Abstract
For efficient ligand binding, integrins must be activated. Specifically, a conformational change has been proposed in a ligand binding domain present within some integrins, the inserted (I) domain [Lee, J., Bankston, L., Arnaout, M. & Liddington, R. C. (1995) Structure (London) 3, 1333–1340]. This proposal remains controversial, however, despite extensive crystal structure studies on the I domain [Lee, J., Bankston, L., Arnaout, M. & Liddington, R. C. (1995) Structure (London) 3, 1333–1340; Liddington, R. & Bankston, L. (1998) Structure (London) 6, 937–938; Qu, A. & Leahy, D. J. (1996) Structure (London) 4, 931–942; and Baldwin, E. T., Sarver, R. W., Bryant, G. L., Jr., Curry, K. A., Fairbanks, M. B., Finzel, B. C., Garlick, R. L., Heinrikson, R. L., Horton, N. C. & Kelly, L. L. (1998) Structure (London) 6, 923–935]. By defining the residues present in the epitope of a mAb against the human Mac-1 integrin (αMβ2, CD11b/CD18) that binds only the active receptor, we provide biochemical evidence that the I domain itself undergoes a conformational change with activation. This mAb, CBRM1/5, binds the I domain very close to the ligand binding site in a region that is widely exposed regardless of activation as judged by reactivity with other antibodies. The conformation of the epitope differs in two crystal forms of the I domain, previously suggested to represent active and inactive receptor. Our data suggests that conformational differences in the I domain are physiologically relevant and not merely a consequence of different crystal lattice interactions. We also demonstrate that the transition between the two conformational states depends on species-specific residues at the bottom of the I domain, which are proposed to be in an interface with another integrin domain, and that this transition correlates with functional activity.
Integrins are heterodimeric adhesion receptors present on cells of multicellular organisms and play key roles in multiple cellular processes. They are type I membrane glycoproteins with short cytoplasmic tails (1, 2). The α-subunit of ≈1,000 aa has seven 60-aa repeats recently predicted to fold into a seven-bladed β-propeller (3). Some α-subunits contain a structurally characterized inserted (I) domain of 200 residues (4–9) that is predicted to be inserted between β-sheets 2 and 3 of the β-propeller (3). The extracellular domain of the β-subunit contains ≈700 residues and includes a conserved domain of 250 residues that may be similar in structure to the I domain (7, 10).
Increased affinity of integrins for ligand can be activated by “inside-out” signals relayed from the cytoplasm; in “outside-in” signaling, ligand binding by integrins transduces signals into the cell (11–13). Earlier experiments have demonstrated gross conformational changes of the extracellular portion of integrins by using fluorescence resonance energy transfer (14), change in protease sensitivity (15), and electron microscopy (16).
In integrins in which it is present, including the leukocyte integrins such as Mac-1 (αMβ2, CD11b/CD8) and LFA-1 (αLβ2, CD11a/CD18), the I domain is important in ligand binding (17, 18). The isolated, recombinant Mac-1 I domain has been shown to interact with ligands (19, 20), and mAbs directed to the I domain can abolish ligand binding to intact integrins (17). Most likely, other domains of both the α-subunit and the β-subunit cooperate to create the complete binding face (21–23). In integrins lacking I domains, the upper face of the putative β-propeller has been shown to be important in ligand binding (3, 24).
Crystal structures of I domains (4–9) reveal a dinucleotide-binding fold, with a metal ion-dependent adhesion site (MIDAS) on the “top” face, that is opposite to the bottom face that connects to the β-propeller domain. The metal ion is believed to ligate directly to an acidic residue in the ligand that completes the metal coordination sphere. Two different crystal forms of the Mac-1 I domain are hypothesized to represent the I domain in active and inactive conformations (4, 25). In both structures, a divalent cation forms a total of five coordinations to the I domain, three of which are direct coordinations; however, the residues that directly coordinate differ, and other nearby residues shift in position. In the putative low-affinity structure, crystallized with Mn2+, one of the three direct coordinations is to an Asp residue, and the sixth coordination is to water. In the putative high-affinity structure, crystallized with Mg2+, none of the three direct coordinations is to a charged residue, but a sixth, direct coordination forms to a Glu in the C-terminal α-helix of a neighboring I domain in the crystal lattice. This C-terminal α-helix undergoes a large 10-Å movement, and it is unclear whether this movement is more closely structurally linked to the rearranged metal coordination in the MIDAS or to the lattice contact between the Mg2+ and Glu of neighboring I domains. Intriguingly, the structurally homologous G protein α subunit undergoes a similar change in metal coordination between the GDP- and GTP-bound forms, which is coupled to long-range structural rearrangements (4). In other studies on LFA-1 and Mac-1 I domains, no significant differences were found between structures in the presence of Mg2+ or Mn2+ or in the absence of bound cation (5, 6). It has been argued that the conformational change, observed in only one of many published structures, is a crystal lattice artifact and is not physiologically relevant (6).
Several activation-dependent mAbs to both α- and β-subunits of integrins are known (26). One such mAb, CBRM1/5 (27), recognizes the I domain of the integrin Mac-1 and is unique in that mAb binding not only depends on activation but also blocks ligand binding. CBRM1/5 mAb saturably binds to only 10 or 30% of total Mac-1 molecules on the surface of neutrophils activated by chemoattractants or phorbol esters, respectively, yet almost completely blocks Mac-1-dependent adhesion to different ligands (27). A priori, the Mac-1 epitope could be exposed as a result of global interdomain rearrangements, or it could be formed as a result of intradomain tertiary conformational changes of the I domain, which could be linked to interdomain, quaternary changes. To what extent such mechanisms operate in integrins currently is not known. Here, using CBRM1/5 and other antibodies that have closely overlapping epitopes, we demonstrate a conformational change in the Mac-1 I domain close to the putative ligand binding site, only three residues from the MIDAS. The epitope of CBRM1/5 is in a region in which major differences are seen between the two crystal forms of the Mac-1 I domain. Further, we show that several species-specific residues in the I domain, near its interface with the β-propeller domain, are important in regulation of integrin activity.
MATERIALS AND METHODS
DNA Constructs and Mutagenesis.
Human αM- and β2-subunit cDNAs were in pCDNA3.1+ as described (28). Mutagenesis of the α-subunit was carried out by overlap extension PCR (29). In brief, outer primers were 5′-TGGTGGAATTCTGCAGATATCCAGC-3′ (5′ end) and 5′-CTGTCTGCGTGTGCTGTTCTTTGTCTC-3′ (3′ end), and internal primers (GIBCO/BRL) with an overlap of ≈22 base pairs were used to generate mutated fragments that were digested with either NotI and BspEI (mutants hm131–136 to hm178) or BspEI and BbsI (mutants hm187–194 to hm313) and were swapped into the wild-type construct. All DNA amplification was with Pfu DNA polymerase (Stratagene), and the final constructs were verified by sequence analysis. Plasmid DNA for transfection was prepared by QIAprep Spin Kit or Maxi Kit (Qiagen, Chatsworth, CA).
Tissue Culture and Transfection.
293T cells (30) were maintained in high glucose DMEM supplemented with 10% fetal bovine serum (JRH Biosciences, Lenexa, KS), 2 mM glutamine (Sigma, nonessential amino acids (GIBCO/BRL), and gentamicin (Sigma). Cells were plated onto 6-cm tissue culture dishes and 18 h later were transfected by calcium phosphate coprecipitation (31) by using 12 μg of α-subunit and 8 μg of β-subunit cDNA. After a further 48 h, transfected cells were detached with 5 mM EDTA in PBS. Approximately 75% of the cells expressed Mac-1 as shown by flow cytometry. For generating stable K562 cell lines that express Mac-1, the wild-type or mutant αM cDNA was subcloned into the XbaI site of pEFpuro (32). One microgram of SspI-linearized pEFpuro containing the αM cDNA was cotransfected with 20 μg of the SfiI-linearized wild-type β2 cDNA contained in plasmid AprM8 by electroporation as described (33). Transfectants were selected for resistance to 4 μg/ml puromycin and were subcloned further by limiting dilution. Clones expressing similar levels of surface Mac-1 were used in binding assays. Stable cell lines were maintained in RPMI medium 1640 (GIBCO/BRL) supplemented with 10% fetal bovine serum and 4 μg/ml puromycin (Sigma).
Analysis of Binding to mAbs.
mAbs to the Mac-1 α-chain I domain were CBRM1/5 (27), LPM19c (Dako), CBRM1/1, CBRM1/2, CBRM1/4, CBRM1/13, CBRM1/21, CBRM1/22, CBRM1/24, CBRM1/27, CBRM1/29, CBRM1/31, CBRM1/33, CBRM1/34, LM2/1, OKM9, TGM-65, and 5A4.C5, as described in ref. 17 or references therein. To monitor expression, mAbs CBRM1/10 and CBRM1/23 to the C-terminal region and mAbs CBRM1/20 and CBRM1/32 to the putative β-propeller domain (17, 28, 34) were used. The activation-dependent mAb M24 (35) and CBRM1/19 (27) also were used. The X63 myeloma IgG1 was used as control. Antibodies were diluted from a 1 mg/ml stock in PBS to 5 μg/ml, except for mAb M24, which was diluted to 10 μg/ml or was diluted 1:200 from ascites. Cells were incubated for 40 minutes in 96-well plates with each mAb and then, after washing twice, for 30 minutes with fluorescein-isothiocyanate-goat anti-mouse IgG (Zymed) diluted 1:50 followed by three washes. All washing and incubation was in L-15 medium with 2% fetal bovine serum (Sigma) at 4°C, except for incubation with M24, which was done at 37°C (35). Cells were fixed with 1% formaldehyde before flow cytometry for measurement of fluorescence intensity. At least 2,500 cells were acquired on a FACScan instrument (Becton–Dickinson).
Binding of Transfectants to Immobilized Ligand.
Human complement component iC3b (Calbiochem) [50 μl/well at 5 or 10 μg/ml in 20 mM Tris, 150 mM NaCl, and 2 mM MgCl2 (pH 9)] was coated in flat-bottomed 96-well plates (ICN) overnight at 4°C and was blocked further with 0.05% polyvinylpyrrolidone (Sigma) at 37°C for 1 h. Cells were labeled at 37°C for 15 minutes with 2′,7′-bis-(carboxyethyl)-5-(and -6)-carboxyfluorescein, acetoxymethyl ester (Molecular Probes) in L-15 medium and were washed and kept at room temperature thereafter. Fifty-thousand cells were pipetted into washed, ligand-coated wells and were spun to the bottom. After incubation for 50 minutes at 37°C, unbound cells were washed off on a microplate autowasher (Bio-Tek, Burlington, VT) with four washes of 250-ml dispense volume and 100-ml volume remaining after aspiration. The pressure applied to the autowasher was 2 psi, and the wash buffer was Hank’s balanced salt solution. The fluorescence of input cells and of cells remaining after each wash was measured with a fluorescence concentration analyzer (Pandex, Idexx Laboratories, Westbrook, ME). Less than 2% of nontransfected cells bound after the final wash.
RESULTS
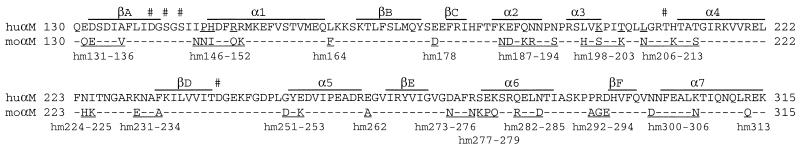
The epitope for the mouse anti-human Mac-1 mAb CBRM1/5 was localized by using 293T cells transfected with human β2 and αM integrin cDNA, in which short segments within the I domain of the αM subunit were substituted with the murine sequence (Fig. 1). Seventeen different chimeras were expressed at wild-type levels, as shown with mAbs that react outside the I domain; however, CBRM1/5 mAb failed to bind to three of the mutants, hm146–152, hm198–203, and hm206–213 (Table 1). The individual amino acid substitutions recognized by CBRM1/5 mAb were defined by using 11 mutants carrying the corresponding single amino acid changes (Table 2). CBRM1/5 is specific for Pro-147, His-148, and Arg-151 in the loop before and at the beginning of α-helix 1 and for Lys-200, Thr-203, and Leu-206 in the loop connecting α-helices 3 and 4 (Fig. 1 and Table 2). Although separated by up to 55 residues in sequence, these six residues are adjacent in the structure (4, 6, 7) (Figs. 2 and 3). All six residues have side chains that are well exposed to solvent (Fig. 3). The residues are present within an area of 17 × 13 Å, which is well within the area typically contacted by an antibody of 600–800 Å2 (36). One of the 11 mutants, I146N, showed increased binding of CBRM1/5 compared with the wild type (Table 2), as discussed below.
Figure 1.
Mouse–human substitutions in the I domain (40) and murine αM (41) I domain sequences are aligned, together with the secondary structure (7). Residue numbers are for the mature proteins. The mouse sequence is only shown when different from the human. Human residues participating in the CBRM1/5 epitope are underlined. Residues engaged in metal coordination are hash-marked (7). The 17 chimeras are shown, with the incorporated mouse sequence underlined.
Table 1.
Antibody staining of human–mouse chimeric αM subunits expressed with human β2 in 293 cells*
| αM variant | CBRM1/5 | CBRM1/21 | CBRM1/22 | CBRM1/1 | CBRM1/34 | CBRM1/2 | 5A4.C5 | CBRM1/29 | OKM9 | CBRM1/27 | CBRM1/31 | CBRM1/4 | TMG-65 | CBRM1/13 | LM2/1 | CBRM1/24 | CBRM1/33 | LPM19c |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mock | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Wild-type | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm131† | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm146 | − | − | − | − | + | − | − | − | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ |
| hm164 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm178 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm187 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm198 | − | − | − | ++ | + | ++++ | ++++ | ++++ | − | + | + | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm206 | − | − | − | ++++ | ++++ | ++++ | ++ | +++ | ++++ | ++++ | +++ | ++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ |
| hm224 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm231 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm251 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | + | + | ++++ | ++++ | +++ | ++++ | ++++ |
| hm262 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm273 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| hm277 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | − | + | ++++ | ++++ | ++++ | ++++ |
| hm282 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | + | + | ++++ | ++++ | ++++ |
| hm292 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | + | − | ++++ | ++++ | ++++ |
| hm300 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| hm313 | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
Fluorescent intensity of transiently transfected 293T cells was normalized to staining with a mAb that was unaffected by the particular mutation, such as LPM19C: ++++, no effect; +++, >60% of wild type; ++, 20–60% of wild type; +, <20% of wild type; −, indistinguishable from staining of mock transfectant. All mAbs are specific for the I domain (17).
The name of each mutant is truncated to show the first altered residue; see Fig. 1 for full names and the identity of substituted residues.
Table 2.
Fine mapping of the CBRM1/5 epitope*
| Mutation | Binding, percent of human wild type ± SD
|
||
|---|---|---|---|
| CBRM1/5 | CBRM1/21 | CBRM1/22 | |
| I146N | 296 ± 5 | 106 ± 14 | 102 ± 8 |
| P147N | 2 ± 3 | 1 ± 1 | 1 ± 1 |
| H148I | 14 ± 2 | 76 ± 9 | 72 ± 4 |
| R151Q | 2 ± 3 | 76 ± 9 | 72 ± 4 |
| R152K | 91 ± 8 | 99 ± 7 | 97 ± 5 |
| L198H | 90 ± 4 | 95 ± 6 | 98 ± 13 |
| K200S | 28 ± 5 | 107 ± 9 | 97 ± 10 |
| T203K | 1 ± 1 | 8 ± 2 | 11 ± 4 |
| L206N | 0 ± 0 | 0 ± 1 | 0 ± 0 |
| H210K | 145 ± 17 | 99 ± 14 | 103 ± 4 |
| T213S | 98 ± 3 | 103 ± 6 | 99 ± 2 |
Transiently transfected 293T cells were stained with mAb as in Table 1; binding of each mAb is expressed as the percent of specifically stained cells relative to staining by the same mAb of wild-type Mac-1. All mutants were expressed equally well to the wild type.
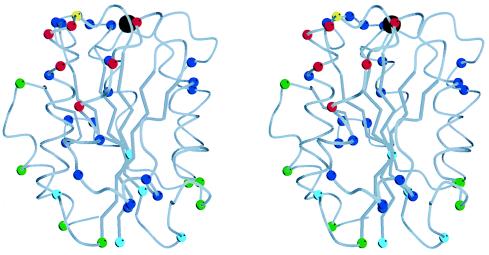
Figure 2.
Stereo view of the Mac-1 I domain (7). In the Cα trace, the Cα atoms of all residues that differ between mouse and human are shown as color-coded spheres. The black sphere at the top is the magnesium ion. The α3 helix is at front center, α-helix 1 is to its left, and the C-terminal α-helix 7 is at the far, lower left, behind α1. The Cα atoms of residues in the CBRM1/5 epitope, as defined by single amino acid substitutions, are red. The Cα of residue I146, which increases CBRM1/5 binding and decreases iC3b binding when substituted, is yellow. The remaining residues in the three human–mouse chimeras that affected CBRM1/5 binding are blue. Residues in the other 14 chimeras are color-coded according to overall effect of the substitutions: blue, decreased binding of one or more mAb and no effect on CBRM1/5 binding; green, no effect on any mAb; and turquoise, enhanced binding of CBRM1/5 mAb and no decrease in binding of any mAb. The figure was made with molscript (42).
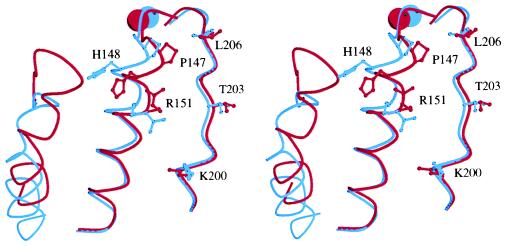
Figure 3.
Stereo view of the CBRM1/5 epitope. The structures of the liganded, Mg2+-bound (7) and nonliganded, Mn2+-bound (4) Mac-1 I domains were superimposed by using Cα atoms of residues 132–140, 167–241, and 245–265 (rms of 0.43 Å) (4). The metal ions and three segments of each structure are shown, with the liganded, putative high-affinity structure in blue and nonliganded, putative low-affinity structure in red. The three segments are left, residues 296–315, including α-helix 7; center, residues 143–163, including α-helix 1; and right, residues 197–209, including α-helix 3. Sidechains of residues in the CBRM1/5 epitope are shown as ball and stick. Metal ions are the spheres at top. The distances between the two structures for N of Pro-147, ND1 of His-148, and NH1 of Arg-151 are 2.6, 4.8, and 5.7 Å, respectively. The figure was made with molscript (42).
Lack of CBRM 1/5 binding to mutants suggests that the residues substituted are the antigenic residues of the epitope. An alternative possibility would be an inability of Mac-1 to assume the active conformation. This possibility can be excluded because the mutants hm146–152, hm198–203, and hm206–213, which abolish CBRM1/5 binding, all reacted as well as the wild type with two other activation-dependent antibodies, M24 (35) and CBRM1/19 (27), and because all six mutants with the antigenic residues substituted one-by-one showed wild-type level binding to immobilized iC3b, a ligand for Mac-1 (data not shown). Therefore, we conclude that the CBRM1/5 binding site is described accurately by the six antigenic residues P147, H148, R151, K200, T203, and L206.
Two other antibodies, CBRM1/21 and CBRM1/22, did not bind mutants hm146–152, hm198–203, and hm206–213 (Table 1). Thus, their binding site must overlap, to a large extent, that of CBRM1/5, although, interestingly, these mAbs react equally well with active and inactive Mac-1 (17, 27). The set of mutants carrying single amino acid substitutions showed that CBRM1/21 and CBRM1/22 could be distinguished from CBRM1/5 (Table 2). Mutant R151Q abolishes CBRM1/5 binding but has very little effect on binding of CBRM1/21 and CBRM1/22. K200S only affects binding of CBRM1/5. Furthermore, the T203K substitution completely abolished CBRM1/5 but left significant albeit greatly reduced binding by CBRM1/21 and CBRM1/22. We conclude that the region of the I domain recognized by these antibodies must be accessible in the inactive as well as in the active state of Mac-1. There might be a shift in position of the I domain relative to other domains in the intact integrin on activation, but the results with CBRM1/21 and CBRM1/22 suggest that this is not the reason for the capability of CBRM1/5 to differentiate between the two states. Rather, the results suggest a change in conformation within the I domain itself.
In the two Mac-1 I domain structures hypothesized to represent active and inactive conformations (4), residues P147, H148, and R151 of the CBRM1/5 epitope occupy markedly different positions and differ in side-chain orientation (Fig. 3), despite low atomic mobilities. Furthermore, these residues are preceded immediately by three of the residues that coordinate the Mg2+ and form the DXSXS motif of the MIDAS, D140, S142, and S144. Thus, movements of P147, H148, and R151 that average 2.4 Å are tightly linked to the 2.0-Å movement of S144 of the MIDAS.
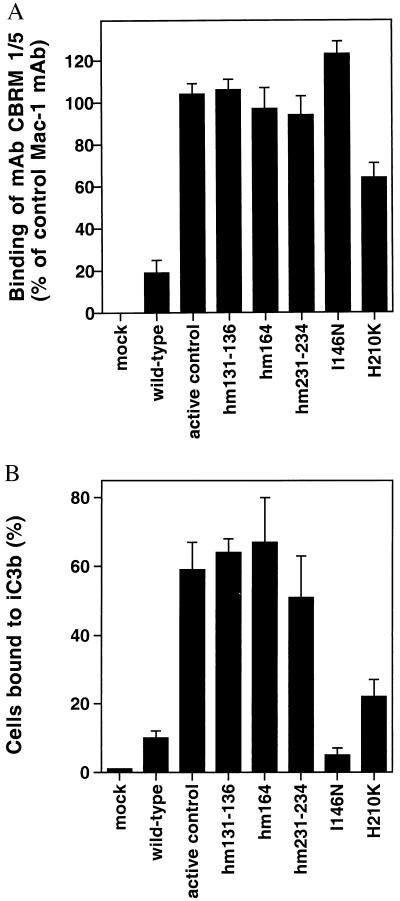
Several mouse–human substitutions activated expression of the CBRM1/5 epitope. Compared with mAbs that are not activation-dependent, CBRM1/5 reacts with ≈30% of Mac-1 molecules expressed on 293T cells (Table 1). Interestingly, the mutations hm131–136, hm164, and hm231–234 increased CBRM1/5 binding; binding was increased to the same level as for non-activation-dependent mAbs both in 293T cells (Table 1) and in K562 cells (Fig. 4A). None of the other 17 initial chimeras increased CBRM1/5 binding. However, when the individual substitutions were studied for the three chimeras that abolished CBRM1/5 binding, two further stimulatory mutations were found, I146N and H210K (Fig. 4A). Notably, all of the mutations that increase expression of CBRM1/5, except for I146N and H210K, mapped to the bottom of the I domain. Furthermore, all of these mutations, except for I146N, activated ligand binding by Mac-1 and resulted in a 2.5- to 6-fold enhancement of binding to the complement component iC3b (Fig. 4B). As a positive control, an activating deletion in the β2 cytoplasmic domain (C.L., unpublished results) resulted in a similar activation of CBRM1/5 mAb and iC3b binding (Fig. 4 A and B). All of these mutants, except for I146N, also increased binding of mAbs M24 and CBRM1/19 (data not shown), both of which bind outside the I domain (27, 35). The I146N mutation decreased binding to iC3b both in K562 cells (Fig. 4B) and in 293T cells in which wild-type Mac-1 was more active (data not shown). These results are consistent with previous observations that Mac-1 binding to iC3b contains a species-specific component (37). Interestingly, residue I146 is located in between the CBRM1/5 mAb epitope and the MIDAS (Fig. 2, yellow sphere) and is in a position in which it may contact ligand. By contrast, the substitutions that activate both CBRM1/5 and ligand binding cluster on the opposite, lower face of the I domain (Fig. 2, turquoise spheres).
Figure 4.
Increased binding of CBRM1/5 and ligand by I domain mutants. K562 cells were transfected stably with vector alone (mock), with wild-type or mutated αM-subunit and wild-type β2-subunit cDNA, or with wild-type αM-subunit and β2-subunit with an activating mutation in the intracellular domain (active control; C.L. and T.A.S., unpublished results). (A) CBRM1/5 binding. Cells were stained with mAbs and fluorescein isothiocyanate-conjugated anti-mouse IgG. Mean fluorescence intensity was calculated after subtraction of control staining with myeloma IgG, and CBRM1/5 staining is shown as a percent of staining with CBRM1/20 mAb to the β-propeller domain. (B) Binding to immobilized iC3b. Bound cells are shown as a percent of the number of input cells.
The location of species-specific residues recognized by the 18 mAbs studied here define surfaces of the I domain that are accessible in the intact integrin. These residues mapped to the upper face of the I domain and to two, opposite faces on the side of the I domain (Fig. 2, blue spheres). One of these faces contains α-helices 1, 2, and 3, and the other contains the top of α-helix 5, α-helix 6, and the prominent loop between α-helix 6 and β-strand 6. None of the activating substitutions on the lower face of the I domain (Fig. 2, turquoise spheres) involved residues that were recognized by any mAb. Other species-specific residues that were not recognized by mAb were confined to the lower face of the I domain and to the C-terminal α-helix 7 (Fig. 2, green spheres).
DISCUSSION
By mapping the epitope of the activation-dependent, function-blocking CBRM1/5 mAb, we provide evidence here for (i) a conformational change in an integrin I domain that is adjacent to and closely linked to the MIDAS and physiologically relevant to ligand binding by an intact integrin and (ii) a linkage between this conformational change and an interface on the lower face of the I domain. Our findings are relevant to a current controversy on whether two conformations observed in crystal structures of I domains are physiologically relevant or represent an artifact induced by crystal lattice interactions (4, 6, 25). The face of the I domain that displays the CBRM1/5 epitope is exposed on both active and inactive forms of Mac-1, as shown by binding of multiple mAb, and in particular CBRM1/21 and CBRM1/22. Thus, of two possible mechanisms for integrin activation, “unmasking” and “shape-shifting” (22), we have evidence for the latter.
We localized the CBRM1/5 epitope to three residues in each of two adjacent polypeptide segments on the I domain. One of these segments immediately follows the DXSXS motif of the MIDAS. When the two crystal forms of the Mac-1 I domain are compared by superposition, the three residues in this segment recognized by CBRM1/5 move 2.4 ± 0.5 Å, and this movement is closely linked by the polypeptide chain to the movement three residues away at S144, the last residue of the DXSXS motif. S144 donates a primary coordination to the metal of the MIDAS, and its 2-Å movement is thus closely linked to the rearrangement of several of the coordinations in the MIDAS between the two crystal forms (4).
Our data provide evidence for conformational change in a specific region of the I domain, adjacent to the MIDAS. Only one of many published I domain crystal structures displays the putative high-affinity conformation; therefore, doubts have been expressed about its physiologic relevance. However, it is highly significant that this is the only structure in which the sixth coordination position in the MIDAS is satisfied by a ligand-like coordination. The crystal studies to date are thus consistent with the hypothesis that fulfillment of the sixth coordination by a negatively charged oxygen increases the stability of the putative high-affinity structure relative to the putative low-affinity structure. This would be consistent with the proposed higher affinity of this structure for ligand, which is suggested by the finding that the only negatively charged oxygen donating a primary coordination comes from the putative ligand-binding position whereas, in the putative low-affinity structure, a negatively charged primary coordination is donated by an I domain residue deeply buried in the Mg2+-binding pocket (4).
Furthermore, our data may be relevant to whether the large movement of the C-terminal α-helix in Mac-1 (4) is physiologically important or instead is related to coordination of a Glu in this helix to the MIDAS in a neighboring I domain in the crystal lattice. Two different LFA-1 I domain structures differ in whether the C-terminal portion of the C-terminal α-helix packs onto the rest of the domain or not, suggesting that its association is loose (5). This movement represents a bend in the helix axis and is not linked to other conformational changes within the I domain. By contrast, in Mac-1 I domain structures, a large movement along the axis of the C-terminal α-helix is observed that is accompanied by a similar 10-Å downward movement of the loop between this α-helix and the preceding β-strand. This loop is adjacent to CBRM1/5 epitope residues H148 and R151 in the putative low-affinity state; its downward movement in the putative high-affinity state leaves H148 and R151 better exposed to solvent and, hence, to binding by CBRM1/5 (Fig. 3). Thus, CBRM1/5 may be recognizing shape-shifting in the C-terminal α-helix and its preceding loop as well as in the adjacent α-helix and loop that bear H148 and R151.
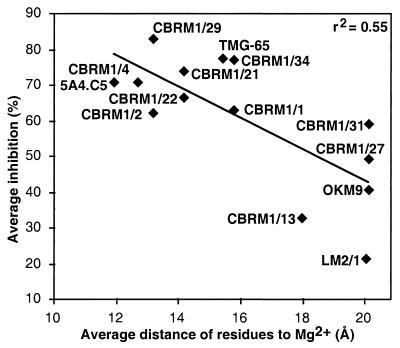
Localization of mAb epitopes supports the importance of the face bearing the MIDAS in ligand binding. The CBRM1/5 mAb epitope is close to the MIDAS, consistent with its ability to block ligand binding (27). The CBRM1/21 and CBRM1/22 mAb that bind an almost identical epitope are also strong function blockers (17). The majority of the mAbs studied here, including all of the CBRM1 series except for CBRM1/5, were selected independently of their ability to block function (27). We tested whether the proximity of mAb epitopes to the Mg2+ ion correlated with how well they blocked function, as determined by the average inhibition of binding to four different ligands (17). Indeed, there was an excellent correlation (Fig. 5), with r2 = 0.55 and a significance of P = 0.002, as determined with an F test. This provides objective support for the use of mAbs to define functionally important sites on proteins.
Figure 5.
Epitope localization on the I domain correlates with inhibition of function. For each mAb, the average distance was calculated between the Mg2+ and the Cα atom of each species-specific residue within human–mouse chimeras that gave ≤++ binding in Table 1. The percent inhibition of iC3b, fibrinogen, ICAM-1 binding, and neutrophil aggregation are averaged from Fig. 6 of ref. 17.
Our results provide information about the inter-relationship between the I domain and the remainder of the integrin molecule in which it is present. The upper surface of the I domain and two of the lateral, α-helix-bearing faces are recognized by mAbs and thus are well exposed in the intact integrin on the cell surface. By contrast, many potentially antigenic residues located near the bottom of the I domain, and on the C-terminal α-helix, were not recognized by any of the 18 mAbs tested. At least a portion of these residues may be shielded from antibody recognition because they are in an interface with another integrin domain. The bottom of the I domain connects to the predicted β-propeller domain in the integrin α-subunit. The N- and C-termini of the I domain are close to one another and are predicted to link to adjacent β-sheets 2 and 3, respectively, in the β-propeller (3). The I-like domain of the β-subunit is also nearby because it interacts with residues in β-propeller sheets 2 and 3, as shown by a shared epitope (Q. Zang, C.L., C. Huang, and T.A.S., unpublished results).
Of relevance to inside-out signaling by integrins are the human–mouse substitutions that activate iC3b binding and CBRM1/5 binding and cluster at the bottom of the I domain (Fig. 2, turquoise spheres). The enhancement of binding to iC3b is marked, ≈6-fold. Of the three substitutions within hm131–136, E131Q and D132E are in the polypeptide chain connection between the β-propeller domain and the I domain and by definition are in the interface between these domains. The other activating substitutions, L164F, K231E, and F234A, are nearby; they are within 6–12 Å of D132, the first residue of the Mac-1 I domain as defined by the crystal structures, and therefore are close to or within the interface between the I domain and the β-propeller domain. Interestingly, L164 is buried by the C-terminal α-helix, and, therefore, the L164F substitution is expected to loosen the packing against the side of the I domain of this α-helix, which connects to the β-propeller domain. Presence within an interface or within the I domain is consistent with a lack of recognition of these species-specific residues by any of the 18 Mac-1 mAbs studied here. Previously, substitutions at the bottom of the von Willebrand factor A1 domain, which is homologous to I domains, have been shown to activate ligand binding near the top of the domain (38). Furthermore, substitutions with LFA-1 for Mac-1 sequence in residues 162–170, and 190–197 of the I domain, have been found to activate Mac-1 (39). These substitutions included loops on the bottom of the I domain but also included residues in α-helices 1, 2, and 3 and involved drastic changes in 9 of 9 and 7 of 8 amino acids, respectively. A total of only three regions were substituted, all on a single face of the I domain. We substituted a total of 17 regions, distributed over all faces of the I domain, and therefore can conclude that the bottom of the I domain, where it interfaces with the β-propeller domain, is especially susceptible to substitutions that activate ligand binding. We speculate that these substitutions weaken the quaternary constraints that hold the I domain in the low-affinity conformation and mimic a physiologic mechanism in which movements in the interface region activate ligand binding. It has been suggested that this could be accomplished by a movement of the C-terminal α-helix of the I domain, which could conformationally couple the MIDAS to the interface between the I domain and the β-propeller domain (3). Because the I-like domain in the β-subunit is near the interface between the I domain and β-propeller domain (Q. Zang, C.L., C. Huang, and T.A.S., unpublished results), it also may participate in the interactions that regulate ligand binding to the MIDAS and expression of the CBRM1/5 epitope. Thus, inside-out signaling may be directed from within the cell by altering the orientation between the integrin α and β subunits, which in turn alters the orientation between the β-propeller domain and the I domain and thus alters the conformation at the MIDAS and its affinity for ligand.
In summary, our results support the hypothesis that, with activation, the integrin I domain undergoes a conformational change near the MIDAS that is tightly linked to the increase in affinity for ligand. Our findings are based on studies of the I domain in the context of the intact Mac-1 molecule. The transition from inactive to active receptor can be provoked with several minor amino acid substitutions located at the bottom of the I domain. This region contacts the β-propeller domain, and we propose that the activation signal is transmitted through the β-propeller domain in association with the I-like domain of the β-subunit.
Acknowledgments
Dr. N. Hogg is thanked for donating mAb M24, and Dr. K. Tan is thanked for help with molscript. We thank Robert Liddington and Martin Humphries for reviewing the manuscript. C.O. was supported by a fellowship from The Danish Natural Science Research Council, and C.L. was supported by The Cancer Research Institute. This work was supported by National Institutes of Health Grant CA31799.
ABBREVIATIONS
- I domain
inserted domain
- MIDAS
metal ion-dependent adhesion site
References
- 1.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 2.Springer T A. Nature (London) 1990;346:425–433. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 3.Springer T A. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J, Bankston L, Arnaout M, Liddington R C. Structure (London) 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 5.Qu A, Leahy D J. Structure (London) 1996;4:931–942. doi: 10.1016/s0969-2126(96)00100-1. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin E T, Sarver R W, Bryant G L, Jr, Curry K A, Fairbanks M B, Finzel B C, Garlick R L, Heinrikson R L, Horton N C, Kelley L L, et al. Structure (London) 1998;6:923–935. doi: 10.1016/s0969-2126(98)00093-8. [DOI] [PubMed] [Google Scholar]
- 7.Lee J-O, Rieu P, Arnaout M A, Liddington R. Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 8.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emsley J, King S L, Bergelson J M, Liddington R C. J Biol Chem. 1997;272:28512–28517. doi: 10.1074/jbc.272.45.28512. [DOI] [PubMed] [Google Scholar]
- 10.Tuckwell D S, Humphries M J. FEBS Lett. 1997;400:297–303. doi: 10.1016/s0014-5793(96)01368-3. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz M A, Schaller M D, Ginsberg M H. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 12.Diamond M S, Springer T A. Curr Biol. 1994;4:506–517. doi: 10.1016/s0960-9822(00)00111-1. [DOI] [PubMed] [Google Scholar]
- 13.Tabbutt S, Nelson D P, Tsai N, Miura T, Hickey P R, Mayer J E, Neufeld E J. Mol Med. 1997;3:600–609. [PMC free article] [PubMed] [Google Scholar]
- 14.Sims P J, Ginsberg M H, Plow E F, Shattil S J. J Biol Chem. 1991;266:7345–7352. [PubMed] [Google Scholar]
- 15.Calvete J J, Mann K, Schäfer W, Fernandez-Lafuente R, Guisán J M. Biochem J. 1994;298:1–7. doi: 10.1042/bj2980001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du X, Gu M, Weisel J W, Nagaswami C, Bennett J S, Bowditch R, Ginsberg M H. J Biol Chem. 1993;268:23087–23092. [PubMed] [Google Scholar]
- 17.Diamond M S, Garcia-Aguilar J, Bickford J K, Corbi A L, Springer T A. J Cell Biol. 1993;120:1031–1043. doi: 10.1083/jcb.120.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loftus J C, Smith J W, Ginsberg M H. J Biol Chem. 1994;269:25235–25238. [PubMed] [Google Scholar]
- 19.Michishita M, Videm V, Arnaout M A. Cell. 1993;72:857–867. doi: 10.1016/0092-8674(93)90575-b. [DOI] [PubMed] [Google Scholar]
- 20.Zhou L, Lee D H S, Plescia J, Lau C Y, Altieri D C. J Biol Chem. 1994;269:17075–17079. [PubMed] [Google Scholar]
- 21.Humphries M J, Newham P. Trends Cell Biol. 1998;8:78–83. [PubMed] [Google Scholar]
- 22.Loftus J C, Liddington R C. J Clin Invest. 1997;99:2302–2306. doi: 10.1172/JCI119408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphries M J. Curr Opin Cell Biol. 1996;8:632–640. doi: 10.1016/s0955-0674(96)80104-9. [DOI] [PubMed] [Google Scholar]
- 24.Irie A, Kamata T, Takada Y. Proc Natl Acad Sci USA. 1997;94:7198–7203. doi: 10.1073/pnas.94.14.7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liddington R, Bankston L. Structure (London) 1998;6:937–938. doi: 10.1016/s0969-2126(98)00094-x. [DOI] [PubMed] [Google Scholar]
- 26.Bazzoni G, Hemler M E. Trends Biochem Sci. 1998;23:30–34. doi: 10.1016/s0968-0004(97)01141-9. [DOI] [PubMed] [Google Scholar]
- 27.Diamond M S, Springer T A. J Cell Biol. 1993;120:545–556. doi: 10.1083/jcb.120.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oxvig C, Springer T A. Proc Natl Acad Sci USA. 1998;95:4870–4875. doi: 10.1073/pnas.95.9.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Heinzel S S, Krysan P J, Calos M P, DuBridge R B. J Virol. 1988;62:3738–3746. doi: 10.1128/jvi.62.10.3738-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotkow K J, Orkin S H. Mol Cell Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, Springer T A. J Immunol. 1997;159:268–278. [PubMed] [Google Scholar]
- 34.Lu C, Oxvig C, Springer T A. J Biol Chem. 1998;273:15138–15147. doi: 10.1074/jbc.273.24.15138. [DOI] [PubMed] [Google Scholar]
- 35.Dransfield I, Cabañas C, Barrett J, Hogg N. J Cell Biol. 1992;116:1527–1535. doi: 10.1083/jcb.116.6.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies D R, Cohen G H. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beller D I, Springer T A, Schreiber R D. J Exp Med. 1982;156:1000–1009. doi: 10.1084/jem.156.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley J, Cruz M, Handin R, Liddington R. J Biol Chem. 1998;273:10396–10401. doi: 10.1074/jbc.273.17.10396. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Plow E F. J Biol Chem. 1996;271:29953–29957. doi: 10.1074/jbc.271.47.29953. [DOI] [PubMed] [Google Scholar]
- 40.Corbi A L, Kishimoto T K, Miller L J, Springer T A. J Biol Chem. 1988;263:12403–12411. [PubMed] [Google Scholar]
- 41.Pytela R. EMBO J. 1988;7:1371–1378. doi: 10.1002/j.1460-2075.1988.tb02953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]