Abstract
Tumor suppressor p53 is an attractive cancer therapeutic target because it can be functionally activated to eradicate tumors. Direct gene alterations in p53 or interaction between p53 and MDM2 proteins are two alternative mechanisms for the inactivation of p53 function. Designing small molecules to block the MDM2-p53 interaction and reactivate the p53 function is a promising therapeutic strategy for the treatment of cancers retaining wild-type p53. This review will highlight recent advances in the design and development of small-molecule inhibitors of the MDM2-p53 interaction as new cancer therapies. A number of these small-molecule inhibitors, such as analogs of MI-219 and Nutlin-3, have progressed to advanced preclinical development or early phase cinical trials.
Keywords: tumor suppressor p53, oncoprotein HDM2, small-molecule inhibitors of HDM2, p53 reactivation
INTRODUCTION
The tumor suppressor p53 plays a central role in the regulation of cell cycle, apoptosis, DNA repair, and senescence (1–4, 4a). The p53 protein was identified in 1979 (5–7) and later the p53 gene was reported to be responsible for most cases of Li-Fraumeni cancer syndrome, a rare inherited condition that leads to the frequent occurrence of several types of cancer in affected families (8–10). In fact, due to its potent tumor suppressor role, p53 is one of the most frequently mutated proteins in human tumors. Indeed, approximately 50% of human cancers have alterations in the p53 gene, resulting in inactivation or loss of p53 protein (2, 11).
Even in cancers retaining wild-type p53, p53 function is effectively inhibited. The inhibition of p53 function is primarily performed by the murine double minute 2 (MDM2; HDM2 in humans). MDM2 is an oncoprotein, discovered by its overexpression in a spontaneously transformed mouse cancer cell line (2, 11–14). MDM2 has both p53-independent and p53-dependent functions. MDM2 directly binds to and forms a complex with p53, inhibiting p53 transactivation (12). A substantial amount of data have confirmed that MDM2 is the central node in the p53 pathway.
The activity and protein levels of p53 are tightly regulated by MDM2 in normal cells (see section below). MDM2 is a ubiquitously expressed protein and plays an important role in tissue development, whereas p53 provides a powerful tumor surveillance mechanism. Deregulation of MDM2/p53 balance leads to malignant transformation of cells. For example, overexpression of MDM2 provides cells with a growth advantage, promotes tumorigenesis, and correlates with worse clinical prognosis and poor response to cancer therapy (15–21). A variety of mechanisms, such as amplification of the MDM2 gene, single nucleotide polymorphism at nucleotide 309 (SNP309) in its gene promoter, increased transcription and increased translation, account for MDM2 overproduction (15, 21–23). Mouse models have also revealed that overexpression of MDM2 at an early stage of differentiation neutralizes p53 tumor suppressor function and predisposes mice to tumorigenesis (24). Analogous to the inherited cancer predisposition Li-Fraumeni syndrome in humans, mice lacking p53 develop normally but are predisposed to develop a variety of tumors (25, 25a). The basic finding that MDM2 binds and inhibits p53 function leads to the prediction that MDM2 overexpression and p53 mutations should be mutually exclusive in tumors. Indeed, a study of MDM2 gene amplification in tumors of 28 different types comprising more than 3000 tumors largely supported this notion and showed a negative correlation between occurrence of p53 mutations and MDM2 amplification (19). MDM2 is thus an important therapeutic target in cancers retaining wild-type p53.
A series of genetic studies in mouse models have shown that loss of p53 induces tumor formation and that restoration of p53 leads to a rapid and impressive regression of established, in situ tumors, providing strong evidence for designing anticancer drugs that restore p53 function (26–28). Several different therapeutic approaches have been attempted with the goal of restoring p53 function (29–34). Among these, targeting the MDM2-p53 interaction by small molecules for the reactivation of p53 has emerged as a promising approach for the treatment of cancers that retain wild-type p53 (4a, 32, 34, 35).
Regulation of p53 and MDM2
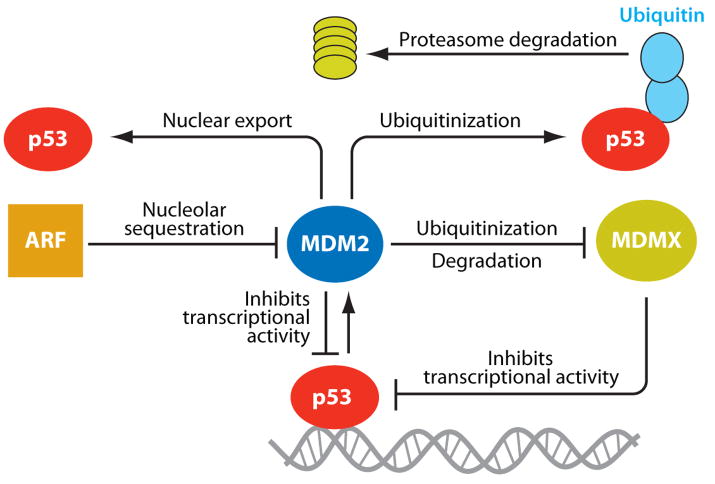
Direct protein-protein interaction between MDM2 and p53 regulates the basal levels and activity of p53 in cells through an autoregulatory feedback loop (Figure 1). Upon activation, p53 binds to the P2 promoter of the MDM2 gene and transcriptionally induces MDM2 protein expression. In turn, MDM2 protein binds to p53 protein and inhibits it through multiple mechanisms: MDM2 (a) inhibits the transactivation function of p53, (b) exports p53 out of the nucleus, and (c) promotes proteosome-mediated degradation of p53 through its E3 ubiquitinization ligase activity (36–38). The physiological relevance of the MDM2-p53 loop has been demonstrated by the genetic evidence that embryonic lethality of MDM2-null mice can be rescued by the simultaneous deletion of the p53 gene (39, 40). In addition, compared with wild-type adult mice, genetically engineered mice expressing reduced levels of MDM2 protein are small in size, have reduced organ weight, and are radiosensitive (41). The p53 dependence was shown by reversal of phenotypes when crossed with p53-null mice. Together, these genetic studies show that MDM2 is critical in the regulation of p53 function during development as well as in adult mice, and that changes in MDM2 levels can dictate tumorigenesis.
Figure 1.
Autoregulatory feedback loop of inhibition of p53 by MDM2. MDM2 directly binds to p53 and inhibits its transcriptional activity, causes ubiquitinization and proteasomal degradation of p53, and exports p53 out of the nucleus. MDMX, a homolog of MDM2, also directly binds to the transactivation domain of p53 and inhibits p53 activity, but does not cause degradation of p53. Tumor suppressor ARF binds to MDM2 and sequesters MDM2 into the nucleolus, leading to stabilization of p53.
DESIGN OF NONPEPTIDIC SMALL-MOLECULE INHIBITORS OF THE MDM2-p53 INTERACTION
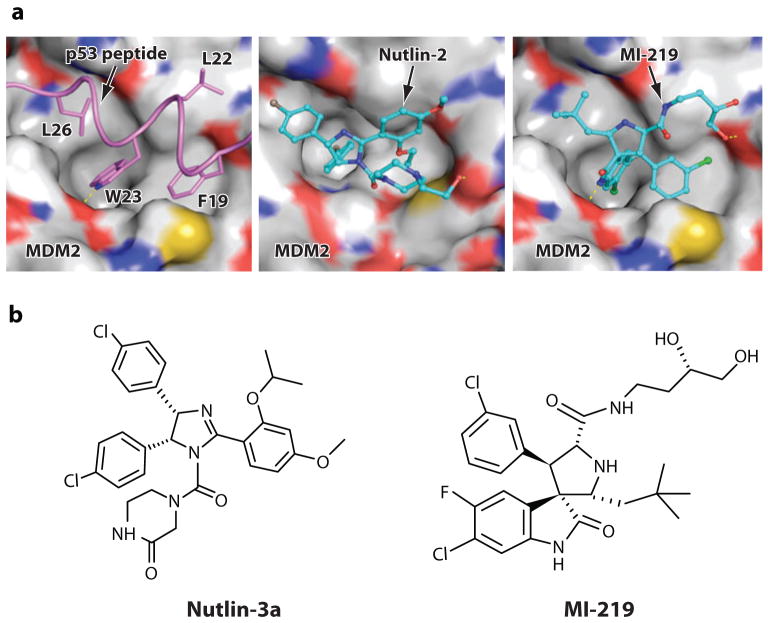
The nature of the interaction between p53 and MDM2 proteins has been firmly established. Genetic and biochemical studies mapped the MDM2-p53 interaction sites to the 106–amino acid-long N terminal domain of MDM2 and the N terminus of the transactivation domain of p53 (42, 43). High-resolution crystal structures of the N-terminal domains of human and Xenopus laevis MDM2, complexed with short peptides derived from the N-terminal domain of p53 (residues 15–29), demonstrated the precise structural requirements for the MDM2-p53 protein-protein interaction (44). The interaction between p53 and MDM2 involves four key hydrophobic residues (Phe 19, Leu 22, Trp 23 and Leu 26) in a short amphipathic helix formed by p53 and a small but deep hydrophobic pocket in MDM2 (Figure 2a). The atomic-level understanding of the MDM2-p53 interaction through X-ray crystallography provided the solid foundation for structure-based design of nonpeptidic, small-molecule antagonists of this interaction.
Figure 2.
Binding mode of a p53 peptide (PDB:1YCR) and the cis-imidazoline analog Nutlin-2 (PDB:1RV1), and the predicted binding model of spiro-oxindole analog MI-219 to MDM2. (a) Side chains of p53 residues involved in the MDM2-p53 interaction are shown in stick representation, whereas Nutlin-2 and MI-219 are shown in ball and stick representation. Nutlin-2 is shown with carbons in cyan, nitrogen in blue, oxygen in red, and bromine in brown. The predicted binding of MI-219 to MDM2 is shown with carbons in cyan, nitrogen in blue, oxygen in red, fluorine in light blue, and chlorine in green. The surface representation of MDM2 in each case is shown with carbons in gray, nitrogen in blue, oxygen in red, and sulfur in yellow. Hydrogen bonds are depicted with yellow lines and hydrogen atoms are excluded for clarity. The PyMOL program was used to generate figures. (b) Chemical structures of Nutlin-3a and MI-219, the two potent and specific small-molecule inhibitors of the MDM2-p53 interaction.
Historically, disruption of protein-protein interaction is a challenging task due to the large binding interface of the protein partners. The well-defined, small interface of MDM2-p53 suggested that design of small-molecule inhibitors to target the MDM2-p53 interaction may be possible (mentioned herein as MDM2 inhibitors; sometimes also called HDM2 inhibitors). Indeed, although very little progress was made in the initial few years after the MDM2-p53 crystal structure became available, several classes of small-molecule inhibitors with distinct chemical structures have now been reported. These are analogs of cis-imidazoline (45), spiro-oxindole (34, 35, 46, 46a), benzodiazepinedione (47–49), terphenyl (50, 51), quilinol (52), chalcone (53) and sulfonamide (54). Below we briefly discuss a number of approaches employed in the design and discovery of MDM2 inhibitors.
Design Approach
Different approaches have been employed to design and identify the small-molecule inhibitors of the MDM2-p53 interaction. These include computational three-dimensional (3D) database screening of large chemical libraries, experimental screening of chemical libraries, and structure-based de novo design.
Computational 3D database screening
Computational 3D screening has been employed for the discovery of novel small-molecule inhibitors of the MDM2-p53 interaction. These include 3D pharmacophore searching, structure-based database searching, and the combination of both approaches.
Pharmacophore searching
A pharmacophore model of the MDM2-p53 interaction was employed for the discovery of nonpeptidic small-molecule MDM2 inhibitors of sulfonamide chemical class (54). A pharmacophore model consists of chemical groups critical for ligand binding to a target protein in their 3D geometrical relationship. The study by Galatin & Abraham (54) examined three separate data sets to draw a pharmacophore model: (a) The p53 mutagenesis data showing measurements of residual MDM2 binding as a percentage of binding observed with wild-type p53 (55), (b) a truncation series based on the best phage display–derived peptide inhibitor to map the prerequisites for activity (56), and (c) p53 peptidic compounds synthesized by No-vartis, incorporating unnatural amino acids to increase binding contacts to MDM2 (57). Various permutations of the MDM2-binding pharmacophore were entered as searches to obtain hit compounds from the database. These compounds were tested in an ELISA-based in vitro MDM2-p53 protein-protein interaction assay. Sulfonamide 1 (NSC 279287) disrupted the interaction between full-length MDM2 and p53 proteins with an IC50 value of 31.8 μM. This compound was thus a fairly weak inhibitor of the MDM2-p53 interaction. When evaluated in a p53 reporter gene assay, only a 20% increase in p53 transcriptional function after treatment with 100 μM of the compound was observed (54).
An integrated approach combining pharmacophore and structure-based screening
An integrated approach utilizes both pharmacophore and structure-based screening of the 3D database. Using this approach, we screened 150,000 drug-like compounds in the NCI 3D database, consisting of both structurally diverse synthetic compounds and a large number of natural products (52, 58, 59). From this database, 110,000 drug-like compounds were filtered and further screened using a pharmacophore model derived from the crystal structure of MDM2 complexed with the p53 peptide and several known nonpeptide small-molecule inhibitors (Nutlin, benzodiazepinedione, and spiro-oxindole). Pharmacophore searching yielded 2599 hits, which were further screened by structure-based screening using the GOLD program (60, 61) to dock each hit to the p53-binding site in MDM2 and to rank their binding affinities. The top 200 compounds ranked by Chemscore (62) and X-score (63) were combined, which yielded a total of 354 nonredundant compounds as potential inhibitors of the MDM2-p53 interaction. Visual inspection of these 354 compounds showed that each compound mimics the key hydrophobic interactions between p53 and MDM2 observed in the MDM2:p53 peptide crystal structure and that each is complementary with MDM2, based on the predicted binding models by GOLD. A total of 67 compounds were obtained from NCI and tested in a competitive fluorescence polarization-based (FP-based) MDM2 binding assay. A total of 10 hits, defined by a Ki value of less than10 μM, were discovered. Compound 4 [NSC 66811, (7-[anilino(phenyl)methyl]-2-methyl-8-quinolinol)] with a Ki value of 120 nM had the highest binding affinity for MDM2. NSC 66811 was shown to activate p53 in the LNCaP prostate cancer cell line with wild-type p53 in a dose-dependent manner (52).
Computational screening using a pharmacophore model incorporating protein flexibility
A pharmacophore model was developed based on the crystal structure of MDM2 and incorporation of the protein flexibility was assessed using molecular dynamics simulation (64). Using this pharmacophore model, a database of approximately 50,000 synthetic compounds was searched. Evaluation of pharmacophore hits in a fluorescence-polarization MDM2 binding assay led to the discovery of five nonpeptidic, small-molecule MDM2 inhibitors, which represent five new scaffolds. The most potent inhibitor exhibited a Ki value of 110 nM. Their cellular activity and mechanism of action have not been reported.
Screening small focused libraries
A fluorescence polarization-based binding assay was used to screen a small library of terphenyl compounds to identify inhibitors of the MDM2-p53 interaction (50). The rationale for screening a terphenyl-based library was their ability to mimic one face of the alpha helical peptide (65). A library of 21 terphenyl derivatives was prepared by substituting the alkyl or aryl group on the three ortho positions of the terphenyl scaffold (50). The side chains in these terphenyl derivatives are projected in a way similar to i, i+4, and i+7 residues of an alpha-helix. An FP-based MDM2 binding assay based on the disruption of MDM2-p53 peptide complex was used to assess the binding affinities of the terphenyls. Screening of the library showed that terphenyl 14 was the most potent inhibitor, with a Ki value of 182 nM for MDM2. Binding to MDM2 was further confirmed using 15N HSQC NMR spectroscopy, which showed that terphenyl 14 targets the same surface area in MDM2 where p53 binds. Terphenyl 14 was thus discovered as an alpha helical mimetic of p53. However, in comparison to its ability to disrupt the MDM2-p53 interaction with a Ki value of 182 nM, terphenyl 14 disrupts the Bcl-2/Bak binding with a Ki value of 15 μM, and Bcl-xL/Bak with a Ki value of 2.5 μM, showing an 82- and 14-fold specificity, respectively, for MDM2 relative to Bcl-2 and Bcl-xL.
An in vitro, quantitative, ELISA-based assay was performed to screen another set of terphenyl derivatives to identify small-molecule inhibitors of the MDM2-p53 protein-protein interaction (51). Compounds were screened for the disruption of the MDM2-p53 interaction. This strategy yielded terphenyl compounds 2 and 6, which disrupted the MDM2-p53 interaction with IC50 values of 10 μM and 15 μM, respectively.
Chalcones are known to have anticancer properties (66). Therefore, Stoll et al. (53) screened a library of 16 chalcones by an ELISA-based assay to identify compounds that disrupt the interaction between MDM2 protein and p53 peptide. Potency of some chalcones to disrupt the MDM2-p53 interaction obtained by ELISA was confirmed by NMR titration experiments. Compound B and B-1, having IC50 values of 49 μM and 117 μM, respectively, were the most potent chalcones. These compounds also disrupted the MDM2-p53 interaction in the in vitro DNA binding electrophoretic mobility supershift assay (EMSA).
High-throughput screening of large chemical libraries
A breakthrough in the design of the potent small-molecule MDM2 inhibitor was obtained through the discovery of small molecules, termed Nutlins by Vassilev and colleagues (32). The Nutlins have a cis-imidazoline core structure. The cis-imidazoline lead compound was discovered by experimental screening of a diverse library of synthetic compounds using a Biacore surface plasmon resonance assay. Through extensive chemical modifications of the lead compound, potent small-molecule MDM2 inhibitors were ultimately obtained. Nutlin-1 and Nutlin-2 are racemic mixtures and Nutlin-3a is an active enantiomer isolated from racemic Nutlin-3 (Figure 2b). Nutlin-1, Nutlin-2, and Nutlin-3 disrupted the MDM2-p53 interaction with IC50 values of 260 nM, 140 nM, and 90 nM, respectively. X-ray crystal structure of the MDM2-Nutlin-2 complex was determined at 2.3 Å resolution. The crystal structure showed that Nutlin-2 binds to the p53 binding site in MDM2 (Figure 2a). Nutlin-2 mimics the interaction of the p53 peptide with MDM2 such that one bromophenyl moiety sits deeply in the Trp 23 pocket, the other bromophenyl group occupies the Leu 26 pocket, and the ethyl ether side chain is directed toward the Phe 19 pocket. The Nutlins are the first examples of potent and specific inhibitors of the MDM2-p53 interaction and one of them, Nutlin-3, has been extensively evaluated for its therapeutic potential and mechanism of action in human cancer.
Parks et al. (48) identified benzodiazepinedione-based MDM2 inhibitors through high-throughput screening of a library of 22,000 benzodiazepinediones, designed using Directed Diversity software, with the ThermoFluor microcalorimetry technology. This technology involves an affinity-based high-throughput screening assay using fluorescent dyes to monitor protein unfolding as a function of temperature for the identification of compounds which bind to MDM2. Detection of compound binding to MDM2 is measured by the resultant increase in thermal stability. Thermal stability is quantified as a change in midpoint transition temperature (ΔTm) in the presence of the compound at a single concentration. The sensitivity of this assay was verified by the shift in Tm on addition of peptides known to bind to MDM2, with higher-affinity peptides generating larger shifts. Based on the initial screening for MDM2 binding, hits were identified which were confirmed in an FP-based peptide displacement binding assay to identify inhibitors of the MDM2-p53 interaction. This study led to the identification of two benzodiazepinedione-based MDM2 inhibitors: Compound 20 and Compound 44 with IC50 value of 420 nM and 490 nM, respectively, in terms of ability to disrupt the MDM2-p53 interaction. Cell activity has not been reported thus far for these compounds.
Another benzodiazepinedione-based MDM2 inhibitor was identified by high-throughput screening of 338,000 compounds from combinatorial libraries using the same ThermoFluor microcalorimetry technology (47). Based on the initial screening, 1216 compounds were selected, which included 116 compounds belonging to a benzodiazepinedione library. The affinity of the selected compounds was confirmed in an FP-based p53 peptide displacement MDM2 binding assay. Benzodiazepinedione compound 1 was the most potent inhibitor, with a binding affinity of 80 nM. A 2.7 Å crystal structure of HDM2 in complex with compound 1 was determined. Compound 1 occupies the same pockets as the peptide side chains Phe 19, Trp 23, and Leu 26 of p53 in the MDM2 binding cleft. The MDM2 interactions with the inhibitor are largely nonspecific van der Waals contacts and, similar to the p53 peptide, the bound conformation of compound 1 is amphipathic. The bound pendant groups in compound 1 are projected towards the MDM2 pocket to, orient themselves such that they mimic the position of the hydrophobic side chains on one face of the helical p53 peptide ligand. Another series of benzodiazepinedione compounds identified by the ThermoFluor microcalorimetry technology and optimized by structure-based design were TDP521252 and TDP665759, which bind to MDM2 with IC50 values of 708 nM and 704 nM, respectively (49).
Structure-based de novo design
Nonpeptidic MDM2 inhibitors based upon the spiro-oxindole core structure were discovered in our laboratory using a structure-based de novo design strategy (34, 35, 46, 46a). The crystal structure shows that the interaction between p53 and MDM2 is primarily mediated by four key hydrophobic residues (Phe 19, Leu 22, Trp 23 and Leu 26) of p53 and a small but deep hydrophobic cleft in MDM2 (44). Because the indole ring of Trp 23 residue of p53 is buried deeply inside a hydrophobic cavity in MDM2 and its NH group forms a hydrogen bond with the backbone carbonyl in MDM2, Trp 23 appears to be most critical for binding of p53 to MDM2. Therefore a search for chemical moieties that can mimic the interaction of Trp 23 with MDM2 was performed. It was found that in addition to the indole ring itself, oxindole can nicely mimic the side chain of Trp 23 for interaction with MDM2. Because many anticancer drugs are natural products or derivatives of natural products, a substructure search technique was used to identify natural products that contain an oxindole substructure. Among the natural products identified were a number of natural alkaloids such as spirotryprostatin A and alstonisine, which both contain a spiro-oxindole substructure. Computational modeling studies predicted that although these compounds fit poorly into the MDM2 cleft (due to steric hindrance), the spiro(oxindole-3,3′-pyrrolidine) core structure may be used as the starting point for the design of a new class of MDM2 inhibitors. The oxindole can closely mimic the Trp 23 side chain in p53 in both hydrogen-bonding formation and hydrophobic interactions with MDM2, and the spiropyrrolidine ring provides a rigid scaffold from which two hydrophobic groups can be projected to mimic the side chain of Phe 19 and Leu 26. We have designed candidate compounds by using different R1, R2, and R3 groups with different configurations and docked them into the MDM2 binding cleft using the GOLD program. Docking studies predicted that compound 1a binds to MDM2 with a good affinity by virtue of its ability to mimic p53 (34). An FP-based binding assay showed that 1a binds to MDM2 with a Ki value of 8.5 μM and provided a starting point for further optimization. Modification of compound 1a yielded spiro-oxindole 1d, which binds to MDM2 with a Ki value of 86 nM. Computational docking predicted that compound 1d binds to MDM2 by mimicking Phe 19, Trp 23, and Leu 26 residues in p53.
X-ray structure of the MDM2-p53 complex showed that a fourth residue, Leu 22, also appears to play an important role in the overall interaction between p53 and MDM2 (44), a suggestion that finds support in results from mutation analysis (55) and alanine scanning of p53 peptides (43). Accordingly, we have designed new analogs to capture the additional interaction between Leu 22 in p53 and MDM2. Among these new compounds, compound 8 (termed MDM2 inhibitor-63 or MI-63) (46) binds to MDM2 with a Ki value of 3 nM.
MI-63 lacked the desirable pharmacological properties in mice and was unsuitable for in vivo evaluations. MI-63 was thus further optimized to obtain new analogs such as MI-219 (Figure 2b) (35). MI-219 mimics all four key residues in p53 and binds to MDM2 with a Ki value of 5 nM (Figure 2a), whereas Nutlin 3 has a Ki value of 36 nM (35). MI-219 has highly desirable pharmacological properties, such as 55% oral bioavailability in mice. (35). MI-219 was greater than10,000-fold selective for MDM2 over its closely related homolog MDMX. In addition, both MI-63 and MI-219 displayed greater than10,000-fold specificity for MDM2 relative to Bcl-2 and Bcl-xL (35, 46). Consistent with the high binding affinity for MDM2 and disruption of the MDM2-p53 complex, MI-63 and MI-219 induced accumulation of p53 in cancer cells with wild-type p53 (35, 46). The compounds inhibited growth of cancer cell lines expressing wild-type p53 with submicromolar IC50 values and showed a 20- to 50-fold-weaker activity in cancer cell lines lacking wild-type p53, indicating a critical role of p53 in cellular activity of the inhibitors (35, 46). Spiro-oxindoles MI-63 and MI-219 are the first examples of nonpeptidic MDM2 small molecules that are more potent and selective than Nutlin-3.
SMALL-MOLECULE MDM2 INHIBITORS AS CANCER THERAPEUTICS
Desirable Properties of a Small-Molecule MDM2 Inhibitor
In order to critically evaluate the mechanism of action and therapeutic potential of a MDM2 inhibitor, it should have the following desirable properties: (a) a high binding affinity and specificity to MDM2, (b) potent cellular activity in cancer cells with wild-type p53, and (c) a highly desirable pharmacokinetic (PK) profile.
To date, two MDM2 inhibitors, Nutlin-3 and MI-219, meet these criteria (32, 35). In addition, benzodiazepinedione compound TDP665759, which binds to MDM2 with a IC50 value of 704 nM, inhibits cell proliferation in cells expressing wild-type p53 (49). Other inhibitors of the MDM2-p53 interaction discovered thus far have either modest binding affinity for MDM2 and/or have only modest activity on cells. Moreover, their cellular selectivity for cancer versus normal cells, in vivo antitumor activity, and mechanism of action have not been reported.
Binding Affinity, Specificity, and Cell Permeability
In addition to achieving high binding affinity for MDM2, it is critical that the MDM2 inhibitors display an excellent specificity over other protein-protein interactions in order to critically evaluate the mechanism of action and therapeutic potential of targeting the MDM2-p53 interaction. In particular, because MDM2 inhibitors mimic the helical p53 peptide for binding to the hydrophobic cleft in MDM2, they may also target other helix-binding proteins such as the Bcl-2 family anti-apoptotic proteins.
To this end, spiro-oxindole inhibitors MI-63 (46) and MI-219 (35) were evaluated for their specificity relative to other protein-protein interactions. Both small molecules display excellent specificity for blocking the MDM2-p53 interaction. For example, MI-63 and MI-219 bind with a greater than 10,000-fold-weaker affinity to Bcl-2 and Bcl-xL proteins, both of which have a deep hydrophobic binding cleft and interact with a helical domain in their binding partners. Moreover, although MDMX is a closely related homolog of MDM2, and both proteins interact with p53 using similar binding clefts, MI-219 shows a greater than10,000-fold selectivity for MDM2 relative to MDMX. These data showed that it is indeed possible to design not only potent, but also highly selective, MDM2 inhibitors.
In addition, excellent cell permeability is essential for employing small molecules as anticancer agents. It is possible that a small molecule might have a high binding affinity to MDM2 but poor cell permeability. This is illustrated by benzodiazepinedione compound 52, which binds to MDM2 with an IC50 value of 810 nM but is poorly cell permeable (67). However, benzodiazepinedione analogs TDP521252 and TDP665759 bind to MDM2 with IC50 values of 708 nM and 704 nM, respectively, and are also cell permeable, as indicated by their IC50 values in inhibiting cell proliferation in a panel of cancer cell lines with wild-type p53 (49).
Potent and selective cis-imidazoline analog (Nutlin-3a) (32) and spiro-oxindole analogs (MI-63 and MI-219) (35, 46) show excellent cell permeability. Small molecules from these two classes have been evaluated for their in vitro and in vivo antitumor activity and mechanism of action (32, 35, 46, 46a). Nutlin-3 and MI-219 represent promising therapeutic candidates for drug development and, by specifically targeting the MDM2-p53 interaction, excellent tools to probe the reactivation of p53.
Reactivation of the p53 Pathway by MDM2 Inhibitors
Studies using the cis-imidazoline analog Nutlin-3 and spiro-oxindole analogs MI-63 and MI-219 have shown the potential of using pharmacological activation of p53 by disrupting the MDM2-p53 interaction as an anticancer strategy for cancers retaining wild-type p53 (4a, 32, 35, 46, 46a). The mechanism of induction of p53 accumulation by MDM2 inhibitors is different from that induced by radiation and traditional chemotherapy drugs. Both radiation and chemotherapy drugs induce p53 accumulation by posttranslational modifications of p53, such as phosphorylation. In contrast, MDM2 inhibitors induce accumulation and activation of p53 in cancer and normal cells without inducing DNA damage or requiring p53 phosphorylation (35, 46, 46a, 68–70, 71–73a). MDM2 inhibitors are thus nongenotoxic agents for the activation of p53.
Furthermore, at submicromolar concentrations such inhibitors decrease the proliferation of a variety of cancer cell lines that retain wild-type p53 with a high degree of specificity over cells that harbor mutated/deleted p53 (32, 35, 46, 46a) Nutlin-3b and MI-61, the respective inactive analogs from each class, bind to MDM2 weakly. These weak MDM2 inhibitors not only have weak cellular activity but also lack cellular specificity between cancer cells with wild-type p53 and those with mutated/deleted p53. The fact that Nutlin-3, MI-63, and MI-219 are able to inhibit cell proliferation in cancer cells lacking wild-type p53 at higher concentrations (greater than 10 μM), together with the observation that at higher concentration inactive analogs inhibit proliferation of cancer cell lines lacking wild-type p53, suggests that the cellular activity of MDM2 inhibitors at higher concentrations is independent of the p53 status and binding to MDM2.
MDM2 Inhibitors Induce Cell Cycle Arrest in Cancer and Normal Cells, but Selective Cell Death in Cancer Cells
It is predicted that activation of the p53 pathway by an MDM2 inhibitor may induce cell cycle arrest and/or apoptosis. Induction of cell cycle arrest by p53 is characterized by depletion of S-phase cells and accumulation at G1/S (74) and/or G2/M (75) phase boundaries of the cell cycle. Studies using Nutlin-3 and MI-219 show that MDM2 inhibitors yield both common and different cellular responses in normal and tumor cells (32, 35). In tumor cell lines, activation of p53 by these inhibitors induces p53- and p21-dependent cell cycle arrest and p53-dependent cell death (32, 35, 46a, 76). However in normal cells, p53 activation by MDM2 inhibitors leads to cell cycle arrest, but not cell death (32, 35, 46a, 77). Evaluation of MI-219 in seven different types of primary human normal cells or cell lines further confirmed that activation of p53 in normal cells by a potent and specific MDM2 inhibitor induces cell cycle arrest but does not cause cell death (32, 35). These results indicate that activation of p53 by an MDM2 inhibitor is nontoxic to normal cells and are thus encouraging from a therapeutic perspective.
Employing Nutlin-3, Tovar et al. (76) tested the effect on cell cycle arrest and apoptosis in 10 randomly selected cancer cell lines of seven different tumor types: colon, breast, lung, prostate, melanoma, osteosarcoma, and renal cancer. Nutlin-3 induced a complete depletion of the S-phase fraction, causing arrest at G1/S and/or G2/M phases in all the cell lines (32, 76). Furthermore, activation of p53 by Nutlin-3 caused transcriptional upregulation of p53 target gene cyclin-dependent kinase inhibitor p21Waf1/Cip1, which is an essential mediator of p53-induced cell cycle arrest (74). Clearly, the p53 pathway is preserved in all cancer cell lines with wild-type p53. Interestingly, it was found that the degree of induction of apoptosis by Nutlin-3 varied among cancer cell lines. SJSA-1 and MHM osteosarcoma cell lines with MDM2 gene amplification were the most sensitive, whereas HCT-116 (colon cancer), A549 (lung cancer), and H460 (lung cancer), which lack the MDM2 gene amplification, were least sensitive. However, U20S, an osteosarcoma cell line lacking the MDM2 gene amplification, markedly resisted apoptosis. LNCaP (prostate cancer), 22Rv1 (prostate cancer), and RKO (colon cancer) with a single copy of MDM2 gene showed intermediate levels of apoptotic response. Thus, amplification of the MDM2 gene in cancer cells may indicate intact p53-dependent apoptosis machinery and such cancer cells are thus likely to be highly sensitive to MDM2 inhibitors. DNA microarray analysis (using Affymetrix microarrays) identified the differential gene expression in cells with high (SJSA-1 and MHM) and low (U2OS and HCT-116) apoptotic index, but did not reveal the identity of the gene set that contributes to differential sensitivity of cells to apoptosis induced by Nutlin-3 (76).
Therapeutic Potential of MDM2 Inhibitors in Blood Malignancies
Because the activity of MDM2 inhibitors depends upon p53 activation in cells expressing wild-type p53, blood malignancies, such as acute myeloid leukemia (AML), B-chronic lymphocytic leukemia (B-CLL), and multiple myeloma, which each have infrequent p53 mutation/deletion, are potentially attractive tumor types for MDM2 inhibitor-based therapy. Ex vivo experiments using AML (78, 79), B-CLL (80–83), and multiple myeloma (84) patient specimens have indeed shown that inhibition of MDM2 by Nutlin-3 and MI-63 effectively triggers apoptosis. Conclusive evidence of the dependence of the activity of MDM2 inhibitors upon p53 status in B-CLL was provided by a recent study using MI-63 and Nutlin-3 in a cohort of more than 100 B-CLL patients (83). Nutlin-3 synergizes with doxorubicin and cytosine arabinoside in killing myeloblasts in AML and with doxorubicin, chlorambucil, and fludarabine in killing leukemic cells in B-CLL patient specimens (80–82). Importantly, both the single agent and the combination effect of Nutlin-3 are selective for cancer versus normal cells, as revealed by the lack of toxicity to peripheral blood mononuclear cells or bone marrow-derived hematopoietic progenitors and bone marrow stromal epithelium cells (78, 80, 84). Ataxia telangiectasia mutated (ATM) protein kinase is a key regulator of p53 activity and a central mediator of cellular responses to DNA double-strand breaks (84a). Low levels of ATM rendered fludarabine ineffective but did not influence Nutlin-3 activity, suggesting that MDM2 inhibitors may retain their activity in cancers missing upstream signals that regulate p53 (78).
MDM2 Inhibitors as Chemoprotective Agents
The ability of MDM2 inhibitors to halt cell cycle progression can be exploited to protect normal cells from the toxic effects of chemotherapy. MDM2 inhibitors halt cell cycle progression at the G1/S and G2/M phases and can thus attenuate the activity of S-phase- and M-phase-specific drugs. For example, taxanes kill cells in M-phase via interfering with the microtubule assembly, whereas gemcitabine and Ara-C kill proliferating cells in S-phase. Treatment of normal proliferating fibroblasts or keratinocytes with Nutlin-3 protects normal cells against these chemotherapy drugs without altering their activity against cancer cells lacking wild-type p53 (85, 86). However, these data from in vitro studies await confirmation in vivo.
MDMX Expression Attenuates the Cellular Activity of MDM2 Inhibitors
MDMX (MDM4 in mouse) is a protein with a high degree of homology to MDM2 and these two proteins have very similar p53-binding sites (87, 88). The MDMX protein binds to the N terminus of p53 and suppresses p53 transcriptional activity but does not cause p53 degradation (88, 89). Unlike MDM2, MDMX is not a transcriptional target of p53 (88). MDM2 and MDMX bind to each other through their respective C terminus RING domains and together with p53 form a p53-MDM2-MDMX trio, which regulates p53 function (90) (Figure 1). Knocking out MDMX in mice causes embryonic lethality, which is rescued by loss of p53 gene (91, 92). Thus, MDMX, like MDM2, is an important regulator of p53 during embryonic development.
Due to close structural similarity between the p53-binding pockets of MDM2 and MDMX (87), it was assumed that an MDM2 inhibitor might have similar affinities in targeting the MDMX-p53 interaction. However, both MI-219 and Nutlin-3 show a high degree of specificity for the MDM2-p53 compared to the MDMX-p53 interaction (35, 68, 69). Direct comparison of the two small-molecule inhibitors showed that MI-219 has a greater than 10,000-fold selectivity, whereas Nutlin-3 has approximately a 250-fold selectivity for the MDM2-p53 compared with the MDMX-p53 interaction (35). Recent availability of the crystal structure of the N-terminal domain of MDMX bound to a 15-residue p53 peptide showed that although basic features of the MDM2-p53 and MDMX-p53 interactions are similar, there are some differences between the binding pockets in MDM2 and MDMX (93). The differences in the p53 binding pockets between MDM2 and MDMX explain the high specificity for MI-219 and Nutlin-3 for MDM2 compared with MDMX.
By targeting the MDM2-p53 interaction, Nutlin-3 efficiently induced apoptosis and decreased long-term survival of MDM2-overexpressing fibroblasts, transformed with E1A/RasV12/hTERT (70). In contrast, due to its inability to efficiently target the MDMX-p53 interaction, Nutlin-3 (68, 70, 71) was ineffective in cells transformed with MDMX. Overexpression of MDMX also decreased the ability of Nutlin-3 to activate the transcriptional function of p53 in MDMX-overexpressing cells, whereas silencing MDMX by RNAi enhanced the activity of Nutlin-3 (68, 70, 71). Interestingly, although Nutlin-3 and MI-219 did not efficiently target the MDMX-p53 interaction, they induced MDMX degradation in some cancer cell lines, implicating an indirect mechanism for inactivating MDMX (35, 68, 71). The inability of MDM2 inhibitors to target the MDMX-p53 interaction or induce MDMX degradation prevents full activation of p53, imparting resistance against MDM2 inhibitors. Nutlin-3 failed to induce apoptosis in cancer cell lines, such as MCF-7 (breast cancer), U2OS (osteosatcoma), BL40 (Burkitt’s lymphoma cells), and JEG3 (choriocarcinoma), in which it failed to induce MDMX downregulation, but induced apoptosis in BL2 (Burkitt’s lymphoma) cells in which it caused downregulation of MDMX protein (68, 71). Silencing MDMX by RNA interference (RNAi) relieves transcriptional repression of p53 and sensitizes MDMX-overexpressing cell lines to apoptosis by Nutlin-3 (70). Thus, the combination of MDM2 inhibitors with chemotheraputic agents such as doxorubicin, which induces MDMX degradation, or with a BH3 mimetic ABT-737, which activates Bax and overcomes MDMX-mediated suppression of p53 function, might have therapeutic value (70a). Downregulation of MDMX by Nutlin-3 is not at the transcriptional level, but perhaps due to ubiquitinization by MDM2 and consequent proteasomal degradation (71). Even in a cancer cell line, such as LNCaP (prostate cancer), which undergoes MDMX degradation by MI-219, silencing MDMX by RNAi further enhances the ability of the MDM2 inhibitor to induce apoptosis and cell cycle arrest, indicating that residual MDMX is sufficient to inhibit p53 function (35).
MDM2 and MDMX dimerize with each other through their RING finger domains. Overex-pression of a RING finger domain–deleted MDMX mutant in transformed fibroblasts did not provide protection to cells against Nutlin-3, indicating that the MDM2-MDMX interaction is required for protective function of MDMX, probably for the MDM2-mediated ubiquitinization of MDMX (70). These studies also suggest the importance of designing small-molecule inhibitors of the MDMX-p53 interaction or dual small-molecule inhibitors of the MDM2-p53/MDMX-p53 interactions for the complete reactivation of p53.
Nutlin-3, despite its selectivity for MDM2-p53 over MDMX-p53 interaction (68, 70, 71), blocks intracellular MDMX-p53 interaction in retinoblastoma cells (69). Nutlin-3 did not induce degradation but disrupted the MDMX-p53 interaction in Weri-1 retinoblastoma cells with MDMX amplification, reduced their viability in a p53-dependent manner, and nullified the growth advantage provided by MDMX, even in the absence of MDM2. In an orthotopic retinoblastoma model, Nutlin-3 alone was inactive at reducing tumor growth (69). However, combined subconjuctival injection of Nutlin-3 with topotecan reduced tumor burden by 82-fold with no systemic ocular toxicity, whereas systemic treatment with Nutlin-3/topotecan caused only a 5-fold decrease in tumor burden with severe side effects, suggesting that the concentrations of Nutlin-3 through subconjuctival injection might be sufficient to disrupt MDMX-p53.
Pharmacological Properties and In Vivo Antitumor Activity of MDM2 Inhibitors
Translating a promising MDM2 inhibitor with excellent in vitro properties into an anticancer drug requires that the inhibitor have excellent pharmacological properties and be nontoxic to normal tissues.
Pharmacokinetic (PK) studies showed that MI-219 has an excellent PK profile and is orally bioavailable (35). In xenograft models of human prostate cancer and osteosarcoma retaining wild-type p53, oral administration of Nutlin-3 and MI-219 induced robust accumulation and activation of p53 in tumor tissues (35, 76). Accumulation of p53 by treatment with MI-219 could be detected at 1 h and 3 h time points, but not at 6 h, which indicated that activation of p53 is transient but correlates with the levels of MI-219 in the plasma (35).
Administration of Nutlin-3 (32, 76, 94) and MI-219 (35) completely inhibited tumor growth in several xenograft models of human cancer with wild-type p53, including SJSA-1 osteosarcoma and LNCaP prostate cancer. MI-219 did not have significant effect on the growth of MDA-MB-231 breast cancer xenograft tumors harboring mutant p53, indicating that the antitumor activity of MDM2 inhibitors correlates with the p53 status (35). Importantly, both Nutlin-3 and MI-219 achieved their antitumor activity without causing visible signs of toxicity in animals, as assessed by necropsy studies and assessment of body weight.
Toxicity of MDM2 Inhibitors to Normal Tissues
Tissue distribution analysis of MI-219 in nude mice bearing SJSA-1 xenografts showed that MI-219 is very well distributed in plasma and normal mouse tissues from different organs (35). The concentrations of MI-219 in SJSA-1 tumor tissues were very similar to those observed in spleen and plasma, but much lower than those in other tissues, such as lung, liver, and kidney. Therefore, the lack of toxicity of MI-219 to animals was not due to low drug exposure in normal tissues.
Radiosensitive tissues, such as small-intestine crypts and thymus, are extremely susceptible to p53-induced apoptosis (95, 96). Restoration of p53 by a genetic approach in the absence of MDM2 results in severe pathological damage to radiosensitive mouse tissues and death of all animals within five days (77), raising concern that p53 activation by MDM2 inhibitors will be toxic to healthy tissues. Both Nutlin-3 (32) and MI-219 (35) show little toxicity to animals at therapeutically efficacious dose schedules. Whereas both γ-radiation and irinotecan chemotherapy induce profound apoptosis in small-intestine crypts and thymus, MI-219, in either single or repeated doses, did not cause apoptosis or damage in either radiosensitive or radioresistant normal mouse tissues (35). Interestingly, in contrast to chemotherapy and radiation, which induce profound accumulation and activation of p53 protein in intestinal crypts and thymus, MI-219 activated p53 in normal tissues with minimal p53 accumulation. Therefore, the higher levels of p53 accumulation induced by radiation and chemotherapeutic agents than by MI-219 could account for their different toxicities.
CONCLUDING REMARKS
Targeting the MDM2-p53 protein-protein interaction using small molecules to reactivate p53 function represents a potentially attractive therapeutic strategy for the treatment of human cancers retaining wild-type p53. Intensive research efforts in the past decade have now yielded Nutlin-3 and MI-219 as potent and specific inhibitors of the MDM2-p53 interaction with desirable pharmacological properties. A number of these small-molecule inhibitors, such as analogs of MI-219 and Nutlin-3, have progressed to advanced preclinical development or early-phase clinical trials. Clinical testing of these new agents will provide the ultimate proof of the usefulness of this therapeutic strategy for the treatment of human cancers.
SUMMARY POINTS
In normal cells, direct binding between the p53 tumor suppressor and MDM2 oncoprotein tightly regulates p53 activity in an autoregulatory feedback manner.
Mutation or deletion of the p53 tumor suppressor gene and overexpression of MDM2 oncoprotein promote tumorigenesis.
Half of human cancers have alterations in the p53 gene, and in the remaining cancers retaining wild-type p53, the function of p53 is inactivated by its interaction with MDM2.
Targeting the MDM2-p53 protein-protein interaction is an attractive therapeutic strategy for the treatment of cancer.
The crystal structure of MDM2 protein in complex with a p53 peptide provides a structural basis for designing small-molecule drugs targeting the MDM2-p53 interaction.
Computational structure-based screening, high-throughput screening and structure-based design are some of the approaches employed in the discovery of small-molecule inhibitors of the MDM2-p53 interaction.
Analogs of MI-219 and Nutlin-3 have progressed to advanced preclinical development or early-phase clinical trials.
FUTURE ISSUES
What tumor types and subsets of tumors retaining wild-type p53 are particularly sensitive to MDM2 inhibitors?
Will treatment of tumors with MDM2 inhibitors lead to mutations in p53 or other defects in the p53 pathway?
What other pathways in addition to p53 are utilized by MDM2 inhibitors for achieving antitumor response?
Can MDM2 inhibitors be used as single agents or in combination regimens for the treatment of cancers with mutated p53?
Acknowledgments
Funding from the National Cancer Institute/National Institutes of Health, the Prostate Cancer Foundation, the Leukemia and Lymphoma Society, and Ascenta Therapeutics, Inc. is greatly appreciated. We thank Dr. Denzil Bernard for the preparation of Figure 2. We apologize in advance to any investigators whose studies are not cited owing to page limitations.
Footnotes
The Annual Review of Pharmacology and Toxicology is online at pharmtox.annualreviews.org
DISCLOSURE STATEMENT
S.W. and the University of Michigan own equity in Ascenta Therapeutics, Inc., which has licensed the technologies related to the MDM2 inhibitors of the spiro-oxindole class. S.W. serves as a consultant for Ascenta and is the principal investigator on a research contract from Ascenta to the University of Michigan.
LITERATURE CITED
- 1.Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–40. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 2.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4a.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14:5318–24. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–63. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 6.Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 7.DeLeo AB, Jay G, Appella E, Dubois GC, Law LW, Old LJ. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc Natl Acad Sci USA. 1979;76:2420–24. doi: 10.1073/pnas.76.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–93. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348:747–49. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 10.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–38. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 11.Feki A, Irminger-Finger I. Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit Rev Oncol Hematol. 2004;52:103–16. doi: 10.1016/j.critrevonc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 13.Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–69. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fakharzadeh SS, Rosenblum-Vos L, Murphy M, Hoffman EK, George DL. Structure and organization of amplified DNA on double minutes containing the mdm2 oncogene. Genomics. 1993;15:283–90. doi: 10.1006/geno.1993.1058. [DOI] [PubMed] [Google Scholar]
- 15.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 16.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M, Gu L, Abshire TC, Homans A, Billett AL, et al. Incidence and prognostic significance of MDM2 oncoprotein overexpression in relapsed childhood acute lymphoblastic leukemia. Leukemia. 2000;14:61–67. doi: 10.1038/sj.leu.2401619. [DOI] [PubMed] [Google Scholar]
- 18.Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. doi: 10.2174/1568009053332636. [DOI] [PubMed] [Google Scholar]
- 19.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–59. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunther T, Schneider-Stock R, Hackel C, Kasper HU, Pross M, et al. Mdm2 gene amplification in gastric cancer correlation with expression of Mdm2 protein and p53 alterations. Mod Pathol. 2000;13:621–26. doi: 10.1038/modpathol.3880107. [DOI] [PubMed] [Google Scholar]
- 21.Bond GL, Hu W, Levine AJ. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 22.Capoulade C, Bressac-de Paillerets B, Lefrere I, Ronsin M, Feunteun J, et al. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene. 1998;16:1603–10. doi: 10.1038/sj.onc.1201702. [DOI] [PubMed] [Google Scholar]
- 23.Momand J, Wu HH, Dasgupta G. MDM2–master regulator of the p53 tumor suppressor protein. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 24.Ganguli G, Abecassis J, Wasylyk B. MDM2 induces hyperplasia and premalignant lesions when expressed in the basal layer of the epidermis. EMBO J. 2000;19:5135–47. doi: 10.1093/emboj/19.19.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–22. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 25a.Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–59. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–65. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 27.Martins CP, Brown-Swigart L, Evan GI. Modeling the th erapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Xue W, Zender L, Miething C, Dickins RA, Hernando E, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–88. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 30.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–28. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 31.Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–48. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 33.Wiman KG. Strategies for therapeutic targeting of the p53 pathway in cancer. Cell Death Differ. 2006;13:921–26. doi: 10.1038/sj.cdd.4401921. [DOI] [PubMed] [Google Scholar]
- 34.Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, et al. Structure-based design of potent nonpeptide MDM2 inhibitors. J Am Chem Soc. 2005;127:10130–31. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 35.Shangary S, Qin D, McEachern D, Liu M, Miller RS, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105:3933–38. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juven-Gershon T, Oren M. Mdm2: the ups and downs. Mol Med. 1999;5:71–83. [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–32. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 39.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 40.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 41.Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. mdm2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23:462–72. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–14. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2–fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–29. [PubMed] [Google Scholar]
- 44.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–53. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 45.Vassilev LT. Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3:419–21. [PubMed] [Google Scholar]
- 46.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2006;49:3432–35. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 46a.Shangary S, Ding K, Qiu S, Nikolovska-Coleska Z, Bauer JA, et al. Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer. Mol Cancer Ther. 2008;7:1533–42. doi: 10.1158/1535-7163.MCT-08-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005;48:909–12. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 48.Parks DJ, Lafrance LV, Calvo RR, Milkiewicz KL, Gupta V, et al. 1,4-Benzodiazepine-2,5-diones as small molecule antagonists of the HDM2-p53 interaction: discovery and SAR. Bioorg Med Chem Lett. 2005;15:765–70. doi: 10.1016/j.bmcl.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Koblish HK, Zhao S, Franks CF, Donatelli RR, Tominovich RM, et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5:160–69. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- 50.Yin H, Lee GI, Park HS, Payne GA, Rodriguez JM, et al. Terphenyl-based helical mimetics that disrupt the p53/HDM2 interaction. Angew Chem Int Ed Engl. 2005;44:2704–7. doi: 10.1002/anie.200462316. [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Yin H, Farooqi B, Sebti S, Hamilton AD, Chen J. p53 alpha-Helix mimetics antagonize p53/MDM2 interaction and activate p53. Mol Cancer Ther. 2005;4:1019–25. doi: 10.1158/1535-7163.MCT-04-0342. [DOI] [PubMed] [Google Scholar]
- 52.Lu Y, Nikolovska-Coleska Z, Fang X, Gao W, Shangary S, et al. Discovery of a nanomolar inhibitor of the human murine double minute 2 (MDM2)-p53 interaction through an integrated, virtual database screening strategy. J Med Chem. 2006;49:3759–62. doi: 10.1021/jm060023+. [DOI] [PubMed] [Google Scholar]
- 53.Stoll R, Renner C, Hansen S, Palme S, Klein C, et al. Chalcone derivatives antagonize interactions between the human oncoprotein MDM2 and p53. Biochemistry. 2001;40:336–44. doi: 10.1021/bi000930v. [DOI] [PubMed] [Google Scholar]
- 54.Galatin PS, Abraham DJ. A nonpeptidic sulfonamide inhibits the p53-mdm2 interaction and activates p53-dependent transcription in mdm2-overexpressing cells. J Med Chem. 2004;47:4163–65. doi: 10.1021/jm034182u. [DOI] [PubMed] [Google Scholar]
- 55.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–46. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 56.Bottger V, Bottger A, Howard SF, Picksley SM, Chene P, et al. Identification of novel mdm2 binding peptides by phage display. Oncogene. 1996;13:2141–47. [PubMed] [Google Scholar]
- 57.Garcia-Echeverria C, Chene P, Blommers MJ, Furet P. Discovery of potent antagonists of the interaction between human double minute 2 and tumor suppressor p53. J Med Chem. 2000;43:3205–8. doi: 10.1021/jm990966p. [DOI] [PubMed] [Google Scholar]
- 58.Milne GW, Nicklaus MC, Driscoll JS, Wang S, Zaharevitz D. National Cancer Institute Drug Information System 3D database. J Chem Inf Comput Sci. 1994;34:1219–24. doi: 10.1021/ci00021a032. [DOI] [PubMed] [Google Scholar]
- 59.Voigt JH, Bienfait B, Wang S, Nicklaus MC. Comparison of the NCI open database with seven large chemical structural databases. J Chem Inf Comput Sci. 2001;41:702–12. doi: 10.1021/ci000150t. [DOI] [PubMed] [Google Scholar]
- 60.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–48. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 61.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–23. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 62.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des. 1997;11:425–45. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 63.Wang R, Lai L, Wang W. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 64.Bowman AL, Nikolovska-Coleska Z, Zhong H, Wang S, Carlson HA. Small molecule inhibitors of the MDM2-p53 interaction discovered by ensemble-based receptor models. J Am Chem Soc. 2007;129:12809–14. doi: 10.1021/ja073687x. [DOI] [PubMed] [Google Scholar]
- 65.Kutzki O, Park HS, Ernst JT, Orner BP, Yin H, Hamilton AD. Development of a potent Bcl-x(L) antagonist based on alpha-helix mimicry. J Am Chem Soc. 2002;124:11838–39. doi: 10.1021/ja026861k. [DOI] [PubMed] [Google Scholar]
- 66.Go ML, Wu X, Liu XL. Chalcones: an update on cytotoxic and chemoprotective properties. Curr Med Chem. 2005;12:481–99. doi: 10.2174/0929867053363153. [DOI] [PubMed] [Google Scholar]
- 67.Leonard K, Marugan JJ, Raboisson P, Calvo R, Gushue JM, et al. Novel 1,4-benzodiazepine-2,5-diones as Hdm2 antagonists with improved cellular activity. Bioorg Med Chem Lett. 2006;16:3463–68. doi: 10.1016/j.bmcl.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 68.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–35. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 69.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 70.Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–76. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- 70a.Wade M, Rodewald LW, Espinosa JM, Wahl GM. BH3 activation blocks Hdmx suppression of apoptosis and cooperates with Nutlin to induce cell death. Cell Cycle. 2008;7:1973–82. doi: 10.4161/cc.7.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–44. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 72.Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, et al. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem. 2004;279:53015–22. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 73.Cheok CF, Dey A, Lane DP. Cyclin-dependent kinase inhibitors sensitize tumor cells to nutlin-induced apoptosis: a potent drug combination. Mol Cancer Res. 2007;5:1133–45. doi: 10.1158/1541-7786.MCR-07-0161. [DOI] [PubMed] [Google Scholar]
- 73a.Jones RJ, Chen Q, Voorhees PM, Young KH, Bruey-Sedano N. Inhibition of the p53 E3 ligase HDM-2 induces apoptosis and DNA damage–independent p53 phosphorylation in mantle cell lymphoma. Clin Cancer Res. 2008;14:5416–25. doi: 10.1158/1078-0432.CCR-08-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.el-Deiry WS, Harper JW, O’Connor PM, Velculescu VE, Canman CE, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–74. [PubMed] [Google Scholar]
- 75.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 76.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–93. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–14. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 78.Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106:3150–59. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Secchiero P, Corallini F, Gonelli A, Dell’Eva R, Vitale M, et al. Antiangiogenic activity of the MDM2 antagonist nutlin-3. Circ Res. 2007;100:61–69. doi: 10.1161/01.RES.0000253975.76198.ff. [DOI] [PubMed] [Google Scholar]
- 80.Secchiero P, Barbarotto E, Tiribelli M, Zerbinati C, di Iasio MG, et al. Functional integrity of the p53-mediated apoptotic pathway induced by the nongenotoxic agent nutlin-3a in B-cell chronic lymphocytic leukemia (B-CLL) Blood. 2006;107:4122–29. doi: 10.1182/blood-2005-11-4465. [DOI] [PubMed] [Google Scholar]
- 81.Kojima K, Konopleva M, McQueen T, O’Brien S, Plunkett W, Andreeff M. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood. 2006;108:993–1000. doi: 10.1182/blood-2005-12-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coll-Mulet L, Iglesias-Serret D, Santidrian AF, Cosialls AM, de Frias M, et al. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood. 2006;107:4109–14. doi: 10.1182/blood-2005-08-3273. [DOI] [PubMed] [Google Scholar]
- 83.Saddler C, Ouillette P, Kujawski L, Shangary S, Talpaz M, et al. Comprehensive biomarker and genomic analysis identifies P53 status as the major determinant of response to MDM2 inhibitors in chronic lymphocytic leukemia. Blood. 2007;111:1584–93. doi: 10.1182/blood-2007-09-112698. [DOI] [PubMed] [Google Scholar]
- 84.Stuhmer T, Chatterjee M, Hildebrandt M, Herrmann P, Gollasch H, et al. Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood. 2005;106:3609–17. doi: 10.1182/blood-2005-04-1489. [DOI] [PubMed] [Google Scholar]
- 84a.Kastan MB. Our cells get stressed too! Implications for human disease. Blood Cells Mol Dis. 2007;39:148–50. doi: 10.1016/j.bcmd.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carvajal D, Tovar C, Yang H, Vu BT, Heimbrook DC, Vassilev LT. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res. 2005;65:1918–24. doi: 10.1158/0008-5472.CAN-04-3576. [DOI] [PubMed] [Google Scholar]
- 86.Kranz D, Dobbelstein M. Nongenotoxic p53 activation protects cells against S-phase-specific chemotherapy. Cancer Res. 2006;66:10274–80. doi: 10.1158/0008-5472.CAN-06-1527. [DOI] [PubMed] [Google Scholar]
- 87.Bottger V, Bottger A, Garcia-Echeverria C, Ramos YF, Van Der Eb AJ, et al. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene. 1999;18:189–99. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 88.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–57. [PMC free article] [PubMed] [Google Scholar]
- 89.Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20:1001–7. doi: 10.1128/mcb.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 91.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 92.Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–38. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Popowicz GM, Czarna A, Rothweiler U, Szwagierczak A, Krajewski M, et al. Molecular basis for the inhibition of p53 by Mdmx. Cell Cycle. 2007;6:2386–92. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- 94.Sarek G, Kurki S, Enback J, Iotzova G, Haas J, et al. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J Clin Invest. 2007;117:1019–28. doi: 10.1172/JCI30945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–49. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 96.Potten CS, Wilson JW, Booth C. Regulation and significance of apoptosis in the stem cells of the gastrointestinal epithelium. Stem Cells. 1997;15:82–93. doi: 10.1002/stem.150082. [DOI] [PubMed] [Google Scholar]