1. Introduction
Ligands of the tumour necrosis factor (TNF) superfamily have diverse roles in the organisation and function of the immune system. Although most TNF family ligands are synthesised as type II transmembrane proteins, many are processed subsequently into soluble forms by limited proteolysis(Bodmer et al., 2002). For some ligands, conversion to the soluble form is essential for biological activity, for example a mutation in the proteolysis motif of ectodysplasin (EDA) is associated with the genetic disorder X-linked hypohidrotic ectodermal dysplasia (XLHED)(Schneider et al., 2001;Chen et al., 2001). In contrast, the cytotoxic activity of FasL (CD95L) is reduced significantly upon cleavage(Tanaka et al., 1998;Shudo et al., 2001) and soluble FasL antagonises killing mediated by membrane bound FasL(Suda et al., 1997). The biological activity of the ligand can be restored by artificially increasing the avidity of the ligand(Holler et al., 2003), suggesting that it is not the solubilisation of the ligand per se that removes the bioactivity but rather its inability to cross-link its cognate receptor. TNF family ligands exist as trimers, both in membrane bound and soluble forms and the engagement of the trimeric ligand with a trimeric signalling competent receptor may be a prerequisite for a secondary multimerisation event and the formation of supramolecular signalling clusters(Siegel et al., 2004;Henkler et al., 2005).
The initial event in the process of viral entry into a target cell is the interaction between the virus and its cellular receptor and the specificity of this interaction determines both viral cell tropism and pathogenicity. The primary receptor for the feline immunodeficiency virus (FIV) is CD134 (OX40), a member of the TNF receptor (TNFR) superfamily and co-stimulatory molecule with a crucial role in the expansion and survival of T cells that have encountered antigen(Gramaglia et al., 1998;Pippig et al., 1999;Kopf et al., 1999). As both CD134(Taylor and Schwarz, 2001) and its ligand CD134L (OX40L, CD252)(Wang et al., 2006) exist in membrane bound and soluble forms in vivo, the process of FIV infection, and ultimately disease pathogenesis, may be sensitive to modulation by soluble receptor and ligand produced endogenously. Further, it may be possible to block the virus-receptor interaction using soluble forms of receptor or ligand administered as therapeutics. Soluble feline CD134 has been produced as an N-terminal immunoglobulin Fc region-fusion protein (sFc-CD134) and used to study the virus-receptor interaction(de Parseval et al., 2006;de Parseval et al., 2004a;de Parseval et al., 2004b;Willett et al., 2007;Willett et al., 2006a). However, when feline CD134L was prepared in soluble forms analogous to the widely used sFc- and FLAG-tagged forms of human and murine CD134L, the ligand exhibited no receptor-binding activity. Similar observations have been made with the related TNF superfamily ligands FasL and TRAIL(Berg et al., 2007) where the soluble murine ligands were completely devoid of Fas and TRAILR2 binding activity respectively, while human FasL or TRAIL prepared under identical conditions bound well to Fas and TRAILR2 respectively. Given that soluble murine CD134L displayed inhibitory activity against FIV(Willett et al., 2007), CD134L may prove a useful tool for the further dissection of the FIV-receptor interaction and its role in viral pathogenesis. In this study, we set out to reconstitute a functional soluble feline CD134L. We demonstrate that enforced trimerisation of feline CD134L via the introduction of a subdomain of the tenascin-C (TNC) oligomerization domain is sufficient to restore full ligand-binding activity. The resulting fusion protein is as effective as anti-CD134 monoclonal antibody at detecting CD134 expression and displays strain-specific blocking of viral entry.
2. Materials and Methods
2.1. Cells and viruses
MYA-1(Miyazawa et al., 1989), MCC(Cheney et al., 1990), CLL(Willett et al., 2006b) and NSO cell lines were cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum (FBS), 2mM glutamine, 0.11mg/ml sodium pyruvate 100 IU/ml penicillin and 100 μg/ml streptomycin (complete RPMI). 293T and NP2 cells were maintained in Dulbecco’s modification of Eagle’s medium (DMEM) supplemented as above. The medium for MYA-1 cells was supplemented with conditioned medium from a murine cell line (L2.3, kindly provided by M. Hattori and T. Miyazawa) transfected with a human IL-2 expression construct (equivalent to 100 U/ml of recombinant human IL-2) and 50μM 2-mercaptoethanol. All media and supplements were obtained from InVitrogen Life Technologies Ltd. (Paisley, UK). Stable cell lines expressing constructs based on the pDONAI (Takara) or PCR3 (InVitrogen) expression vectors were maintained in G418-supplemented media (InVitrogen, Paisley, UK) while those based on pTorsten(Spiller et al., 2003) were maintained in hygromycin-supplemented media as previous(Willett et al., 2007). Chimeric viruses derived from the GL8 molecular were prepared by transfection of the molecular clone into 293T cells followed by passage of the 0.45μm-filtered supernatant onto MYA-1 cells. The infected cells were cultured until cytopathicity was evident (normally ~1 to 2 weeks) at which time the supernatant was clarified by centrifugation, 0.45μm-filtered and stored at −80°C.
2.2. Antibodies and flow cytometry
Anti-feline CD134 has been described previously(Willett et al., 2007). Fluorescein isothiocyanate (FITC)-conjugated anti-feline CD134 was obtained from AbD Serotec, Oxford, UK. Anti-human CD134 (BerACT35) was obtained from Alexis Biochemicals (Axxora (UK) Ltd., Nottingham), anti-mouse B220 (CD45R) was from BD Biosciences (Pharmingen, Oxford, United Kingdom), while anti-human CXCR4 (#44701) was obtained from R&D Systems, Abingdon, Oxford, United Kingdom. Cells to be processed for flow cytometry were resuspended in phosphate buffered saline (PBS) supplemented with 1.0% (w/v) bovine serum albumin, 0.1% (w/v) sodium azide (PBA). Cells were incubated with 1μg of primary antibody for 30 minutes at 4°C and then washed twice with PBA by centrifugation at 1000 rpm for 5 minutes. Bound primary antibody was detected with the appropriate anti-mouse IgG secondary antibody (Serotec, Oxford, United Kingdom) corresponding to the isotype of the primary antibody and conjugated to either fluorescein isothiocyanate (FITC) or R-phycoerythrin (PE). Cells were incubated with secondary antibody for 30 minutes at 4°C and then washed twice with PBA by centrifugation at 1000rpm for 5 minutes and resuspended in 1ml of PBA for analysis. All samples were analysed on a Beckman Coulter EPICS MCS-XL flow cytometer, 10,000 events being collected for each sample in LIST mode. Analysis gates were set using isotype-matched negative control antibodies such that <1.0% of events fell within the gate. Data were analysed using EXPO 32 ADC Analysis (Advanced Cytometry Systems).
2.3. Production of soluble feline CD134-ligand
The cloning of a cDNA encoding feline CD134L (fCD134L) has been described previously(Willett et al., 2007). In order to generate soluble fCD134L, the extracellular domain (aa 51–183) of fCD134L was cloned as a PstI-EcoRI fragment into both PS261(Bossen et al., 2006) (generating FLAG-fCD134L) and PS1297(Bossen et al., 2006) (generating Fc-fCD134L), similar constructs bearing murine and human CD134L have been described previously(Bossen et al., 2006). Chimeric feline x human and human x feline CD134L fusions were prepared by exchanging PstI-SacI (aa 1–113) and SacI-EcoRI (aa 114–183) fragments between feline and human CD34L in the PS261 vector (although only the fCD134L cDNA has an internal SacI, a similar site was introduced into hCD134L by PCR-directed mutagenesis using the oligonucleotide 5′-AATGTTGAGCTCCTGGGAGAAGTAGCCCTT-3′). The trimerisation domain of murine ACRP30 (aa 18–111) was introduced into the expression vector for FLAG-tagged murine and feline ligands(Bossen et al., 2006) as an NsiI-PstI fragment. The trimerisation domain of chicken tenascin (aa 110–139)(Berg et al., 2007) was introduced into the expression vectors for the FLAG and Fc-tagged ligands(Bossen et al., 2006) as an NsiI-PstI fragment. The resulting Fc-tagged vectors encode the hemagglutinin signal peptide, the hinge and Fc domain of human IgG1 (excluding the Stop codon, aa 104–330 of GenBank:P01857), a linker with sequence RSPQPQPKPQPKPEPEGSLH, aa 110–139 of chicken tenascin-C, a linker with sequence GTGGGSGGRGLQ, and the TNF homology domain of CD134L (aa 52–183 of human CD134L, aa 51–198 of mouse CD134L or aa 52–183 of feline CD134L). The expression vector for FLAG-tagged ligands encodes the hemagglutinin signal peptide, the FLAG sequence (DYKDDDDKG), a linker with sequence GPGQVQLH, aa 110–139 of chicken tenascin-C, a linker with sequence GTGGGSGGRGLQ, and the TNF homology domain of feline OX40L (aa 52–183).
Soluble ligands were prepared by transient transfection of 293T cells with expression vector using SuperFect, QIAgen (Crawley, UK) and culturing in OptiMEM (InVitrogen) for 72 hours. Supernatants were collected, filtered at 0.45μm, concentrated 10-fold using an Amicon Ultra centrifugal filter (Ultracel 10K, Millipore Corporation, Billerica, MA, USA) and stored at −80°C prior to use.
2.4. Preparation of HIV (FIV) pseudotypes
FIV env gene expression constructs GL8, B2542, CPG41 and PPR have been described previously(Shimojima et al., 2004;Willett et al., 2006b;Willett et al., 2006a). 5μg of each VR1012-env and 7.5μg of pNL4-3-Luc-E−R− were co-transfected into HEK-293T cells using SuperFect activated dendrimer (QIAgen) as per manufacturer’s instructions. The nomenclature “HIV(FIV)” denotes an FIV Env protein on an HIV particle. Culture supernatants were collected at 48 hours post-transfection, filtered at 0.45μm and frozen at −80°C until required. Target cell lines were seeded at 5×104 cells per well of a CulturPlate™-96 assay plate (Perkin Elmer, Life and Analytical Sciences, Beaconsfield, UK) and used immediately. The cells were then infected with 50μl of HIV (FIV) luciferase pseudotypes, cultured for 72 hours and then luciferase activity quantified by the addition of 50μl of Steadylite HTS™ (Perkin Elmer) luciferase substrate prior to measurement by single photon counting on a MicroBeta TriLux luminometer (Perkin Elmer).
2.5. Large scale production of recombinant IgG-Fc fusion proteins
Fc-TNC-mCD134L, fCD134L and hCD134L were produced from CHO cells stably transfected with the respective construct in CELLine AD1000 bioreactor flasks (Integra Biosciences (Scientific Laboratory Supplies, Nottingham, U.K.)) in medium supplemented with low IgG serum (Integra Biosciences). Culture supernatants were filtered at 0.45μm and frozen at −80°C prior to use in order to preserve optimal bioactivity. Where appropriate, Fc-tagged ligands were purified by affinity chromatography using protein A-Sepharose (HiTrap™ Protein A HP, GE Healthcare, Chalfont St. Giles, U.K) and bound protein was eluted with 0.1M glycine pH 3.0, neutralising the eluate immediately to pH 7.0 by collection into 1.5M Tris pH8.6. The integrity of the purified proteins was assessed by immunoblotting, fusion protein being detected with goat anti-human IgG-Fc horseradish peroxidase (HRP) conjugate (Sigma, Poole, U.K.) and visualised with amino-9-ethylcarbazole (Vectastain AEC substrate kit, Vector Laboratories Ltd., Peterborough, U.K.).
2.6. Binding of soluble CD134Ls to CD134
MCC cells expressing feline, human or chimeric CD134s(Willett et al., 2007) were incubated with soluble CD134Ls for 30 minutes on ice in complete RPMI supplemented with 10mM N-(2-Hydroxyethyl) piperazine-N′-(2-ethanesulfonic acid) (HEPES). Cells were then washed twice by centrifugation through phosphate buffered saline supplemented with 1%bovine serum albumin and 0.1% sodium azide (PBA) at 1000rpm and incubated with i) PE-conjugated anti-human IgG Fc (eBioscience, San Diego, USA) for 30 minutes at 4°C or ii) M2 anti-FLAG (Sigma, Poole, Dorset, U.K.) for 30 minutes at 4°C followed by PE-anti-mouse IgG (AbD Serotec, Oxford, U.K.). Cells were then washed twice with PBA by centrifugation at 1000rpm for 5 minutes and resuspended in 1ml of PBA for analysis and analysed by flow cytometry as above.
2.7. Inhibition of viral entry
1×105 MYA-1 or CLL-CD134 cells were incubated with ligand or inhibitor in complete medium + HEPES for 30 minutes at room temperature. Viral pseudotypes were added, incubated for a further hour before the cells were washed once by centrifugation and then plated in 96-well culture-treated luciferase assay plates (CulturPlate™ 96) in complete medium in the presence of maintenance ligand at a 1:4 dilution of the blocking concentration. Cultures were maintained for 72 hours post-infection at which point glow luciferase assay substrate was added and luminescence quantified.
The effect of the ligands on productive infection was assayed by incubating cells with ligand at 12.5μg/ml for 30 mins followed by the addition of virus for 1 hour. Cells were then washed twice with complete RPMI by centrifugation at 1000rpm, 5 mins. and plated into 48 well culture plates in the presence of 1.25 μg/ml ligand. The culture wells were sampled (every 2–3 days), the supernatants clarified by centrifugation (13.5K, 5 mins.) and stored at −80°C for reverse transcriptase (RT) assay. RT was quantified in the thawed samples by using a non-isotopic assay bromo-deoxyuridine-triphosphate (BrdU) incorporation test kit (Lenti RT, Cavidi AB, Uppsala, Sweden).
2.8. Polyacrylamide gel electrophoresis (PAGE) analyses
Approximately 1μg of affinity-purified proteins were separated using reducing and non-reducing Laemmli sample buffer(Laemmli, 1970) on 12% polyacrylamide gels. Blue-native electrophoresis(Schagger, 2001) used 4–16% NativePAGE™ Bis-Tris gels (Invitrogen) according to the manufacturer’s instructions in comparison with NativeMark™ unstained molecular weight markers (Invitrogen). Separated proteins were transferred to nitrocellulose by electroblotting (iBlot™, Invitrogen); molecular weight markers were visualised using Ponceau stain and bound ligands were detected using goat anti-human IgG-Fc horseradish peroxidase (HRP) conjugate followed by visualisation with amino-9-ethylcarbazole.
3. Results
3.1. Production of soluble feline CD134L
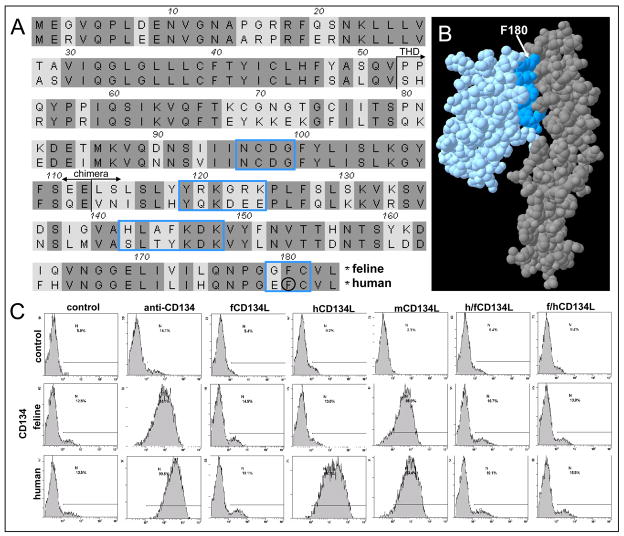
Previous studies have demonstrated that the TNF homology domain (THD) of human and murine CD134L (OX40L) can be produced in soluble forms that retain full receptor-binding activity(Bossen et al., 2006). In contrast, when the THD of feline CD134L (aa 52–183) was expressed in mammalian cells as N-terminal FLAG and Fc-tagged proteins, feline CD134L bound to neither feline nor human CD134, irrespective of the tag(Willett et al., 2007). As expression of both feline and human CD134 was detected readily by anti-CD134 antibody, the data suggested that the ligand binding capacity of feline CD134L was disrupted by expression in soluble form(Willett et al., 2007) (previously we had shown that surface expressed fCD134L bound soluble feline CD134(Willett et al., 2007)). The amino acid sequences of feline and human CD134L were compared in order to assess the degree of sequence divergence and to investigate the likely cause of the failure to bind CD134 (Fig. 1a). FCD134L and hCD134L shared 69% amino acid identity and when the 3D-structure of feline CD134L and CD134 were modelled using the SWISS-MODEL modelling server in “First Approach” mode(Schwede et al., 2003;Peitsch, 1996), based on the published structure of the human CD134L:CD134 complex(Compaan and Hymowitz, 2006) (Fig. 1b), divergent residues were identified in the region predicted to form the receptor-binding interface (Fig. 1a). The contact residues at the binding interface were predicted to be derived from four regions; 96NCDG99, 119YRKGRK124, 142HLAFKDK148 and 179GFC181. 96NCDG99 was conserved between feline and human CD134L, thus we postulated that a chimeric human x feline CD134L bearing residues 114–183 of feline CD134L may retain the ligand-binding specificity of fCD134L but the stability of human CD134L when expressed in soluble form. Two FLAG-tagged chimeras were prepared consisting of aa 52–113 of human CD134L with aa 114–183 of feline CD134L (h/fCD134L) and, as a control, the converse chimera with aa 52–113 of feline CD134L and aa 114–183 of human CD134L. Binding of the chimeric ligands was assessed in comparison with feline, human and murine CD134L on cells expressing feline or human CD134 (Fig. 1c). While human CD134L bound specifically to human CD134-expressing cells, murine CD134L bound to both feline and human CD134L-expressing cells, consistent with the promiscuity of binding observed with mCD134L previously(Bossen et al., 2006;Willett et al., 2007). In contrast, neither fCD134L nor the h/fCD134L and f/hCD134L chimeric ligands bound to either receptor. Thus, expression of the receptor binding domain of feline CD134L in the context of human CD134L was insufficient to generate a functional soluble CD134L.
Fig. 1.
Binding properties of chimeric CD134L fusion proteins. (A) The amino acid sequences of feline and human CD134L were compared and regions of homology (dark shading) identified. Start of the TNF homology domain (THD) and chimera junction are arrowed. Residues predicted to lie at the CD134 binding face of CD134L are in blue boxes while aa F180, critical for the CD134:CD134L interaction(Compaan and Hymowitz, 2006) is circled. (B) Predicted 3D-structure of the feline CD134L (blue) and CD134 (gray) complex illustrating residues at the binding face of CD134L (dark blue) and residue F180(Compaan and Hymowitz, 2006) (arrowed). (C) Binding of FLAG-tagged fCD134L, hCD134L, mCD134L and the human/feline and feline/human chimeric CD134Ls (h/fCD134L and f/hCD134L) to MCC cells expressing feline or human CD134. Bound ligands were detected with anti-FLAG followed by PE-anti-mouse IgG. Cells were processed for flow cytometry and histograms represent 10,000 events collected in LIST mode, bars illustrate percentage positive. Histograms are representative of two independent analyses.
3.2. Restoration of ligand binding by enforced covalent trimerisation
Ligands of the TNF family are expressed at the cell surface as trimeric type II transmembrane proteins that may then be cleaved to yield soluble trimeric forms(Bodmer et al., 2002). The soluble trimers interact with three molecules of their respective receptors, triggering secondary multimerisation into supramolecular clusters that are capable of transducing signals(Holler et al., 2003;Henkler et al., 2005;Siegel et al., 2004). The loss of specific binding properties by TNF family ligands when expressed in a soluble form may be the result of inappropriate aggregation, as has been observed previously with the related members of the TNF family mFasL and mLIGHT(Bossen et al., 2006). In order to restore bioactivity to soluble feline CD134L, novel CD134L expression constructs were generated into which synthetic trimerisation domains from either the collagen domain of murine adiponectin/ACRP30 (ACRP, aa 18–111)(Holler et al., 2003) or chicken tenascin-C (TNC, aa 110–139) had been inserted (Berg et al., 2007). ACRP:FasL assembles into hexamers (of two trimers) that trigger Fasmediated apoptosis(Holler et al., 2003) while the trimerisation domain of tenascin conferred receptor binding activity upon TRAIL and FasL (bioactivity required further cross-linking of the bound TNC-FasL trimers) (Berg et al., 2007). Binding studies comparing murine and feline FLAG-tagged ACRP-CD134L revealed that while the murine ligand retained full binding activity, the ACRP trimerisation domain did not restore binding activity to the feline ligand (data not shown). Accordingly, no further studies with the FLAG-ACRP-fCD134L were undertaken. In contrast, the introduction of a TNC trimerisation domain into either the FLAG or Fc-tagged fCD134L vectors generated soluble fCD134Ls capable of binding feline CD134 (Fig. 2c). Ligand binding was first assessed on MCC cells expressing fCD134 or hCD134 ectopically (in comparison with vector-only control cells). Fc-TNC-fCD134L bound to the majority of MCC cells expressing feline CD134 (99.8%, mean fluorescence intensity(MFI)=221) but only weakly to those expressing human CD134 (45.5%, MFI=9.9). Conversely, Fc-TNC-hCD134L recognised 100% of human CD134-expressing cells (MFI=608) but showed no reactivity with feline CD134 (0.4%). Fc-TNC-mCD134L bound both feline and human CD134 with similar efficiency (99.8%, MFI=300 and 99.8%, MFI=367 respectively). No binding of the Fc-TNC-mFasL control protein was detected on either cell line, confirming the specificity of the CD134L:CD134 interactions. Thus the species-specificity of feline and human CD134L binding to their respective receptors was replicated when the ligands were expressed in soluble forms bearing TNC trimerisation domains. Binding of the Fc-TNC ligands to native, endogenously expressed, feline CD134 was assessed using the IL2-dependent T cell line MYA-1. MYA-1 cells are non-transformed feline CD4+ T cells(Miyazawa et al., 1989) and in cell culture the population is comprised of a mixture of cells at various stages of blastogenesis. Thus CD134 expression can vary between analyses but is essentially bi-modal(Willett et al., 2007) with larger blast cells being 100% CD134-positive, consistent with CD134 being up-regulated upon activation(Willett et al., 2007). Anti-feline CD134 mAb 7D6 recognised 83.3% of MYA-1 cells (MFI=19) (Fig. 2c). In comparison, Fc-TNC-fCD134L and Fc-TNC-mCD134L bound to 82.4% (MFI=14) and 78.2% (MFI=7) of MYA-1 cells respectively. The bi-modal expression pattern observed with anti-CD134 mAb was reproduced by the soluble feline and murine Fc-TNC ligands, confirming sensitivity and specificity. In contrast, Fc-TNC-hCD134L did not bind to MYA-1 T cells (0.5%).
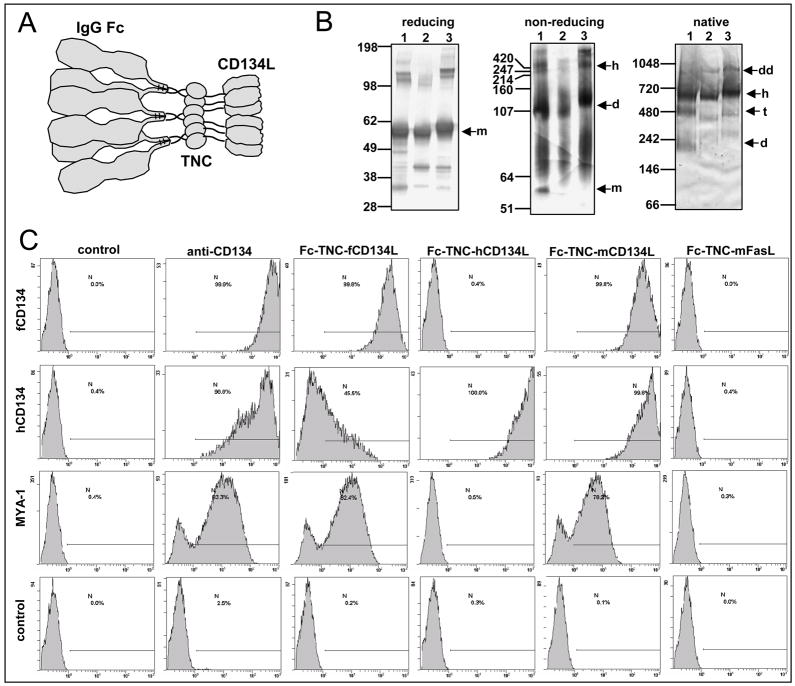
Fig. 2.
Enforced trimerisation of feline CD134L rescues ligand-specific binding. (A) Predicted hexameric structure of the Fc-TNC ligands, comprising three IgG Fc dimers and two CD134L trimers. (B) Polyacrylamide gel electrophoretic analyses of the Fc-TNA-ligands on reducing SDS-PAGE, non-reducing SDS-PAGE or blue NativePAGE™. 1-Feline, 2-Murine, 3- Human; m-monomer, d-dimer, t-tetramer, h-hexamer, dd-dodecamer (C) Binding of Fc-TNC ligands to MCC cells expressing feline or human CD134 (or vector only control), or to MYA-1 cells. Fc-TNC-fCD134 was compared with Fc-TNC-mCD134, Fc-TNC-hCD134 or Fc-TNC-mFasL. Bound ligands were detected with phycoerythrin (PE)-conjugated anti-IgG Fc. CD134 expression was assessed using mAb 7D6, bound antibody being detected with PE-anti-mouse IgG. Cells were processed for flow cytometry and histograms represent 10,000 events collected in LIST mode, bars illustrate percentage positive. Histograms are representative of two independent analyses.
The biochemical properties of the Fc-TNC ligands were confirmed by polyacrylamide gel electrophoresis (PAGE). Under reducing conditions, the ligands migrated as a single dominant species with an apparent molecular mass of ~50–60kDa, consistent with the predicted molecular masses of 48,333, 50,237 and 48,986 for the feline, murine and human monomeric ligands respectively (Fig 3b). Fc-TNC-hCD134L displayed a greater apparent size, this likely due to the presence of 5 predicted sites for N-linked glycosylation (compared with 4 and 2 in the feline and murine ligands respectively). Under non-reducing conditions, the ligands migrated primarily at ~100–130 kDa, consistent with dimeric forms of the ligand driven by dimerisation of the Fc-domain. However higher molecular weight forms of ~420 kDa were also evident (Fig. 2b) as would be predicted for the hexameric form of the ligand due to covalent trimerisation mediated by the TNC domain. This high molecular weight form was most evident under native PAGE where the dominant species migrated at ~590kDa (feline and murine) and ~600kDa (human). Recent studies using a trimerizing isoleucine zipper (ILZ) domain to stabilise human CD134L found that a hexamer with predicted molecular weight of 240kDa and confirmed size of 336kDa (sedimentation analysis) eluted at ~540kDa using molecular sieve chromatography. It is likely that such discrepancies are the result of sizing estimates being made between globular size standards and asymmetrical/non-globular species(Morris et al., 2007). Here, although the hexameric form of Fc-TNC-fCD134L dominated, there was also evidence of both smaller forms (dimer and trimer/tetramer) and single larger form ~940–980kDa (dodecamer).
Fig. 3.
Binding of anti-CD134 and Fc-TNC-CD134L to primary CD4+ T cells. Popliteal lymph node cells were stained with Fc-TNC-fCD134L, anti-CD134 and as a control for non-specific binding, Fc-TNC-hCD134. Bound ligands were detected with phycoerythrin (PE)-conjugated anti- human IgG Fc. CD134 expression was assessed using mAb 7D6, bound antibody being detected with PE-anti-mouse IgG. Cells were processed for flow cytometry and dotplots represent 10,000 events collected in LIST mode and analysis gates show percentages of total cells analysed. Dotplots are representative of two independent analyses.
Given the similar binding profiles of Fc-TNC-fCD134L and anti-CD134 on IL2-dependent T cells, the binding specificity of the Fc-TNC-fCD134L was confirmed on primary lymph node CD4+ T cells (CD134 expression is restricted predominantly to CD4+ T cells in blood and lymphoid tissues(Willett et al., 2007)). Anti-CD134 antibody stained 5.2% of CD4+ T cells while Fc-TNC-fCD134L labelled 6.6% of CD4+ T cells (the difference of 0.8% corresponding to the non-specific binding of Fc-TNC-hCD134L) (Fig. 3). Thus, Fc-TNC-CD134L and anti-CD134 appear to identify similar populations of cells on primary CD4+ T cells, underlining the potential for the use of Fc-TNC-ligands to assess receptor expression where monoclonal antibodies are not available.
Introduction of the TNC trimerisation domain into the FLAG-CD134L construct restored functionality to the soluble fCD134L (Fig. 4), the resulting protein binding selectively to feline CD134 (66.2%, MFI=5) but not human CD134. However, binding was weak in comparison with FLAG-mCD134L which recognised 99.7% (MFI=309) and 99.2% (MFI=291) of cells expressing feline and human CD134 respectively. The weak binding of the FLAG-TNC-fCD134L was confirmed on MYA-1 cells where 44.9% (MFI=3) of cells were stained while FLAG-mCD134L recognised 86.5% (MFI=12), consistent with the binding patterns of mAb 7D6 and the Fc-TNC ligands. Given the poor binding of the FLAG-TNC-CD134L, subsequent studies focussed on the Fc-TNC ligands.
Fig. 4.
Specific binding of FLAG-tagged TNC feline CD134L to feline CD134. Specificity of binding of FLAG-tagged TNC feline CD134L was examined on MCC cells expressing human or feline CD134, vector only control cells (control), or MYA-1 T cells. Binding was compared with FLAG-tagged murine CD134L. Bound ligands were detected with anti-FLAG followed by PE-anti-mouse IgG. Cells were processed for flow cytometry and histograms represent 10,000 events collected in LIST mode, bars illustrate percentage positive. Histograms are representative of two independent analyses.
3.3. Modulation of FIV infection by soluble feline CD134L
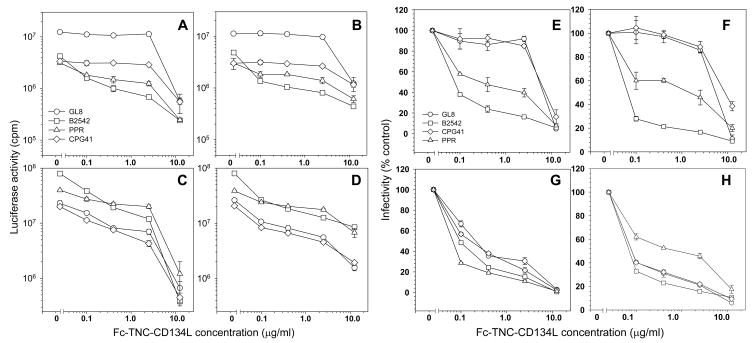
Soluble murine CD134L inhibited FIV infection of MYA-1 cells with the PPR and B2542 strains but had little effect on infection with strains such as GL8 and CPG41(Willett et al., 2007). The effect of the Fc-TNC-CD134L ligands on infection with viruses at extremes of sensitivity to murine CD134L was examined on MYA-1 cells (MYA-1 cells are IL2-dependent T cells expressing low, but biologically relevant, levels of CD134 and CXCR4). Infection of MYA-1 cells with B2542 and PPR was markedly reduced in a dose dependent manner by both the feline (Fig. 5a., e.) and murine (Fig. 5b., f.) Fc-TNC ligands, significant inhibitory activity being revealed with ligand concentrations as low as 0.1μg/ml (2nM). Fc-TNC-hCD134L had no activity (not shown), consistent with the species-specificity of the CD134L:CD134 interaction. GL8 and CPG41 pseudotypes were more resistant to inhibition by the Fc-TNC ligands compared to B2542 and PPR pseudotypes, significant inhibitory activity not being revealed until a concentration of 12.5μg/ml (250nM) had been achieved. The reduction in PPR and B2542 infectivity by both the Fc-TNC-fCD134L and Fc-TNC-mCD134L at 0.1, 0.4 and 2.5μg/ml was significant (t-test, P<0.001) relative to either GL8 or CPG41 at the equivalent concentrations. The data suggest that when assayed on IL2-dependent T cells, the presumed primary target of the virus, Fc-TNC ligands display potent but strain-specific inhibitory activity.
Fig. 5.
Strain-specific inhibition of viral entry by Fc-TNC ligands. Inhibitory activities of Fc-TNC-fCD134L (A, C, E, G) and Fc-TNC-mCD134L (B, D, F, H) were compared on MYA-1 T cells (A, B, E, F) and CLL-CD134 (C, D, G, H) against HV(FIV) pseudotypes bearing GL8, B2542, PPR and CPG41 Envs. (A–D) Luciferase activity versus Fc-TNC-CD134L concentration, (E–H) Infectivity (percent infection relative to no antagonist) versus Fc-TNC-CD134L concentration. Each point represents the mean +/− standard error (n=3).
Next, the inhibitory activity of the ligands was examined on cells expressing high levels of both CD134 and CXCR4, CLL-CD134 cells (Fig. 5c., d., g., h.). On these cells, a dose dependent inhibition of all viral strains was observed. GL8, CPG41, PPR and B2542 were inhibited with similar efficiency with the feline Fc-TNC ligand. PPR appeared more resistant to murine CD134L on CLL cells, this may reflect PPRs previously described ability to utilise high levels of CXCR4 (see Fig. 7a) for CD134-independent infection(de Parseval et al., 2004c). Given that GL8 and CPG41 were more resistant to the inhibitory activity of the Fc-TNC ligands on MYA-1 cells, the data suggest that GL8 and CPG41 may either bind to MYA-1 cells with a higher affinity than CLL-CD134 cells, or have the capacity to utilise a distinct entry pathway on MYA-1 cells that is relatively insensitive to perturbation by Fc-TNC CD134L. Conversely, it is notable that with matched doses of input virus, PPR and B2542 pseudotypes gave higher values on CLL-CD134 compared to MYA-1 and that PRR was the least sensitive pseudotype to antagonism by Fc-TNC CD134L on CLL-CD134 cells.
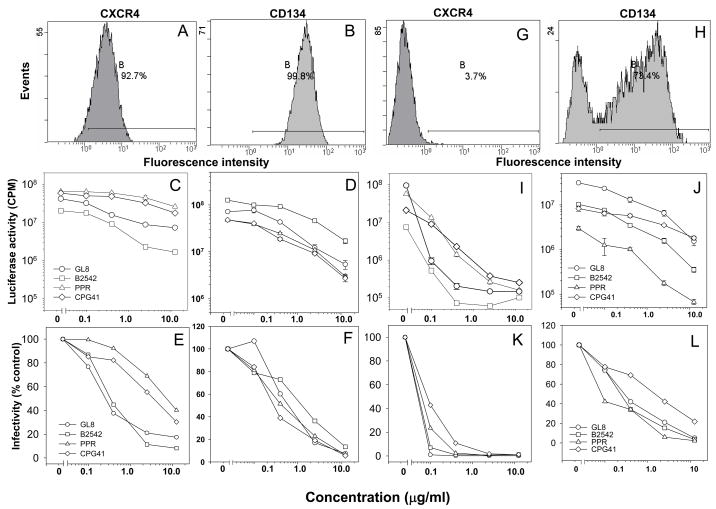
Fig. 7.
Strain-specificity of receptor antagonism. CXCR4 (A., G.) and CD134 (B., H.) were quantified on CLL-CD134 (A., B) and MYA-1 (G., H.) cells by flow cytometry. Bound anti-CXCR4 antibody was detected with PE-anti-mouse IgG. Histograms show 10,000 events collected in LIST mode. Inhibitory activity of AMD3100 (C., E., I., K) and soluble Fc-CD134 (D., F., J., L.) were then compared on CLL-CD134 (C., D., E., F.) and MYA-1 (I., J., K., L.) cells. (C., D., I., J.) Luciferase activity versus antagonist concentration, (E., F., K., L.) Infectivity (percent infection relative to no antagonist) versus antagonist concentration. Each point represent the mean +/− standard error (n=3).
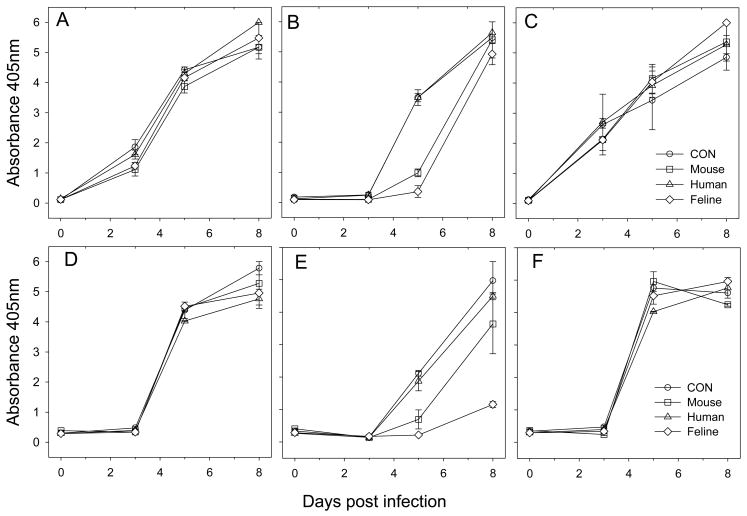
The strain specificity of the antiviral effect of the Fc-TNC-ligands translated from the inhibition of viral entry to suppression of viral replication in vitro. MYA-1 T cells were infected with chimaeric viruses derived from the GL8 molecular clone and bearing the GL8 (Fig. 6a, d), B2542 (Fig. 6b, e) and CPG41 (Fig. 6c, f) env genes at either 1000 TCID50 (a–c) or 100 TCID50 (d–f). While neither the human, feline nor murine ligands altered the kinetics of infection of MYA-1 cells with chimaeras bearing the GL8 or CPG41 envs, the feline and murine TNC-ligands significantly suppressed viral replication with B2542 at 5 days post-infection although this inhibitory effect was overwhelmed by 8 days post-infection (Fig. 6b). When the MOI of the input virus was reduced (Fig. 6e), the inhibitory effect was more stark with feline CD134L suppressing viral replication to the end point of the assay. Thus, infection with B2542, the viral strain for which entry was blocked most efficiently on MYA-1 cells (Fig. 5e. & f.), was inhibited in the assay for productive infection (Fig. 6c.), confirming the sensitivity of this strain to antagonism by Fc-CD134-CD134L. These data may suggest that viral variants that interact with CD134 in a manner similar to B2542 may be susceptible to suppression in vivo by endogenously produced CD134L while viruses such as GL8 and CPG41 would be resistant to antagonism by CD134L and would thus have a replicative advantage.
Fig. 6.
Effect of Fc-TNC-CD134L on productive infection. MYA-1 cells were infected with 1000 (A–C) or 100 (D–F) TCID50 of GL8 chimaeras bearing the (A, D) GL8, (B, E) B2542 and (C, F) CPG41 envs in the presence of 12.5μg/ml Fc-TNC-CD134L (mouse, human, or feline) or saline control. Cultures were maintained for one week and supernatants analysed for virus production by non-isotopic reverse transcriptase assay (Absorbance 405nm). Infection with B2542 was inhibited with the murine and feline but not human ligands. Results are mean of two replicates and are typical of two analyses.
Previous studies have demonstrated that over-expression of human CXCR4 alone is sufficient to facilitate entry mediated by the PPR Env(de Parseval et al., 2004c). Accordingly, where CXCR4 is abundant, infection mediated by PPR may be more efficient (and resistant to antagonism of CD134 binding). CXCR4 expression on CLL-CD134 (Fig. 7a.) and MYA-1 (Fig. 7g.) was examined by flow cytometry; 92.7% of CLL-CD134 compared with 3.7% of MYA-1 were CXCR4-positive. Simultaneously, we infected CLL-CD134 (Fig. 7c., e.) and MYA-1 (Fig. 7i., k.) with HIV(FIV) pseudotypes in the presence of increasing concentrations of the CXCR4 antagonist AMD3100. Infection of MYA-1 cells, where CXCR4 is limiting, was blocked efficiently by AMD3100 in a dose-dependent manner. In contrast, infection of CLL-CD134 cells, where CXCR4 is abundant, was less sensitive to modulation by AMD3100 but all viruses did exhibit a dose-dependent reduction in infectivity. On both cellular substrates, PPR and CPG41 were least sensitive to antagonism by AMD3100, suggesting that these viruses may have a higher affinity for CXCR4. In contrast with the differential effect of the Fc-TNC-CD134Ls, infection with all four viruses was blocked efficiently by soluble Fc-CD134 on either or CLL-CD134 (7d., f.) MYA-1 (7j., l.). CPG41 alone displayed a lower sensitivity to inhibition by Fc-CD134 on MYA-1 cells, perhaps indicating a higher affinity for CD134 than GL8, PPR and B2542. However, the findings with Fc-CD134 are in marked contrast to those observed with Fc-TNC-CD134L on MYA-1 cells (Fig. 5e., f.) where both GL8 and CPG41 displayed significant resistance to inhibition.
4. Discussion
The aim of this study was to produce a functional soluble feline CD134L with which the role of the virus-receptor interaction in the pathogenesis of AIDS in FIV infected cats could be studied. Previously, it was observed that soluble feline CD134L was non-functional when expressed in identical expression constructs to those used for the production of fully-functional soluble murine and human CD134L(Willett et al., 2007), either as a FLAG-tagged monomer or as an Fc-tagged dimer. Thus while the TNF homology domain of murine and human CD134L is sufficient to drive trimerisation and receptor binding, this does not extend to feline CD134L. Electrophoretic analyses suggested that soluble feline CD134L may have been forming into misfolded aggregates as had been observed previously with murine FasL(Berg et al., 2007). We predicted that preparation of a chimeric feline/human CD134L in which the ligand binding face of human CD134L was exchanged with that of feline CD134L would retain the binding properties of feline CD134 and the stability of human CD134L. However, once again, both the feline/human and human/feline ligands were non-functional. As CD134L engages CD134 as a trimer(Compaan and Hymowitz, 2006), we introduced a hexamerising domain from ACRP-30 (adiponectin) to bring together two CD134L trimers, an approach that has proved successful in the generation of functional CD95L(Holler et al., 2003). Our rationale was that assuming the TNF homology domain of fCD134L was driving trimerisation of the ligand, then the additional ACRP-30 domain would facilitate dimerisation of two trimers to from a stable ligand with higher binding avidity. However, while this approach generated a functional murine CD134L it did not restore receptor-binding activity to feline CD134L. We next examined the trimerisation domain of tenascin-C, postulating that the spatial fixation of the TNF homology domain may be critical missing element for receptor binding(Berg et al., 2007) and that the ACRP-30 domain may not have recapitulated the arrangement of the trimer found in the native membrane-bound form of CD134L that we had shown previously to bind CD134 well(Willett et al., 2007). TNC-fCD134L fusion proteins tagged with either immunoglobulin (Ig) Fc or FLAG were functional and bound feline CD134. Further, ligand binding specificity was retained in the TNC-ligands in that both feline and human CD134L bound their respective receptors specifically. The specificity and sensitivity of feline Fc-TNC-CD134L for CD134 mirrored that of the anti-CD134 MAb 7D6(Willett et al., 2007) staining IL2-dependent feline T cells with similar intensity. Electrophoretic analyses indicated that the predominant form of the native Fc-TNC-ligands was likely to be a hexamer; each monomer dimerising through the Fc-tag and then trimerising through TNC domain (thus the Fc-TNC ligands would comprise two ligand trimers and three Fc dimers). That soluble feline CD134L required the introduction of a TNC domain suggests that the spatial distribution of the CD134L monomers is critical for the correct presentation of the receptor binding face to CD134. Soluble CD134L is likely trimeric however in the absence of the “stalk” region, the receptor binding residues may be misaligned. Adding the TNC domain may fix the receptor-binding region in a spatial distribution closer to the native membrane anchored-ligand, however this arrangement is still sub-optimal (given the relatively modest increase in binding observed with the FLAG-tagged ligand). Addition of the Fc domain would appear to increase the receptor binding affinity of the TNC-CD134L fusion further, presumably by increasing avidity for CD134.
Whether cleavage of the membrane bound form in vivo releases a bioactive soluble mediator will be of importance to the regulation of T cell survival during antigen-specific immune responses. Elevated levels of soluble CD134L have been detected in the sera of patients with Graves’ disease(Wang et al., 2006), suggesting a possible role in the immune-dysregulation associated with this autoimmune response. Therefore, if the FIV Env protein competes with CD134L for binding to CD134, either as a competitive antagonist or as a surrogate agonist, then FIV infection may also modulate T cell survival indirectly.
The activity of the Fc-TNC-ligands as antagonists of FIV infection was investigated on primary (non-transformed) IL2-dependent T cells (MYA-1) and on a cell line engineered to express high levels of CD134 (CLL-CD134). Feline and murine Fc-TNC-CD134L had similar activities against HIV(FIV) pseudotypes in assays of viral entry, inhibiting infection with the B2542 and PPR strains by 60 and 40% respectively at a concentration of 0.1μg/ml (2nM). The difference in sensitivity to inhibition between B2542/PPR and GL8/CPG41 was striking, GL8 and CPG41 requiring a ligand concentration of 12.5μg/ml (250nM) to achieve significant inhibition of viral entry. However, when the assays were repeated on a cellular substrate on which CD134 was expressed (ectopically) at high levels, the differential effect disappeared, suggesting that the differential sensitivity that we observed may be dependent on the level of CD134 expression. However, it is possible that the IL2-dependent T cells, express (endogenous) CD134 in a different conformation to that on CLL-CD134 cells and that the GL8 and CPG41 Envs are capable of recognising this endogenously expressed CD134 conformation with a high affinity. It is notable therefore that infection with both the GL8 and CPG41 strains was highly sensitive to perturbations of CRD2(Willett et al., 2006a), the region of CD134 that forms a component of the CD134L binding site(Willett et al., 2006b;Willett et al., 2006a). Previously, we hypothesised that GL8 and CPG41 may represent early isolates of FIV and that such viral strains may target cell types efficiently where CD134 is limiting. With time post-infection, and with developing immunodeficiency, viruses may acquire mutations that enable a higher affinity interaction with the co-receptor CXCR4, evolving towards CD134-independence. However, when sensitivity to the CXCR4 antagonist AMD3100 was compared, the PPR and CPG41 strains were most resistant to inhibition, suggesting a higher affinity interaction with CXCR4. The PPR strain is known to use CXCR4 efficiently, to the extent that over-expression of CXCR4 alone can confer susceptibility to PPR infection(de Parseval et al., 2004c). That CPG41 also displayed a substantial resistance to AMD3100 indicates that sensitivities to CXCR4 and CD134 antagonism are not mutually exclusive. Our findings are reminiscent of previous studies comparing infection of primary macrophages with cell culture adapted and primary X4 strains of HIV where efficiency of infection was governed largely by differential requirements for expression CD4 and/or CXCR4(Tokunaga et al., 2001). While the previous study compared cell culture adapted with primary X4 strains of virus, here, we compare four distinct primary strains of FIV (all CXCR4-dependent) and show that they differ in their requirements for CD134 and CXCR4.
The efficiency of CD134 and CXCR4 usage may alter with time post infection; early in infection viruses may dominate that have a high affinity for CD134 and either a high or low affinity for CXCR4. As the CXCR4 binding site is concealed from potentially neutralising antibodies, the affinity for CXCR4 may be less critical in early infection. With time post-infection, as the neutralising antibody response diminishes, the affinity of the interaction with CD134 may decline, leading to the emergence of CD134-independent viruses that are capable of spreading into compartments rich in CXCR4-expressing cells. Thus, although FIV does not appear to switch chemokine receptor usage from CCR5 to CXCR4 with disease progression as has been described in HIV infection(Connor et al., 1997) (there are no data at present supporting a role for chemokine receptors other than CXCR4 in FIV infection), by modulating affinity for CD134 binding and a gradual progression towards usage of CXCR4 alone, the virus may evolve in a similar manner. Accordingly, “late” viruses may be more sensitive to virus neutralising antibodies and less transmissible due to their low affinity for CD134. Soluble CD134L produced in vivo may suppress both the replication and spread of such variants. Thus, viruses such as CPGammer(Diehl et al., 1995;de Rozieres et al., 2004) (a virus isolated following acute phase passage(Diehl et al., 1995)) and GL8 (isolated from a cat displaying no clinical signs of infection at the time of sampling) may represent the most robust (and biologically relevant) challenge strains for FIV vaccine studies. If candidate vaccines can be designed that protect against experimental challenge with these viruses, then it is likely that they will protect cats in the field.
Acknowledgments
We thank Andrew Lane (AbD-Serotec Limited, U.K.) for the provision of reagents and Harald Wajant (University of Wuerzburg, Germany) for supplying the cDNA encoding the tenascin-C trimerisation domain. This work was supported by Public Health Service grant AI049765 to B.J.W & M.J.H. from the National Institute of Allergy and Infectious Diseases and by grants from the Swiss National Science Foundation to PS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berg D, Lehne M, Muller N, Siegmund D, Munkel S, Sebald W, Pfizenmaier K, Wajant H. Enforced covalent trimerization increases the activity of the TNF ligand family members TRAIL and CD95L. Cell Death Differ. 2007 doi: 10.1038/sj.cdd.4402213. [DOI] [PubMed] [Google Scholar]
- Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- Bossen C, Ingold K, Tardivel A, Bodmer JL, Gaide O, Hertig S, Ambrose C, Tschopp J, Schneider P. Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J Biol Chem. 2006;281:13964–13971. doi: 10.1074/jbc.M601553200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Molloy SS, Thomas L, Gambee J, Bachinger HP, Ferguson B, Zonana J, Thomas G, Morris NP. Mutations within a furin consensus sequence block proteolytic release of ectodysplasin-A and cause X-linked hypohidrotic ectodermal dysplasia. Proc Natl Acad Sci USA. 2001;98:7218–7223. doi: 10.1073/pnas.131076098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney CM, Rojko JL, Kociba GJ, Wellman ML, Di Bartola SP, Rezanka LJ, Forman L, Mathes LE. A feline large granular lymphoma and its derived cell line. In Vitro Cell Dev Biol. 1990;26:455–463. doi: 10.1007/BF02624087. [DOI] [PubMed] [Google Scholar]
- Compaan DM, Hymowitz SG. The crystal structure of the costimulatory OX40-OX40L complex. Structure. 2006;14:1321–1330. doi: 10.1016/j.str.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV- 1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval A, Chatterji U, Morris G, Sun P, Olson AJ, Elder JH. Structural mapping of CD134 residues critical for interaction with feline immunodeficiency virus. Nat Struct Mol Biol. 2004a;12:60–66. doi: 10.1038/nsmb872. [DOI] [PubMed] [Google Scholar]
- de Parseval A, Chatterji U, Sun P, Elder JH. Feline immunodeficiency virus targets activated CD4+ T cells by using CD134 as a binding receptor. Proc Natl Acad Sci USA. 2004b;101:13044–13049. doi: 10.1073/pnas.0404006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval A, Grant CK, Sastry KJ, Elder JH. Sequential CD134-CXCR4 interactions in feline immunodeficiency virus (FIV): soluble CD134 activates FIV Env for CXCR4-dependent entry and reveals a cryptic neutralization epitope. J Virol. 2006;80:3088–3091. doi: 10.1128/JVI.80.6.3088-3091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval A, Ngo S, Sun P, Elder JH. Factors that increase the effective concentration of CXCR4 dictate feline immunodeficiency virus tropism and kinetics of replication. J Virol. 2004c;78:9132–9143. doi: 10.1128/JVI.78.17.9132-9143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rozieres S, Mathiason CK, Rolston MR, Chatterji U, Hoover EA, Elder JH. Characterization of a highly pathogenic molecular clone of feline immunodeficiency virus clade C. J Virol. 2004;78:8971–8982. doi: 10.1128/JVI.78.17.8971-8982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl LJ, Mathiason-Dubard CK, O’Neil LL, Obert LA, Hoover EA. Induction of accelerated feline immunodeficiency virus disease by acute-phase virus passage. J Virol. 1995;69:6149–6157. doi: 10.1128/jvi.69.10.6149-6157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramaglia I, Weinberg AD, Lemon M, Croft M. Ox-40 ligand: a potent costimulatory molecule for sustaining primary CD4 T cell responses. J Immunol. 1998;161:6510–6517. [PubMed] [Google Scholar]
- Henkler F, Behrle E, Dennehy KM, Wicovsky A, Peters N, Warnke C, Pfizenmaier K, Wajant H. The extracellular domains of FasL and Fas are sufficient for the formation of supramolecular FasL-Fas clusters of high stability. J Cell Biol. 2005;168:1087–1098. doi: 10.1083/jcb.200501048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler N, Tardivel A, Kovacsovics-Bankowski M, Hertig S, Gaide O, Martinon F, Tinel A, Deperthes D, Calderara S, Schulthess T, Engel J, Schneider P, Tschopp J. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol Cell Biol. 2003;23:1428–1440. doi: 10.1128/MCB.23.4.1428-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Ruedl C, Schmitz N, Gallimore A, Lefrang K, Ecabert B, Odermatt B, Bachmann MF. OX40-deficient mice are defective in Th cell proliferation but are competent in generating B cell and CTL Responses after virus infection. Immunity. 1999;11:699–708. doi: 10.1016/s1074-7613(00)80144-2. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Miyazawa TM, Furuya T, Itagaki S, Tohya Y, Takahashi E, Mikami T. Establishment of a feline T-lymphoblastoid cell line highly sensitive for replication of feline immunodeficiency virus. Arch Virol. 1989;108:131–135. doi: 10.1007/BF01313750. [DOI] [PubMed] [Google Scholar]
- Morris NP, Peters C, Montler R, Hu HM, Curti BD, Urba WJ, Weinberg AD. Development and characterization of recombinant human Fc:OX40L fusion protein linked via a coiled-coil trimerization domain. Mol Immunol. 2007;44:3112–3121. doi: 10.1016/j.molimm.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch MC. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- Pippig SD, Pena-Rossi C, Long J, Godfrey WR, Fowell DJ, Reiner SL, Birkeland ML, Locksley RM, Barclay AN, Killeen N. Robust B cell immunity but impaired T cell proliferation in the absence of CD134 (OX40) J Immunol. 1999;163:6520–6529. [PubMed] [Google Scholar]
- Schagger H. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 2001;65:231–244. doi: 10.1016/s0091-679x(01)65014-3. [DOI] [PubMed] [Google Scholar]
- Schneider P, Street SL, Gaide O, Hertig S, Tardivel A, Tschopp J, Runkel L, Alevizopoulos K, Fergusonα BM, Zonana J. Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A. J Biol Chem. 2001;276:18819–18827. doi: 10.1074/jbc.M101280200. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M, Miyazawa T, Ikeda Y, McMonagle EL, Haining H, Akashi H, Takeuchi Y, Hosie MJ, Willett BJ. Use of CD134 as a primary receptor by thefeline immunodeficiency virus. Science. 2004;303:1192–1195. doi: 10.1126/science.1092124. [DOI] [PubMed] [Google Scholar]
- Shudo K, Kinoshita K, Imamura R, Fan H, Hasumoto K, Tanaka M, Nagata S, Suda T. The membrane-bound but not the soluble form of human Fas ligand is responsible for its inflammatory activity. Eur J Immunol. 2001;31:2504–2511. doi: 10.1002/1521-4141(200108)31:8<2504::aid-immu2504>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Muppidi JR, Sarker M, Lobito A, Jen M, Martin D, Straus SE, Lenardo MJ. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J Cell Biol. 2004;167:735–744. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller OB, Robinson M, O’Donnell E, Milligan S, Morgan BP, Davison AJ, Blackbourn DJ. Complement regulation by Kaposi’s sarcoma-associated herpesvirus ORF4 protein. J Virol. 2003;77:592–599. doi: 10.1128/JVI.77.1.592-599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Hashimoto H, Tanaka M, Ochi T, Nagata S. Membrane Fas ligand kills human peripheral blood T lymphocytes, and soluble Fas ligand blocks the killing. J Exp Med. 1997;186:2045–2050. doi: 10.1084/jem.186.12.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Itai T, Adachi M, Nagata S. Downregulation of Fas ligand by shedding. Nat Med. 1998;4:31–36. doi: 10.1038/nm0198-031. [DOI] [PubMed] [Google Scholar]
- Taylor L, Schwarz H. Identification of a soluble OX40 isoform: development of a specific and quantitative immunoassay. J Immunol Methods. 2001;255:67–72. doi: 10.1016/s0022-1759(01)00424-0. [DOI] [PubMed] [Google Scholar]
- Tokunaga K, Greenberg ML, Morse MA, Cumming RI, Lyerly HK, Cullen BR. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J Virol. 2001;75:6776–6785. doi: 10.1128/JVI.75.15.6776-6785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chen Y, Xie F, Ge Y, Chen L, Wu H, Qu Q, Wang X, Zhang X. Development of a sandwich ELISA for evaluating soluble OX40L (CD252) in human sera of different ages or with Graves’ disease. Cytokine. 2006;36:23–28. doi: 10.1016/j.cyto.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Willett BJ, McMonagle EL, Bonci F, Pistello M, Hosie MJ. Mapping the domains of CD134 as a functional receptor for feline immunodeficiency virus. J Virol. 2006a;80:7744–7747. doi: 10.1128/JVI.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett BJ, McMonagle EL, logan N, Spiller OB, Schneider P, Hosie MJ. Probing the interaction between the feline immunodeficiency virus and CD134 using a novel monoclonal antibody 7D6 and CD134L (OX40L) J Virol. 2007;81:9665–9679. doi: 10.1128/JVI.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett BJ, McMonagle EL, Ridha S, Hosie MJ. Differential utilization of CD134 as a functional receptor by diverse strains of feline immunodeficiency virus (FIV) J Virol. 2006b;80:3386–3394. doi: 10.1128/JVI.80.7.3386-3394.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]