Abstract
Tumor suppressor p53 is the most frequently mutated gene in human tumors. Meanwhile, under stress conditions, p53 also acts as a transcription factor, regulating the expression of a series of target genes to maintain the integrity of genome. The target genes of p53 can be classified into genes regulating cell cycle arrest, genes involved in apoptosis, and genes inhibiting angiogenesis. p53 protein contains a transactivation domain, a sequence-specific DNA binding domain, a tetramerization domain, a non-specific DNA binding domain that recognizes damaged DNA, and a later identified proline-rich domain. Under stress, p53 proteins accumulate and are activated through two mechanisms. One, involving ataxia telangiectasia-mutated protein (ATM), is that the interaction between p53 and its down-regulation factor murine double minute 2 (MDM2) decreases, leading to p53 phosphorylation on Ser15, as determined by the post-translational mechanism; the other holds that p53 increases and is activated through the binding of ribosomal protein L26 (RPL26) or nucleolin to p53 mRNA 5′ untranslated region (UTR), regulating p53 translation. Under hypoxia, p53 decreases transactivation and increases transrepression. The mutations outside the DNA binding domain of p53 also contribute to tumor progress, so further studies on p53 should also be focused on this direction. The subterranean blind mole rat Spalax in Israel is a good model for hypoxia-adaptation. The p53 of Spalax mutated in residue 172 and residue 207 from arginine to lysine, conferring it the ability to survive hypoxic conditions. This model indicates that p53 acts as a master gene of diversity formation during evolution.
Keywords: p53, Transcription factor, Hypoxia, Adaptation
INTRODUCTION
Tumor suppressor p53 is a transcription factor involved in maintaining genomic integrity by regulating genes involved in cell cycle arrest, DNA repair, and programmed cell death, in response to various stress stimuli such as DNA damage and hypoxia (Asker et al., 1999). p53 protein was first discovered in 1979, during the study that simian virus 40 (SV40), a large T-antigen, was found able to bind to a protein of 53 000 Da to form a complex (Lane and Crawford, 1979). p53 was defined as an oncogene until 1989 when it was shown to be a tumor suppressor by Baker et al.(1989). p53 is the most frequently mutated gene in human cancers. Its functions as a transcription factor are crucial for tumor suppression (Miled et al., 2005; Pietenpol et al., 1994), because p53 suppresses tumor progress mainly by inducing cell cycle arrest and apoptosis (Avivi et al., 2005).
Dozens of genes have been identified as the targets of p53, which are classified into three groups: (1) down-stream mediators of p53-dependent cell cycle arrest, including p21 (WAF1), 14-3-3σ, GADD45 (growth arrest and DNA-damage inducible gene #45), and B99; (2) down-stream mediators of p53-dependent apoptosis, including p21(Bax), Fas/APO1 (factor associated suicide/apoptosis-1), Killer/DR5 (death receptor 5), PIGs (p53-induced genes), p85, PAG608 (p53-activated gene 608), and IGF-Bp3 (insulin-like growth factor-binding protein #3); and (3) p53 targets that inhibit angiogenesis, including Tsp1 (thrombospondin 1), BAI1 (brain-specific angiogenesis inhibitor 1), and GD-AiF (glioma-derived angiogenesis inhibitory factor). Another group of p53-upregulated targets has unknown functions, including Cyclin G, GML (glycosyl phosphatidyl inositol (GPI)-anchored molecule-like protein), Wip1, EI24 (etoposide-induced #24), EF-1α (elongation factor-1α), HIC-1 (hypermethylated in cancer #1), and RTP/rit42 (reduced in tumor, 42 kDa) (el-Deiry, 1998).
Human p53 has 393 residues and 4 domains (Fig.1): (1) a transcriptional activation domain that contacts TAF (TATA-binding protein and associated factors) components of TFIID (transcription factor IID), residues 1~40 (Cho et al., 1994; Thut et al., 1995); (2) a sequence-specific DNA binding domain, residues 120~290 (Cho et al., 1994; Bargonetti et al., 1993); (3) a tetramerization domain, residues 310~360, including the nuclear localization signal (NLS) region (Clore et al., 1995); and (4) a domain that recognizes and binds to damaged DNA nonspecifically, residues 364~390 (Wang et al., 1993; Lee et al., 1995). A new fifth functional domain was identified, and is defined approximately by amino acids 61~94 (Walker and Levine, 1996). This region is rich in the amino acid proline (12/34 residues), contains five repeats of the amino acid sequence PXXP (P designating proline and X designating any amino acid), and plays an important role in efficient growth suppression (Walker and Levine, 1996).
Fig. 1.
Structural features of p53 protein
Since the p53 transcription factor regulates transcription by sequence-specific DNA binding, the conventional p53 consensus binding site is composed of two copies of the 10-bp motif 5′-PuPuPuC (A/T)(T/A)GPyPyPy-3′, where Pu is G or A, Py is C or T (Miled et al., 2005; Avivi et al., 2005).
CRYSTAL STRUCTURE OF p53 CORE DNA-BINDING DOMAIN
p53 acts as a tumor repressor mainly by binding to its target genes to regulate their expression. Thus the structure of the p53 DNA-binding domain is critical for its function. The transcriptional activity of p53 is mediated by a tetrameric form of the protein that binds to DNA in a sequence-specific fashion to activate the transcription of target genes (Zhao et al., 2001).
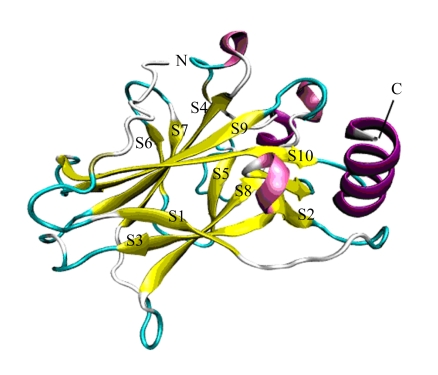
The overall crystal structure of mouse p53 contains three molecules of the core domain. The core domain forms a central region that adopts an immunoglobulin-like β sandwich architecture of two long twisted antiparallel β sheets of four (S1, S3, S8, and S5) and five (S10, S9, S4, S7, and S6) strands (Zhao et al., 2001). The overall computerized structure (http://swissmodel.expasy.org/workspace/index.php?func=modelling_simple1&userid=USERID&token=TOKEN) of the mouse p53 core domain based on the mouse p53 sequence (GenBank accession No. NM_011640) is shown in Fig.2.
Fig. 2.
The overall structure of the mouse p53 core domain
p53 ACTIVATION
Generally, p53 is activated by stress through two mechanisms, one post-translational and the other before translation.
Post-translational mechanism
Under normal conditions, p53 is expressed at a low level, and has a short half-life. When cells are exposed to a stressful environment, p53 is activated and stabilized.
p53 is activated in two ways, an N-terminal mechanism and a C-terminal mechanism (el-Deiry, 1998; Waterman et al., 1998; Sakaguchi et al., 1998). The N-terminus of p53 ranging from residues 1 to 42 is the transactivation region, including the site binding to murine double minute 2 (MDM2). The Mdm2 gene is transcriptionally regulated by p53, and the product of the Mdm2 gene is a p53 binding protein. When MDM2 binds to the p53 N-terminus, p53 activity is inhibited due to the obscuring of the transactivation region. On the other hand, p53 is degraded through the ubiquitination pathway (Shieh et al., 1997; Woo et al., 1998; Canman et al., 1998). Proteins like ataxia telangiectasia-mutated protein (ATM) and DNA-dependent protein kinase (DNA-PK) can be activated under conditions of DNA damage, inducing p53 Ser15 phosphorylation, thus reducing the ability of p53 to bind to MDM2, stabilizing p53 protein, and increasing its activity (Canman et al., 1998; Banin et al., 1998).
The C-terminus of p53 contains a tetramerization domain (TD) and a nuclear localization signal (NLS). The tetramerization of p53 plays an important role in its DNA binding and prevents p53 from being transported out of the nucleus. NLS is involved in p53 nuclear retention (Shaulsky et al., 1990; Stommel et al., 1999). The dephosphorylation of Ser376 induces p53 binding to a 14-3-3 protein, enhancing p53 DNA binding ability (Waterman et al., 1998).
Mechanism before translation
It is known that the presence of p53 in the cell is in synthesis/degradation balance. The increase of p53 under stress is due to the inhibition of degradation, similar to some other transcription factors, such as hypoxia-inducible factor (HIF) (Nikinmaa and Rees, 2005). Recent studies show that the accumulation of p53 under stress is not solely by the inhibition of p53 degradation. Rather, two proteins, ribosomal protein L26 (RPL26) and nucleolin, increase p53 synthesis under stress (Takagi et al., 2005). The studies demonstrated that both RPL26 and nucleolin bind to the 5′ untranslated region (UTR) of p53 mRNA. In normal cells, nucleolin binds to p53 mRNA 5′UTR, inhibiting p53 translation, while in stressed cells, RPL26 binds competitively to the 5′UTR, increasing the p53 protein level. The increase is independent of Ser15 phosphorylation induced by ATM, indicating that under stress RPL26 induces p53 translation directly to increase its protein level (Takagi et al., 2005). Through the two mechanisms, p53 can be accumulated and activated efficiently under stress, preserving genomic integrity.
p53 AND HYPOXIA
Following DNA damage, p53 binds to DNA in a sequence-dependent manner to induce transcription that ultimately influences cell cycle arrest or apoptosis. The transcription needs the interaction between p53 and p300 (also called cAMP-response-element-binding protein (CREB) binding protein (CBP)), the transcription coactivator of p53. Besides transactivation, p53 also has the ability to induce transrepression of several genes, mainly through the interaction between p53 and the corepressor mSin3A (Murphy et al., 1999).
Accumulating evidence shows that under hypoxia, p53 level increases without the induction of its target gene p21 (Koumenis et al., 2001; Hammond et al., 2006). In the human colorectal carcinoma RKO, in which expression of p21 is p53-dependent, when cells are exposed to a series of hypoxic environments (oxygen concentrations ranging from 20% to 0.2%), p53 increases (Koumenis et al., 2001). This increase is evident at an oxygen concentration of 0.2%. However, this increase in p53 does not increase p21 protein level (Koumenis et al., 2001). Similar results were found in the human breast carcinoma MCF-7, suggesting that the inability of hypoxia-induced p53 to induce p21 accumulation is not cell line specific (Koumenis et al., 2001). MDM2, another target of p53, binds to the N-terminus of p53 protein, inhibiting its transactivation and inducing proteolysis (Haupt et al., 1997; Kubbutat et al., 1997). Under hypoxia, MDM2 level decreases, and this may be related to the p53 increase under hypoxia. In the MCF-7 cell line, p53 mRNA does not increase after exposure to hypoxia, indicating that the increase of p53 level after hypoxia is through a post-transcriptional mechanism. Similar results were also found in experiments on cervical epithelial cells, suggesting that the mechanism is not cell line specific (Koumenis et al., 2001). Different from DNA damage, hypoxia fails to induce the acetylation of Lys382 of p53 and the association between p53 and p300, preventing p53 transactivation. Meanwhile, the ability of p53 to bind to mSin3A increases, enhancing p53 transrepression (Koumenis et al., 2001; Hammond et al., 2006).
The decrease in p53 transactivation and increase in p53 transrepression under hypoxia is a strategy for the cell to adapt to hypoxia. Inducing a series of genes to synthesize new mRNA costs much energy, and thus consumes more oxygen. In cells under hypoxia, it is much easier for p53 to inhibit a series of genes’ expressions than to transactivate them. This is partly attributed to the transactivation of HIF, which requires the presence of the transcriptional coactivator CBP/p300. HIF may attract CBP/p300 away from p53, inhibiting p53 transactivation under hypoxia.
HIF was first discovered in cancer cells, playing roles in promoting vessel and blood formation and preventing cells from being damaged in the hypoxic environment of tumor tissues. HIF induces the expressions of vascular endothelial growth factor (VEGF) and erythropoietin (EPO), which promote angiogenesis and hemocytogenesis, respectively.
During hypoxia, HIF-1α accumulates in cells, and when cells are reoxygenated, HIF-1α protein is rapidly degraded. The fluctuation of HIF-1α with the changes of reactive oxygen species (ROS) levels may attribute to the interaction between p53 and HIF-1α. Earlier studies demonstrated an indirect interaction between p53 and HIF-1α, probably through MDM2 (Chen et al., 2003). However, a later study presented evidence of a direct interaction between p53 and HIF-1α (Fels and Koumenis, 2005; Hansson et al., 2002; Sánchez-Puig et al., 2005). Two p53-binding sites have been identified within the oxygen-dependent degradation (ODD) domain of HIF-1α, suggesting that one HIF-1α interacts with one p53 dimer. Under normoxia, both p53 and HIF-1α are low because of proteasome-mediated degradation. Mild or moderate hypoxia activates HIF-1-dependent angiogenesis but is not stringent enough to induce p53 accumulation. Under severe hypoxic or anoxic conditions, p53 also accumulates and, when it reaches a threshold level, it binds to the ODD domain of HIF-1α (Fels and Koumenis, 2005; Sánchez-Puig et al., 2005). The binding of p53 inhibits HIF-1-dependent transactivation or facilitates MDM2-dependent degradation of the HIF-1α subunit (Fels and Koumenis, 2005; Sánchez-Puig et al., 2005). A recent study using Mdm2/p53 double knockout mouse embryonic fibroblasts (MEFs) and p53−/− MEFs indicated that MDM2, either alone or in cooperation with p53, does not promote HIF-1α protein degradation during hypoxia (Nieminen et al., 2005).
HIF-1 protein level increases under mild hypoxia, while p53 increases only in severe hypoxia (close to anoxia). The degree of hypoxia that induces p53 accumulation is also enough to facilitate the inhibition of duplication, which induces phosphorylation of p53 and other molecules involved in DNA repair and cell cycle regulation. The tolerance of p53 to mild hypoxia and the induction of apoptosis under severe hypoxia play significant roles in the adaptation of cells and tissues to hypoxia. HIF functions to protect cells under mild hypoxia; under severe hypoxia such as in tumors, p53 is induced to promote apoptosis, inhibiting tumor progress and protecting the normal physiological functions of tissues and organs.
p53 shows specificity in some hypoxia-adaptive animals. The Israeli blind subterranean mole rat Spalax has four allospecies. Spalax spends its entire life underground in subterranean burrows at decidedly low oxygen tension (Ashur-Fabian et al., 2004). Since its origin some 40 million years ago, Spalax has adopted multiple adaptations to the underground habitat. A series of genes relate to hypoxia change in their structure and function (Ashur-Fabian et al., 2004). p53 in the four allospecies of Spalax has mutated during evolution, substituting arginine (R) to lysine (K) in residue 172 (Arg174 in human) and residue 207 (Arg209 in human) (Ashur-Fabian et al., 2004). The two sites, especially Arg174, are highly conserved during evolution. In the evolution of Spalax, p53 underwent a change that mimics the human tumor mutation (Ashur-Fabian et al., 2004). In Spalax p53, the mutation of Arg174 to Lys reduces its DNA binding due to reduction of dimer stability (Ashur-Fabian et al., 2004). The Spalax p53 favors cell cycle arrest rather than apoptosis targets, preventing cells from damage induced by p53 under hypoxic conditions; arrest is beneficial to normal/healthy individuals but harmful to individuals who suffer from tumor mutation. Thus we can speculate that, during evolution, Spalax adopted a strategy by which normal individuals survived under stressful conditions (hypoxia) but in the cost of a high mortality rate of tumor-bearing individuals. Additionally, several animals living in hypoxic aquatic or muddy bottoms carry the R174K mutation of p53. These aquatic animals include Xenopus (African frog), Xiphophorus (tropical fish), Loligo forbesi (squid), and Mya arenaria (softshell clam).
CLINICAL MUTATIONS OF p53 OUTSIDE THE DNA-BINDING DOMAIN
p53 is the most frequently mutated gene in various cancers. The mutation of p53 in cancer occurs mainly within the highly-conserved DNA-binding domain, in which changes of amino acids may affect its DNA binding ability, hence decreasing the transactivation of p53 targets that induce cell cycle arrest or apoptosis. Until recently, little was understood about the mutation outside of the DNA-binding domain that also contributes to cancers.
Recently, a family suffering from Li-Fraumeni syndrome (LFS) was found, bearing a mutation of p53 outside the DNA-binding domain (Gu et al., 2001). In this syndrome, mutation was found in residues 108~111, where a change from Gly-Phe-Arg-Leu to Ile-Gln has occurred. The mutation localized within the region that is necessary for MDM2-mediated p53 degradation. Mutation in this region is also associated with an impaired response to DNA damage. In addition, p53 (LFS) mutant is defective in its transactivation function, due to its predominantly cytoplasmic localization, probably caused by a faulty nuclear import mechanism. An Arg337His mutation was found in several adrenocortical tumor patients (Latronico et al., 2001). Two p53 mutants, Arg342Pro and Leu344Pro, are inactive in vitro; three mutants, Leu330His, Arg337Cys, and Arg337Leu, show no activity in vitro at low expression levels but become active at higher expression levels (Davison et al., 1998). p53 lacking parts of the oligomerization domain and NLS was found in SK-N-AS (human, bone marrow, neuroblastoma) cells. The protein is expressed largely in the cytoplasm and has lost its transactivation function (Nakamura et al., 2007). Thus, in clinical investigations, more attention should be paid to mutations outside the p53 DNA-binding domain, especially exons 9 and 10.
Hypoxia is a stressful environmental condition harmful to many organisms. However, due to their adaptability, many special species live under different and unusual conditions. Adapting to hypoxia requires the species to change (mutation), and that is how the diversity occurs. The site of mutation in Spalax p53 differentially changes the expression and function of a series of target genes, suggesting p53 as a master gene of diversity. The functional mutations of p53 and the variations in expression contribute to a vast number of phenotype outcomes (Resnick and Inga, 2003). This hypothesis leads to a new understanding of upstream transcription factor p53.
CONCLUSION
p53 is an important tumor suppressor and a transcription factor, inhibiting tumor progression by transcriptionally regulating many target genes. p53 is sensitive to stress and is activated under stress like DNA damage or irradiation. Meanwhile, p53 is also sensitive to hypoxia. The manner of expression and transactivation of p53 is different under hypoxia from other stresses, showing increase in protein level, reduction of transcriptional activity, and enhancement of transrepression, and promoting apoptosis. The manner of expression and activity of p53 under hypoxia corresponds to its induction of tumors when mutated, because in tumor tissues, the microenvironment is hypoxic. Thus, further studies on the understanding of the relationship between p53 and hypoxic adaptation will provide new strategies for cancer therapy.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 30393130 and 30870300) and the National Basic Research Program (973) of China (No. 2006CB504100)
References
- 1.Ashur-Fabian O, Avivi A, Trakhtenbrot L, Adamsky K, Cohen M, Kajakaro G, Joel A, Amariglio N, Nevo E, Rechavi G. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc Natl Acad Sci USA. 2004;101(33):12236–12241. doi: 10.1073/pnas.0404998101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asker C, Wiman KG, Selivanova G. p53-induced apoptosis as a safeguard against cancer. Biochem Biophys Res Commun. 1999;265(1):1–6. doi: 10.1006/bbrc.1999.1446. [DOI] [PubMed] [Google Scholar]
- 3.Avivi A, Ashur-Fabian O, Amariglio N, Nevo E, Rechavi G. p53—a key player in tumoral and evolutionary adaptation: a lesson from the Israeli blind subterranean mole rat. Cell Cycle. 2005;4(3):367–372. doi: 10.4161/cc.4.3.1534. [DOI] [PubMed] [Google Scholar]
- 4.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, van Tuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244(4901):217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 5.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 6.Bargonetti J, Manfredi JJ, Chen X, Marshak DR, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7(12B):2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 7.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281(5383):1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 8.Chen D, Li M, Luo J, Gu W. Direct interaction between HIF-1α and Mdm2 modulate p53 function. J Biol Chem. 2003;278(16):13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- 9.Cho Y, Gorina S, Jeffrey PD. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265(5170):346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 10.Clore GM, Ernst J, Clubb R, Omichinski JG, Kennedy WM, Sakaguchi K, Appella E, Gronenborn AM. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat Struct Biol. 1995;2(4):321–333. doi: 10.1038/nsb0495-321. [DOI] [PubMed] [Google Scholar]
- 11.Davison TS, Yin P, Nie E, Kay C, Arrowsmith CH. Characterization of the oligomerization defects of two p53 mutants found in families with Li-Fraumeni and Li-Fraumeni-like syndrome. Oncogene. 1998;17(5):651–656. doi: 10.1038/sj.onc.1202062. [DOI] [PubMed] [Google Scholar]
- 12.el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8(5):345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 13.Fels DR, Koumenis C. HIF-1α and p53: the ODD couple? TRENDS in Biochemical Sciences. 2005;30(8):426–429. doi: 10.1016/j.tibs.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Gu J, Kawai H, Wiederschain D, Yuan ZM. Mechanism of functional inactivation of a Li-Fraumeni syndrome p53 that has a mutation outside of the DNA-binding domain. Cancer Res. 2001;61(4):1741–1746. [PubMed] [Google Scholar]
- 15.Hammond EM, Mandell DJ, Salim A, Krieg AJ, Johnson TM, Shirazi HA, Attardi LD, Giaccia AJ. Genome-wide analysis of p53 under hypoxic conditions. Mol Cell Biol. 2006;26(9):3492–3504. doi: 10.1128/MCB.26.9.3492-3504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansson LO, Friedler A, Freund S, Rudiger S, Fersht AR. Two sequence motifs from HIF-1α bind to the DNA-binding site of p53. Proc Natl Acad Sci USA. 2002;99(16):10305–10309. doi: 10.1073/pnas.122347199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, Derr J, Taya Y, Lowe SW, Kastan M, et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol. 2001;21(4):1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 20.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 21.Latronico AC, Pinto EM, Domenice S, Fragoso MC, Martin RM, Zerbini MC, Lucon AM, Mendonca BB. An inherited mutation outside the highly conserved DNA-binding domain of the p53 tumor suppressor protein in children and adults with sporadic adrenocortical tumors. J Clin Endocrinol Metab. 2001;86(10):4970–4973. doi: 10.1210/jc.86.10.4970. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, Elenbaas B, Levine A, Griffith J. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell. 1995;81(7):1013–1020. doi: 10.1016/S0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 23.Miled C, Pontoglio M, Garbay S, Yaniv M, Weitzman JB. A genomic map of p53 binding sites identifies novel p53 targets involved in an apoptotic network. Cancer Res. 2005;65(12):5096–5104. doi: 10.1158/0008-5472.CAN-04-4232. [DOI] [PubMed] [Google Scholar]
- 24.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3A. Genes Dev. 1999;13(19):2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura Y, Ozaki T, Niizuma H, Ohira M, Kamijo T, Nakagawara A. Functional characterization of a new p53 mutant generated by homozygous deletion in a neuroblastoma cell line. Biochem Biophys Res Commun. 2007;354(4):892–898. doi: 10.1016/j.bbrc.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 26.Nieminen A, Qanungo S, Schneider EA, Jiang B, Agani FH. Mdm2 and HIF-1α interaction in tumor cells during hypoxia. J Cell Physiol. 2005;204(2):364–369. doi: 10.1002/jcp.20406. [DOI] [PubMed] [Google Scholar]
- 27.Nikinmaa M, Rees BB. Oxygen-dependent gene expression in fishes. Am J Physiol Regul Integr Comp Physiol. 2005;288(5):R1079–R1090. doi: 10.1152/ajpregu.00626.2004. [DOI] [PubMed] [Google Scholar]
- 28.Pietenpol JA, Tokino T, Thiagalingam S, el-Deiry WS, Kinzler KW, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91(6):1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci USA. 2003;100(17):9934–9939. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12(18):2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez-Puig N, Veprintsev DB, Fersht AR. Binding of natively unfolded HIF-1α ODD domain to p53. Mol Cell. 2005;17(1):11–21. doi: 10.1016/j.molcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Shaulsky G, Goldfinger N, Ben-Ze′ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10(12):6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325–334. doi: 10.1016/S0092-8674(00)80416-X. [DOI] [PubMed] [Google Scholar]
- 34.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18(6):1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123(1):49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267(5194):100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 37.Walker KK, Levine AJ. Identification of a novel p53 function domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93(26):15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang BQ, Kostrub CF, Finkelstein A, Burton ZF. Production of human RAP30 and RAP74 in bacterial cells. Protein Expr Purif. 1993;4(3):207–214. doi: 10.1006/prep.1993.1027. [DOI] [PubMed] [Google Scholar]
- 39.Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19(2):175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 40.Woo RA, McLure KG, Lees-Miller SP, Rancourt DE, Lee PW. DNA-dependent protein kinase acts upstream of p53 in response to DNA damage. Nature. 1998;394(6694):700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- 41.Zhao K, Chai X, Johnston K, Clements A, Marmorstein R. Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution. J Biol Chem. 2001;276(15):12120–12127. doi: 10.1074/jbc.M011644200. [DOI] [PubMed] [Google Scholar]