Abstract
Mutations in the ELG1 gene of yeast lead to genomic instability, manifested in high levels of genetic recombination, chromosome loss, and gross chromosomal rearrangements. Elg1 shows similarity to the large subunit of the Replication Factor C clamp loader, and forms a RFC-like (RLC) complex in conjunction with the 4 small RFC subunits. Two additional RLCs exist in yeast: in one of them the large subunit is Ctf18, and in the other, Rad24. Ctf18 has been characterized as the RLC that functions in sister chromatid cohesion. Here we present evidence that the Elg1 RLC (but not Rad24) also plays an important role in this process. A genetic screen identified the cohesin subunit Mcd1/Scc1 and its loader Scc2 as suppressors of the synthetic lethality between elg1 and ctf4. We describe genetic interactions between ELG1 and genes encoding cohesin subunits and their accessory proteins. We also show that defects in Elg1 lead to higher precocious sister chromatid separation, and that Ctf18 and Elg1 affect cohesion via a joint pathway. Finally, we localize both Ctf18 and Elg1 to chromatin and show that Elg1 plays a role in the recruitment of Ctf18. Our results suggest that Elg1, Ctf4, and Ctf18 may coordinate the relative movement of the replication fork with respect to the cohesin ring.
Introduction
Genomic instability is a hallmark of cancer cells. Most human cancer cells show signs of genome instability, ranging from elevated mutation rates, to gross chromosomal rearrangements including deletions and translocations. Several surveillance and repair mechanisms operate in eukaryotic cells to ensure the stability of the genome. The current view is that most spontaneous chromosomal rearrangements arise during DNA replication. The activity of the DNA polymerases may be impaired by the presence of secondary structures, bound proteins or DNA lesions; this may lead to stalling or even collapse of replication forks. In response, cellular mechanisms are activated that arrest cell cycle progression, induce DNA repair, and restore replication [1], [2]. These repair mechanisms act on lesions to promote their repair and to prevent them from being converted into fatal genomic rearrangements. Pathways for DNA repair and genome stability are highly conserved across species. Because of this conservation, simple organisms, such as the budding yeast Saccharomyces cerevisiae, are extremely useful for studying the basic principles of genome stability and maintenance.
The ELG1 gene was identified in yeast as a mutant that causes enhanced levels of genomic instability [3], [4], [5]. Deletion of ELG1 in yeast leads to increased recombination levels [4], [6], as well as elevated levels of chromosome loss [4], [7] and gross chromosomal rearrangements [7]. elg1 mutants also exhibit elongated telomeres [8] and increased levels of Ty transposition [9]. Elg1 function is thus clearly required for maintaining genome stability during normal growth, and its absence has severe genetic consequences. The Elg1 protein shares sequence homology with the large subunit of RFC clamp loader and with the product of two additional genes involved in checkpoint functions and genome maintenance: RAD24 and CTF18. Elg1, Rad24 and Ctf18 form three alternative RFC-like (RLC) protein complexes, by interacting with the four small subunits of RFC [3], [4], [5]. The Rad24 RLC (Rad17 in S. pombe and hRad17 humans) [10] is predicted to load an alternative DNA sliding clamp (9-1-1) composed of checkpoint proteins [Ddc1/Rad17/Mec3 in S. cerevisiae, Rad9/Rad1/Hus1 in S. pombe and humans] [11]. Genetic data indicates that the Elg1, Ctf18 and Rad24 RLCs work in three separate pathways important for maintaining the integrity of the genome and for coping with various genomic stresses. Mutations in each of the genes cause a mild sensitivity to DNA damage or DNA replication block. Double mutants show increased sensitivity, whereas the triple mutant is extremely sensitive and shows impaired growth [4].
Genetic screens in yeast have shown that elg1 mutants have synthetic growth defects when combined with genes that are involved in homologous recombination, DNA repair, replication fork restart, checkpoint response and sister chromatid cohesion [2], [3], [12]. These genetic interactions emphasize the pivotal role of Elg1 as a guardian of genomic stability. Among the strongest synthetic interactions is the one with ctf4: double mutant spores elg1 ctf4 germinate, but fail to undergo cell divisions [3], [4], [13]. In addition, Δctf4 and Δelg1 mutant cells show partially overlapping phenotypes. Both mutants have high levels of recombination, chromosome loss, as well as synthetic growth defects with rad52, mec1 [14], [15] and other genes that are involved in DNA metabolism.
CTF4 was found, together with CTF18, in two independent screens for mutants that affect chromosome transmission fidelity [16], [17]. Both ctf4 and ctf18 exhibit elevated levels of recombination [18] and share genetic interactions with genes involved in DNA replication [14]. The proposed role for these proteins is in sister chromatid cohesion [19], [20]. Sister chromatids are held close to each other with the help of a complex called cohesin. Cohesin is composed of four subunits and forms a ring, although the precise topology of the ring relative to the replicated DNA is still debated [e.g.: [21], [22]]. Cohesin complexes are loaded on the DNA during late G1 at specific sites along the chromosomes. Although they are present along the whole chromosome arm with an average distance of 11 kb, they are especially enriched near the centromeres [23]. Sister chromatids remain associated from the beginning of S phase throughout G2, until metaphase. During mitosis the microtubules are bound to the kinetochore, establishing bipolar attachment to the spindle apparatus. Cohesin complexes oppose the tension that is implemented by the microtubules until all the chromosomes are bound to the microtubules properly and are arranged in the metaphase plate. Only then, a tightly regulated cascade is activated to trigger the cleavage of Mcd1/Scc1 (a cohesin subunit, hereafter referred to as Scc1 for simplicity), enabling the separation of sister chromosomes to opposite poles of the nucleus. Defects in cohesin loading, establishment, maintenance or cleavage lead to chromosome loss and cell death.
A clear linkage exists between replication progression and sister chromatid cohesion. First, Ctf7/Eco1, encoded by another gene that was found in the ctf screen for increased chromosome loss [17], is required for cohesion establishment solely during S phase. Mutations in this gene are synthetic lethal (SL) with mutations in the DNA replication clamp PCNA and overexpression of PCNA partially suppresses temperature sensitive ctf7 mutants, allowing them to grow at a higher temperature before arresting [24], [25], [26]. Second, loading of cohesin at the beginning of S phase is essential for proper sister chromatid cohesion. If cohesin subunits are expressed after DNA replication, they are successfully loaded onto the chromatin but fail to establish sister chromatid cohesion [27]. Finally, in the absence of Ctf4 or Ctf18, which are proteins that act close to the replication fork [28], a strong phenotype of precocious sister chromatid separation is observed. In addition, Ctf4 was shown to bind physically to the catalytic subunit of polymerase α [14].
Although ELG1 has tight genetic interactions with CTF4 and CTF18, it is not known how it affects genomic stability. Here we performed a genetic screen in order to find high copy number suppressors of the SL interaction between elg1 and ctf4. We found a cohesin subunit and a cohesin loader, pointing to the fact that the synergistic genetic interaction between elg1 and ctf4 is due to enhanced defects in sister chromatid cohesion. Consistent with this possibility we also show that elg1 mutants exhibit defects in cohesion. Our results show that both the Elg1 and the Ctf18 clamp loaders affect sister chromatin cohesion and establish a link between Elg1 activity and cohesin loading.
Results
A screen for high copy number suppressors of the synthetic lethal phenotype of elg1 ctf4
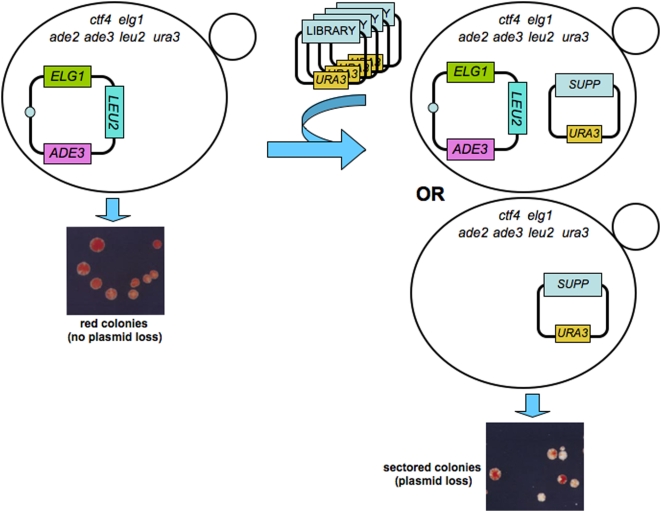
Mutations in ELG1 and CTF4 are synthetic lethal (SL), namely, despite the fact that each single-mutant grows well, the double mutant is inviable [2], [4]. In order to further investigate the interaction between these genes, we carried out a genetic screen for high-copy-number suppressors of the synthetic lethality (Figure 1). First, we constructed a double mutant strain carrying a ELG1 centromeric plasmid enabling the strain to stay alive. This plasmid has two marker genes: LEU2 and ADE3. In the appropriate genetic background, the presence of the ADE3 gene causes accumulation of a red pigment. Thus, elg1 ctf4 strains carrying the LEU2/ADE3/ELG1 plasmid grow well, and form uniformly red colonies, as they are unable to lose the ELG1-containing plasmid. These cells were then transformed with a yeast genomic library cloned in a high copy number plasmid (containing the URA3 marker). Cells that received a plasmid with a gene that, when over-expressed, can suppress the SL phenotype of elg1 ctf4 are now able to lose the ELG1-ADE3-LEU2 plasmid, and therefore show white/red sectored colonies (Figure 1).
Figure 1. Schematic explanation of the genetic screen carried out.
A double mutant elg1 ctf4 strain is kept alive by the presence of a plasmid carrying the ELG1 gene. The ADE3 marker on the plasmid confers a red pigment to the cells carrying it. As these cells are unable to lose the plasmid during colony formation, all colonies are uniformly red. This strain was transformed with a high copy number library carrying random fragments of the yeast genome. Cells that received a plasmid that suppresses the synthetic lethality phenotype can now lose the ELG1-containing plasmid, becoming white. These cells create white or red/white sectored colonies.
After transformation with the high copy number library, we screened about 30,000 colonies; 62 of them showed some degree of sectoring and were further analyzed. After re-introduction of the plasmid to naïve yeast strains, only 29 clones gave a positive result. Twenty six carried the CTF4 gene, two clones carried a genomic DNA fragment containing the SCC1 gene and an additional clone carried the SCC2 gene. SCC1 encodes a subunit of the Cohesin complex. This complex is composed of four core subunits: Smc1 and Smc3, which are members of the structural maintenance of chromosomes (SMC) protein family, and two non-SMC subunits, Scc1, which is a member of the kleisin family, and Scc3. Smc1, Smc3 and Scc1 form a ring large enough to contain two dsDNA molecules [21], [22]. Scc3 binds Scc1 and has an unknown function [27], [29]. The second gene isolated in our screen, SCC2, encodes a protein that forms, together with Scc4, a complex that helps loading cohesin on the DNA [30].
All the known phenotypes of mutants in SCC1 are related to its function as a structural component of cohesin. This raises the question of whether Scc1 is the only subunit of the complex able to suppress the synthetic lethal phenotype of elg1 ctf4 strains. The fact that CTF4 was isolated numerous times but ELG1 was not, suggest that the screen may not have been saturated enough. Alternatively, only Scc1 and Scc2 may be the limiting factors that determine the amount of cohesin complexes loaded onto the DNA. Scc1 may be the limiting component in cohesin formation, and Scc2 availability may be limiting the amount of Scc1 loaded. According to this possibility ctf4 elg1 strain is inviable due to a low level of the cohesin ring, for example, at the centromeres.
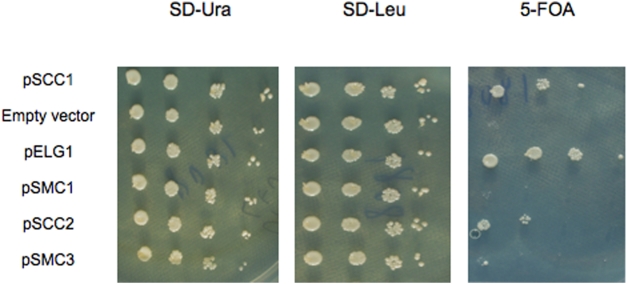
We therefore directly tested the ability of other cohesin subunits to suppress the SL phenotype by overexpressing SCC1, SCC2, SMC1 or SMC3 from LEU2-marked plasmids in a ctf4 elg1 strain carrying a ELG1/URA3 plasmid. If overexpression of any of these genes can suppress the SL phenotype, the ELG1/URA3 plasmid can be lost, allowing the cells to grow on 5-FOA medium (which selects for Ura- cells). Figure 2 shows that SCC1and SCC2, (and of course ELG1), but not SMC1 or SMC3, can rescue the SL phenotype of the ctf4 elg1 double mutant strain. Thus, in agreement with the second hypothesis expressed above, Scc1 and Scc2 are the only limiting components able to suppress the lethality when overexpressed. Scc1 seems to be a slightly better suppressor than Scc2 (Figure 2).
Figure 2. Overexpression of SCC1 or SCC2 suppress the synthetic lethality between elg1 and ctf4.
A elg1 ctf4 double mutant maintained alive by the presence of a URA3-marked plasmid carrying the ELG1 gene was transformed with LEU2-marked plasmids overexpressing SCC1, SCC2, SMC1 or SMC3, or, as controls, with an empty vector or a plasmid carrying ELG1. Cells were successively diluted ten-fold and plated on plates lacking Uracil (SD-Ura), Leucine (SD-Leu), or on plates containing 5-fluoroorotic acid (5-FOA), which select for cells that became Ura- (i.e., lost the URA3-marked, ELG1-containing plasmid). Only plasmids overexpressing SCC1 and SCC2 allowed cells to grow on 5-FOA plates.
One of the most prominent phenotypes of both elg1 mutants and ctf4 mutants is their increased levels of recombination. We tested whether the suppression of the SL phenotype by Scc1 overexpression was related to the hyper-recombination phenotypes observed. Table 1 shows that this was not the case, as overexpression of SCC1 or SCC2 failed to affect the hyper-recombination phenotype of either elg1 or ctf4 strains.
Table 1. Overexpression of Scc1 and Scc2 does not affect the hyper-recombination phenotype of ctf4 or elg1.
| Strain | His+ recombinants | Ty recombinants |
| MK166/vector | 5.8×10−6 (×1) | 1.1×10−6 (×1) |
| MK166/pSCC1 | 6.2×10−6 (×1.1) | 1.2×10−6 (×1.1) |
| MK166/pSCC2 | 6.0×10−6 (×1.0) | 0.97×10−6 (×0.9) |
| elg1/vector | 30.6×10−6 (×5.3) | 12.1×10−6 (×11) |
| elg1/pSCC1 | 37.7×10−6 (×6.5) | 8.9×10−6 (×8) |
| elg1/pSCC2 | 33.1×10−6 (×5.7) | 11.1×10−6 (×10) |
| ctf4/vector | 20.2×10−6 (×3.5) | 8.6×10−6 (×7.8) |
| ctf4/pSCC1 | 26.1×10−6 (×4.5) | 9.1×10−6 (×8.3) |
| ctf4/pSCC2 | 22.1×10−6 (×3.8) | 7.6×10−6 (×6.9) |
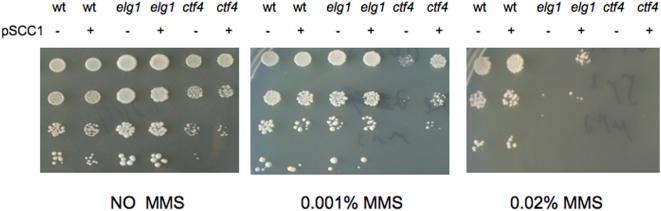
To investigate whether the suppression by Scc1 was due to bypass of the elg1 phenotype, the ctf4 phenotype, or the combination of both, we tested whether Scc1 overexpression could suppress the sensitivity of elg1 or ctf4 mutants to DNA damage. Figure 3 shows that Scc1 (and Scc2, data not shown) overexpression is able to partially suppress the sensitivity to MMS of both ctf4 and elg1 mutants, suggesting that the synthetic lethality, as well as the DNA damage sensitivity, are due to shared or overlapping functions involving sister chromatin cohesion. No changes in the cell cycle profile (as analyzed by FACS) were detected in these experiments (data not shown).
Figure 3. Overexpression of SCC1 suppresses the sensitivity of both elg1 and ctf4 to DNA damaging agents.
Isogenic wild type, elg1 or ctf4 strains were serially diluted and plated on plates without, and with methyl methanesulfonate (MMS) at two different concentrations (0.001% and 0.02%). Although ctf4 strains are more sensitive than elg1 strains, SCC1 overexpression suppresses the sensitivity of both to MMS.
Genetic interactions between ELG1 and Cohesin subunits
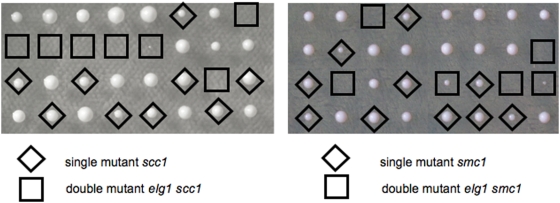
In order to determine the genetic relationship between Elg1 and the Cohesin complex, we attempted to create double mutants between elg1 and mutants defective in each of the components of cohesin. Figure 4 shows the results of tetrad dissection of elg1/ELG1 SCC1/scc1-73 and elg1/ELG1 SMC1/smc1-259 double heterozygous diploids. The double mutants elg1 scc1 and elg1 smc1 were very sick being either completely unable to form colonies or forming tiny, sick colonies that failed to develop.
Figure 4. Mutations in SCC1 and in ELG1 show synthetic lethality.
Tetrad analysis of double heterozygotes elg1/ELG1 SCC1/scc1-73 and elg1/ELG1 SMC1/smc1-259. Plates were incubated at 25C, a permissive temperature for the single scc1 and smc1 mutants. The double mutants were unable to form viable colonies.
We used the above method to examine the genetic relationship between elg1 and mutations in other components of the cohesin complex such as Scc3 and Smc3, and additional genes that interact with cohesin, such as the loaders Scc2 and Scc4 [30], Esp1, the protease that cleaves Scc1 at the metaphase transition [31], Pds1, the securin that prevents cleavage until the right cell cycle stage is reached [32], and Pds5, a protein of unknown function recruited by cohesin and important for cohesion maintenance [33]. Results are presented in Table 2: elg1 exhibited synthetic fitness phenotypes already at the permissive temperature when combined with any member of the cohesin complex or with the Scc2-Scc4 loaders, but not with mutations in the separin Esp1, in the securin Pds1 or in Pds5. These synthetic genetic interactions show that Elg1 activity is important in the early stages of cohesin establishment and not in the maintenance or the cleavage of cohesin, strengthening the possibility that both Elg1 and Ctf4 play an important role in the coordination between replication fork progression and cohesin loading.
Table 2. Synthetic genetic interactions between elg1 and cohesin subunits/accessory factors.
| Allele | Predicted Role | Synthetic effect with elg1Δ |
| smc1-259 | Subunit of cohesin | Very sick |
| smc3-42 | Subunit of cohesin | Very sick |
| scc1-73 | Subunit of cohesin | Very sick |
| scc3-1 | Subunit of cohesin | sick |
| scc2-4 | Cohesin loader – acts during late G1 | Very sick |
| scc4-4 | Cohesin loader – forms a complex with Scc2 | sick |
| esp1-1 | The protease that cleavages Scc1 during mitosis | Normal growth |
| pds1-1 | Securin that inhibits Esp1 (protease) activity until metaphase | Normal growth |
| pds5-1 | Recruited by cohesin, role unknown | Normal growth |
The Elg1 protein plays a role in sister chromatid cohesion
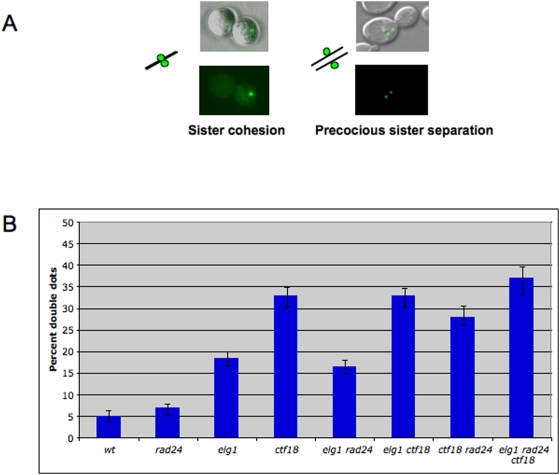
The results presented above suggest that Elg1 may have a direct role in sister chromatid cohesion. In order to directly measure this potential role we used a cohesion assay in which a TetO array is integrated approximately 40 kb from the centromere of chromosome V. These strains constitutively express a GFP-tagged TetR protein, which binds the arrays and can be visualized as a single GFP “dot”. In G2-arrested cells, lack of chromatid cohesion is seen as a “double dot” [13] (Figure 5A). Previous analysis, using a similar system, showed that the Elg1 homolog Ctf18 plays a role in sister chromatid cohesion [20]. Here we confirm these results and show that Elg1 has also a role in cohesion: whereas only 5% of wild type cells showed a “double dot” phenotype, elg1 mutants showed this phenotype in 18.5% of the cells. Mutations in CTF18 exhibited a stronger phenotype (33%). Interestingly, double mutants elg1 ctf18 exhibit the same phenotype as the ctf18 single, indicating epistatic interactions. In contrast, mutations in the gene encoding the third RFC-like protein, Rad24, had only a modest effect, if at all, in sister chromatid cohesion, and no aggravating phenotype was observed when the rad24 mutation was combined with the elg1 or ctf18 mutations, or with the double mutant (Figure 5B). We thus conclude that both Ctf18 and Elg1 play a role in chromatid cohesion, with Ctf18 having a stronger contribution.
Figure 5. Elg1 and Ctf18, but not Rad24, play a role in sister chromatid cohesion.
(A) Cohesion test. Isogenic strains carrying TetO tandem repeats on chromosome V were arrested in G2 with nocodazole and the percent of cells exhibiting two GFP dots (indicating separated sister chromatids) was counted. (B) Percentage of cells exhibiting precocious sister chromatid separation. At lest 400 cells were counted for each strain.
Elg1 localizes to chromatin and plays a role in the recruitment of Ctf18
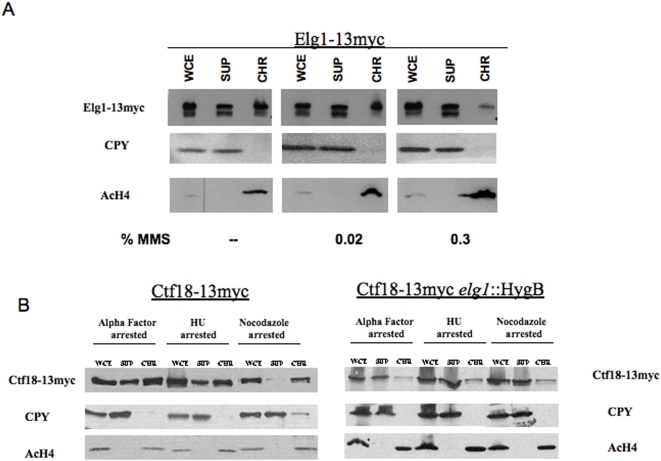
To further examine the interactions between Elg1 and Ctf18, we analyzed the recruitment of these proteins to chromatin in vivo, using chromatin fractionation assays (Figure 6). Cells were spheroplasted, lysed, and the chromatin-containing fraction was separated from the soluble fraction. Appropriate fractionation was verified in all experiments by assessing the enrichment of acetylated histone H4 in the chromatin fraction and the enrichment of Carboxypeptidase Y (CPY) in the soluble fraction. Figure 6 shows that Elg1 was present both in the chromatin and in the soluble fraction. The protein was almost always seen as a double band; it is not clear yet what kind of modification or processing is responsible for this phenomenon. The amount of Elg1 at the chromatin fraction did not seem to fluctuate greatly during the cell cycle (data not shown) (Figure 6A). We also monitored the recruitment of Ctf18 to the chromatin fraction. Figure 6B shows that, similar to Elg1, Ctf18 is present both in the soluble fraction, as well as in the chromatin. Similar results were obtained in cycling cells (not shown), as well as in cells arrested in G1 with alpha factor or in early S with hydroxyurea. Interestingly, in cells arrested in G2 with nocodazole, most of the Ctf18 protein is present in the chromatin fraction. When the experiment was repeated in an elg1 strain, the localization of Ctf18 changed, from being mainly in the chromatin, to being mainly at the soluble fraction (Figure 6B). Thus, Elg1 plays a role in determining Ctf18's localization to chromatin.
Figure 6. Elg1 and Ctf18 are present both at the chromatin and soluble fractions.
Whole cell extracts from cells at mid-logarithmic growth phase were fractionated into a chromatin and a soluble fraction, and the presence of Myc- tagged Elg1 was followed with anti-Myc specific antibodies. As controls, anti-Carboxypeptidase Y and anti- Acetylated histone H4 antibodies were used. (A) Elg1 is present in both fractions, but its abundance decreases with DNA damage. Cells were untreated, or incubated in the presence of methyl methanesulfonate (MMS) at two different concentrations (0.02% and 0.3%). (B) Ctf18 is present in both fractions, and its localization in G2 depends on Elg1. Wild type or elg1 cells were arrested with either alpha factor (G1), hydroxyurea (early S) or nocodazole (G2) before being fractionated.
Discussion
Elg1 and Ctf4 are two proteins that where found to interact genetically and physically with the replication fork. Elg1 binds the small subunits of the RFC complex [4] and, similarly to the RFC complex, is assumed to act during replication to load or unload the processivity clamp PCNA. Accordingly, physical contacts have been detected between these proteins and replication factors: Elg1 interacts with PCNA [[5] and data not shown], while Ctf4 interacts with polymerase α [14]. In a previous study we found that mutations in the ELG1 and CTF4 genes show a synthetic lethal (SL) interaction [4], suggesting that they jointly participate in an essential process in the cells. In order to uncover this essential process we performed a high copy number screen to find suppressors of the SL interaction. We found two proteins involved in sister chromatid cohesion: the cohesin subunit Scc1 and the cohesin loader complex subunit Scc2. These results suggest that the defective process common to these mutants is sister chromatid cohesion. Although it is well known that ctf4 exhibits defects in sister chromatin cohesion [16], [17], [19], [20], Elg1 was not found previously to be involved in this process. The fact that high copy number plasmids carrying Scc1 or Scc2 are able to suppress the known phenotypes of the elg1 mutant such as its sensitivity to MMS (Figure 3) indicates that elg1 has a sister chromatid cohesion phenotype by itself, independently of its interactions with Ctf4. Using a direct sister chromatid cohesion assay, we show that elg1 mutants indeed have a sister chromatid separation phenotype (Figure 5B), and have a 3.5-fold higher level of precocious separation, compared to wild type cells. This level is comparable to that observed in bona fide cohesion mutants, such as chl1, bim1 or kar3 [34].
Moreover, we also show that the activity of Elg1 becomes essential when there are defects in cohesin subunits or in its loading (Table 2 and Figure 4). The SL phenotype of elg1 with cohesin components takes place in cells that have functional Ctf4 protein, again supporting a direct role for Elg1 in sister chromatid cohesion. Importantly, elg1 exhibits synthetic genetic interactions with scc2, which was found to be important for cohesin loading [30] and with the subunits of the cohesin complex, but not with genes that are involved in late stages of cohesin maintainance or cohesin cleavage, such as PDS1 [32], ESP1 [31] and PDS5 [33] (Table 2). These results point to an early role of Elg1 in sister chromatid cohesion, possibly during cohesion establishment or during DNA replication.
Our results show that out of the three non-essential RLCs, Ctf18 and Elg1 affect sister chromatid cohesion, whereas Rad24 seems to play at most a minor role in this process. This is consistent with the fact that the Rad24 RLC loads the 9-1-1 alternative clamp [11] whereas both Ctf18 and Elg1 have been shown to interact physically with PCNA, the replication clamp [3], [4], [19], [20]. Moreover, it has been recently found that ctf18 mutants, as elg1 mutants, are SL with ctf4 [35].
This result is consistent with those presented in Figure 5, where we show that Elg1 affects cohesion via the same genetic pathway as Ctf18 (as the double mutant elg1 ctf18 shows a similar phenotype to that of the single ctf18 mutant). Moreover, we show that both proteins localize to chromatin, and that Elg1 is required in order to direct Ctf18 to the chromatin fraction (Figure 6B).
In previous studies, different interactions between elg1 and ctf18 were observed, depending on the phenotype examined. In response to DNA damage (MMS, UV light, hydroxyurea) the elg1 ctf18 double mutant showed a higher sensitivity compared to the single mutants, suggesting that these proteins work in different pathways in the presence of DNA damage [4]. Contrasting phenotypes were seen in elg1 and ctf18 mutants of S. pombe [36] suggesting that the two RLCs act antagonistically to each other. Opposite effects were also observed at telomeres: elg1 mutants exhibit elongated telomeres, whereas ctf18 mutants have short telomeres [2], [8], [37]. Finally, it has recently been shown that mutations in either ELG1 or CTF18 have the same effect in suppressing the hst3 hst4 mutant that contains hyperacetylated histone H3 at position K56 [38]. Interestingly, the same effect could be achieved by deletion of CTF4, consistent again with a model in which Elg1, Ctf18 and Ctf4 carry out related functions.
While this work was in progress, Maradeo and Skibbens [39] reported that mutations in ELG1 can partially suppress the temperature sensitivity and the cohesion defects of an ctf7/eco1-1 mutant, whereas Elg1 overexpression exacerbates its conditional growth defect. Ctf7 is a pivotal sister chromatid cohesion factor that was found to be important for cohesion establishment [25], [26] and for sister chromatid cohesion in response to DNA damage [40]. The reported genetic interaction between ELG1 and ctf7 strengthen our characterization of Elg1 as a cohesion factor. We further extend these conclusions by analyzing the interactions between elg1 and all known cohesion factors. In addition, we show that Elg1 plays a possible role in recruiting the Ctf18 RLC to the chromatin, thus providing a mechanistic explanation for the epistatic genetic interactions between elg1 and ctf18 in sister chromatid cohesion. Consistent with our results (Figure 5B), no genetic interactions could be detected between ctf7/eco1-1 and the third RLC, containing Rad24 [39].
The physical interactions of Ctf18 and Elg1 with the small RFC subunits and with PCNA and the genetic interactions with CTF4, a gene encoding a Polymerase α -interacting protein, suggest a model in which the two RLC subunits and Ctf4 mediate the interaction between the DNA replication machinery and the loading of cohesin. Cohesin is loaded onto DNA at the end of G1 [30]. Thus, during DNA replication, it is expected that the progressing fork would encounter cohesin molecules on its path. A possible model of action of the RLCs and of Ctf4 would be that passage through a cohesin loop may require momentary unloading, then re-loading, of PCNA. This would explain the physical and genetic interactions of these proteins with DNA replication components, as well as with cohesin and its associated proteins.
Materials and Methods
Yeast strains and plasmids
A list of all strains used in this paper is presented as Table 3. Plasmids pSB418 (URA3-ELG1-ADE3) and pSBA419 (LEU2-ELG1-ADE3) have been described [41]. High copy number URA3-marked and LEU2-marked plasmids pSCC1, pSCC2, pSMC1, pSMC3 and pELG1 were created by cloning the respective genes in YEplac181 (LEU2) or YEplac195 (URA3).
Table 3. List of strains used in this study.
| Strain | Genotype | Reference |
| MK166 | MATα lys2:: Ty1Sup ade2-1(o) can1-100(o) ura3-52 leu2-3, 112 his3del200 trp1del901 HIS3 :: lys2 :: ura3 his4 :: TRP1 :: his4 | [42] |
| BY7471elg1 | MATa elg1 :: KanMX his3-1 leu2-0 met15-0 ura3-0 | [45] |
| BY7471ctf4 | MATa ctf4 :: KanMX his3-1 leu2-0 met15-0 ura3-0 | [45] |
| MK1001 | MATa elg1 ::KanMX ctf4:KanMX lys2:: LTR ade2-1(o) ade3 del can1-100(o) ura3-52 leu2-3, 112 his3del200/pSBA419 (LEU2-ELG1-ADE3) | [4] |
| DD282 | MATa his3_1 leu2_0 met15_0 ura3_0 pep4Δ::KAN ELG1::13myc-his5. | [5] |
| 6745 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3::3xURA3tetO112 leu2::LEU2 tetR-GFP | [46] |
| 6745elg1 | MATα elg1::LEU2 | This study |
| 6745ctf18 | MATα ctf18::HygB | This study |
| 6745rad24 | MATα rad24::KanMX | This study |
| 6745rad24ctf18 | MATα rad24::KanMX ctf18::HygB | This study |
| 6745elg1 rad24 | MATa elg1 :: LEU2 rad24::KanMX | This study |
| 6745elg1 ctf18 | MATa elg1 :: LEU2 ctf18 :: HygB | This study |
| 6745elg1 ctf18 rad24 | MATa elg1 :: LEU2 ctf18 :: HygB rad24::KanMX | This study |
| W303Ascc1-73 | MATa ura3-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 scc1-73 | [46] |
| W303Asmc1-259 | MATa ura3-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15smc1 -259 | [46] |
| W303Asmc3-42 | MATa ura3-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 smc3-42 | [46] |
| W303Ascc3-1 | MATa ura3-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 scc3-1 | [46] |
| W303A scc2-4 | MATa ura3-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 scc2-4 | [30] |
| W303A scc4-4 | MATa ura3-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 scc4-4 | [30] |
| SY96 | MATa esp1-1 leu2-3, 112 ura3-52 lys2 can1 | B. Byers collection |
| VG986-5B | Mata trp1 ura3 bar1 pds5-1 | [33] |
| RA 2806-2b | MATa pds1-38-HA::URA3 ESP1-Myc::TRP1 ura3-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 | [47] |
Genetic screen
Strain MK1001 (elg1 ctf4 ura3 ade2 ade3 leu2/pSBA419) was transformed with a high copy number URA3 library as described [41]. After re-streaking colonies suspected of carrying suppressor plasmids, these were isolated and re-transformed into fresh MK1001 cultures. The plasmids in positive transformed colonies were sequenced.
Recombination assay
The rate of recombination of wild type, elg1 and ctf4 mutants carrying various plasmids was measured as described [4]in derivatives of strain MK166 [42].
Chromatin fractionation assay
Logarithmically growing cultures were arrested in G1 with α factor, at the beginning of S-phase with hydroxyurea or in G2 with nocodazole and incubated for one hour before being cropped by centrifugation [43]. Fractionation was carried out as in [44]. In brief, cells were incubated for 10 min at 37°C with 20 µg/µl of zymolase T-100, then lysed with 0.25% Triton X-100. Lysates were separated to supernatant and chromatin fractions by sucrose gradient. Whole cell extracts (WCE), supernatant (Sup) and chromatin pellets (CHR) were subjected to SDS-page Western blot using anti myc (SC-789, Santa Cruz Biotechnology), anti acetylated histone H4 (SC-06-946) or anti-CPY (SC-0998) antibodies. 30–50 µg of total proteins were loaded in each lane. Acetylated histone H4 serves as a loading control as well as marker for chromatin fraction and CPY serves as a marker for the soluble fraction.
Two dot assay
This experiment was carried out as described [30]. At least 400 cells were counted for each strain.
Acknowledgments
We thank Philip Hieter, Kim Nasmyth, Vincent Guacci, and Orna Cohen-Fix for generous sharing of yeast strains and plasmids. We thank Robert Skibbens for providing information before publication and all of the members of the Kupiec laboratory for helpful comments and support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants to MK from the Association for International Cancer Research, the German-Israeli Research fund (GIF), and the Israel Cancer Research Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 2.Aroya SB, Kupiec M. The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Repair (Amst) 2005;4:409–417. doi: 10.1016/j.dnarep.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Bellaoui M, Chang M, Ou J, Xu H, Boone C, et al. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. Embo J. 2003;22:4304–4313. doi: 10.1093/emboj/cdg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci U S A. 2003;100:9906–9911. doi: 10.1073/pnas.1633757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- 6.Ogiwara H, Ui A, Enomoto T, Seki M. Role of Elg1 protein in double strand break repair. Nucleic Acids Res. 2007;35:353–362. doi: 10.1093/nar/gkl1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, et al. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smolikov S, Mazor Y, Krauskopf A. ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc Natl Acad Sci U S A. 2004;101:1656–1661. doi: 10.1073/pnas.0307796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 11.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci U S A. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee S, Sikdar N, Myung K. Suppression of gross chromosomal rearrangements by a new alternative replication factor C complex. Biochem Biophys Res Commun. 2007;362:546–549. doi: 10.1016/j.bbrc.2007.07.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 14.Miles J, Formosa T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol Cell Biol. 1992;12:5724–5735. doi: 10.1128/mcb.12.12.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouprina N, Kroll E, Bannikov V, Bliskovsky V, Gizatullin R, et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouprina N, Pashina OB, Nikolaishwili NT, Tsouladze AM, Larionov VL. Genetic control of chromosome stability in the yeast Saccharomyces cerevisiae. Yeast. 1988;4:257–269. doi: 10.1002/yea.320040404. [DOI] [PubMed] [Google Scholar]
- 17.Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouprina N, Kroll E, Kirillov A, Bannikov V, Zakharyev V, et al. CHL12, a gene essential for the fidelity of chromosome transmission in the yeast Saccharomyces cerevisiae. Genetics. 1994;138:1067–1079. doi: 10.1093/genetics/138.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 21.Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 23.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, et al. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, et al. Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 28.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, et al. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 30.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 31.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 32.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, et al. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 33.Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, et al. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu H, Boone C, Brown GW. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics. 2007;176:1417–1429. doi: 10.1534/genetics.107.072876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Robertson K, Mylonas KJ, Gray FC, Charapitsa I, et al. Contrasting effects of Elg1-RFC and Ctf18-RFC inactivation in the absence of fully functional RFC in fission yeast. Nucleic Acids Res. 2005;33:4078–4089. doi: 10.1093/nar/gki728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiraga S, Robertson ED, Donaldson AD. The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. Embo J. 2006;25:1505–1514. doi: 10.1038/sj.emboj.7601038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celic I, Verreault A, Boeke JD. Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics. 2008;179:1769–1784. doi: 10.1534/genetics.108.088914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maradeo ME, Skibbens RV. The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS ONE. 2009;4:e4707. doi: 10.1371/journal.pone.0004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- 41.Koren A, Ben-Aroya S, Steinlauf R, Kupiec M. Pitfalls of the synthetic lethality screen in Saccharomyces cerevisiae: an improved design. Curr Genet. 2003;43:62–69. doi: 10.1007/s00294-003-0373-8. [DOI] [PubMed] [Google Scholar]
- 42.Liefshitz B, Steinlauf R, Friedl A, Eckardt-Schupp F, Kupiec M. Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat Res. 1998;407:135–145. doi: 10.1016/s0921-8777(97)00070-0. [DOI] [PubMed] [Google Scholar]
- 43.Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 44.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giaever G, Chu AM, Ni L, Connelly C, Riles L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 46.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 47.Agarwal R, Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]