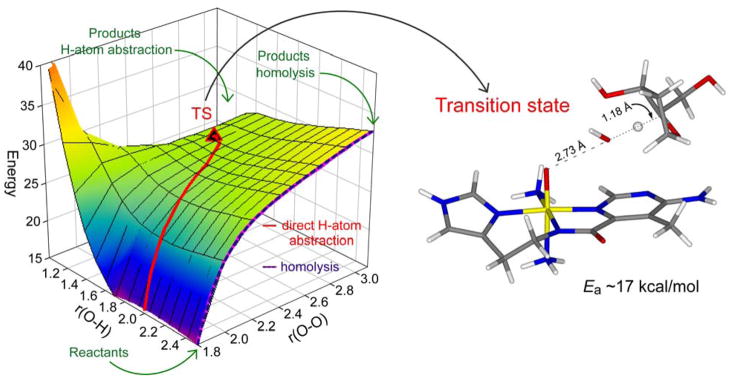
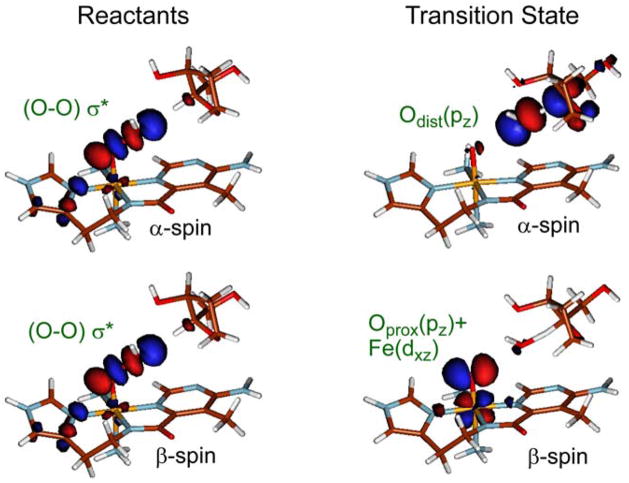
Figure 9 shows the 2-dimensional reaction coordinate, where the O–O bond is elongated going from front to rear on the right. This edge corresponds to O–O homolysis. On the left from front to rear the H-atom is transferred from the H–C bond of the sugar substrate to the distal O of the hydroperoxide to form H2O and an FeIV=O S = 1 species. The H-atom abstraction reaction is given by the red trajectory and the transition state is given to the right of Figure 9. The calculated activation energy is somewhat higher than observed experimentally but is dependent on the hydrogen bonds included in the model. Interestingly, the transition state is late along O–O bond cleavage but early in H–C cleavage, consistent with the relatively low primary KIE. Figure 10 gives the FMOs involved in this reaction and elucidates their nature at the transition state. On the left are the α and β holes in the σ* orbital of the hydroperoxide. Elongation of the O–O bond in going to the transition state (Figure 10, right) shows that the O–O σb/σ* interaction is greatly weakened and the holes now localize: one on the distal O (α-spin) which strongly activates it for the H-atom abstraction and the second on the proximal O (β-spin) which results in an FeIV=O S = 1 species. We have shown that this resultant FeIV=O S = 1 BLM species is also capable of an H-atom abstraction from the DNA sugar with an even lower barrier [33]. This is important as it provides a mechanism for ABLM, tethered to DNA through its bithiazole tail, to effectively perform a double-strand cleavage which is not readily repaired and is associated with the effectiveness of BLM as an anticancer drug.
Figure 9.
2D potential energy surface for the reaction pathway of ABLM: the right edge shows O–O homolysis and the red trajectory represents direct H-atom abstraction from DNA, the transition state of which is shown on the right.
Figure 10.
FMO interactions of ABLM with DNA before reaction (left) and at the transition state of H-atom abstraction (right). Note that the transition state is late along O–O cleavage and early along O–H formation.
FeIV=O Intermediates
High-spin FeIV=O S = 2 intermediates have now been confirmed to be present in TauD, P4H, TryH and the halogenase CytC3 [3–8]. Model complexes with low-spin FeIV=O S = 1 sites have been structurally defined for two complexes, (TMC)FeIV=O and (N4Py)FeIV=O, but only (N4Py)FeIV=O is reactive in H-atom abstraction [36–38]; trends in the reactivities of these and related complexes have been studied [39–41]. However, no well-defined FeIV=O S = 2 model complex exists at this point. In this section we combine experiment and calculations to understand the nature of the FeIV=O bond in the S = 1 models and the contribution of its FMOs to reactivity. We then computationally extend these results to the FeIV=O S = 2 enzyme intermediates.
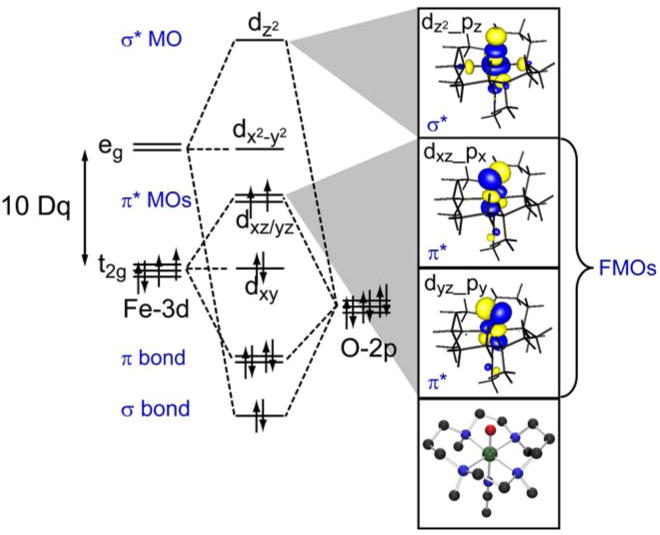
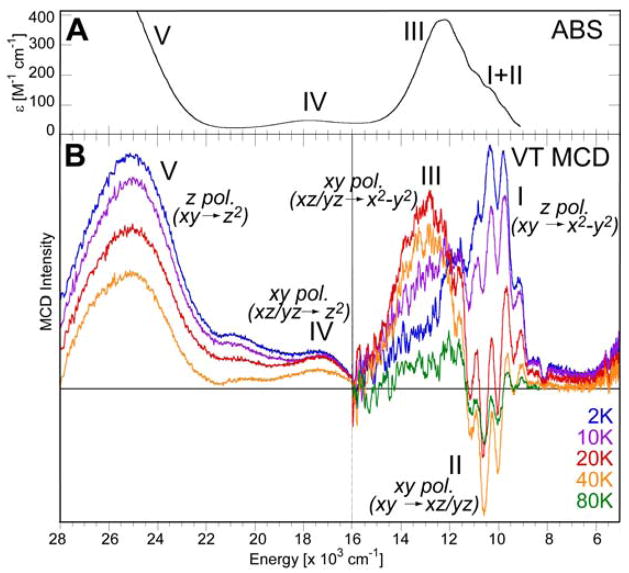
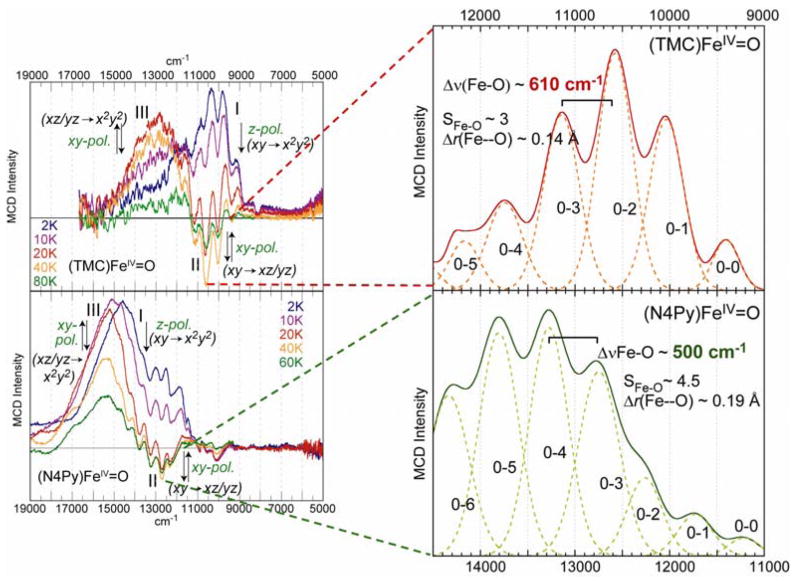
The (TMC)FeIV=O(NCCH3) complex (shown at the bottom right of Figure 11) of Que and colleagues was the first to be structurally defined [36]. As shown in Figure 11, the Fe–O bonding interactions dominate its electronic structure. Starting from the octahedral limit on the left, the O pz will undergo a strong bonding/antibonding interaction with the Fe dz2 orbital. In addition, the O px,py orbitals experience strong bonding/antibonding interactions with the dxz,yz orbitals of the Fe center. Inclusion of the six valence O p electrons with the four FeIV d electrons leads to a (dxy)2(dxz,yz)2 configuration for the S = 1 ground state. The filled dxy is nonbonding while the dxz,yz set is half-filled and π*, the σ* dz2 is empty; thus in addition to the strong σ bond there are also 2 × ½ π bonds contributing to the Fe–O bond. Importantly, the π* β orbitals are the lowest-energy unoccupied MOs and should be the frontier orbitals involved in electrophilic attack. These frontier orbitals can be studied experimentally using electronic spectroscopy, through the excitation of an electron into these unoccupied orbitals. However, the electronic absorption spectrum of (TMC)FeIV=O(NCCH3) is rather uninformative (Figure 12A) in having a broad, weak absorption band in the near-IR region at around 12,000 cm−1. However, the LT MCD spectrum in this region (Figure 12B) is rich in information content and allows a detailed experimental study of the FMOs of the Fe–O bond [42,43]. The variable-temperature MCD data in Figure 12B show that there are in fact three electronic transitions contributing to this one absorption envelope. The intensities of these bands exhibit different temperature-dependent behaviors, allowing resolution and assignment of each individual transition.
Figure 11.
Key orbital interactions defining the Fe–O bond in (TMC)FeIV=O(CH3CN).
Figure 12.
(A) Absorption and (B) variable-temperature MCD spectra of (TMC)FeIV=O(NCCH3).
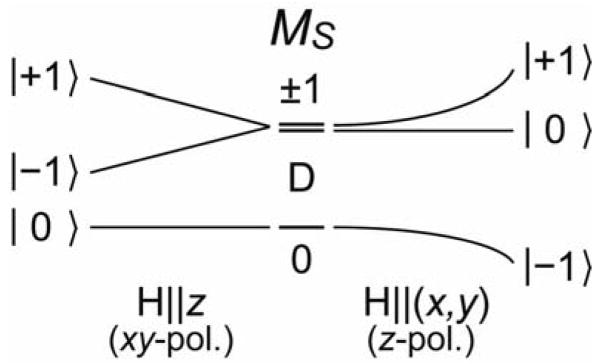
The FeIV=O S = 1 ground state undergoes a positive zero-field splitting with a D = +29 cm−1 [36]. As shown in Scheme 2 left, an (x, y)-polarized electronic transition requires the magnetic field to be along the z axis for MCD intensity. In this case, the lowest Zeeman-split sublevel has MS = 0 and thus no MCD activity, which is proportional to the spin-expectation value of the sublevel. Thermal population of the MS = −1 sublevel produces MCD intensity which then decreases as the temperature further increases, due to the population of the MS = +1 sublevel. Alternatively, for a z-polarized transition, the magnetic field must be in the (x, y) plane for MCD intensity. In this configuration, the lowest-energy Zeeman-split sublevel behaves as an MS = −1 with MCD intensity; this decreases as temperature increases due to the population of the MS = 0, +1 sublevels. Thus, the temperature dependence of the MCD intensity allows the determination of the polarizations of electronic transitions even for a frozen solution of randomly-oriented molecules.
Scheme 2.
Zeeman splitting of an S = 1 system with positive zero-field splitting (H||z on left, H⊥z on right.) The selection rule for C-term MCD intensity requires that the field be perpendicular to the polarization directions of the electronic transition.
Examining the temperature dependence of the band intensities in Figure 12B, Band III has little intensity at low temperature, increases in intensity up to 20 K and then decreases as the temperature is further increased. Band III can thus be assigned as an (x, y)-polarized transition. Band I, at lowest energy, is intense in MCD at low temperature with positive intensity, and then decreases in intensity as the temperature increases. It can be assigned as the lowest-energy z-polarized transition, which is the dxy → dx2 − y2 transition that reflects the strength of the equatorial ligand field. Interestingly, while the energy level diagram in Figure 11 indicates that it should be a higher-energy transition, it is instead at lowest energy due to the change in e−–e− repulsion; this emphasizes the limitation of a 1e− MO diagram. Importantly, band II exhibits sharp structure which is negative and overlaps with band I. It initially increases in intensity as the temperature is increased, reaches a maximum in negative structured intensity at 40K and then decreases in intensity as the temperature is further increased. It can thus be assigned as the lowest-energy (x, y)-polarized transition, which is the dxy→ dxz/yz ligand-field transition. This involves the excitation of an electron from a nonbonding MO to the π* manifold of the FeIV=O unit and thus directly, experimentally reflects the FMO involved in reactivity.
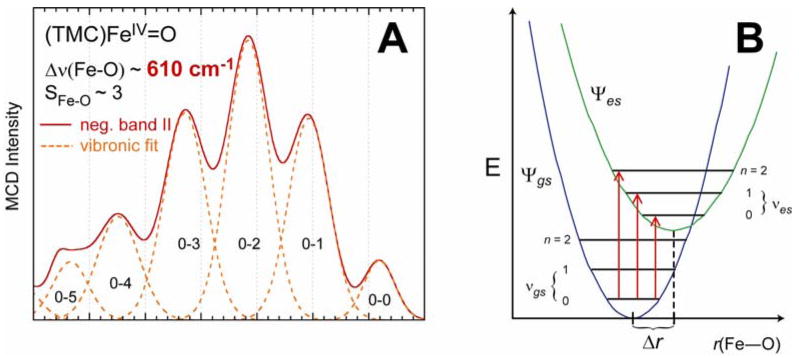
The energy of this transition at ~11,000 cm−1 reflects the strong π bond which is also probed by its vibronic structure. The latter has been deconvoluted and plotted as a positive progression in Figure 13A. This Franck-Condon progression reflects an elongation of the Fe–O bond in the excited state relative to the ground state due to the excitation of a dxy nonbonding electron into a π antibonding orbital (Figure 13B). A fit of this progression shows that the Fe–O bond length increases from 1.65 Å in the (dxy)2(dxz,yz)2 ground state to 1.79 Å in the (dxy)1(dxz,yz)3 excited state. The energy spacing of the progression is 610 cm−1; this is the Fe–O stretching vibration in this excited state which has gone down in energy from 830 cm−1 in the ground state (vide infra) due to the loss of ½ a π bond.
Figure 13.
(A) Franck-Condon progression in the n → π* (dxy → dxz/yz) excited state (band II) of the FeIV=O unit due to (B) vibronic transitions between the ground state and distorted excited state.
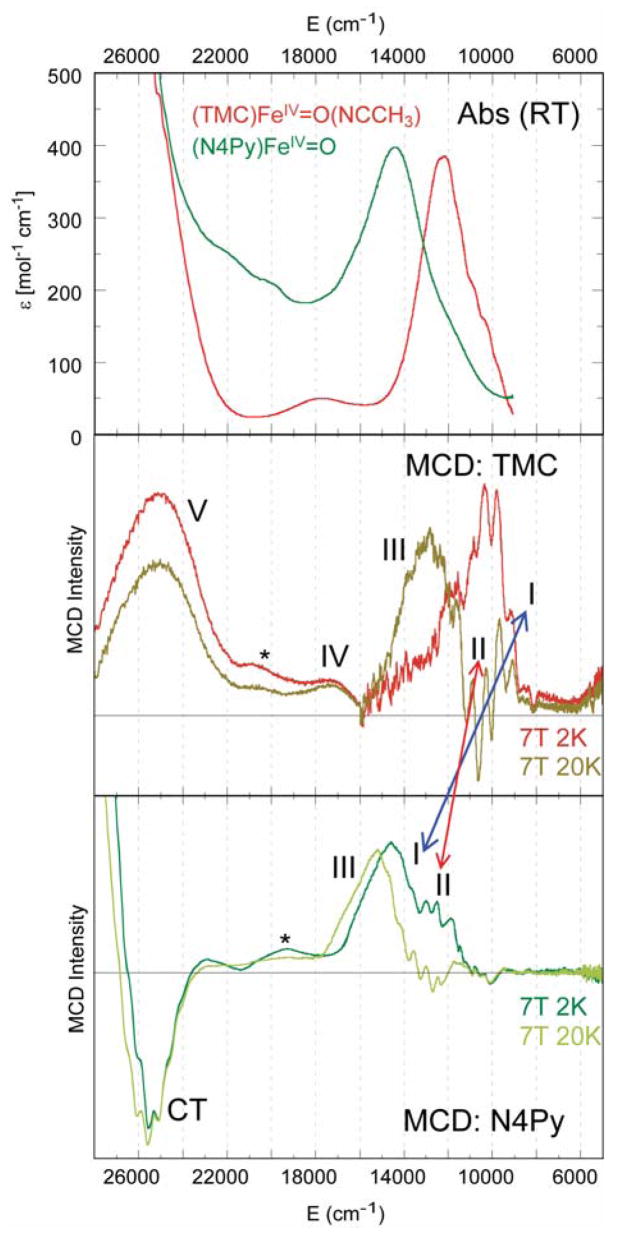
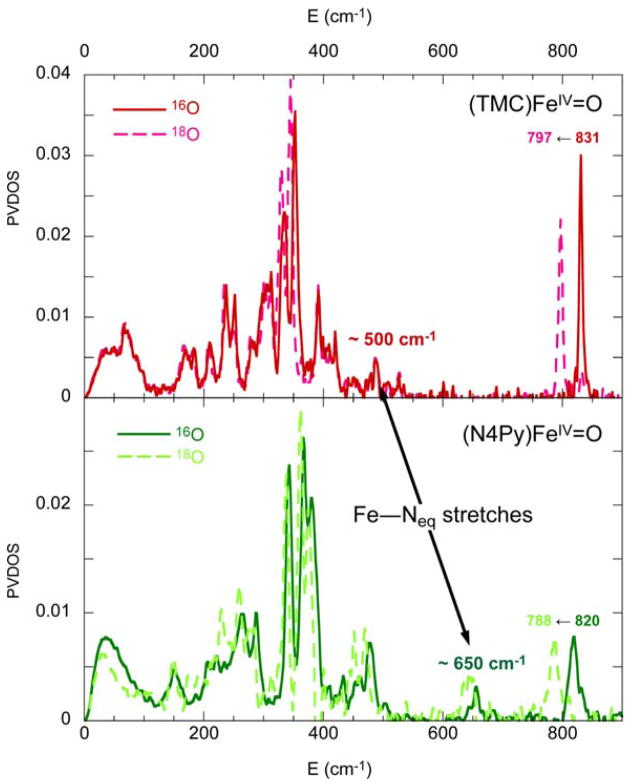
The quality of these experimental data also allows a quantitative comparison of (N4Py)FeIV=O with (TMC)FeIV=O to define differences in their excited states related to their difference in reactivity. In (N4Py)FeIV=O, the four equatorial tertiary amine ligands of (TMC)FeIV=O are replaced by four pyridine ligands, and in contrast to (TMC)FeIV=O, (N4Py)FeIV=O is able to H-atom abstract from inert C–H bonds, such as from cyclohexane [37]. From the absorption/MCD data in Figure 14, in going from (TMC)FeIV=O to (N4Py)FeIV=O, band I and band II both shift up in energy, indicating that both the equatorial ligand field (band I) and the Fe–O π bond strength have increased in the (N4Py)FeIV=O complex. Figure 15 shows the Nuclear Resonance Vibrational Spectroscopy (NRVS) data for both complexes, where the vibrations involving Fe motion are excited by recoil in a Mössbauer transition [44–46]. From Figure 15 and DFT calculations of the NRVS partial-vibrational-density-of-states (PVDOS) data, a peak at ~500 cm−1 in the (TMC)FeIV=O complex shifts up to ~650 cm−1 in the (N4Py)FeIV=O complex [47]. This peak is assigned as originating from the equatorial Fe–N stretches, and its shift to higher energy for (N4Py)FeIV=O also indicates stronger equatorial bonding interactions. Since the Fe–O stretch at 831 cm−1 in the (TMC)FeIV=O complex goes slightly down in energy to 816 cm−1 in the (N4Py)FeIV=O complex and the Fe–O π bond has become stronger (vide supra), the Fe–O σ bond must be weaker in the (N4Py)FeIV=O complex. The strong σ donation by N4Py into the equatorial lobe of the dz2 orbital weakens its interaction with the σ (pz) orbital of the oxo which is compensated by stronger π-donor interactions.
Figure 14.
Comparison of absorption and MCD spectra of (TMC)FeIV=O and (N4Py)FeIV=O.
Figure 15.
Comparison of NRVS ground-state vibrational spectra of (TMC)FeIV=O and (N4Py)FeIV=O.
This can be experimentally quantified from a comparison of the vibronic data in the LT MCD spectra of Band II in the two complexes in Figure 16, right [43]. In going from the (TMC)FeIV=O to the (N4Py)FeIV=O complex, the Franck-Condon progression shifts in intensity to a higher vibrational excitation, indicating a larger Fe–O bond length increase in the (dxy)1(dxz,yz)3 excited state. Analysis shows that the Fe–O bond lengthens by 0.19 Å in the excited state of (N4Py)FeIV=O, relative to the 0.14 Å increase for the (TMC)FeIV=O complex. Additionally, the Fe–O vibrational frequency in (N4Py)FeIV=O decreases to 500 cm−1 in the π* excited state. Thus, in (N4Py)FeIV=O the π bonding is stronger and so excitation of a nonbonding electron into a π* orbital leads to a longer, weaker Fe–O bond relative to the (TMC)FeIV=O complex.
Figure 16.
Vibronic structure comparison of band II in (TMC)FeIV=O and (N4Py)FeIV=O.
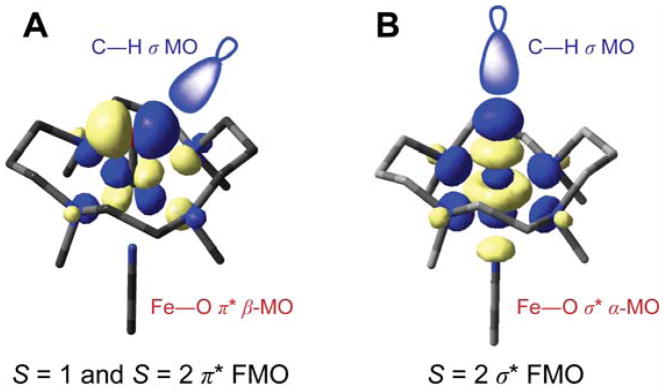
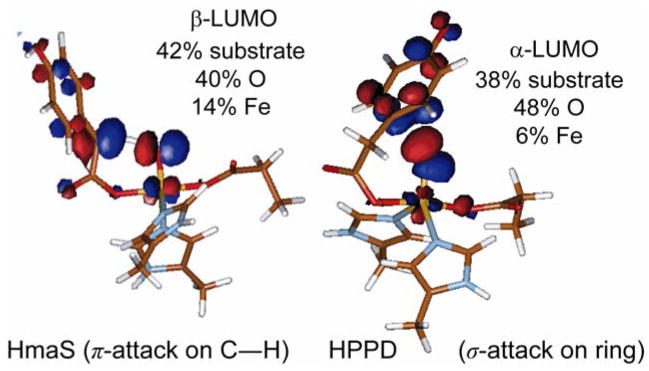
Because of this strong π bond in the (N4Py)FeIV=O complex, the amount of oxo px, py character in this low-lying, unoccupied MO increases, which activates it for reactivity. From the FMO in Figure 17A, the ability of the Fe d π* MO to H-atom abstract from a C–H bond directly reflects the amount of oxo p character mixed into this low-lying orbital due to the strength of the oxo π bond. Consequently, the stronger equatorial donor interactions of the N4Py ligand compete with the oxo σ donation which is compensated by the oxo π donor character that activates the π* FMO for reactivity.
Figure 17.
Reactive FMOs of (TMC)FeIV=O involved in H-atom abstraction by (A) π attack (both S = 1 and S = 2) and (B) σ attack (S = 2 only.)
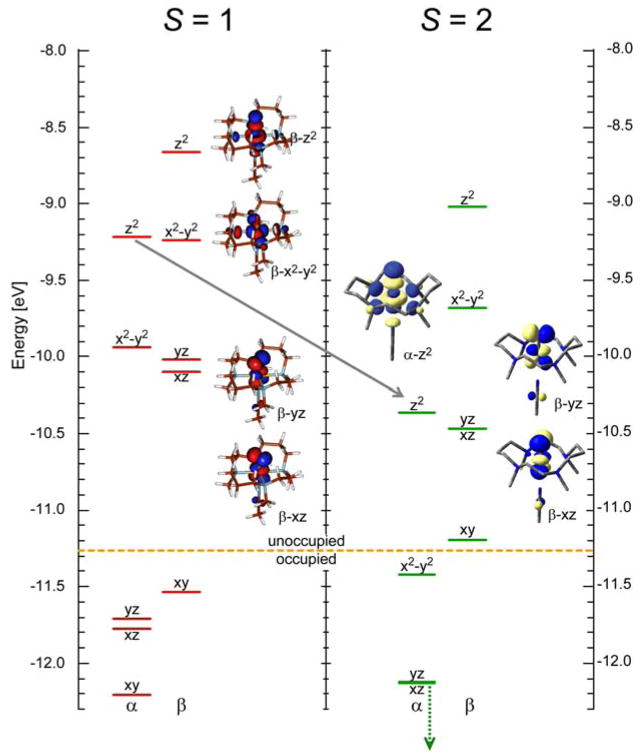
In going from the FeIV=O models to enzyme intermediates, the S = 1 to S = 2 spin-state change involves the promotion of an electron from a dxy to a dx2 − y2 orbital (with a spin flip, Scheme 3). This does not affect the strength or nature of the Fe–O bond in the ground state because only a change in the equatorial ligand field is involved [42]. However, this change in spin does strongly affect the excited states [48]. As shown in Figure 18, for the same complex, in going from an S = 1 to an S = 2 ground state, the α-spin manifold is polarized such that the dz2 unoccupied orbital now shifts down and becomes comparable in energy with the dxz/yz β LUMOs. Thus, both σ and π FMOs must be considered in the reactivity of FeIV=O S = 2 intermediates in non-heme iron enzymes (Figure 17B as well as A.)
Scheme 3.
Orbital occupation diagrams for S = 1 and S = 2 FeIV=O systems in the C4v point group.
Figure 18.
Molecular orbital diagrams for (TMC)FeIV=O in the S = 1 and S = 2 spin states.
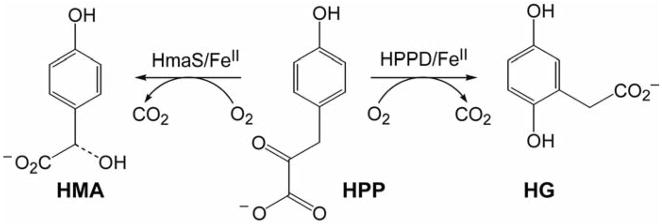
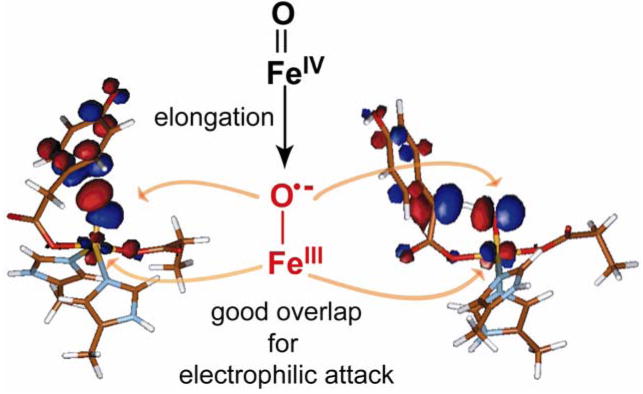
The enzymes (4-hydroxyphenyl)pyruvate dioxygenase (HPPD) and (4-hydroxy)mandelate synthase (HmaS) provide an interesting comparison set for extending these model studies to biological systems. Both react with the same substrate HPP (an α-keto acid covalently bound to the substrate) yet undergo different reactions: HmaS performs H-atom abstraction while HPPD performs an electrophilic attack on the aromatic ring of HPP [9] (Scheme 4.) From spectroscopic studies, the anaerobic substrate-bound complexes are are equivalent, but the protein pocket induces a conformational change in the ring (See Figure 19, where the ligand-field CD data for the resting and pyruvate-bound sites are the same but the HPP band signs are opposite, reflecting a local conformational change associated with the presence of the substrate’s aromatic ring.) As the aromatic substrate is covalently linked to the α-keto acid group bound to the iron, a difference in ring orientation would lead to differences at the FeIV=O intermediate level of the reaction pathway. From Figure 20, the decarboxylated substrate-intermediate bound to the Fe center in HmaS has the benzylic C–H oriented for good overlap with the π* FMO of the FeIV=O intermediate, activating it for H-atom abstraction. Alternatively for HPPD, the decarboxylated substrate-intermediate bound to the Fe center would have the occupied p π orbital of its ring oriented for σ* FMO attack by the oxo character in the dz2 orbital. It is also important to note that, as with the hydroperoxide FMOs at the O–O elongated transition state in Figure 10, elongation of the Fe–O bond of the FeIV=O S = 2 unit at the transition state also leads to strong spin polarization such that the unoccupied FMO greatly increases in oxo character (Figure 21), leading to a species best described as an FeIII–O•−. This high oxo character in a low-lying FMO increases electrophilicity and would make a major contribution to reactivity.
Scheme 4.
Reactivity differences of HPPD and HmaS for the same substrate.
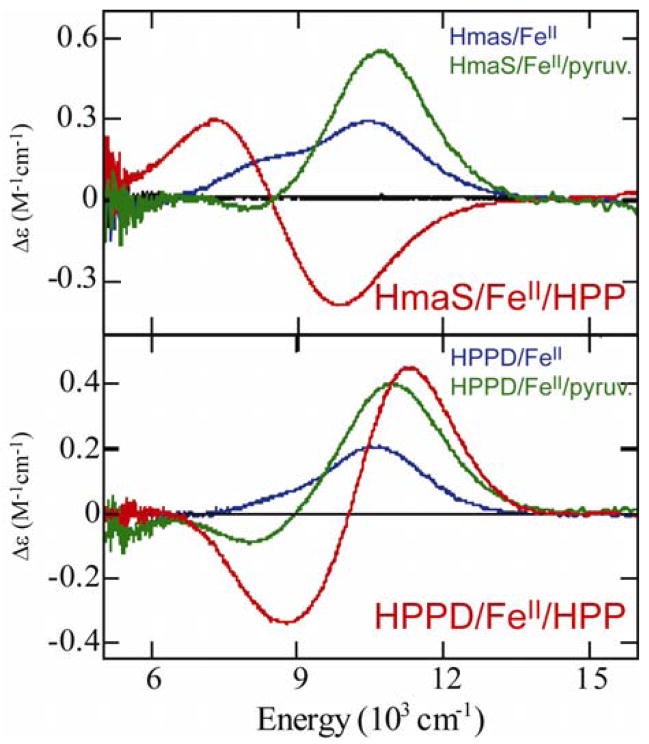
Figure 19.
298K CD spectra of HPPD and HmaS and HPPD coordinated with pyruvate (substrate analog) and HPP (natural substrate.)
Figure 20.
Nature of FMOs at the transition state along the reaction pathways of HPPD and HmaS.
Figure 21.
Activation of FeIV=O S = 2 species at the transition state: radical character of elongated FeIII–O•− bond increases electrophilicity.
Concluding comments
The reactivity of the low-spin FeIII–hydroperoxide in a non-heme relative to a heme environment is different and this difference is reasonably well-understood. Alternatively for the Rieske dioxygenases, the active species appears to involve a high-spin non-heme FeIII–peroxy complex; the nature of this complex and whether it directly reacts with the substrate or undergoes heterolytic cleavage to produce an (HO)FeV=O species which then reacts with substrate is an open issue. For the FeIV=O S = 2 enzyme intermediates, the abundance of computational data overshadows the paucity of experimental data, and it is important to generate data comparable to those for the FeIV=O S = 1 species examined here, to experimentally correlate the nature of this species with its electronic structure contributions to reactivity.
Acknowledgments
E.I.S. thanks Prof. Lawrence Que, Jr. for FeIV=O model complexes which were analyzed by MCD and NRVS spectroscopies and the National Institute of Health (GM40392) for research funding.
References
- 1.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem Rev. 2000;100:235–349. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 2.Neidig ML, Solomon EI. Structure-function correlations in oxygen activating non-heme iron enzymes. Chem Comm. 2005:5843–5863. doi: 10.1039/b510233m. [DOI] [PubMed] [Google Scholar]
- 3.Price JC, Barr EW, Tirupati B, Bollinger JM, Krebs C. The first direct characterization of a high-valent iron intermediate in the reaction of an α-ketoglutarate-dependent dioxygenase: A high-spin Fe(IV) complex in taurine/α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochem. 2003;42:7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 4.Riggs-Gelasco PJ, Price JC, Guyer RB, Brehm JH, Barr EW, Bollinger JM, Krebs C. EXAFS Spectroscopic Evidence for an Fe=O Unit in the Fe(IV) Intermediate Observed during Oxygen Activation by Taurine: α-Ketoglutarate Dioxygenase. J Am Chem Soc. 2004;126:8108–8109. doi: 10.1021/ja048255q. [DOI] [PubMed] [Google Scholar]
- 5.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eser BE, Barr EW, Frantom PA, Saleh L, Bollinger JM, Krebs C, Fitzpatrick PF. Direct Spectroscopic Evidence for a High-Spin Fe(IV) Intermediate in Tyrosine Hydroxylase. J Am Chem Soc. 2007;129:11334–11335. doi: 10.1021/ja074446s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galonic Fujimori DP, Barr EW, Walsh CT, Bollinger JM, Krebs C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat Chem Biol. 2007;3:113–116. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- *8.Galonic Fujimori D, Barr EW, Matthews ML, Koch GM, Yonce JR, Walsh CT, Bollinger JM, Krebs C, Riggs-Gelasco PJ. Spectroscopic Evidence for a High-Spin Br-Fe(IV)-Oxo Intermediate in the α-Ketoglutarate-Dependent Halogenase CytC3 from Streptomyces. J Am Chem Soc. 2007;129:13408–13409. doi: 10.1021/ja076454e. The most recently-characterized mononuclear non-heme FeIV=O S = 2 intermediate is shown to have the ability to activate an inert C-H bond and replace the H with either Cl or Br, via a proposed H-atom abstraction mechanism. [DOI] [PubMed] [Google Scholar]
- *9.Neidig ML, Decker A, Choroba OW, Huang F, Kavana M, Moran GR, Spencer JB, Solomon EI. Spectroscopic and electronic structure studies of aromatic electrophilic attack and hydrogen-atom abstraction by non-heme iron enzymes. Proc Natl Acad Sci USA. 2006;103:12966–12973. doi: 10.1073/pnas.0605067103. This study uses DFT in combination with MCD spectroscopy to examine the reactivities of the α-keto-acid-dependent enzymes HPPD and HmaS, the FeIV=O intermediates of which perform two different organic transformations (aromatic electrophilic attack and H-atom abstraction, respectively) on the same substrate. The different geometries of their transition states determine the key FMOs involved in reactivity. However, the activation mechanism is the same in both cases: elongation of the FeIV=O bond results in the development of FeIII–O•− character, increasing the electrophilicity of the oxygen moiety and thus its reactivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worth L, Frank BL, Christner DF, Absalon MJ, Stubbe J, Kozarich JW. Isotope Effects on the Cleavage of DNA by Bleomycin - Mechanism and Modulation. Biochem. 1993;32:2601–2609. doi: 10.1021/bi00061a018. [DOI] [PubMed] [Google Scholar]
- 11.Hecht SM. Bleomycin: New perspectives on the mechanism of action. J Nat Prod. 2000;63:158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 12.Burger RM. Cleavage of nucleic acids by bleomycin. Chem Rev. 1998;98:1153–1169. doi: 10.1021/cr960438a. [DOI] [PubMed] [Google Scholar]
- 13.Petering DH, Mao QK, Li WB, DeRose E, Antholine WE. Metallobleomycin-DNA interactions: Structures and reactions related to bleomycin-induced DNA damage. Metal Ions Biol Syst. 1996;33:619–648. [PubMed] [Google Scholar]
- 14.Stubbe J, Kozarich JW, Wu W, Vanderwall DE. Bleomycins: A structural model for specificity, binding, and double strand cleavage. Acc Chem Res. 1996;29:322–330. [Google Scholar]
- 15.Wu JC, Kozarich JW, Stubbe J. Mechanism of Bleomycin - Evidence for a Rate-Determining 4′-Hydrogen Abstraction from Poly(Da-Du) Associated with the Formation of Both Free Base and Base Propenyl. Biochem. 1985;24:7562–7568. doi: 10.1021/bi00347a009. [DOI] [PubMed] [Google Scholar]
- 16.Burger RM. Nature of Activated Bleomycin. Struct Bonding. 2000;97:287–303. [Google Scholar]
- 17.Burger RM, Peisach J, Horwitz SB. Activated Bleomycin - a Transient Complex of Drug, Iron, and Oxygen That Degrades DNA. J Biol Chem. 1981;256:1636–1644. [PubMed] [Google Scholar]
- 18.Sugiura Y. Bleomycin-Iron Complexes - Electron-Spin Resonance Study, Ligand Effect, and Implication for Action Mechanism. J Am Chem Soc. 1980;102:5208–5215. [Google Scholar]
- 19.Veselov A, Sun HJ, Sienkiewicz A, Taylor H, Burger RM, Scholes CP. Iron Coordination of Activated Bleomycin Probed by Q- and X-Band ENDOR - Hyperfine Coupling to Activated 17O Oxygen, 14N and Exchangeable 1H. J Am Chem Soc. 1995;117:7508–7512. [Google Scholar]
- 20.Burger RM, Kent TA, Horwitz SB, Münck E, Peisach J. Mössbauer Study of Iron Belomycin and Its Activation Intermediates. J Biol Chem. 1983;258:1559–1564. [PubMed] [Google Scholar]
- 21.Veselov A, Burger RM, Scholes CP. Q-band electron nuclear double resonance of ferric bleomycin and activated bleomycin complexes with DNA: Fe(III) hyperfine interaction with P-31 and DNA-induced perturbation to bleomycin structure. J Am Chem Soc. 1998;120:1030–1033. [Google Scholar]
- 22.Sam JW, Tang X-J, Peisach J. Electrospray Mass Spectrometry of Iron Bleomycin: Demonstration That Activated Bleomycin Is a Ferric Peroxide Complex. J Am Chem Soc. 1994;116:3250–5256. [Google Scholar]
- 23.Westre TE, Loeb KE, Zaleski JM, Hedman B, Hodgson KO, Solomon EI. Determination of the Geometric and Electronic Structure of Activated Bleomycin Using X-Ray-Absorption Spectroscopy. J Am Chem Soc. 1995;117:1309–1313. [Google Scholar]
- 24.Neese F, Zaleski JM, Loeb Zaleski K, Solomon EI. Electronic Structure of Activated Bleomycin: Oxygen Intermediates in Heme versus Non-Heme Iron. J Am Chem Soc. 2000;122:11703–11724. [Google Scholar]
- 25.Kovaleva EG, Lipscomb JD. Crystal Structures of Fe2+ Dioxygenase Superoxo, Alkylperoxo, and Bound Product Intermediates. Science. 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S. Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron. Science. 2003;299:1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 27.Pau MYM, Lipscomb JD, Solomon EI. Substrate activation for O2 reactions by oxidized metal centers in biology. Proc Natl Acad Sci USA. 2007;104:18355–18362. doi: 10.1073/pnas.0704191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon EI, Decker A, Lehnert N. Non-heme iron enzymes: Contrasts to heme catalysis. Proc Natl Acad Sci USA. 2003;100:3589–3594. doi: 10.1073/pnas.0336792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheesman M, Greenwood C, Thomson A. Magnetic Circular Dichroism of Hemoproteins. Adv Inorg Chem. 1991;36:201–255. [Google Scholar]
- 30.Lehnert N, Ho RYN, Que L, Jr, Solomon EI. Electronic Structure of High-Spin Iron(III)-Alkylperoxo Complexes and Its Relation to Low-Spin Analogues: Reaction Coordinate of O-O Bond Homolysis. J Am Chem Soc. 2001;123:12802–12816. doi: 10.1021/ja011450+. [DOI] [PubMed] [Google Scholar]
- 31.Lehnert N, Ho RYN, Que L, Jr, Solomon EI. Spectroscopic Properties and Electronic Structure of Low-Spin Fe(III)-Alkylperoxo Complexes: Homolytic Cleavage of the O-O Bond. J Am Chem Soc. 2001;123:8271–8290. doi: 10.1021/ja010165n. [DOI] [PubMed] [Google Scholar]
- 32.Lehnert N, Ho RYN, Neese F, Que L, Jr, Solomon EI. Electronic Structure and Reactivity of Low-spin Fe(III)-Hydroperoxo Complexes: Comparison to Activated Bleomycin. J Am Chem Soc. 2002;124:10810–10822. doi: 10.1021/ja012621d. [DOI] [PubMed] [Google Scholar]
- *33.Decker A, Chow MS, Kemsley JN, Lehnert N, Solomon EI. Direct Hydrogen-Atom Abstraction by Activated Bleomycin: An Experimental and Computational Study. J Am Chem Soc. 2006;128:4719–4733. doi: 10.1021/ja057378n. This paper presents comprehensive kinetic studies and a computational evaluation of the two-dimensional potential energy surface of ABLM reacting with DNA. These studies confirm that ABLM is thermodynamically and kinetically competent for direct H-atom abstraction, and that this route is favored over homolytic and heterolytic O–O cleavage. Furthermore, the resulting FeIV=O species is capable of second-strand DNA cleavage, providing a reasonable mechanism for DNA cleavage by ABLM. [DOI] [PubMed] [Google Scholar]
- *34.Kumar D, Hirao H, Shaik S, Kozlowski PM. Proton-Shuffle Mechanism of O-O Activation for Formation of a High-Valent Oxo-Iron Species of Bleomycin. J Am Chem Soc. 2006;128:16148–16158. doi: 10.1021/ja064611o. Based on DFT calculations, this paper proposes a heterolytic cleavage pathway for ABLM involving a proton shuffle mechanism, generating a Compound-I-like intermediate, (BLM•)FeIV=O. See however Reference 35. [DOI] [PubMed] [Google Scholar]
- *35.Chow MS, Liu LV, Solomon EI. Further insights into the mechanism of the reaction of activated bleomycin with DNA. Proc Natl Acad Sci USA. 2008;105:13241–13245. doi: 10.1073/pnas.0806378105. CD spectroscopy is used to directly monitor the kinetics of ABLM decay with and without DNA present, and the accelerated rate of the ABLM+DNA reaction establishes the DNA-dependence of ABLM decay. It is thus experimentally determined that DNA reacts directly with ABLM and not its heterolytic decay product. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde J-U, In J-H, Lim MH, Brennessel WW, Bukowski MR, Stubna A, Munck E, Nam W, Que L., Jr Crystallographic and spectroscopic characterization of a nonheme Fe(IV)=O complex. Science. 2003;299:1037–1039. doi: 10.1126/science.299.5609.1037. [DOI] [PubMed] [Google Scholar]
- 37.Kaizer J, Klinker EJ, Oh NY, Rohde J-U, Song WJ, Stubna A, Kim J, Munck E, Nam W, Que L. Nonheme FeIVO Complexes That Can Oxidize the C-H Bonds of Cyclohexane at Room Temperature. J Am Chem Soc. 2004;126:472–473. doi: 10.1021/ja037288n. [DOI] [PubMed] [Google Scholar]
- 38.Klinker E, Kaizer J, Brennessel W, Woodrum N, Cramer C, Que L., Jr Structures of Nonheme Oxoiron(IV) Complexes from X-ray Crystallography, NMR Spectroscopy, and DFT Calculations. Angew Chem Int Ed. 2005;44:3690–3694. doi: 10.1002/anie.200500485. [DOI] [PubMed] [Google Scholar]
- 39.Hirao H, Que L, Jr, Nam W, Shaik S. A Two-State Reactivity Rationale for Counterintuitive Axial Ligand Effects on the C-H Activation Reactivity of Nonheme FeIV=O Oxidants. Chem Eur J. 2008;14:1740–1756. doi: 10.1002/chem.200701739. [DOI] [PubMed] [Google Scholar]
- 40.Hirao H, Kumar D, Que JL, Shaik S. Two-State Reactivity in Alkane Hydroxylation by Non-Heme Iron-Oxo Complexes. J Am Chem Soc. 2006;128:8590–8606. doi: 10.1021/ja061609o. [DOI] [PubMed] [Google Scholar]
- *41.Sastri CV, Lee J, Oh K, Lee YJ, Lee J, Jackson TA, Ray K, Hirao H, Shin W, Halfen JA, et al. Axial ligand tuning of a nonheme iron(IV)-oxo unit for hydrogen atom abstraction. Proc Natl Acad Sci USA. 2007;104:19181–19186. doi: 10.1073/pnas.0709471104. This study on the reactivity of (TMC)FeIV=O with a series of different axial ligands demonstrates that the more electron-donating the axial ligand, the slower the rate of oxo-atom transfer, following the electrophilicity trend. For H-atom abstraction however, the trend is opposite; it is postulated that the quintet excited state participates in this reaction pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Decker A, Rohde JU, Que L, Jr, Solomon EI. Spectroscopic and Quantum Chemical Characterization of the Electronic Structure and Bonding in a Non-Heme FeIV=O Complex. J Am Chem Soc. 2004;126:5378–5379. doi: 10.1021/ja0498033. [DOI] [PubMed] [Google Scholar]
- *43.Decker A, Rohde JU, Klinker EJ, Wong SD, Que L, Jr, Solomon EI. Spectroscopic and Quantum Chemical Studies on Low-Spin FeIV=O Complexes: Fe-O Bonding and Its Contributions to Reactivity. J Am Chem Soc. 2007;129:15983–15996. doi: 10.1021/ja074900s. MCD and DFT analyses elucidate the nature of iron–oxo bonding in S = 1 FeIV=O model complexes and show that a stronger equatorial ligand field leads to stronger iron–oxo π-bonding interactions and consequently greater oxo contribution to the reactive π* FMOs, activating the FeIV=O unit for H-atom abstraction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rüffer R, Chumakov AI. Nuclear inelastic scattering. Hyperfine Interact. 2000;128:255–272. [Google Scholar]
- 45.Scheidt WR, Durbin SM, Sage JT. Nuclear resonance vibrational spectroscopy - NRVS. J Inorg Biochem. 2005;99:60–71. doi: 10.1016/j.jinorgbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Sturhahn W. Nuclear resonant spectroscopy. J Phys Condens Matt. 2004;16:S497–S530. [Google Scholar]
- *47.Bell CB, III, Wong SD, Xiao Y, Klinker EJ, Tenderholt AL, Smith MC, Rohde J-U, Que L, Jr, Cramer SP, Solomon EI. A Combined NRVS and DFT Study of FeIV=O Model Complexes: a Diagnostic Method for the Elucidation of Non-Heme Iron Enzyme Intermediates. Angew Chem Int Ed (Early View) 2008 doi: 10.1002/anie.200803740. Combined Nuclear Resonance Vibrational Spectroscopy (NRVS) and DFT studies confirm that the equatorial nitrogens of (N4Py)FeIV=O have stronger σ-bonding interactions with Fe. In addition, it is shown that the peaks with the highest intensities in NRVS result from the O=FeIV-Nax bending modes, allowing the prediction of the geometry of high-spin FeIV=O enzyme intermediates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *48.Decker A, Clay MD, Solomon EI. Spectroscopy and electronic structures of mono- and binuclear high-valent non-heme iron-oxo systems. J Inorg Biochem. 2006;100:697–706. doi: 10.1016/j.jinorgbio.2006.01.013. A theoretical comparison of non-heme FeIV=O species demonstrates that going from the S = 1 to S = 2 spin state does not change the Fe–O bonding interactions but spin-polarizes the α dz2 FMO, allowing it to participate in the reactivity of the FeIV=O unit (see for example Reference 9.) [DOI] [PubMed] [Google Scholar]