Abstract
Staphylococcus aureus has emerged as a significant pathogen causing severe invasive disease in otherwise healthy people. Despite considerable advances in understanding the epidemiology, resistance mechanisms, and virulence factors produced by the bacteria, there is limited knowledge of the in vivo host immune response to acute, invasive S. aureus infections. Herein, we report that peripheral blood mononuclear cells from patients with severe S. aureus infections demonstrate a distinctive and robust gene expression profile which is validated in a distinct group of patients and on a different microarray platform. Application of a systems-wide modular analysis framework reveals significant over-expression of innate immunity genes and under-expression of genes related to adaptive immunity. Simultaneous flow cytometry analyses demonstrated marked alterations in immune cell numbers, with decreased central memory CD4 and CD8 T cells and increased numbers of monocytes. CD14+ monocyte numbers significantly correlated with the gene expression levels of genes related to the innate immune response. These results demonstrate the value of applying a systems biology approach that reveals the significant alterations in the components of circulating blood lymphocytes and monocytes in invasive S. aureus infections.
Introduction
Staphylococcus aureus has emerged as a leading cause of both community-associated and nosocomial invasive bacterial infections in the United States, causing a wide spectrum of clinical illnesses ranging from superficial soft tissue infections to severe, invasive disease leading to considerable morbidity and mortality in otherwise healthy children and adults [1]–[5]. As such, the US Centers for Disease Control and Prevention has identified S. aureus as a major public health problem in a recent population-based surveillance study [6]. The increasing magnitude of S. aureus disease burden, changes in bacterial susceptibility, and continued poor patient outcomes with available antimicrobial therapies clearly demonstrate a need to improve our understanding of its pathogenesis in order to develop more effective therapeutic strategies. To date, most efforts have been focused on advancing our knowledge of the pathogen and its remarkable repertoire of virulence factors [7]. To complement this progress, it is critical that we also gain new insight into the host immune response to S. aureus infection in a clinically relevant context.
Genome-wide analyses of blood leukocytes provide a global and comprehensive assessment of the complexity of the immune network and response to disease [8]. Previous work has demonstrated the applicability of blood leukocyte microarray data into clinical relevance, allowing for biomarker discovery leading to improved diagnostic and prognostic indicators and innovative therapeutics in autoimmune diseases [9]–[11] and cancer [12]–[15]. With advances in gene expression profiling and application of microarray technology to the field of infectious diseases, we can now more clearly define the host's immune response to a pathogen and identify a unique biosignature that is not limited by traditional diagnostic or microbiologic techniques [16]–[18]. Peripheral blood mononuclear cells (PBMCs) provide an accessible source of molecular phenotypic information as they traffic within the systemic circulation, to and from sites of infection and inflammation. When a pathogen infects the host, specific pattern-recognition receptors expressed on leukocytes recognize unique pathogen-associated molecular patterns displayed on microbes and initiate the host's innate and pro-inflammatory response to the infection while simultaneously activating the adaptive immune response [19], [20]. The microbe-induced changes in host cells demonstrate significant and stereotypical changes at the gene expression level that are unique to the pathogen and can be objectively measured [21], [22]. Genome-wide arrays have been used to advance our understanding of the human host response to febrile illnesses such as malaria [17], acute dengue hemorrhagic fever [17], HIV [23], and Kawasaki syndrome [17]. Our previous work has demonstrated that PBMCs of children with different acute infections carry distinct gene expression patterns that allow discrimination between viral and bacterial pathogens and even among subtypes of bacteria [24]. Despite different bacterial strains and diverse clinical manifestations, we hypothesized that gene expression analysis of PBMCs from children with acute invasive S. aureus infections would demonstrate a characteristic and unique host immune response and provide a novel and broad insight into the pathogenesis of these infections. The present study was designed to obtain a comprehensive characterization of the host immune response to invasive S. aureus infections in children using a combination of gene expression and flow cytometry analyses.
Results
Patient characteristics
Over a period of 4 years, samples from 53 previously healthy patients hospitalized with invasive S. aureus infections and 24 healthy control subjects were analyzed. Patients were chosen representing the clinical spectrum of acute severe S. aureus disease including bacteremia, osteomyelitis, suppurative arthritis, pyomyositis, and pneumonia with empyema. Patients with a diagnosis of staphylococcal toxic shock syndrome or polymicrobial infections were excluded. Patient demographic data, clinical characteristics, analysis group, and microarray platform are summarized in Table 1 . There were no statistical differences between the S. aureus-infected children and their respective healthy controls with regards to age, sex, or race in the training and test sets ( Table 2 ). Patients were enrolled only after a bacteriologic diagnosis was established; the median time from patient hospitalization to procurement of study blood sample was 4 days [IQ range 3–8 days]. Viral direct fluorescent antibody testing or culture of the nasopharynx was obtained on 68 subjects (88%, 46 patients, 22 controls) and did not reveal the presence of a concomitant viral infection.
Table 1. Subject characteristics.
| SUBJECT # | AGE (yrs) | RACE | SEX | CONDITION | PATHOGEN | ANALYSIS | PLATFORM |
| 3N | 6 | H | M | Control | Healthy | Training/Test | A |
| 5 | 10 | H | M | Osteomyelitis | MSSA | Training | A |
| 7N | 1.6 | B | F | Control | Healthy | Training/Test | A |
| 8N | 10 | B | M | Control | Healthy | Training/Test | A |
| 11N | 4 | B | F | Control | Healthy | Training/Test | A |
| 23N | 7 | H | F | Control | Healthy | Training/Test | A |
| 24 | 3 | B | M | Bacteremia, Osteomyelitis, Myositis | MRSA | Test | A |
| 24N | 3 | W | M | Control | Healthy | Training/Test | A |
| 30 | 15 | B | M | Bacteremia | MRSA | Test | A |
| 40 | 13 | W | M | Bacteremia, Osteomyelitis | MSSA | Training | A |
| 43 | 7 | B | M | Bacteremia, Osteomyelitis, SArthritis, Pyomyositis, Emboli | MRSA | Test | A |
| 62 | 2 | W | M | Osteomyelitis, Pyomyositis | MRSA | Training | A |
| 66 | 0.25 | B | F | Pneumonia | MRSA | Test | A |
| 67 | 7 | W | F | Bacteremia, Osteomyelitis | MRSA | Training | A |
| 88 | 0.92 | H | M | Bacteremia, Osteomyelitis, Pneumonia, Emboli | MRSA | Test | A |
| 90 | 0.67 | B | M | Bacteremia, SArthritis | MSSA | Test | A |
| 109 | 0.67 | H | F | Bacteremia, SST Abscess | MRSA | Training | A |
| 150 | 9 | B | F | Bacteremia, Osteomyelitis, SArthritis, Myositis | MRSA | Test | A |
| 179 | 12 | W | M | Bacteremia, Endocarditis, Emboli | MSSA | Training | A |
| 205 | 7 | H | M | Bacteremia, Pneumonia, SST Abscess | MRSA | Test | A |
| 208 | 10 | W | F | Bacteremia, Osteomyelitis, CNS abscess, Pneumonia, Emboli | MRSA | Test | A |
| 216 | 10 | H | F | Bacteremia, Osteomyelitis | MRSA | Training | A |
| 220 | 11 | H | M | Bacteremia, Osteomyelitis | MSSA | Test | A |
| 221 | 6 | B | F | Bacteremia, Osteomyelitis | MRSA | Training | A |
| 224 | 10 | W | M | Bacteremia, Osteomyelitis | MSSA | Test | A |
| 230 | 20 | B | M | Bacteremia, Endocarditis, SST Abscess | MRSA | Test | A |
| 241 | 0.92 | B | F | Bacteremia, Osteomyelitis, Pneumonia | MRSA | Training | A |
| 242 | 1.2 | B | M | Bacteremia, Osteomyelitis, Pyomyositis | MRSA | Test | A |
| 258 | 8 | W | F | Bacteremia, Osteomyelitis, Cellulitis | MSSA | Training | A |
| 262 | 13 | H | M | Bacteremia, SST Abscess | MRSA | Training | A |
| 264 | 13 | B | M | Bacteremia, Osteomyelitis, SArthritis, SST Abscess, Myositis, Emboli | MSSA | Test | A |
| 271 | 13 | B | M | Osteomyelitis, pyomyositis | MSSA | Training | A |
| 294 | 12 | B | F | Control | Healthy | Training/Test | A |
| 301 | 8 | W | M | Control | Healthy | Training/Test | A |
| 303 | 6 | W | F | Control | Healthy | Training/Test | A |
| 304 | 6 | W | M | Control | Healthy | Training/Test | A |
| 305 | 4 | H | F | Bacteremia, Osteomyelitis, SArthritis | MSSA | Training | A |
| 308 | 12 | B | F | Bacteremia, Pneumonia, Pyomyositis, SST Ab | MSSA | Training | A |
| 328 | 0.38 | B | F | Bacteremia, Osteomyelitis, Pneumonia, SST Abscess | MSSA | Test | A |
| 329 | 0.58 | O | F | Bacteremia, Lymphadenitis | MRSA | Training | A |
| 330 | 11 | B | F | Bacteremia, Pneumonia | MRSA | Test | A |
| 354 | 2 | H | F | Bacteremia, Osteomyelitis | MRSA | Training | A |
| 366 | 11 | B | F | Osteomyelitis, SST Abscess | MRSA | Test | A |
| 369 | 14 | B | M | Bacteremia, SArthritis, SST Abscess, Emboli | MRSA | Test | A |
| 372 | 14 | W | M | Bacteremia, Osteomyelitis, Myositis | MRSA | Test | A |
| 412 | 1.75 | W | M | Bacteremia, SArthritis | MSSA | Test | A |
| 418 | 8 | H | F | Osteomyelitis | MSSA | Training | A |
| 423 | 8 | W | M | Bacteremia, Osteomyelitis, Pyomyositis | MSSA | Training | A |
| 434 | 5 | W | M | Osteomyelitis, Pyomyositis | MSSA | Test | A |
| 440 | 0.5 | B | M | Bacteremia, Osteomyelitis, Pneumonia, Emboli | MRSA | Training | A |
| 450 | 0.83 | B | M | Bacteremia, Osteomyelitis, SArthritis, Pyomyositis | MSSA | Test | A |
| 451 | 4 | H | M | Bacteremia, Osteomyelitis, SArthritis, Pyomyositis, Emboli | MRSA | Training | A |
| 894 | 8 | W | F | Osteomyelitis, SST Abscess | MSSA | Validation, FACS | I |
| 903 | 13 | W | F | Control | Healthy | Validation, FACS | I |
| 904 | 16 | W | F | Control | Healthy | FACS | I |
| 905 | 16 | W | F | Control | Healthy | FACS | I |
| 907 | 1.3 | H | F | Bacteremia | MRSA | FACS | I |
| 908 | 8 | B | M | Bacteremia, Pleural effusion, Pyomyositis, SST Abscess, Emboli | MSSA | FACS | I |
| 909 | 6 | H | F | Bacteremia, Osteomyelitis, SArthritis, Pneumonia | MRSA | Validation, FACS | I |
| 910 | 5 | B | M | Osteomyelitis, SArthritis | MSSA | Validation, FACS | I |
| 922 | 3 | H | F | Control | Healthy | FACS | I |
| 926 | 3 | H | M | Control | Healthy | FACS | I |
| 927 | 2 | H | M | Control | Healthy | Validation, FACS | I |
| 929 | 11 | O | F | Control | Healthy | Validation, FACS | I |
| 930 | 8 | O | M | Control | Healthy | Validation | I |
| 931 | 12 | W | F | Control | Healthy | FACS | I |
| 933 | 9 | W | M | Control | Healthy | Validation, FACS | I |
| 935 | 9 | W | F | Control | Healthy | Validation, FACS | I |
| 939 | 8 | W | M | Control | Healthy | Validation, FACS | I |
| 940 | 7 | W | M | Control | Healthy | Validation, FACS | I |
| 941 | 9 | W | F | Control | Healthy | Validation, FACS | I |
| 943 | 9 | W | M | Osteomyelitis, SArthritis, Pyomyositis | MRSA | Validation, FACS | I |
| 944 | 6 | B | F | Bacteremia, Osteomyelitis, SArthritis, Pyomyositis | MRSA | Validation, FACS | I |
| 949 | 11 | W | M | Bacteremia, Osteomyelitis, SArthritis | MSSA | Validation | I |
| 952 | 15 | B | M | Bacteremia, Osteomyelitis, Pneumonia | MRSA | Validation, FACS | I |
| 954 | 13 | W | M | Bacteremia, Osteomyelitis, Pyomyositis | MSSA | Validation, FACS | I |
| 960 | 3 | H | M | Pneumonia | MRSA | Validation, FACS | I |
H = Hispanic, W = White, B = Black, O = Other; M = Male, F = Female; SST Ab = Skin/soft tissue abscess, SArthritis = Suppurative arthritis, CNS = central nervous system; MSSA = methicillin-susceptible Staphylococcus aureus, MRSA = methicillin-resistant Staphylococcus aureus; Platform: A = Affymetrix U133 A&B I = Illumina Sentrix Hu6 BeadChips.
Table 2. Demographic and Laboratory Characteristics of Patients and Controls in Training and Test Sets.
| Parameter | TRAINING SET | TEST SET | ||||
| Patients | Controls | p a | Patients | Controls | p a | |
| Age (years) | 7.5 [2–11] | 6 [3.5–9] | 0.86 | 7 [1–12] | 6 [3.5–9] | 0.93 |
| Race | 5B,8H,6W,1O | 4B,2H,4W | 1 | 14B,3H,5W | 4B,2H,4W | 0.25 |
| Gender | 9M:11F | 5M:5F | 0.57 | 16M:6F | 5M:5F | 0.45 |
| WBC (thousand/mm3) | 8.9 [7.5–17.4] | 7.2 [5.2–8.2] | 0.03 | 11 [7.7–16] | 7.2 [5.2–8.2] | 0.009 |
| Neutrophils (%) | 60 [48–73] | 43 [27–50] | 0.007 | 61 [40–66] | 43 [27–50] | 0.022 |
| Lymphocytes (%) | 24 [13–38] | 47 [39–55] | 0.002 | 24 [12.7–47] | 47 [39–55] | 0.038 |
| Monocytes (%) | 8 [7–11] | 7 [6–10] | 0.37 | 9 [6.3–12.5] | 7 [6–10] | 0.38 |
| Hematocrit (%) | 31.6 [29.8–36] | 37.7 [34.9–40] | 0.007 | 30 [27–33.9] | 37.7 [34.9–40] | 0.002 |
| Platelets (thousand/mm3) | 382 [297–459] | 310 [264–329] | 0.07 | 353 [273–445] | 310 [264–329] | 0.373 |
| CRP (mg/dL) | 6.65 [2.1–16.5] | 0.4 [0.4–0.7] | <0.001 | 7.4 [2.3–16.9] | 0.4 [0.4–0.7] | 0.001 |
Median values [25–75% range]; B = Black, H = Hispanic, W = White, O = Other; M = Male, F = Female; CRP = C-reactive protein.
p values calculated within each individual subject set and respective, matched controls (Mann-Whitney).
Patients with culture-proven invasive S. aureus infections were divided into 3 groups for analysis: training, test, and validation sets. The training set of subjects composed of 20 children with invasive S. aureus infections (median age 7.5 years; 11 methicillin-resistant S. aureus, MRSA, and 9 methicillin-susceptible S. aureus, MSSA) and 10 healthy controls (median age 6 years) matched for age, sex, and race, were initially analyzed to identify the gene expression profile in PBMCs from S. aureus-infected patients. As expected, there were statistical differences in laboratory parameters with higher total peripheral white blood cell count and percent neutrophil count, but lower percent lymphocyte count and hematocrit values in patients with S. aureus infections compared with healthy controls ( Table 2 ). The test set of subjects included an independent group of 22 patients with S. aureus infection (median age 7 years; 8 MSSA, 14 MRSA) and 10 healthy controls (median age 6 years) and was used to validate the gene expression profile in PBMCs from S. aureus-infected patients. As in the training set, there were differences in laboratory values between patients with S. aureus infection and healthy controls ( Table 2 ).
A third independent group of 25 subjects was included to validate our initial findings using (1) a second microarray platform (Illumina) and (2) flow cytometry. This validation set was comprised of 11 patients with S. aureus infection (median age 8 years; 5 MSSA, 6 MRSA) and 13 healthy controls (median age 9 years). PBMCs from 23 subjects were evaluated by flow cytometry to determine the relative abundance of different immune cell populations. Simultaneous flow cytometry evaluation and gene expression analysis was conducted with the same PBMC samples in 18 (9 with S. aureus infection and 9 healthy controls, matched for age, sex, race) of these 23 subjects. There were no statistical differences in extent of disease severity, antimicrobial therapy, or demographic and laboratory data between the training, test, and validation sets ( Table 3 ).
Table 3. Demographic and Laboratory Characteristics of Patients in Training, Test, and Validation Sets.
| S. aureus patients | Training Set | Test Set | Validation Set | p-valuea |
| Age (years) | 7.5 [2–11] | 7 [1–12] | 8 [5–11] | 0.82 |
| Race | 5B,8H,6W,1O | 14B,3H,5W | 4B,3H,4W | 0.08 |
| Gender | 9M:11F | 16M:6F | 7M:4F | 1 |
| WBC (thousand/mm3) | 8.9 [7.5–17.4] | 11 [7.7–16] | 8.5 [5.7–15.5] | 0.52 |
| Neutrophils (%) | 60 [48–73] | 61 [37–66] | 55 [45–71] | 0.73 |
| Lymphocytes (%) | 24 [13–38.3] | 24 [12.7–47.5] | 29 [19.5–44.5] | 0.70 |
| Monocytes (%) | 8 [7–11] | 9 [6.3–12.5] | 7 [3.5–11.5] | 0.58 |
| Hematocrit (%) | 31.6 [29.8–36.2] | 30 [27.2–33.9] | 32 [29–34.1] | 0.08 |
| Platelets (thousand/mm3) | 382 [297–459] | 353 [273–445] | 364 [313–473] | 0.61 |
| ESR (mm/hr) | 70 [34–98] | 46 [36–72] | 72 [46–87] | 0.28 |
| CRP (mg/dL) | 6.7 [2.1–16.5] | 7.4 [2.4–16.9] | 7.7 [3.7–21.3] | 0.69 |
Median values [25–75% range]; B = Black, H = Hispanic, W = White, O = Other; M = Male, F = Female; CRP = C-reactive protein.
p values calculated within each individual subject set and respective to matched controls (Kruskal-Wallis).
Gene expression signature in patients with invasive S. aureus infections
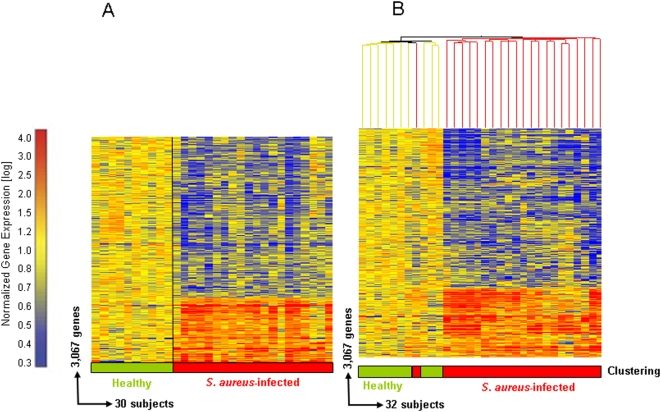
Statistical group comparison (Mann-Whitney, p<0.01) was applied to the class comparisons on the quality control (QC) genes present in the training set revealing 3,168 genes differentially expressed between S. aureus-infected patients and healthy controls. These genes were then filtered to include those transcripts with a 1.25-fold or greater change in expression level relative to the healthy control group, for a total of 3,067 genes. A hierarchical clustering algorithm was applied to these 3,067 genes in order to visualize the transcriptional pattern ( Figure 1a ). For purposes of validation, the 3,067 gene list comprising the gene expression profile in PBMC of S. aureus-infected patients was then evaluated in an independent test set of 22 new patients. The samples were organized into a condition tree utilizing the 3,067 genes and correctly classified 31 of 32 samples as either healthy or S. aureus infection based on the gene expression patterns ( Figure 1b ).
Figure 1. Gene expression biosignature in PBMCs from S. aureus patients and healthy controls.
(a) Statistical group comparisons between 10 healthy subjects and 20 patients with acute S.aureus infections yielded 3,067 genes expressed at statistically different levels (Mann-Whitney p<0.01 and 1.25 fold change) between the two groups. Significant genes were organized by hierarchical clustering to reveal differential expression, each row representing a single gene and each column an individual subject. Transformed expression levels are indicated by color scale: red representing relatively high and blue relatively low gene expression compared to the median expression for each gene across all patients compared to healthy controls. (b) The same 3,067 gene list was used to perform a condition tree on 22 new subjects with S. aureus infections and correctly grouped 21 of the 22 patients based solely on gene expression.
Genes represented in the expression profile in PBMCs from S. aureus-infected patients were then ranked according to differences in both fold-change and significance in gene expression levels (p<0.05) compared with healthy controls. The top 50 genes that were significantly over-expressed in patients with S. aureus infection versus healthy controls are shown in Table 4 . Over-expressed genes included those with microbicidal functions (lactotransferrin, alpha-defensins 1 and 4, bactericidal/permeability-increasing protein), involved in coagulation (thrombomodulin), hemoglobin synthesis (hemoglobin D and G), and pro-inflammatory and immune-related genes related to pathways of cellular growth, proliferation, and apoptosis (ADM, ARG1, CLU, EGR1, IL8, HBEGF, ITGA2B, MMP9) and involved in cell to cell signaling such as CEACAM6 and CEACAM8.
Table 4. Top 50 over-expressed genes in invasive S. aureus infections.
| Rank | Common Name | Gene Bank Symbol | Description | Fold Change | Significance |
| 1 | LTF | NM_002343 | Lactotransferrin | 46.266 | 0.000029 |
| 2 | CEACAM8 | M33326 | Carcinoembryonic antigen-related cell adhesion molecule 8 | 31.985 | 0.000052 |
| 3 | AW337833 | Transcribed locus | 32.904 | 0.000029 | |
| 4 | IL8 | NM_000584 | Interleukin 8 | 21.581 | 0.000075 |
| 5 | HBG1 | NM_000559 | Hemoglobin, gamma A | 21.543 | 0.000705 |
| 6 | DEFA4 | NM_001925 | Defensin, alpha 4, corticostatin | 20.559 | 0.000075 |
| 7 | DEFA1 | NM_004084 | Defensin, alpha 1, myeloid-related sequence | 17.011 | 0.000043 |
| 8 | HBG2 | NM_000184 | Hemoglobin, gamma G | 15.804 | 0.001540 |
| 9 | LCN2 | NM_005564 | Lipocalin 2 (oncogene 24p3) | 13.696 | 0.000090 |
| 10 | HBD | NM_000519 | Hemoglobin, delta | 13.327 | 0.000432 |
| 11 | ARG1 | NM_000045 | Arginase, liver | 14.386 | 0.000366 |
| 12 | BPI | NM_001725 | Bactericidal/permeability-increasing protein | 12.791 | 0.000129 |
| 13 | HPR | NM_020995 | Haptoglobin-related protein | 12.218 | 0.000155 |
| 14 | S100P | NM_005980 | S100 calcium binding protein P | 11.571 | 0.000052 |
| 15 | THBD | NM_000361 | Thrombomodulin | 9.005 | 0.000309 |
| 16 | ADM | NM_001124 | Adrenomedullin | 8.670 | 0.000020 |
| 17 | CA1 | NM_001738 | Carbonic anhydrase I | 8.627 | 0.000432 |
| 18 | ERAF | NM_016633 | Erythroid associated factor | 7.890 | 0.000155 |
| 19 | MSCP | H69701 | Mitochondrial solute carrier protein | 7.635 | 0.000043 |
| 20 | MYL9 | NM_006097 | Myosin, light polypeptide 9, regulatory | 7.509 | 0.000510 |
| 21 | MMP9 | NM_004994 | Matrix metalloproteinase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) | 7.023 | 0.00580 |
| 22 | RETN | NM_020415 | Resistin | 6.900 | 0.000108 |
| 23 | ANXA3 | M63310 | Annexin A3 | 6.465 | 0.000705 |
| 24 | CEACAM6 | M18728 | 5.918 | 0.002400 | |
| 25 | MGAM | NM_004668 | Maltase-glucoamylase (alpha-glucosidase) | 5.823 | 0.000432 |
| 26 | PBEF1 | AI681868 | Pre-B-cell colony enhancing factor 1 | 5.630 | 0.001540 |
| 27 | CYP4F3 | NM_000896 | Cytochrome P450, family 4, subfamily F, polypeptide 3 | 5.451 | 0.000432 |
| 28 | PBEF | AA873350 | Pre-B-cell colony enhancing factor 1 | 5.407 | 0.000600 |
| 29 | ITGA2B | AF098114 | Integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41B) | 5.238 | 0.000827 |
| 30 | HP | NM_005143 | Haptoglobin | 5.138 | 0.000035 |
| 31 | SNCA | BG260394 | Synuclein, alpha (non A4 component of amyloid precursor) | 4.891 | 0.000016 |
| 32 | FCGR3A | J04162 | Fc fragment of IgG, low affinity IIIb, receptor for (CD16) | 4.877 | 0.000029 |
| 33 | EGR1 | AI459194 | Early growth response 1 | 4.422 | 0.003200 |
| 34 | LOC199675 | BF433657 | Hypothetical protein LOC199675 | 4.420 | 0.000309 |
| 35 | SOCS3 | AI244908 | qj98g11.x1 NCI_CGAP_Kid3 Homo sapiens cDNA clone IMAGE:1867556 3′, mRNA sequence. | 4.312 | 0.000013 |
| 36 | EREG | NM_001432 | Epiregulin | 4.192 | 0.002070 |
| 37 | DTR | M60278 | Heparin-binding EGF-like growth factor | 4.087 | 0.002400 |
| 38 | TCN1 | NM_001062 | Transcobalamin I (vitamin B12 binding protein, R binder family) | 3.975 | 0.007280 |
| 39 | MS4A3 | L35848 | Membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | 3.966 | 0.002780 |
| 40 | GLUL | NM_002065 | Glutamate-ammonia ligase (glutamine synthase) | 3.932 | 0.000366 |
| 41 | N63920 | FP15737 | 3.883 | 0.000062 | |
| 42 | FLJ31978 | AI041543 | Hypothetical protein FLJ31978 | 3.879 | 0.000261 |
| 43 | CLU | M25915 | Clusterin (complement lysis inhibitor, SP-40,40, sulfated glycoprotein 2, testosterone-repressed prostate message 2, apolipoprotein J) | 3.733 | 0.000827 |
| 44 | MAD | AW071793 | MAX dimerization protein 1 | 3.715 | 0.000016 |
| 45 | H1F0 | BC000145 | H1 histone family, member 0 | 3.713 | 0.000043 |
| 46 | FCGR1A | X14355 | Fc fragment of IgG, high affinity Ia, receptor for (CD64) | 3.712 | 0.000029 |
| 47 | SOCS3 | BG035761 | Suppressor of cytokine signaling 3 | 3.708 | 0.000013 |
| 48 | MS4A4A | NM_024021 | Membrane-spanning 4-domains, subfamily A, member 4 | 3.648 | 0.001790 |
| 49 | PBEF | NM_005746 | Pre-B-cell colony enhancing factor 1 | 3.611 | 0.000155 |
| 50 | ORF1-FL49 | AL522667 | Putative nuclear protein ORF1-FL49 | 3.594 | 0.000155 |
Module-level analysis reveals over-expression of innate and under-expression of adaptive immune response genes
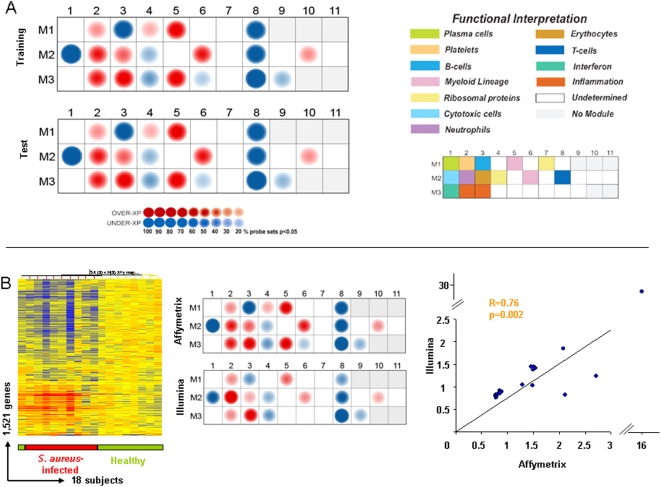
To better characterize the biological significance of the gene expression profiles seen in the PBMCs of patients with S. aureus infections, gene expression levels between patients and healthy controls were mapped using a modular analysis framework that we recently described [25]. A key to the functional interpretation of each transcriptional module is detailed in Table S1 . Gene expression levels were compared between patients and healthy controls on a module-by-module basis. The percentage of genes with a significant change (Mann-Whitney p<0.05) within each module are graphically displayed on a module map, with over-expressed genes represented in red and under-expressed genes in blue ( Figure 2a ). Following the approach described previously with the class comparisons analysis, module analysis was applied initially to the training set of patients. Patients with S. aureus infection demonstrated significant over-expression of genes in modules related to innate immunity including myeloid (M1.5, M2.6), neutrophil (M2.2), and inflammation (M3.2, M3.3) modules and under-expression of genes regulating adaptive immunity such as B cell module M1.3, cytotoxic cell module M2.1, and T cell specific module M2.8. The levels of expression of each transcriptional gene profile significantly changed (Mann Whitney p<0.05) in the PBMCs of patients with S. aureus infection versus controls are detailed in Figure S1 .
Figure 2. Module analysis identifies a specific gene expression profile in the PBMCs of S. aureus-infected patients.
(a) Gene expression levels were compared between patients with S. aureus infections and healthy controls on a module-by-module basis. Colored spots represent the percentage of significantly over-expressed (red) or under-expressed (blue) transcripts (p<0.05, Mann Whitney) within a module in patients with S. aureus infections; spot intensity represents the magnitude of the gene expression change, blank modules demonstrate no significant differences between groups (p>0.05). Information is displayed on a grid, with the coordinates corresponding to one of 28 modules with the key (upper right) representing the functional interpretation of modules. Nineteen modules are shown to be significantly different between healthy subjects and patients with S. aureus infection in the training set (a, upper module map). The same gene list applied to the independent test set of patients reveals the same modular map (a, lower module map). (b) Transcript sequences (RefSeq) were used to map the corresponding 3,067 significant gene probe sets comprising the gene expression profile of PBMC in patients with S. aureus infection on the Affymetrix platform to their corresponding 1,521 gene probes on the Illumina platform (b, far left panel) and tested in new S. aureus patients (n = 9) and controls (n = 9). Module analysis (b, middle panel) and correlation analyses (Spearman) between the average gene expression levels per module in each platform (b, right panel) were performed.
Significantly over-expressed genes included those in: M1.5 and M2.6 (“Myeloid”), genes related to cells of the myeloid lineage which are involved in bacterial pathogen recognition such as TLR 2 and CD14, IL10 signaling (CD32, BLVRA) and leukocyte extravasation signaling (CTNNA1, NCF2, PECAM1, ITGB2), also in M2.6 genes related to the inflammatory response (calgranulin B, NFKB inhibitor, TNF superfamily members, and metalloproteinase inhibitors); in M2.2 (“Neutrophils”) genes encoding innate molecules including LTF, DEFA 1 and 4, BPI, CEACAM 8; in modules M3.2 and M3.3 (“Inflammation I and II”) genes involved in inflammatory and endothelial cell processes. Significant genes found over-expressed in M3.2 (“Inflammation II”) included those involved in coagulation (THBD), endovascular inflammation, TLR signaling (IRAK 3, Ly96), and transcriptional regulation (zinc finger proteins) while M3.3 included genes encoding antigens to scavenger receptor proteins (CD36), coagulation (factor V), and lysosomal functions (LAMP2). A number of genes were significantly over-expressed in patients compared to controls in module M2.3 (“erythrocytes”) related to hemoglobin (hemoglobin alpha and gamma, erythrocyte membrane protein, erythroid factors) and module M3.5 (“undetermined”) including hemoglobin alpha and gamma proteins.
Conversely, there was significant under-expression of genes related to the adaptive immune response including those in M1.3 (“B cells”) such as genes encoding cell surface molecules CD19, CD22, CD72, CD79 and genes involved in immunoglobulin production, and in M2.8 (“T cells”) including CD6, CD96, ITK, and M2.1 (“cytotoxic cells”) (KLR subfamilies, Granulysin, Granzyme B) and genes encoding TNF family members. Module M1.8 (“undetermined”) included under-expression of factors involved in DNA replication and transcription (zinc finger protein genes) and cytokines (IL16); significant under-expressed genes in M3.8 (“undetermined”) also included multiple zinc finger proteins and TNF receptor-associated factors (TRAF5).
The PBMC gene list from S. aureus-infected patients was then analyzed in the test set of patients and module analysis validated the initial findings. Both the actual gene probes and the percentage of over or under-expressed genes per module in patients with S. aureus infection in the test set matched what had been demonstrated in the training set of patients, confirming the value and consistency of the PBMC transcriptional signature in patients with S. aureus infection ( Figure 2a ).
Confirming the robustness of the gene expression signature in S. aureus-infected patients
Twenty-five additional subjects were enrolled to further validate the PBMC gene expression profile of S. aureus-infected patients by confirming its reproducibility across microarray platforms. This validation set was comprised of 11 patients with invasive S. aureus infections and 14 age and sex-matched healthy controls. Transcript sequences from RefSeq were used to match corresponding valid gene probe sets on each platform. This allowed mapping of the 3,067 significant genes comprising the gene expression profile of PBMC of S. aureus-infected patients based on the Affymetrix gene probes to their corresponding 1,521 gene probes on the Illumina platform. This 1,521 gene list was then applied to the independent validation set of 9 patients and 9 controls using an unsupervised scheme that allowed clustering of samples based solely on intrinsic gene expression levels ( Figure 2b ). Despite technical differences between the two platforms, all 9 patients with S. aureus infection clustered together (red horizontal bar) based on similarities in PBMC gene expression patterns alone.
Module level analyses of the 1,521 gene probes on the Illumina platform demonstrate similarities between the Affymetrix and the Illumina data. As illustrated in Figure 2b , 16 of 19 modules yielded concordant results in both the Illumina and Affymetrix platforms. Only modules M1.4, M3.5, and M3.6 did not show significant differences in Illumina and may be a result of representation of less gene probes in those modules. Correlation analyses of the average normalized values among the differentially expressed genes for each given module between both platforms was statistically significant (Spearman R = 0.76, p = 0.002). Thus, both unsupervised hierarchical clustering and modular analyses confirmed the robustness of the gene expression profile in PBMC of S. aureus-infected patients across both the Affymetrix and Illumina BeadChip microarray platforms in distinct groups of subjects.
Decreased number of central memory CD4+ and CD8+ T cells in patients with S. aureus infections
To understand how the changes in blood transcriptional profiles in patients with S. aureus infection relate to changes in the numbers of the different immune cell populations, a detailed flow cytometry analysis was simultaneously conducted in PBMCs of a subset of patients with S. aureus (n = 11) and appropriate age-matched healthy controls (n = 13).
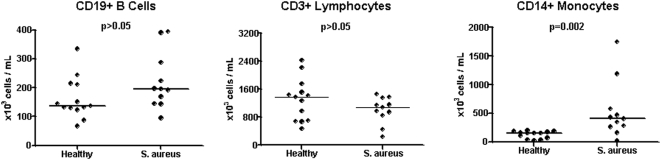
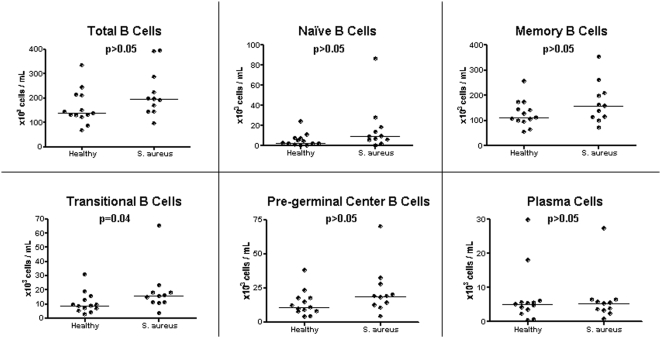
As illustrated in Figure 3 , there were no significant differences in the total number of B cells and T cells between patients with S. aureus infection and healthy controls (p>0.05). Given the significant under-expression of genes related to these cell populations observed in the modular analysis (M1.3, M2.1, M2.8), further detailed flow analysis was performed in each lymphocyte compartment. Characterization of B cell subpopulations revealed no significant differences in the absolute number of naïve (CD19+/CD20+, IgD+, CD27−), memory (CD19+/CD20+/IgD+&−/CD27+), and plasma cells (CD19+/CD20−/CD27+&++/CD38++) between patients with S. aureus infections and healthy controls ( Figure 4 ). Transitional B cells (CD19+/CD20+/CD24++/CD38++) were increased in patients with S. aureus infection compared with healthy controls (p = 0.04); there was also a trend (p = 0.06) toward increased numbers of pre-germinal B cells (CD19+/CD20+/CD27+/CD38++) in patients versus control subjects.
Figure 3. Peripheral blood monocytes are significantly expanded in patients with invasive S. aureus infections.
PBMCs obtained from age-matched healthy donors (n = 13) and patients with S. aureus infection (n = 11) were analyzed by flow cytometry for the expression of CD19 (left panel), CD3 (middle), and CD14 (right) markers. Results are expressed as absolute number of cells per mL of blood. Bars represent median values. Mann-Whitney test was applied for statistical analysis.
Figure 4. Analysis of the B cell compartment.
PBMCs obtained from age-matched healthy donors (n = 13) and patients with S. aureus infection (n = 11) were stained by multicolor panel staining according to the expression of specific B cell markers. Results are expressed as absolute number of cells per mL of blood. Bars represent median values. Mann-Whitney test was applied for statistical analysis.
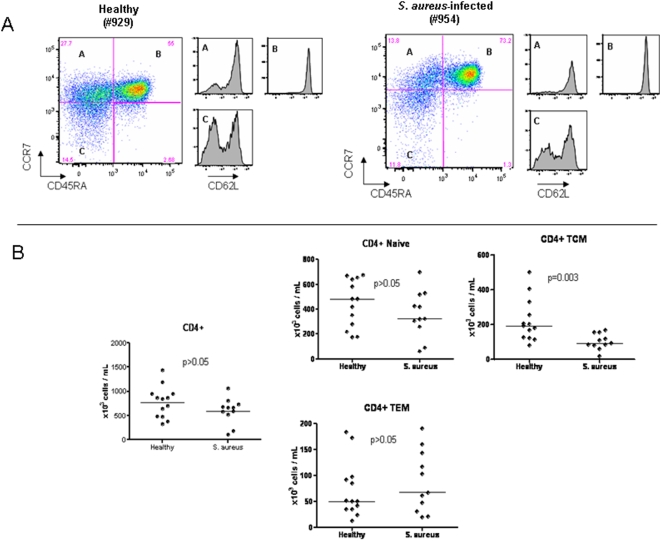
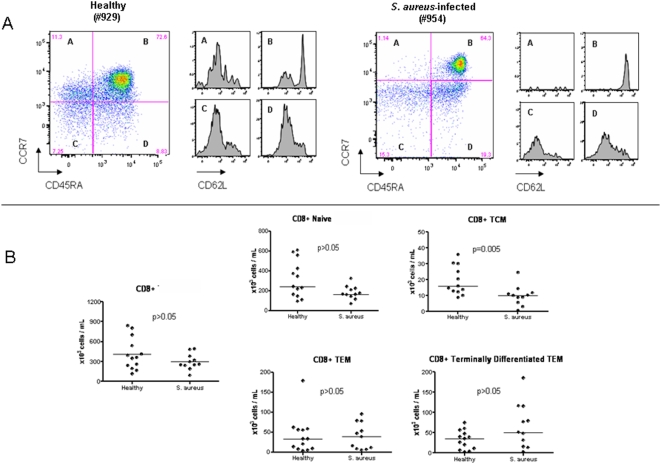
We then analyzed both CD4+ and CD8+ T cell and subpopulations of naïve and memory cells based on the expression of CD45RA, CD62L, and CCR7 as previously described [26]. As illustrated in Figure 5 there were no differences in the absolute numbers of naïve CD4+ T cells and effector memory CD4+ T cells between the S. aureus patients and healthy controls. However, the absolute number of central memory CD4+ T cell was significantly reduced in S. aureus patients compared with controls (p = 0.003). With respect to CD8+ T cells, there were no differences between S. aureus patients and controls in the number of naïve, effector memory, and terminally differentiated CD8+ T cells ( Figure 6 ). However, as was observed in the central memory CD4+ T cells, central memory CD8+ T cells were also significantly reduced in S. aureus patients compared with controls (p = 0.005). Thus, PBMCs of patients with invasive S. aureus infections demonstrate a significant reduction in the numbers of circulating central memory T cells.
Figure 5. Analysis of the CD4+ T cell compartment.
PBMCs obtained from age-matched healthy controls (n = 13) and patients with S. aureus infections (n = 11) were stained with CD3, CD4, CCR7, CD45RA, and CD62L antibodies and analyzed by flow cytometry. CD4 T cell subsets are labeled as (A) central memory T cells (TCM), (B) naïve T cells, and (C) effector memory T cells (TEM). (a) Flow cytometry plots of CD4 T cell subsets in a representative S. aureus patient and healthy control. CD62L expression is shown for each CD4 T cell subset. (b) Graphs show the absolute numbers of CD4 T cells and CD4 T cell subsets expressed as absolute number of cells per mL of blood. Horizontal lines represent median values. Statistical analysis was performed using a Mann-Whitney test.
Figure 6. Analysis of the CD8+ T cell compartment.
PBMCs obtained from patients with S. aureus infections (n = 11) and age-matched healthy controls (n = 13) were stained with CD3, CD8, CCR7, CD45RA, and CD62L antibodies and analyzed by flow cytometry. CD 8 T cells and CD 8 T cell subsets are labeled as: (A) central memory T cells (TCM); (B) naïve T cells, (C) effector memory T cells (TEM), and (D) terminally differentiated effector T cells (TEM). (a) Flow cytometry plots show CD8 T cell subsets in a representative S. aureus patient and healthy control. CD62L expression is shown for each CD8 T cell subset. (b) Graphs show the absolute numbers of CD8 T cells and CD8 T cell subsets. Results are expressed as absolute number of cells per mL of blood. Horizontal lines represent median values. Statistical analysis was performed using a Mann-Whitney test.
Monocyte expansion in patients with acute S. aureus infection
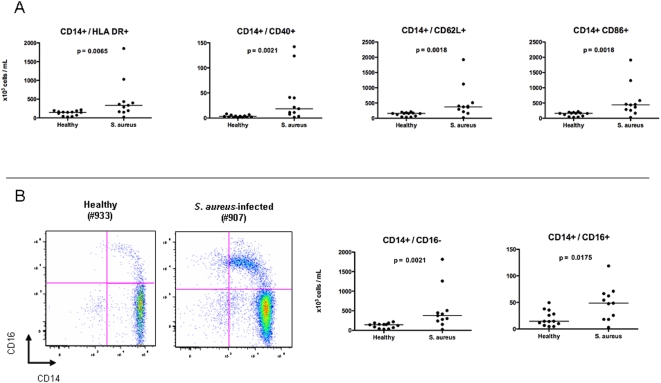
As shown in Figure 3 , there was a significant increase in the median total number of monocytes in PBMCs of patients with S. aureus infection when compared with healthy controls (p = 0.002). Detailed characterization of CD14+ subpopulations showed a significant increase of absolute number of monocytes expressing activation markers such as CD86, CD40, HLA-DR, and CD62L homing lymphocyte molecule in patients with S. aureus infection compared with controls ( Figure 7a ). Reanalysis of the CD14+ subpopulations was performed after removing patients with extreme values (patients 908, 952, or 960) in each subpopulation; S. aureus patients still demonstrated statistically significant increased numbers of CD14+ HLADR+ (p = 0.023), CD14+ CD40+ (p = 0.0076), CD14+ 62L+ (p = 0.0062), and CD14+86+ (p = 0.0062) when compared to healthy controls.
Figure 7. Analysis of monocyte compartment.
PBMCs obtained from patients with S. aureus infections (n = 11) and age-matched healthy controls (n = 13) were stained with Monocytes expressing HLA DR, CD40, CD62L, CD16, and CD86 antibodies and analyzed by flow cytometry. (a) Graphs show the absolute monocyte cell numbers. Results are expressed as absolute number of cells per mL of blood. Horizontal lines represent median values. Statistical analysis was performed using a Mann-Whitney test. (b) There was a significant expansion of both the CD14+ CD16− and the CD14+ CD16+ monocyte populations as shown in a flow cytometry plot of a representative S. aureus patient (907) and healthy control (933).
Based on the CD16 expression, circulating monocytes can be divided into functionally distinct subpopulations: CD14+CD16− monocytes and CD14+16+ monocytes. Patients with S. aureus infection showed a significant expansion of both CD14+16− (p = 0.0021) and CD14+16+ (p = 0.0175) monocytes compared with healthy controls ( Figure 7b ). These differences remained significant even when outliers were removed (CD14+16−, p = 0.0076; CD14+16−, p = 0.0324). Thus, PBMCs of patients with invasive S. aureus infections demonstrate a significant increase in the numbers of circulating monocytes.
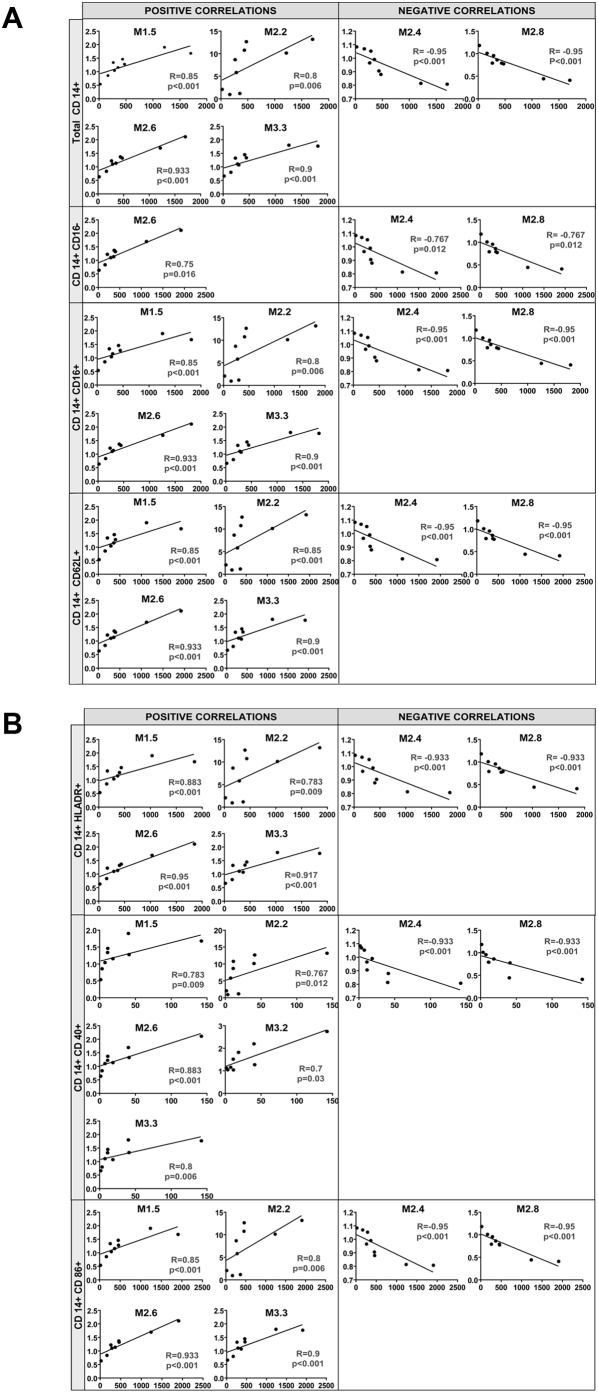
Gene expression levels correlate with specific immune cell populations
To further characterize the relationship between the gene expression profiles in PBMCs of patients with S. aureus infections and the changes of the different immune cell populations, analyses were conducted on a group of patients (n = 9) and healthy controls (n = 9) in whom simultaneous gene expression and flow cytometry analyses were performed. To this end, the per module PBMC average gene expression obtained from patients with S.aureus infection relative to the healthy controls, were run on the Illumina platform and correlated with the absolute number of immune cells in each subject as measured by flow cytometry ( Table 5 ). The most significant statistical correlations observed in patients with S. aureus infections were between the total number of CD14+ monocytes and CD14+ monocyte subpopulations and several gene expression modules ( Figure 8 ). Absolute number of CD14+ monocytes significantly positively correlated with myeloid cell (M1.5 and M2.6), neutrophil (M2.2), and inflammation II (M3.3) modules (Spearman R≥0.8, p≤0.006). Total CD14+ monocyte cell numbers inversely correlated with the modules encoding genes for ribosomal proteins (M2.4) and T cells (M2.8). The number of CD14+16− cells positively correlated with M2.6 (“Myeloid”); CD14+16+ cells correlated with M2.6 (“Myeloid”). M1.5 (“Myeloid”), M2.2 (“Neutrophils”), and M3.3 (“Inflammation II”). The absolute number of CD14+CD16− and CD14+CD16+ cells inversely correlated with the ribosomal protein (M2.4) and T cell (M2.8) modules. Further correlations performed in the monocyte compartment of CD14+CD62L+, CD14+ HLA-DR+, CD14+CD40+, and CD14+CD86+ revealed significant positive correlations between these subpopulations and the myeloid module M2.6 and inverse correlations with ribosomal protein (M2.6) and T cell (M2.8) modules. Gene expression levels in modules related to neutrophils (M2.2) and inflammation module II (M3.3) positively correlated with CD14+CD62L+, CD14+ HLA-DR+, CD14+CD40+, and CD14+CD86+ cell numbers.
Table 5. Correlation of gene expression levels and absolute number of immune cells in patients with S. aureus infections.
| FACS Cell Marker | Module | Assigned Immune Function | Correlation (Spearman) | p-value |
| CD19 | none | |||
| CD3 | none | |||
| CD4 | M1.5 | Myeloid | −0.683 | 0.0361 |
| M2.6 | Myeloid | −0.650 | 0.05 | |
| M3.3 | Inflammation II | −0.683 | 0.0361 | |
| CD8 | M3.1 | Interferon | 0.65 | |
| NK | none | |||
| CD14 | M1.3 | B cells | −0.633 | 0.0583 |
| M1.5 | Myeloid | 0.85 | <0.001 | |
| M2.2 | Neutrophils | 0.8 | 0.006 | |
| M2.4 | Ribosomal | −0.95 | <0.001 | |
| M2.6 | Myeloid | 0.933 | <0.001 | |
| M2.8 | T cells | −0.95 | <0.001 | |
| M3.3 | Inflammation II | 0.9 | <0.001 |
FACS = fluorescence-activated cell sorting; NK = Natural Killer cell.
Figure 8. Significant correlations between absolute and subpopulations of monocytes and modular gene expression in patients with S. aureus infection.
Correlation analyses were performed between the significant modules comprising the gene expression profile of PBMCs from S. aureus-infected patients and monocyte populations. Graphs represent the correlation (Spearman) between the average normalized average gene expression [log] significantly changed (Mann Whitney p<0.05) in the PBMCs of patients with S. aureus infection relative to the median gene expression of PBMCs in healthy controls in each significant module (y axis) and the corresponding absolute monocyte number and CD14+ subpopulations per mL of blood in S. aureus-infected patients (x axis).
With respect to the T cell compartment, the number of total CD4+ T cells inversely correlated with the myeloid (M1.5 and M2.6) and inflammation (M3.3) modules (p = 0.0361) ( Table 5 ). Numbers of naïve CD4+ T cells positively correlated with the modules related to ribosomal proteins (M2.4) and T cells (M3.8), while inversely correlating with the M2.6 (Myeloid) and inflammation (M3.2 and M3.3) modules. The number of CD4+ T central memory cells (TCM) correlated inversely with myeloid (M1.5 and M2.6), neutrophil (M2.2), and inflammation (M3.3) modules, but positively with the ribosomal protein (M2.4), T cell (M2.8), and interferon (M3.1) modules. CD8+ T central memory cell numbers correlated inversely with myeloid modules (M1.5 and M2.6) and inflammation (M3.3) modules; conversely, CD8+ T effector memory cells correlated positively with cytotoxic cell module (M2.1) and undetermined (M1.8 and M3.7) modules.
There were no significant correlations between modular gene expression levels and CD4+ T effector memory, CD8+ naïve, and CD8+ terminally differentiated T cells. There were no significant correlations between the total number of CD19+ B cells, nor B cell subpopulations such as naïve B cells, memory B cells, transitional B cells, pre-germinal center B cells, and plasma cells B cell and modular gene expression patterns ( Table S2 ).
Discussion
The versatility of S. aureus has allowed emergence of highly resistant and virulent bacterial strains in the community [27]. Its pathogenicity is in part due to its repertoire of virulence factors and proclivity for tissue and endovascular invasion, destruction, and dissemination while simultaneously evading multiple components of the innate immune system and secreting immunomodulatory proteins that compromise both humoral and cell-mediated immunity [7], [27], [28]. Very little is known about the relationship between the human host and S. aureus during invasive infections, leading us to carry out the present study. We chose to apply a systems biology approach utilizing both gene expression microarray profiling and corresponding flow cytometry analyses to allow for a comprehensive assessment of the immunopathogenesis of the disease and interaction with the host.
By conducting several different step-wise analyses, the gene expression profile of patients with invasive S. aureus infections was defined and its robustness validated among distinct patient populations and across microarray platforms. Modular analysis demonstrated significant activation of host genes related to the innate immune response with increased expression of genes related to inflammatory processes and cells of the myeloid lineage and significant under-expression of genes related to the adaptive immune response. Although the over-expression of the innate immune response genes was not unexpected [28], [29], the striking and consistent decreased expression of B and T cell-related genes was less anticipated.
There is limited and conflicting information regarding the numbers of lymphocyte populations in patients with acute S. aureus infections [30]–[33]. In one study, patients with S. aureus and S. pneumoniae sepsis showed significantly decreased numbers of CD4+ and CD8+ T cells, and NK cells [34] while in another, patients with MRSA superantigen-associated glomerulonephritis showed increased numbers of DR+ CD4+ and CD8+ T cells and NK cells [35]. Flow cytometry analyses were performed in our study subjects to better understand whether the changes observed in the gene expression patterns simply reflected alterations in immune cell numbers. Despite significant under-expression of T and B cell-related genes observed in our patients with S. aureus infections, there were no differences in the absolute numbers of total B and T cells between infected patients and healthy controls. No consistent significant differences were seen between patients and controls across B cell subpopulations evaluated, however there were possible trends in transitional and pre-germinal B cells that will require analysis in a larger sample size.
Detailed analysis of the T cell sub-populations revealed decreased numbers of both central memory CD4+ T cells and CD8+ T cells, but no differences in the other T cell subsets in patients with S. aureus infection. Central memory T cells have been shown to have a high proliferative potential and to demonstrate in vivo persistence [36], [37]. Our results demonstrate a decreased number of central memory T cells, suggesting a possible reorganization of the circulating T cell compartment that may also explain the reduced expression of T cell-related genes. With increased expression of CCR7 and CD62L, central memory T cells are programmed to preferentially migrate to lymphoid tissues to interact with other T and B cells in establishing a repertoire of effector functions against the invading pathogen [26]. One possible explanation then, for the significant reduction in the numbers of circulating central memory T cells seen in our subjects with S. aureus infection may be central memory T cell homing to these secondary lymph organs. S. aureus expresses factors that promote its immune evasion and could also elicit this cellular imbalance. In vitro observations have demonstrated a shift from central memory to effector memory T cells in the presence of S. aureus enterotoxin [26]. Secretion of other superantigens by S. aureus, such as Map/Eap (MHC Class II analogous protein/extracellular adherence protein), bind to T-cell receptors and directly thwart T cell responses by reducing T cell proliferation, altering effector functions, and stimulating apoptosis [7].
Alternatively, the under-expression of T cell genes may be due to an increase in other cell populations. Indeed, the monocyte compartment, total number and subpopulations, was significantly expanded in patients with S. aureus infection and correlated with gene expression levels, providing the cellular component to the increased expression of innate immune genes. Based on CD16 expression, circulating monocytes can be divided into functionally distinct subpopulations CD14+CD16− and CD14+16+ monocytes, which may have distinct roles in the innate immune response [38]. In the S. aureus patients, there was a significant expansion of both of these monocyte subsets. Furthermore, the high levels of expression of genes generally associated with neutrophil functions, such as defensins, lactotransferrins, CAMP, and elastase, as seen in module M2.2, may originate from the CD14+16+ (CD62L−) monocytes subset, as has been recently described [38]. Although significant numbers of neutrophils are not generally present in PBMCs, low-density neutrophils have been demonstrated and accounted for over-expression of neutrophil genes in patients with SLE [39]. Despite using similar technical approaches, we were not able to detect neither these low-density immature neutrophils nor high-density neutrophils that could result from contamination in our samples. Studies with purified cell subsets will be necessary to understand the distinct contribution from each cell subtype.
These results demonstrate the value of gene expression profiling in combination with flow cytometry as a new strategy to study disease pathogenesis within the clinical context. This study provides information on the profound dysregulation of both the innate and adaptive immune responses induced by invasive S. aureus infections and opens new avenues for improving our understanding of this infection that may lead to improved biomarker discovery and therapeutic interventions.
Materials and Methods
Ethics Statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Boards of the University of Texas Southwestern Medical Center and Children's Medical Center of Dallas (IRB #0802-447) and Baylor Institute of Immunology Research (BIIR, IRB # 002-141). Informed consent was obtained from legal guardians and informed assent was obtained from patients 10 years of age and older prior to any study-related procedure.
Patient information
Blood samples were collected from 77 children: 53 patients with S. aureus infection and 24 healthy controls. Children with suspected or proven polymicrobial infections, underlying chronic disease, immunodeficiency, or those who received steroids or other immunomodulatory therapies were excluded. Control samples were obtained from healthy children undergoing elective surgical procedures and at healthy outpatient clinic visits. Nasopharyngeal viral cultures were obtained in both patients and controls to exclude viral co-infections. Children hospitalized with acute S. aureus infections were offered participation in the study after microbiologic confirmation of the diagnosis by standard bacterial culture of blood or tissue specimens. Patients were analyzed in 3 groups: training (20 S. aureus, 10 healthy), test (22 S. aureus, 10 healthy), and validation sets (11 S. aureus, 14 healthy).
Sample collection
Blood samples (3–8 mL) were collected in acid-citrate-dextrose tubes (ACD tubes, BD Vacutainer, Franklin Lakes, NJ) and delivered to the laboratory at room temperature for processing. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-hypaque technique and lysed in RLT reagent (Qiagen, Valencia, CA) with β-mercaptoethanol (BME) and stored at -80°C until processing. Samples were run in batches by the same laboratory team to ensure standardization of quality and handling of samples. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA) per the manufacturer's instructions and RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
Microarray procedures
Affymetrix
From 2–5 micrograms of total RNA, double-stranded cDNA was generated as a template for single-round in vitro transcription with biotin-labeled nucleotides using the Affymetrix cDNA Synthesis and In Vitro Transcription kits (Affymetrix Inc., Santa Clara, CA). Biotinylated cRNA targets were then purified (Sample Cleanup Module, Affymetrix) and hybridized to the Affymetrix HG-U133A and B GeneChip arrays (Affymetrix Inc., Santa Clara, CA) according to the manufacturer's standard protocols.
Affymetrix Gene Chips
Arrays were scanned using a laser confocal scanner (Agilent). Global gene expression analysis was carried out using the Affymetrix HG-U133A and U133B GeneChips. The HG-U133 set contains 44,760 probe sets representing >39,000 transcripts derived from ∼33,000 human genes. Raw signal intensity values were normalized to the mean intensity of all measurements per gene chip and scaled to a target intensity value of 500 using the MAS 5.0 global scaling method to adjust for possible chip-to-chip variations in hybridization intensities (GeneChip Operating System version 1.0). Data was imported into GeneSpring software (version 7.3.1, Agilent) to perform the gene expression analyses, statistical testing, hierarchical clustering, and classification of samples.
Illumina: Double-stranded cDNA was obtained from 200 ng of total RNA and after in vitro transcription underwent amplification and labeling steps according to the manufacturer's instructions. 1.5 µg of amplified biotin-labeled cRNA was hybridized to the Illumina Sentrix Hu6 BeadChips according to the sample labeling procedure recommended by Illumina. (Ambion, Inc, Austin, TX).
Illumina BeadChips: The Sentrix Hu6 BeadChips consist of 50mer oligonucleotide probes attached to 3-µm beads within microwells on the surface of the glass slide representing 48,687 probes. Slides were scanned on Illumina BeadStation 500 and Beadstudio software was used to assess fluorescent hybridization signals.
Microarray Data Analysis
Using the Genespring™ software program (Agilent), the expression value for each gene per individual subject's sample was normalized to the median expression value of the same gene in samples from healthy controls. Class comparison analyses were performed on probe sets present in at least 75% of samples in each group (quality control (QC) probes). Non-parametric statistical testing (Wilcoxon-Mann-Whitney U-test; p<0.01 for class comparisons; p<0.05 for modular analyses with no multiple test corrections) was used to rank genes based on their ability to discriminate among pre-specified groups of patients. Final lists of significantly changed genes used in class comparisons were filtered to include only those transcripts that showed a 1.25-fold or greater fold change in expression level relative to the control group. Hierarchical clustering was applied to order genes according to expression levels. The list of most highly ranked genes from the S. aureus biosignature was created by ranking genes with the highest fold change difference and the most significantly different genes (by p value) between healthy controls and patients with infection.
For the cross microarray platform validation part of the analysis we performed a two-step procedure. First, we examined the genes that defined the S. aureus biosignature obtained on the test and training set of subjects analyzed on the Affymetrix platform in a new set of 18 subjects (validation set) and run on the lllumina microarray platform. Transcript sequences from RefSeq were used to perfectly match corresponding valid gene probe sets on each platform; Affymetrix probe set data encoded by Mage-ML files in XML format were matched by GenBank accession numbers and reference sequence transcripts to the Illumina manifest probe mapping file (http://www.switchtoi.com/probemapping.ilmn) allowing for mapping of the significant Affymetrix gene probes to their corresponding gene probes on the Illumina platform. In the second step of the analysis, the Illumina gene list was applied to an independent validation set of subjects using an unsupervised scheme that allowed clustering of samples based solely on intrinsic gene expression levels.
Transcriptional Module Analysis
A detailed account of this module-based data mining analysis strategy has been reported elsewhere [25]. Briefly, this is a systems-scale strategy for microarray analysis that has identified transcriptional modules formed by genes coordinately expressed across multiple disease data sets thus allowing functional interpretation of the microarray data into biologically useful information. Analysis of linear correlations between gene expression levels per module in the different microarray platforms (Affymetrix vs. Illumina) and between the average gene expression levels per module (from the validation set run on Illumina) and the individual immune cell numbers per patient (calculated by Flow cytometry) were performed using a non-parametric statistical correlation analysis test (Spearman).
Flow cytometry
PBMCs were isolated by density gradient centrifugation using Ficoll-hypaque technique from blood samples (3–8 mL) collected in ACD tubes. One million PBMCs were washed and incubated with conjugated antibodies for 15 minutes at room temperature, in dark conditions according to multi-color staining panels described below. Cells were washed with phosphate buffer and then fixed with 2% paraformaldehyde. Samples were run on a BD LSRII flow cytometer (BD BioScience, San Jose, CA); 50,000 events were acquired with the BD FACSDiva™ software according to the lymphocyte gate and analyzed using FlowJo™ software (Tree Star, Inc).
Multicolor staining panels included: B cell panel: CD19 (ECD, Beckman-Coulter), CD20 (Pe-Cy5, BD Pharmingen), CD24 (PE, BD Pharmingen), CD27 (APC, BD Pharmingen), CD38 (Pe-Cy7, BD Biosciences) and IgD (FITC, SouthernBiotech). T cell panel: CD3 (Alexa700, BD Pharmingen), CD4 (Pacific Blue, Invitrogen), CD8 (APC-Cy7, BD Pharmingen), CD45RA (ECD, Beckman-Coulter), CD62L (Pe-Cy5, BD Pharmingen), CCR7 (Pe-Cy7, BD Pharmingen). Monocyte panel: CD14 (Pacific Blue, BD Pharmingen), CD16 (APC, Invitrogen), CD40 (PE, BD Pharmingen), CD86 (FITC, BD Pharmingen), HLA-DR (APC-Cy7, BD Pharmingen) and CD62L (ECD, Beckman-Coulter). Isotype controls: IgG1 (FITC, BD Biosciences), IgG1 (PE, BD Biosciences), IgG1 (ECD, Beckman-Coulter), IgG1 (Pe-Cy5, BD Biosciences), IgG1 (Pe-Cy7, BD Biosciences), IgG1 (APC, BD Biosciences), IgG1 (Alexa 700, BD Biosciences), IgG1 (APC-Cy7, BD Biosciences), IgG1 (Pacific Blue, Invitrogen).
Cell populations were analyzed based on the following markers: B cells: CD19; Naïve B cells: CD19+/CD20+, IgD+, CD27−; Memory B cells: CD19+/CD20+/IgD+&−/CD27+; Plasma cells: CD19+/CD20−/CD27+&++/CD38++; Transitional B cells: CD19+/CD20+/CD24++/CD38++; Pre-germinal center B cells: CD19+/CD20+/CD27+/CD38++; T cells: Naïve T cells: CD3+/CD4+ or CD8+/CD45RA+/CCR7+; Central memory T cells: CD3+/CD4+ or CD8+/CD45RA−/CCR7+; Effector memory T cells: CD3+/CD4+ or CD8+/CD45RA−/CCR7−; CD8+ terminally differentiated T cells: CD3+/CD8+/CD45RA+/CCR7−.
Online Supporting Information
Table S1 provides the functional interpretation of the transcriptional modules including module, number of probe tests evaluated, key words included in data mining, and immune interpretation. Table S2 details the results of the non-parametric correlation analyses performed between B and T cell populations and module analyses. Figure S1 provides graphic representation and detailed information regarding significantly differently expressed genes in patients with S. aureus infection versus healthy control per module.
Accession Numbers
The microarray data used in this study is deposited in NCBI Gene Expression Omnibus (GEO).
Supporting Information
Functional interpretation of transcriptional modules.
(0.06 MB DOC)
Correlations of B and T cell subsets with transcriptional modules.
(0.05 MB DOC)
Module analysis of the gene expression profile in patients with S. aureus infections. Graphs represent the average gene expression level (y axis) significantly changed (Mann Whitney p<0.05) in patients with S. aureus infections (x axis, red horizontal bar) versus healthy controls (x axis, green horizontal bar) in each of the 19 significant modules (M) comprising the gene expression profile seen in PBMCs of S. aureus-infected patients. A detailed list citing the gene probe, pvalue, and description of the significant transcript is noted.
(0.57 MB DOC)
Acknowledgments
Special thanks to our patients and their guardians for participating in this study and to Evelyn Torres, RN, MS for actively recruiting subjects.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National Institutes of Health (NIH) grants U19 AIO57234-02 (JB and OR), P01 CA084512, R01 CA078846 and R01 I068842 (JB); the Center for Lupus Research P50 AR054083 and R01 AR050770-01 (VP); The Baylor Health Care System Foundation. JB holds the W.W. Caruth, Jr Chair in Organ Transplantation Immunology. MIA was supported in part by NIH grants KL2RR024983 and UL1RR024982. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Francis JS, Doherty MC, Lopatin U, Johnston CP, Sinha G, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–107. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 2.Fridkin SK, Hageman JC, Morrison M, Sanza LT, Como-Sabetti K, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 3.Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. Jama. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan SL, Hulten KG, Gonzalez BE, Hammerman WA, Lamberth L, et al. Three-year surveillance of community-acquired Staphylococcus aureus infections in children. Clin Infect Dis. 2005;40:1785–1791. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- 5.Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis. 2005;5:275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 6.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 7.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 8.Benoist C, Germain RN, Mathis D. A plaidoyer for ‘systems immunology’. Immunol Rev. 2006;210:229–234. doi: 10.1111/j.0105-2896.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- 9.Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204:2131–2144. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med. 2005;201:1479–1486. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes MG, Aronow BJ, Luyrink LK, Moroldo MB, Pavlidis P, et al. Gene expression in juvenile arthritis and spondyloarthropathy: pro-angiogenic ELR+ chemokine genes relate to course of arthritis. Rheumatology (Oxford) 2004;43:973–979. doi: 10.1093/rheumatology/keh224. [DOI] [PubMed] [Google Scholar]
- 12.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 13.Jeffrey SS, Fero MJ, Borresen-Dale AL, Botstein D. Expression array technology in the diagnosis and treatment of breast cancer. Mol Interv. 2002;2:101–109. doi: 10.1124/mi.2.2.101. [DOI] [PubMed] [Google Scholar]
- 14.Clement S, Vaudaux P, Francois P, Schrenzel J, Huggler E, et al. Evidence of an intracellular reservoir in the nasal mucosa of patients with recurrent Staphylococcus aureus rhinosinusitis. J Infect Dis. 2005;192:1023–1028. doi: 10.1086/432735. [DOI] [PubMed] [Google Scholar]
- 15.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubins KH, Hensley LE, Jahrling PB, Whitney AR, Geisbert TW, et al. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc Natl Acad Sci U S A. 2004;101:15190–15195. doi: 10.1073/pnas.0405759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–1107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medzhitov R, Janeway C., Jr Innate immunity. N Engl J Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 20.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 21.Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, et al. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci U S A. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 23.Montano M, Rarick M, Sebastiani P, Brinkmann P, Russell M, et al. Gene-expression profiling of HIV-1 infection and perinatal transmission in Botswana. Genes Immun. 2006;7:298–309. doi: 10.1038/sj.gene.6364297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 27.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 28.Fournier B, Philpott DJ. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev. 2005;18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadley JS, Wang JE, Foster SJ, Thiemermann C, Hinds CJ. Peptidoglycan of Staphylococcus aureus upregulates monocyte expression of CD14, Toll-like receptor 2 (TLR2), and TLR4 in human blood: possible implications for priming of lipopolysaccharide signaling. Infect Immun. 2005;73:7613–7619. doi: 10.1128/IAI.73.11.7613-7619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baran J, Pajdak E, Isendorf M, Szmyd D. [Evaluation of interactions between strains of S. aureus isolated from different clinical specimens with peripheral blood phagocytic cells]. Med Dosw Mikrobiol. 1999;51:37–46. [PubMed] [Google Scholar]
- 31.Bueno C, Almeida J, Alguero MC, Sanchez ML, Vaquero JM, et al. Flow cytometric analysis of cytokine production by normal human peripheral blood dendritic cells and monocytes: comparative analysis of different stimuli, secretion-blocking agents and incubation periods. Cytometry. 2001;46:33–40. [PubMed] [Google Scholar]
- 32.Sundstedt A, Grundstrom S, Dohlsten M. T cell- and perforin-dependent depletion of B cells in vivo by staphylococcal enterotoxin A. Immunology. 1998;95:76–82. doi: 10.1046/j.1365-2567.1998.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viau M, Longo NS, Lipsky PE, Zouali M. Staphylococcal protein a deletes B-1a and marginal zone B lymphocytes expressing human immunoglobulins: an immune evasion mechanism. J Immunol. 2005;175:7719–7727. doi: 10.4049/jimmunol.175.11.7719. [DOI] [PubMed] [Google Scholar]
- 34.Holub M, Kluckova Z, Helcl M, Prihodov J, Rokyta R, et al. Lymphocyte subset numbers depend on the bacterial origin of sepsis. Clin Microbiol Infect. 2003;9:202–211. doi: 10.1046/j.1469-0691.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 35.Yoh K, Kobayashi M, Yamaguchi N, Hirayama K, Ishizu T, et al. Cytokines and T-cell responses in superantigen-related glomerulonephritis following methicillin-resistant Staphylococcus aureus infection. Nephrol Dial Transplant. 2000;15:1170–1174. doi: 10.1093/ndt/15.8.1170. [DOI] [PubMed] [Google Scholar]
- 36.Lanzavecchia A, Sallusto F. Understanding the generation and function of memory T cell subsets. Curr Opin Immunol. 2005;17:326–332. doi: 10.1016/j.coi.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Lanzavecchia A, Sallusto F. Dynamics of T lymphocyte responses: intermediates, effectors, and memory cells. Science. 2000;290:92–97. doi: 10.1126/science.290.5489.92. [DOI] [PubMed] [Google Scholar]
- 38.Mobley JL, Leininger M, Madore S, Baginski TJ, Renkiewicz R. Genetic evidence of a functional monocyte dichotomy. Inflammation. 2007;30:189–197. doi: 10.1007/s10753-007-9036-0. [DOI] [PubMed] [Google Scholar]
- 39.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional interpretation of transcriptional modules.
(0.06 MB DOC)
Correlations of B and T cell subsets with transcriptional modules.
(0.05 MB DOC)
Module analysis of the gene expression profile in patients with S. aureus infections. Graphs represent the average gene expression level (y axis) significantly changed (Mann Whitney p<0.05) in patients with S. aureus infections (x axis, red horizontal bar) versus healthy controls (x axis, green horizontal bar) in each of the 19 significant modules (M) comprising the gene expression profile seen in PBMCs of S. aureus-infected patients. A detailed list citing the gene probe, pvalue, and description of the significant transcript is noted.
(0.57 MB DOC)