Abstract
High-level expression of Bcl-2 associated athanogene (BAG-1) protects cancer cells from stress-induced cell death and growth inhibition. These protective effects of BAG-1 are dependent on interactions with the HSC70 and HSP70 chaperones. However, the key stress-response molecules that are regulated by a BAG-1/chaperone mechanism have not been identified. In this study, we investigated the effects of BAG-1 overexpression on the function of p53 family proteins, p53, p63 and p73. Overexpression of BAG-1 isoforms interfered with the transactivating activity of p73 and p63, but had modest and variable effects on p53-dependent transcription. p73 and BAG-1 interacted in intact cells and overexpression of BAG-1 decreased the expression of p73. siRNA-mediated ablation of endogenous BAG-1 increased the activity of a p73-responsive promoter and this was reversed by knock-down of p73. The ability of BAG-1 to modulate p73 activity and expression, and to interact with p73 were dependent on amino acid residues required for the interaction of BAG-1 with HSC70 and HSP70. These results show that BAG-1 inhibits the transactivating functions of p73 and provide new insight into the mechanisms that control the expression of p73. Inhibition of p73 function may be one mechanism that contributes to the pro-survival activity of BAG-1.
Keywords: BAG-1, HSC70, HSP70, p73, transcription, apoptosis
Bcl-2 associated athanogene (BAG-1) is a multifunctional protein that interacts with multiple cellular targets and modulates a wide range of cellular processes (Alberti et al, 2003; Townsend et al, 2003a, 2005; Gehring, 2004). Overexpression of BAG-1 protects cells from various apoptotic stimuli, enhances proliferation and metastasis, and modulates the transcriptional activity of a variety of nuclear hormone receptors. BAG-1 is essential for the survival and differentiation of haemopoietic and neuronal cells in mice (Gotz et al, 2005). Functional and expression studies suggest that overexpression of BAG-1 may play an important role in diverse cancer types (Cutress et al, 2002; Tang, 2002; Sharp et al, 2004). For example, BAG-1 is frequently overexpressed in breast cancer and can correlate with important clinical parameters (Tang et al, 1999, 2004; Turner et al, 2001; Townsend et al, 2002; Cutress et al, 2003; Pusztai et al, 2004; Sirvent et al, 2004).
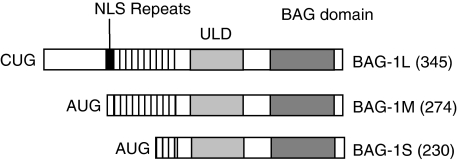
In human cells BAG-1 exists as three major isoforms (BAG-1S, BAG-1M and BAG-1L) derived by alternate translation initiation from a single mRNA (Figure 1). All BAG-1 isoforms contain a C-terminal, evolutionary conserved BAG domain (Takayama and Reed, 2001) and a central ubiquitin-like domain (ULD), but the larger isoforms have unique N-terminal extensions. In general, the functional significance of these variable N-terminal regions is poorly understood. However, BAG-1L possesses a nuclear localisation sequence and is a predominantly nuclear protein, whereas the other isoforms partition between the cytoplasm and nucleus (Packham et al, 1997; Takayama et al, 1998; Yang et al, 1998; Brimmell et al, 1999).
Figure 1.
Human Bcl-2 associated athanogene (BAG-1) isoforms. The structures of the three major human BAG-1 isoforms are shown, along with their size (amino acid residues). Translation of BAG-1L initiates at an upstream CUG codon, whereas BAG-1M and BAG-1S are AUG-derived. The position of the nuclear localisation sequence (NLS), acidic repeats, ubiquitin-like domain (ULD) and BAG domain are shown.
The C-terminal BAG domain is comprised of a bundle of three α-helices of which helices 2 and 3 mediate electrostatic interactions with the ATPase domain of the 70 kDa heat shock proteins, HSC70 and HSP70 (Briknarova et al, 2001; Sondermann et al, 2001). BAG-1 acts as a cochaperone and stimulates nucleotide exchange of HSC70/HSP70 (Hohfeld and Jentsch, 1997; Takayama et al, 1997; Luders et al, 2000b; Brehmer et al, 2001; Nollen et al, 2001). HSC70 and HSP70 play important roles in multiple cell processes, for example, by effects on protein (re)folding and degradation, and on the expression and activity of nuclear hormone receptors (Mayer and Bukau, 2005; Daugaard et al, 2007; Grad and Picard, 2007). Binding to these multifunctional proteins may explain, at least in part, the multiple effects associated with BAG-1 overexpression. The BAG-1 ULD is required for the interaction of BAG-1 with the proteasome (Luders et al, 2000a), and substantial evidence shows that BAG-1 can act to coordinate the function of chaperones and the proteasome in the degradation of specific proteins (Arndt et al, 2007). BAG-1 can interact simultaneously with HSC70 and the proteasome (Luders et al, 2000a; Alberti et al, 2002), and its ability to influence chaperone function may facilitate the unloading of chaperone clients in the vicinity of the proteasome to enhance degradation. BAG-1 also interacts with CHIP, an E3 ubiquitin ligase, which plays a key role in protein triage (i.e., degradation versus refolding) decisions (McDonough and Patterson, 2003). BAG-1 and CHIP cooperate to target the glucocorticoid receptor for proteasomal degradation (Demand et al, 2001).
Functional studies show that BAG-1 isoforms promote the survival, proliferation and metastasis of cancer cells. For example, overexpression of BAG-1S or BAG-1L increases breast cancer cell survival in vitro and tumour growth in vivo (Kudoh et al, 2002). Our own studies (Townsend et al, 2003b) showed that overexpression of BAG-1 isoforms provided robust protection from cell death and long-term growth inhibition induced by heat shock, and other cellular stress, including hypoxia, radiation and certain cytotoxic agents. RNAi-mediated knock-down of BAG-1 is also sufficient to promote apoptosis (Sawitzki et al, 2002; Clemo et al, 2008). The survival-promoting function of BAG-1 was dependent on the BAG domain as a BAG-1 mutant lacking this region failed to promote cell survival in breast cancer cells (Kudoh et al, 2002; Townsend et al, 2003b). In addition to HSC70/HSP70, the BAG domain also acts as a docking site for c-Raf; BAG-1 activates c-Raf independent of Ras (Wang et al, 1996; Song et al, 2001). However, the critical functional requirement for the BAG domain seemed to be chaperone binding as the introduction of mutations that specifically ablated HSC70/HSP70 interaction interfered with BAG-1-mediated survival in breast cancer cells (Townsend et al, 2003b) and other cell systems (Townsend et al, 2004). Thus, the survival function of BAG-1 is dependent on HSC70/HSP70. However, the specific molecular regulators of stress-induced apoptosis that are targeted by BAG-1 remain to be identified.
Members of the p53 family of transcription factors (p53, p63 and p73 and their splice variants) are critical regulators of stress-induced apoptosis (Murray-Zmijewski et al, 2006; McKeon and Melino, 2007; Stiewe, 2007). These proteins accumulate or are activated following cellular stress (including DNA damaging treatments and oncogenic stress) and transactivate target genes to induce cell cycle arrest (e.g., GADD45, p21cip1), apoptosis (e.g., Bax, Puma, IGFBP3, PIG3) or activate negative-feedback control loops (e.g., Mdm2). The function of p53 family proteins is required for normal stress-induced apoptosis (Agami et al, 1999; Gong et al, 1999; Yuan et al, 1999; Flores et al, 2002; Gressner et al, 2005; Stiewe, 2007) and they are frequently inactivated in cancer cells by mutations (p53 in particular) or alternate mechanisms. A number of proteins that have been shown to be overexpressed in cancer cells act, at least in part, by inhibiting the function of p53 family proteins. These proteins may be attractive therapeutic targets, as interfering with their function can lead to a reactivation of tumour suppressor function. For example, Mdm2 is overexpressed in many cancers with wild type p53 and inhibits p53 function by inhibition of transcriptional activity and targeting for degradation (Toledo and Wahl, 2006). Inhibitors of the p53:Mdm2 interaction induce p53-dependent apoptosis and are being developed as anti-cancer drugs (Dudkina and Lindsley, 2007).
Our earlier study showed that BAG-1 overexpression suppressed stress-induced apoptosis in MCF7 breast cancer cells (Townsend et al, 2003b), despite the presence of wild type p53, p63 and p73 in these cells (Gudas et al, 1995; Toh et al, 2005). This suggested that BAG-1 could interfere with the normal function of these proteins. As transcriptional regulation plays a major role in the function of p53, p63 and p73, we investigated the effects of BAG-1 on transactivation by p53 family proteins.
Materials and methods
Cell lines and culture
SaOs2 (human osteosarcoma), HEK293 (human embryonic kidney) and NIH3T3 (mouse fibroblast) cell lines were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) and maintained in Dulbecco's Modified Eagle's medium (Gibco, Paisley, UK) supplemented with 10% (v/v) foetal calf serum (FCS) (PAA Laboratories, Yeovil, UK), 1 mM l-glutamine and penicillin/streptomycin (Gibco). H1299 (human non-small lung carcinoma) cells were obtained from ATCC and maintained in RPMI 1640 medium (Gibco) supplemented with 10% (v/v) FCS, 1 mM l-glutamine and penicillin/streptomycin.
Plasmids
The reporter plasmids Bax-lux, GADD45-luc, Mdm2-luc, Pig3-luc, IGFBP3B-luc and the p53 expression plasmid (Rowan et al, 1996; Hsieh et al, 1999) were kindly provided by Prof. Xin Lu, (Ludwig Institute for Cancer Research, Oxford, UK). The p63, p73α and p73β expression plasmids (De Laurenzi et al, 1998, 2000) were a kind gift of Prof. Gerry Melino (Medical Research Council, Toxicology Unit, Leicester, UK). Human BAG-1S, BAG-1M and BAG-1L isoform specific expression constructs and point mutations have been described earlier (Townsend et al, 2003b, 2004). pcDNA3 plasmid was from Invitrogen Life Technologies (Paisley, UK). pGL2-Basic (Promega, Southampton, UK) and the human Bcl-X IB promoter construct pBcl-XIB (MacCarthy-Morrogh et al, 2000) were used as control reporter plasmids.
Transfections and reporter gene assays
For luciferase assays, SaOs2 cells (5 × 104) were plated in 24-well tissue culture plate one day before transfection. Cells were transfected using FuGene 6 transfection reagents (Roche Applied Science, Burgess Hill, UK) according to the manufacturer's instructions. Empty vector pcDNA3 was used to maintain equal quantity of total DNA per transfection. At 48 h after transfection, cells were washed with cold phosphate-buffered saline (PBS), collected by centrifugation and resuspended in 100 μl cell lysis buffer (0.65% (v/v) IGEPAL CA-630, 10 mM Tris(hydroxymethyl)methylamine (Tris)-HCl, 1 mM ethylenediaminetetraacetic acid (EDTA) disodium salt, 150 mM NaCl, pH 8.0) and incubated on ice for 5 min. The cell lysate was clarified by centrifugation and luciferase activity was measured using the Luciferase Assay System reagents (Promega) and a Sirius luminometer (Berthold Detection System, Oak Ridge, TN, USA). In the experiments to determine the effect of BAG-1 on p73α expression levels, H1299 cells were plated in 10-cm tissue culture dishes and co-transfected with 1 μg of p73α expression construct in the presence or absence of 7 μg of BAG-1S expression construct or pcDNA3. After 24 h, expression of p73α, BAG-1 and PCNA (loading control) were analysed by immunoblotting.
Immunoblotting
Immunoblots were carried out as described earlier (Brimmell et al, 1999) using the following primary antibodies: rabbit polyclonal anti-BAG-1 (TB3, (Brimmell et al, 1999)), mouse monoclonal anti-BAG-1 (3.10 G3E2; (Brimmell et al, 1999)), mouse monoclonal anti-p73 antibody E4 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal anti-p73 antibody R26 (generated by immunisation of rabbits with purified GST-p73α1−131 fusion protein), rabbit polyclonal anti-β-actin antibody (Sigma, Poole, UK) and mouse monoclonal anti-PCNA antibody (Santa Cruz Biotechnology). Horseradish peroxidase-conjugated secondary antibodies were from Amersham (GE Healthcare UK, Amersham, UK) and bound immunocomplexes were detected using SuperSignal West Pico Chemiluminescent reagents (Perbioscience UK Ltd, Pierce, Northumberland, UK). To quantify the effects of overexpression of BAG-1 on p73α expression, immunoblots were analysed using Quantity One program (BioRad, Hemel Hempstead, UK). The expression of p73α was normalised to the expression of PCNA and the relative expression of p73α in the absence of BAG-1 overexpression was set at 1.0.
RNA interference
Control siRNA (control 1), siRNA against Bcl-w (control 2), siRNA against human BAG-1 and siRNA against human p73 were obtained from Ambion Ltd (Huntingdon, UK) as annealed double-stranded RNA-DNA hybrids. Their sequences are: Bcl-w sense 5′-r(GCUGGAGAUGAGUUCGAGA)d(tt)-3′ and antisense 5′-r(UCUCGAACUCAUCUCCAGC)d(tg), BAG-1 sense 5′-r(GGUUGUUGAAGAGGUCAUA)d(tt)-3′ and antisense 5′-r(UAUGACCUCUUCAACAACC)d(tg)-3′, hp73 sense 5′-r(CGGAUUCCAGCAUGGACGU)d(TT)-3′ and antisense 5′-r(ACGUCCAUGCUGGAAUCCG)d(TT)-3′. H1299 cells were co-transfected with siRNA oligonucleotides at a final concentration of 75 nM, together with a reporter plasmid (pig3-luc at 400 ng) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 72 h, cells were harvested and lysed for western blotting and luciferase assay.
Quantitative-reverse transcription-polymerase chain reaction (Q–RT–PCR)
Total RNA was isolated using Trizol (Invitrogen) and the quantity and quality of RNA was analysed using a Agilent 2100 Bioanalyser (Agilent Technologies Inc., South Queensferry, UK). cDNA was synthesised using oligo(dT) and MMLV reverse transcriptase (Promega) according to the manufacturer's instructions. Q–RT–PCR was carried out in 20-μl reactions containing 5 μl cDNA, 10 μl Universal Taqman PCR master mix (Applied Biosystems, Warrington, UK) and 1 μl of the Taqman Gene Expression Assay of interest (Applied Biosystems). Expression assays used for this study were p73 (Hs00232088_m1) and β-actin (Hs99999903_m1). All reactions were carried out in duplicate using the ABI PRISM 7500 Sequence Detection System (Applied Biosystems) according to the following thermal cycle protocol: 94°C for 10 min, followed by 40 cycles at 94°C for 15 s and 60°C for 1 min. Control reactions with no cDNA were run on each plate for each Taqman gene Expression Assay used and no amplification was detected in any control reaction. All expression values were normalised using expression of β-actin as a control.
Co-Immunoprecipitations
H1299 cells were transfected (FuGene 6) in 4 × 100 mm tissue culture plates, each with 5 μg of p73α expression plasmid together with 5 μg of BAG-1S expression plasmids, or 5 μg of pcDNA3 empty vector. Cells were washed and harvested in cold PBS and resuspended in 1.5 ml of HMKEN buffer (10 mM n-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.2), 5 mM MgCl2, 142 mM KCl, 2 mM ethylene glycol-bis(b-aminoethylether)N,N,N′,N′-tetra-acetic acid, 0.2% (v/v) IGEPAL, and Protease Inhibitor Cocktail (Sigma)) by passing repeatedly through 21-gauge and 25-gauge needles, followed by incubation on ice for 30 min. The lysate was clarified by centrifugation. A portion (50 μl) of the resultant cell lysate was retained as a whole cell lysate. The remaining sample was pre-cleared using protein G-Sepharose beads (pre-blocked with 5% (w/v) skimmed milk overnight) for 30 min at 4°C on a Spiramixer. Protein G-Sepharose beads were removed by centrifugation. To immunoprecipitate BAG-1, the lysate was divided into two parts and incubated with the BAG-1-specific rabbit polyclonal antibody TB3 or with pre-immune control serum (both at 4 μl/500 μl lysate), respectively. After overnight incubation at 4°C, the lysate was incubated with protein G-Sepharose beads at 4°C for 4 h, and immunocomplexes were removed by centrifugation. The beads were washed four times using HMKEN buffer, re-suspended in 50 μl SDS-PAGE sample buffer and heated at 95°C for 5 min before immunoblot analysis.
Results
BAG-1 isoforms interfere with the transactivating function of p73α
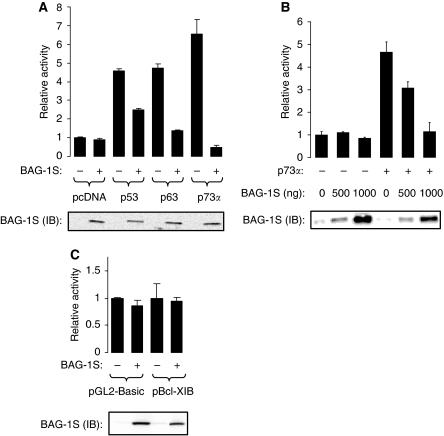
We carried out transient transfection assays to determine whether the major BAG-1 isoform, BAG-1S, modulates the transcriptional activities of p53 family proteins. p53-null SaOs2 cells were selected for this study as they have been widely used for investigations of p53 family protein function (Jost et al, 1997; Mantovani et al, 2004; Melino et al, 2004). SaOs2 cells were transfected with a Bax promoter–reporter construct and p53, p63 or p73α expression plasmids, in the presence or absence of a human BAG-1S expression plasmid (Figure 2A). BAG-1S overexpression did not have a significant effect on the basal expression of the Bax promoter but did interfere with the ability of p53, p63 and p73α to increase promoter expression. Whereas the ability of BAG-1S to modulate p53 function was modest and variable between experiments (mean inhibition from two separate experiments each carried out in duplicate (±s.d.) was 22±33%), p63 and p73α functions were strongly inhibited by BAG-1S (mean inhibition 76±7% and 91±1%, respectively). Co-expression of p53, p63 or p73α did not significantly alter the levels of BAG-1S (Figure 2A).
Figure 2.
Effect of Bcl-2 associated athanogene (BAG-1)S overexpression on transcriptional regulation by p53 family proteins. (A) SaOs2 cells were transfected with 100 ng of Bax-luc reporter construct and 100 ng of p53, 100 ng of p63 or 50 ng of p73α expression plasmids, respectively, in the presence or absence of BAG-1S expression construct (2500 ng). Transfected cells were analysed for luciferase activity (top) and BAG-1S expression (by immunoblotting (IB); bottom) 48 h after transfection. Data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells transfected with Bax-luc only (set to 1.0). (B) SaOs2 cells were transfected with 100 ng of Bax-luc reporter construct in the absence or presence of p73α expression plasmid (50 ng) and increasing amount of BAG-1S expression construct (0, 500, 1000 ng). Transfected cells were analysed for luciferase activity (top) and BAG-1S expression (bottom) 48 h after transfection. Data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells transfected with Bax-luc only (set to 1.0). (C) SaOs2 cells were transfected with 200 ng of pGL-Basic or Bcl-XIB reporter constructs in the absence or presence of 1000 ng of BAG-1S expression plasmid and luciferase activity (top), and BAG-1S expression (by immunoblotting (IB); bottom) was measured after 48 h. Data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells without BAG-1S overexpression (set to 1.0). The experiments shown in (A–C) are representative of two similar experiments.
As the effects of BAG-1S on p73α function were most dramatic, we focused our analysis on this interaction. Plasmid-titration experiments showed that the effects of BAG-1S overexpression were concentration dependent (Figure 2B) and were specific, because BAG-1S overexpression did not interfere with the activity of control promoters not regulated by p73α (Figure 2C). The inhibitory effects of BAG-1S were also observed in all the cell lines tested (HEK293, NIH3T3 and H1299; Figure 3A) and using a range of p73-responsive promoter constructs (IGFBP3, GADD45, Pig3 and MDM-2; Figure 3B). All three BAG-1 isoforms inhibited p73α-mediated transcription when overexpressed in SaOs2 cells (Figure 3C). The expression of BAG-1 isoforms was not altered by co-expression of p73α.
Figure 3.
Regulation of p73α transcriptional activity by Bcl-2 associated athanogene (BAG-1) isoforms. (A) Human embryonic kidney (HEK)293, NIH3T3 and H1299 cells were transfected with the Bax-luc reporter (100 ng) and the indicated amounts (ng) of p73α and BAG-1S expression plasmids. Luciferase activity (top) and BAG-1S expression (by immunoblotting (IB); bottom) was measured after 48 h. Data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells transfected with Bax-luc only (set to 1.0). Experiments shown are representative of at least three similar experiments. (B) SaOs2 cells were transfected with various promoter–reporter constructs with or without p73α (25 ng for transfections with the Pig3 promoter, 50 ng for transfections with the Bax promoter, IGFBP3 or MDM-2 promoter or 100 ng for transfections with the GADD45β promoter) and in the presence (closed bars) or absence (open bars) of BAG-1S (1000 ng). The following amounts of reporter constructs were used in each transfection; Bax, 100 ng; GADD45β, 200 ng; IGFBP3, 50 ng; MDM-2, 50 ng; or Pig3, 100 ng. Luciferase activity (top) and BAG-1S expression (by immunoblotting (IB); bottom) was measured after 48 h. Data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells transfected with each reporter construct in the absence of BAG-1S or p73α (set to 1.0). Experiments shown are representative of at least two similar experiments. (C) SaOs2 cells were transfected with the Bax-luc reporter construct (100 ng) and the p73α expression plasmid (50 ng) and BAG-1S, BAG-1M or BAG-1L expression plasmids (1000 ng), as indicated. Luciferase activity (top panel) was measured after 48 h. Data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells transfected with the Bax reporter construct in the absence of BAG-1 or p73α (set to 1.0). Experiment shown is representative of four similar experiments. The bottom panel shows BAG-1 protein expression in transfected cells analysed by immunoblotting. Arrows indicate the BAG-1L, BAG-1M and BAG-1S isoforms. Note that some faster migrating BAG-1 forms are detected, especially in cells transfected with the BAG-1M expression plasmid. These may derive from internal translation initiation or degradation (Lee et al, 2007).
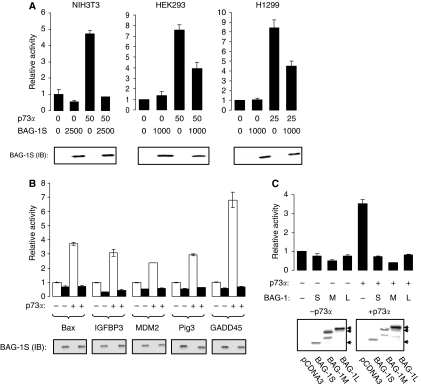
BAG-1S is a more effective inhibitor of p73α compared with p73β
p73 is expresssed as multiple isoforms (Murray-Zmijewski et al, 2006). p73β is generated by alternative splicing and has a truncated C-terminus compared with p73α. p73β is transcriptionally active and we therefore compared the ability of BAG-1S to inhibit transcriptional activation by p73α and p73β (Figure 4A). Whereas BAG-1S overexpression substantially reduced p73α-mediated transcription (mean inhibition 84±5%, mean derived from three identical experiments, ±s.d.), the activity of p73β was relatively modestly affected (mean inhibition 32±9%). The difference between the effects of BAG-1S on p73α and p73β was statistically significant (Student's t-test, P=0.003). p73 isoforms were expressed at approximately equivalent levels (Figure 4B). In these experiments, we used 100 ng of p73β expression plasmid (compared with 50 ng of p73α expression plasmid) to achieve approximately equivalent levels of activation of the Bax reporter construct. However, BAG-1S also failed to effectively repress p73β activity when cells were co-transfected with 50 ng of p73β expression plasmid (data not shown).
Figure 4.
Effect of Bcl-2 associated athanogene (BAG-1) S on the activity of p73α and p73β. SaOs2 cells were transfected with Bax-luc reporter construct (100 ng) and the indicated amounts of p73α, p73β and BAG-1S expression constructs. After 48 h luciferase activity was measured (top) and BAG-1 expression analysed by immunoblotting (bottom). In (A), data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells transfected with the Bax reporter construct in the absence of BAG-1 or p73α (set to 1.0). Experiment shown is representative of three similar experiments. (B) SaOs2 cells were transfected with BAG-1S (500 ng), p73α (50 ng) or p73β (100 ng) expression constructs and analysed by immunoblottting using the R26 antibody. BAG-1S and PCNA were analysed as controls.
BAG-1 knockdown reactivates p73 function
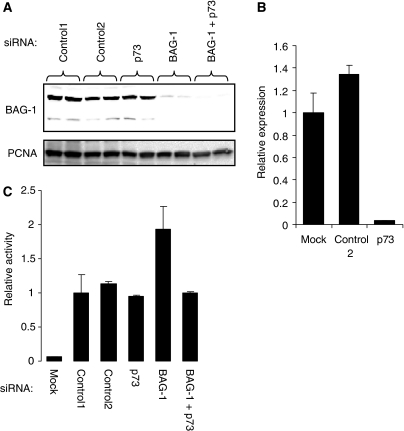
As overexpression studies showed that BAG-1 inhibited the transcription-activating function of p73, we used RNA interference to determine whether similar functional interactions occur between endogenous BAG-1 and p73 proteins. H1299 cells were selected for these studies because of the very high efficiency of siRNA-mediated knock-down obtained. H1299 cells were transfected with the Pig3 promoter–reporter construct to monitor p73 activity and BAG-1 and p73 were depleted by siRNA. The BAG-1 siRNA has been validated in earlier studies (Clemo et al, 2008), and immunoblot analysis confirmed the effective knock-down of BAG-1 expression in H1299 cells (94±6% reduction in BAG-1 siRNA transfected cells, mean±s.d. derived from four experiments), which predominantly express BAG-1 L (Figure 5A). Because of the absence of suitable antibodies to reliably detect endogenous p73, we were unable to confirm knockdown of p73 at the protein level. However, this siRNA has been validated in earlier studies (Basu et al, 2003) and Q-RT-PCR analysis showed a clear knock-down of p73 RNA (Figure 5B). Depletion of BAG-1 resulted in a 1.9±0.3 fold increase in the activity of the Pig3 promoter (mean±s.d. of four experiments), compared with cells transfected with control siRNA (Figure 5C). Knock-down of p73 reversed the activation of the Pig3 promoter observed in cells transfected with the BAG-1 siRNA, but had no effect on Pig3 promoter activity when tested alone (Figure 5C).
Figure 5.
Effect of Bcl-2 associated athanogene (BAG-1) knock-down on endogenous p73 activity. H1299 cells were co-transfected in duplicate with control, p73 or BAG-1-specific siRNAs and the Pig3-luc promoter–reporter plasmid (400 ng). BAG-1 protein levels (A) and luciferase activity (C) were analysed 72 h after transfection. (C) shows mean luciferase activity (±s.d.) of duplicate transfections normalised to luciferase activity in cells transfected with control 1 siRNA (set to 1.0). Mock-transfected cells are the cells without any siRNA/reporter plasmid. In (A), PCNA expression was also analysed as a loading control. Experiment shown is representative of three similar experiments. (B) H1299 cells were transfected with control or p73-specific siRNAs, or mock transfected in the absence of any siRNAs, and expression of p73 RNA was analysed by quantitative-reverse transcription (Q–RT)–PCR after 72 h.
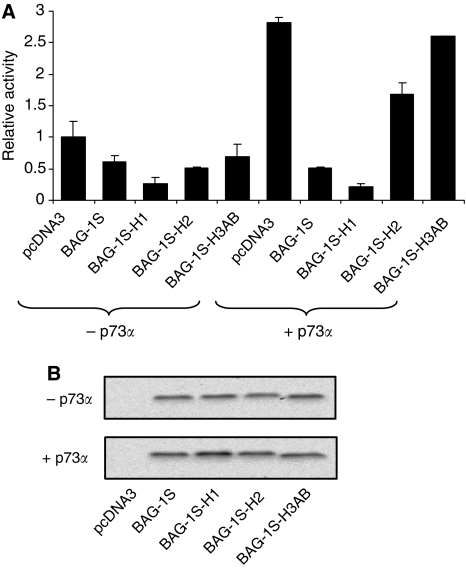
Inhibition of p73α function by BAG-1S requires helix 2 and 3 of the BAG domain
We showed earlier that suppression of apoptosis by BAG-1S requires amino acids within helix 2 and 3 of the BAG domain important for interaction with HSC70/HSP70 (Townsend et al, 2003b, 2004). We therefore analysed the effect of BAG-1 C-terminal mutations on the ability of BAG-1S to inhibit p73α activity. Mutations within helix 2 or 3, in BAG-1S-H2 (Q169A, K172A) and BAG-1S-H3AB (Q201A, D208A, Q212A) significantly reduced the ability of BAG-1S to inhibit p73α-mediated transcription (Figure 6A). By contrast, mutations within helix 1 in BAG-1S-H1 (E112A and K116A) did not interfere with p73α-mediated transcription. Immunoblot analysis showed that the wild type and mutant BAG-1S proteins were expressed at approximately equivalent levels (Figure 6B). As shown earlier (Briknarova et al, 2001; Townsend et al, 2004; Lee et al, 2007), mutation of helix 2 and 3, but not helix 1, prevented interaction of BAG-1S with HSC70 in co-immunoprecipitation assays (Figure 8A).
Figure 6.
Effect of Bcl-2 associated athanogene (BAG-1) C-terminal point mutations on p73α activity. SaOs2 cells were transfected with 100 ng of Bax-luc reporter construct and p73α expression plasmid (100 ng) and wild type/mutant BAG-1S expression constructs (2500 ng) as indicated. After 48 h luciferase activity was measured (A) and BAG-1 expression analysed by immunoblotting (B). In (A), data shown are the mean luciferase activity (±s.d.) of duplicate transfections normalised to cells transfected with the Bax reporter construct in the absence of BAG-1 or p73α (set to 1.0). Experiment shown is each representative of three similar experiments.
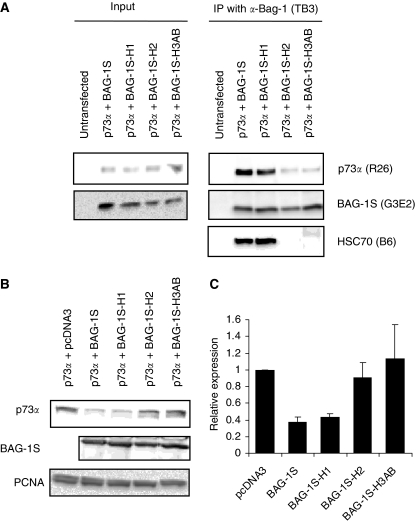
Figure 8.
Effect of Bcl-2 associated athanogene (BAG-1) C-terminal point mutations on p73α interaction and modulation of p73α expression levels. (A) H1299 cells were transfected with the indicated p73α and wild type/mutant BAG-1S expression constructs. After 24 h, immunoprecipitations were carried out using the BAG-1-specific TB3 antibody. Immunoprecipitates were analysed by immunoblotting using antibodies specific for p73 (R26), BAG-1 (G3E2) or HSC70 (B6). ‘Input’ is the lysate from transfected cells before immunoprecipitation. (B) H1299 cells were transfected with the p73α expression construct in the presence or absence of wild type/mutant BAG-1S expression constructs. After 24 h, expression of p73α, BAG-1S and PCNA (loading control) were analysed by immunoblotting. (C) Quantitation of p73α expression in wild type and mutant BAG-1S transfected cells. The expression of p73α in control cell lysates was set to 1.0. Data are mean (±s.d.) expression levels derived from two identical experiments.
BAG-1S interacts with p73α and decreases p73α expression levels
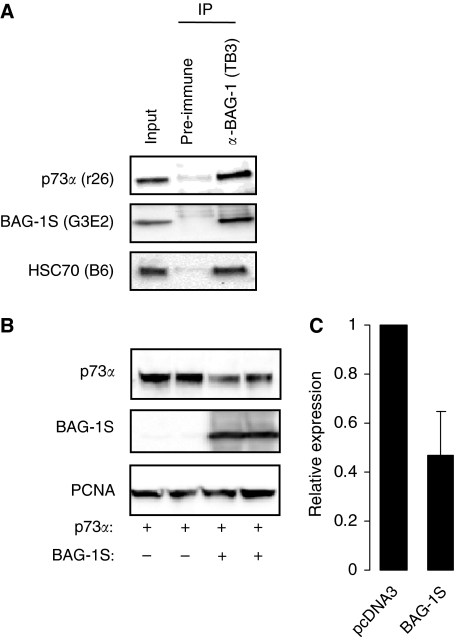
We carried out immunoprecipitation experiments to determine whether BAG-1S and p73α associate in intact cells. H1299 cells were transfected with BAG-1S and p73α expression plasmids, and BAG-1 complexes immunoprecipitated. In addition to the expected association with HSC70, there was a clear interaction between BAG-1S and p73α (Figure 7A). We also determined whether overexpression of BAG-1S altered the levels of p73α. In H1299 cells co-overexpressing BAG-1S, the levels of p73α were significantly reduced (Figure 7B). On an average, co-expression of BAG-1S reduced p73α expression by ∼50% (mean of eight experiments; Student's t-test, P=6 × 10−5) compared with control cells (Figure 7C). Therefore, when overexpressed in cells, BAG-1S and p73α interact and BAG-1S expression leads to a reduction in p73α levels.
Figure 7.
Bcl-2 associated athanogene (BAG-1) S interacts with p73α and decreases the expression levels of co-expressed p73α. (A) H1299 cells were transfected with p73α and BAG-1S expression constructs. After 24 h, immunoprecipitations were carried out using the BAG-1-specific TB3 antibody or pre-immune serum as a control. Immunoprecipitates were analysed by immunoblotting using antibodies specific for p73 (R26), BAG-1 (G3E2) or HSC70 (B6). ‘Input’ is the lysate from transfected cells before immunoprecipitation. (B) H1299 cells were transfected with a p73α expression construct in the presence or absence of a BAG-1S expression construct. After 24 h, expression of p73α, BAG-1S and PCNA (loading control) were analysed by immunoblotting. (C) Quantitation of p73α expression in BAG-1S and control (pcDNA3 transfected) cells. The expression of p73α in control cell lysates was set to 1.0. Data are mean (±s.d.) expression levels derived from eight similar experiments.
p73α binding and regulation of expression is dependent on helix 2 and 3 of the BAG-1S BAG domain
As regulation of p73α activity is dependent on residues within helix 2 and 3 of the BAG domain, we analysed the effects of these mutations on the ability of BAG-1S to interact with p73α and to decrease the levels of p73α. Mutations within helix 2 and 3, but not helix 1, decreased the interaction between BAG-1S and p73α (Figure 8A) and reduced the ability of BAG-1S to decrease the expression of p73α (Figures 8B and C). Therefore the ability of BAG-1S to inhibit p73α activity, to interact with p73α and to decrease p73α expression levels are all dependent on the residues with helix 2 and 3 of the BAG-1S BAG domain. As these residues are also required for interaction with HSC70/HSP70, chaperones are likely to be critical mediators of the regulation of p73α by BAG-1S.
Discussion
Earlier study has shown that BAG-1 contributes to the inappropriate survival of malignant cells. However, the specific molecular targets of BAG-1 that mediate survival remain to be identified. Although originally identified as a Bcl-2 interacting protein (Takayama et al, 1995) the significance of this interaction remains unclear and at present data showing a role for Bcl-2 modulation in BAG-1-mediated survival are lacking. We have shown earlier that endogenous BAG-1 enhances the function of NF-κB in colorectal cancer cells (Clemo et al, 2008). BAG-1 also interacts with and interferes with the function of the stress-responsive, pro-apoptotic GADD34 protein (Hung et al, 2003). However, the requirements for cochaperone binding in these activities are not clear.
Data presented here show that overexpression of BAG-1 inhibits the transcriptional activating functions of p53 family proteins (p73 in particular) and this may represent one mechanism by which BAG-1 interferes with stress-induced apoptosis. The absence of robust reagents to reliably detect endogenous p73 forced us to focus on overexpression studies, and this is one of the major limitations of our study. However siRNA-mediated ablation showed similar functional interactions between endogenous BAG-1 and p73 because BAG-1 knock-down leads to an increase in p73-dependent transcription. p73 is activated in response to a variety of chemotherapeutic drugs and γ-irradiation, and is an important determinant of cellular sensitivity to apoptosis (Agami et al, 1999; Gong et al, 1999; Yuan et al, 1999; Flores et al, 2002; Bergamaschi et al, 2003). Although p73 is rarely mutated in cancer cells, the expression of alternate spliced isoforms (e.g., ΔNp73) might interfere with p73 function in a dominant negative manner (Melino et al, 2002; Murray-Zmijewski et al, 2006). p73 activity can also be regulated by interaction with several cellular partners such as Mdm-2, Yap, ASPP family proteins and these are often altered in cancer cells. Our data suggest that overexpression of BAG-1 might be an additional mechanism that limits p73 function in malignant cells.
The inhibitory function of BAG-1 was more pronounced for p73α, compared with p73β, a splice variant that is transcriptionally active and is frequently co-expressed in cancer cells. Compared with p73α, p73β lacks a C-terminal SAM domain. The molecular function of the SAM domain is not known, but is considered to act as a negative control domain (Liu and Chen, 2005). It is interesting that, p63 also contains an N-terminal SAM domain whereas p53 does not. Thus, the ability of BAG-1 to modulate p53-family proteins may involve, but not absolutely require, the SAM domain. Although the function of the SAM domain is likely to be multifunctional, including effects of co-activator recruitment (Liu and Chen, 2005) and DNA binding, our data further suggest that the SAM domain may also act to confer chaperone-dependent negative regulation. However, it is important to note that under our experimental conditions we did not detect enhanced transcriptional activation by p73β, compared with p73α, as shown earlier (De Laurenzi et al, 1998; Ueda et al, 2001; Liu and Chen, 2005).
The ability of BAG-1S to inhibit p73α function seems to be mediated through physical association. Both the ability of BAG-1S to modulate p73α-transcriptional activity and to associate with p73α was dependent on specific residues within the BAG-1 BAG domain that are also required for binding to HSC70 and for promotion of cell survival (Townsend et al, 2004; Lee et al, 2007). The data are consistent with a model in which the binding of BAG-1 to p73α is mediated by HSC70/HSP70, but studies with purified components are required to test this. One consequence of formation of this complex may be a reduction in the steady state levels of p73α, as co-expression of BAG-1S and p73α reduced p73α levels by ∼50%. Decreased expression of p73α is not an artefact of co-expression (e.g., competition for transcription/translation machinery), as certain mutants of BAG-1S did not show this activity although expressed at equivalent levels. Moreover, this function of BAG-1S was dependent on residues required for binding to HSC70, again strongly implicating chaperones in this effect. One mechanism by which BAG-1S may decrease p73α function is by increasing proteolytic degradation. However, the decrease in p73 levels (50%) did not fully account for the inhibition of p73α activity observed in transfection studies (80%). Thus, alternate mechanisms are also likely to contribute, and chaperone-dependent changes in the conformation of p73 and its association with co-regulatory molecules may also be important.
A key question is to what extent the ability of BAG-1S to decrease p73α levels is dependent on the ability of BAG-1 to coordinate the activity of the proteasome and chaperones in protein triage decisions (Arndt et al, 2007). We have not shown that the reduction in p73α levels are proteasome-mediated, but there is substantial evidence that p73α levels can be controlled by ubiquitination and proteasomal degradation. Several E3-ligase have been shown to modulate p73 turnover, including Itch and UFD2a (Hosoda et al, 2005; Oberst et al, 2005; Rossi et al, 2005). Proteasome binding of BAG-1 is dependent on the ULD (Luders et al, 2000a; Demand et al, 2001). Our analysis of BAG-1 ULD mutants did not clarify the role of this domain in controlling p73 function because deletion of the entire ULD or mutation of a conserved lysine (K80 in BAG-1S) required for cell survival (Townsend et al, 2003b) resulted in significant destabilisation and stabilisation of BAG-1S, respectively, making interpretation of data obtained for these proteins unclear.
Although BAG-1 very effectively inhibited p73 function, its effects on p53 were modest and variable between experiments. These results are probably consistent with those of Matsuzawa et al who showed that BAG-1S did not interfere with p53-dependent transcription in HEK293 cells (Matsuzawa et al, 1998). However, in addition to the direct effects of BAG-1 on p73-transactivating function that we have described, it remains possible that BAG-1 inhibits p53 function independent of effects in transcription. BAG-1 participates in complexes with wild type and mutant p53 (King et al, 2001), and although the functional implications of BAG-1 are not known, CHIP targets p53 for proteasomal degradation (Esser et al, 2005). BAG-1 overexpression interferes with p53-induced growth arrest and apoptosis, perhaps by interfering with downstream effector molecules, such as Siah (Danen-van Oorschot et al, 1997; Matsuzawa et al, 1998). Thus, we believe that the inhibitory effects of BAG-1 on p53 family proteins as a whole are likely to involve both direct modulation of transactivating function and indirect effects. However, further study is required to dissect the molecular details of these interactions.
Acknowledgments
We thank Profs Xin Lu and Gerry Melino for the kind gift of reporter and expression plasmids. This work was supported by the Cancer Research UK.
References
- Agami R, Blandino G, Oren M, Shaul Y (1999) Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399: 809–813 [DOI] [PubMed] [Google Scholar]
- Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J (2002) Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J Biol Chem 277: 45920–45927 [DOI] [PubMed] [Google Scholar]
- Alberti S, Esser C, Hohfeld J (2003) BAG-1—a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones 8: 225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V, Rogon C, Höhfeld J (2007) To be, or not to be--molecular chaperones in protein degradation. Cell Mol Life Sci 64: 2525–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J (2003) Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 11: 11–23 [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, Attard M, Reelfs O, Gusterson B, Bell AK, Heath V, Tavassoli M, Farrell PJ, Smith P, Lu X, Crook T (2003) p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 3: 387–402 [DOI] [PubMed] [Google Scholar]
- Brehmer D, Rüdiger S, Gässler C, Klostermeier D, Packschies L, Reinstein J, Mayer M, Bukau B (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol 8: 427–432 [DOI] [PubMed] [Google Scholar]
- Briknarova K, Takayama S, Brive L, Havert ML, Knee DA, Velasco J, Homma S, Cabezas E, Stuart J, Hoyt DW, Satterthwait AC, Llinas M, Reed JC, Ely KR (2001) Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol 8: 349–352 [DOI] [PubMed] [Google Scholar]
- Brimmell M, Burns JS, Munson P, McDonald L, O'Hare MJ, Lakhani SR, Packham G (1999) High level expression of differentially localized BAG-1 isoforms in some oestrogen receptor-positive human breast cancers. Br J Cancer 81: 1042–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemo N, Collard T, Southern S, Edwards K, Moorghen M, Packham G, Hague A, Paraskeva C, Williams A (2008) BAG-1 is up-regulated in colorectal tumour progression and promotes colorectal tumour cell survival through increased NF-kappaB activity. Carcinogenesis 29: 849–857 [DOI] [PubMed] [Google Scholar]
- Cutress RI, Townsend PA, Brimmell M, Bateman AC, Hague A, Packham G (2002) BAG-1 expression and function in human cancer. Br J Cancer 87: 834–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutress RI, Townsend PA, Sharp A, Maison A, Wood L, Lee R, Brimmell M, Mullee MA, Johnson PW, Royle GT, Bateman AC, Packham G (2003) The nuclear BAG-1 isoform, BAG-1 L, enhances oestrogen-dependent transcription. Oncogene 22: 4973–4982 [DOI] [PubMed] [Google Scholar]
- Danen-van Oorschot AA, den Hollander AI, Takayama S, Reed JC, van der Eb AJ, Noteborn MH (1997) BAG-1 inhibits p53-induced but not apoptin-induced apoptosis. Apoptosis 2: 395–402 [DOI] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jaattela M (2007) The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett 581: 3702–3710 [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G (1998) Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med 188: 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Rossi A, Terrinoni A, Barcaroli D, Levrero M, Costanzo A, Knight RA, Guerrieri P, Melino G (2000) p63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem Biophys Res Commun 273: 342–346 [DOI] [PubMed] [Google Scholar]
- Demand J, Alberti S, Patterson C, Hohfeld J (2001) Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol 11: 1569–1577 [DOI] [PubMed] [Google Scholar]
- Dudkina AS, Lindsley CW (2007) Small molecule protein-protein inhibitors for the p53-MDM2 interaction. Curr Top Med Chem 7: 952–960 [DOI] [PubMed] [Google Scholar]
- Esser C, Scheffner M, Hohfeld J (2005) The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem 280: 27443–27448 [DOI] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T (2002) p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416: 560–564 [DOI] [PubMed] [Google Scholar]
- Gehring U (2004) Biological activities of HAP46/BAG-1. The HAP46/BAG-1 protein: regulator of HSP70 chaperones, DNA-binding protein and stimulator of transcription. EMBO Rep 5: 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin Jr WG, Levrero M, Wang JY (1999) The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399: 806–809 [DOI] [PubMed] [Google Scholar]
- Gotz R, Wiese S, Takayama S, Camarero GC, Rossoll W, Schweizer U, Troppmair J, Jablonka S, Holtmann B, Reed JC, Rapp UR, Sendtner M (2005) Bag1 is essential for differentiation and survival of hematopoietic and neuronal cells. Nat Neurosci 8: 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad I, Picard D (2007) The glucocorticoid responses are shaped by molecular chaperones. Mol Cell Endocrinol 275: 2–12 [DOI] [PubMed] [Google Scholar]
- Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, Lena AM, Candi E, Terrinoni A, Catani MV, Oren M, Melino G, Krammer PH, Stremmel W, Muller M (2005) TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. Embo J 24: 2458–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudas J, Nguyen H, Li T, Hill D, Cowan KH (1995) Effects of cell cycle, wild-type p53 and DNA damage on p21CIP1/Waf1 expression in human breast epithelial cells. Oncogene 11: 253–261 [PubMed] [Google Scholar]
- Hohfeld J, Jentsch S (1997) GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J 16: 6209–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda M, Ozaki T, Miyazaki K, Hayashi S, Furuya K, Watanabe K, Nakagawa T, Hanamoto T, Todo S, Nakagawara A (2005) UFD2a mediates the proteasomal turnover of p73 without promoting p73 ubiquitination. Oncogene 24: 7156–7169 [DOI] [PubMed] [Google Scholar]
- Hsieh JK, Chan FS, O'Connor DJ, Mittnacht S, Zhong S, Lu X (1999) RB Regulates the Stability and the Apoptotic Function of p53 via MDM2. Molecular Cell 3: 181–193 [DOI] [PubMed] [Google Scholar]
- Hung WJ, Roberson RS, Taft J, Wu DY (2003) Human BAG-1 proteins bind to the cellular stress response protein GADD34 and interfere with GADD34 functions. Mol Cell Biol 23: 3477–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost CA, Marin MC, Kaelin Jr WG (1997) p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature 389: 191–194 [DOI] [PubMed] [Google Scholar]
- King FW WA, Höhfeld J, Zylicz M (2001) Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J 20: 6297–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh M, Knee DA, Takayama S, Reed JC (2002) Bag1 proteins regulate growth and survival of ZR-75-1 human breast cancer cells. Cancer Res 62: 1904–1909 [PubMed] [Google Scholar]
- Lee SS, Crabb SJ, Janghra N, Carlberg C, Williams AC, Cutress RI, Packham G, Hague A (2007) Subcellular localisation of BAG-1 and its regulation of vitamin D receptor-mediated transactivation and involucrin expression in oral keratinocytes: implications for oral carcinogenesis. Exp Cell Res 313: 3222–3238 [DOI] [PubMed] [Google Scholar]
- Liu G, Chen X (2005) The C-terminal sterile alpha motif and the extreme C terminus regulate the transcriptional activity of the alpha isoform of p73. J Biol Chem 280: 20111–20119 [DOI] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J (2000a) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem 275: 4613–4617 [DOI] [PubMed] [Google Scholar]
- Luders J, Demand J, Papp O, Hohfeld J (2000b) Distinct isoforms of the cofactor BAG-1 differentially affect Hsc70 chaperone function. J Biol Chem 275: 14817–14823 [DOI] [PubMed] [Google Scholar]
- MacCarthy-Morrogh L, Wood L, Brimmell M, Johnson PW, Packham G (2000) Identification of a novel human BCL-X promoter and exon. Oncogene 19: 5534–5538 [DOI] [PubMed] [Google Scholar]
- Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, Blandino G, Del Sal G (2004) Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell 14: 625–636 [DOI] [PubMed] [Google Scholar]
- Matsuzawa S, Takayama S, Froesch BA, Zapata JM, Reed JC (1998) p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J 17: 2736–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Bukau B (2005) Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H, Patterson C (2003) CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon F, Melino G (2007) Fog of war: the emerging p53 family. Cell Cycle 6: 229–232 [DOI] [PubMed] [Google Scholar]
- Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, Vousden KH (2004) p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem 279: 8076–8083 [DOI] [PubMed] [Google Scholar]
- Melino G, De Laurenzi V, Vousden KH (2002) p73: Friend or foe in tumorigenesis. Nat Rev Cancer 2: 605–615 [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC (2006) p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ 13: 962–972 [DOI] [PubMed] [Google Scholar]
- Nollen EA, Kabakov AE, Brunsting JF, Kanon B, Hohfeld J, Kampinga HH (2001) Modulation of in vivo HSP70 chaperone activity by Hip and Bag-1. J Biol Chem 276: 4677–4682 [DOI] [PubMed] [Google Scholar]
- Oberst A, Rossi M, Salomoni P, Pandolfi PP, Oren M, Melino G, Bernassola F (2005) Regulation of the p73 protein stability and degradation. Biochem Biophys Res Commun 331: 707–712 [DOI] [PubMed] [Google Scholar]
- Packham G, Brimmell M, Cleveland JL (1997) Mammalian cells express two differently localized Bag-1 isoforms generated by alternative translation initiation. Biochem J 328: 807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusztai L, Krishnamurti S, Perez Cardona J, Sneige N, Esteva FJ, Volchenok M, Breitenfelder P, Kau SW, Takayama S, Krajewski S, Reed JC, Bast Jr RC, Hortobagyi GN (2004) Expression of BAG-1 and BcL-2 proteins before and after neoadjuvant chemotherapy of locally advanced breast cancer. Cancer Invest 22: 248–256 [DOI] [PubMed] [Google Scholar]
- Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, Cesareni G, Melino G (2005) The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J 24: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Ludwig RL, Haupt Y, Bates S, Lu X, Oren M, Vousden KH (1996) Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBO J 15: 827–838 [PMC free article] [PubMed] [Google Scholar]
- Sawitzki B, Lehmann M, Vogt K, Seifert M, Risch K, Brock J, Kupiec-Weglinski JW, Volk HD (2002) Bag-1 up-regulation in anti-CD4 mAb-treated allo-activated T cell confers resistance to activation-induced cell death (AICD). Transpl Immunol 9: 83–91 [DOI] [PubMed] [Google Scholar]
- Sharp A, Crabb SJ, Townsend PA, Cutress RI, Brimmell M, Wang XH, Packham G (2004) BAG-1 in carcinogenesis. Expert Rev Mol Med 6: 1–15 [DOI] [PubMed] [Google Scholar]
- Sirvent JJ, Aguilar MC, Olona M, Pelegri A, Blazquez S, Gutierrez C (2004) Prognostic value of apoptosis in breast cancer (pT1-pT2). A TUNEL, p53, bcl-2, bag-1 and Bax immunohistochemical study. Histol Histopathol 19: 759–770 [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl F, Moarefi I (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291: 1553–1557 [DOI] [PubMed] [Google Scholar]
- Song J, Takeda M, Morimoto RI (2001) Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat Cell Biol 3: 276–282 [DOI] [PubMed] [Google Scholar]
- Stiewe T (2007) The p53 family in differentiation and tumorigenesis. Nat Rev Cancer 7: 165–168 [DOI] [PubMed] [Google Scholar]
- Takayama S, Bimston DN, Matsuzawa S, Freeman BC, Aime-Sempe C, Xie Z, Morimoto RI, Reed JC (1997) BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J 16: 4887–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Krajewski S, Krajewska M, Kitada S, Zapata JM, Kochel K, Knee D, Scudiero D, Tudor G, Miller GJ, Miyashita T, Yamada M, Reed JC (1998) Expression and location of Hsp70/Hsc-binding anti-apoptotic protein BAG-1 and its variants in normal tissues and tumor cell lines. Cancer Res 58: 3116–3131 [PubMed] [Google Scholar]
- Takayama S, Reed JC (2001) Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol 3: E237–E241 [DOI] [PubMed] [Google Scholar]
- Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan JA, Reed JC (1995) Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell 80: 279–284 [DOI] [PubMed] [Google Scholar]
- Tang SC (2002) BAG-1, an anti-apoptotic tumour marker. IUBMB Life 53: 99–105 [DOI] [PubMed] [Google Scholar]
- Tang SC, Beck J, Murphy S, Chernenko G, Robb D, Watson P, Khalifa M (2004) BAG-1 expression correlates with Bcl-2, p53, differentiation, estrogen and progesterone receptors in invasive breast carcinoma. Breast Cancer Res Treat 84: 203–213 [DOI] [PubMed] [Google Scholar]
- Tang SC, Shehata N, Chernenko G, Khalifa M, Wang X (1999) Expression of BAG-1 in invasive breast carcinomas. J Clin Oncol 17: 1710–1719 [DOI] [PubMed] [Google Scholar]
- Toh WH, Kyo S, Sabapathy K (2005) Relief of p53-mediated telomerase suppression by p73. J Biol Chem 280: 17329–17338 [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Townsend PA, Cutress RI, Carroll CJ, Lawrence KM, Scarabelli TM, Packham G, Stephanou A, Latchman DS (2004) BAG-1 proteins protect cardiac myocytes from simulated ischemia/reperfusion-induced apoptosis via an alternate mechanism of cell survival independent of the proteasome. J Biol Chem 279: 20723–20728 [DOI] [PubMed] [Google Scholar]
- Townsend PA, Cutress RI, Sharp A, Brimmell M, Packham G (2003a) BAG-1: a multifunctional regulator of cell growth and survival. Biochim Biophys Acta 1603: 83–98 [DOI] [PubMed] [Google Scholar]
- Townsend PA, Cutress RI, Sharp A, Brimmell M, Packham G (2003b) BAG-1 prevents stress-induced long-term growth inhibition in breast cancer cells via a chaperone-dependent pathway. Cancer Res 63: 4150–4157 [PubMed] [Google Scholar]
- Townsend PA, Dublin E, Hart IR, Kao RH, Hanby AM, Cutress RI, Poulsom R, Ryder K, Barnes DM, Packham G (2002) BAG-i expression in human breast cancer: interrelationship between BAG-1 RNA, protein, HSC70 expression and clinico-pathological data. J Pathol 197: 51–59 [DOI] [PubMed] [Google Scholar]
- Townsend PA, Stephanou A, Packham G, Latchman DS (2005) BAG-1: a multi-functional pro-survival molecule. Int J Biochem Cell Biol 37: 251–259 [DOI] [PubMed] [Google Scholar]
- Turner BC, Krajewski S, Krajewska M, Takayama S, Gumbs AA, Carter D, Rebbeck TR, Haffty BG, Reed JC (2001) BAG-1: a novel biomarker predicting long-term survival in early-stage breast cancer. J Clin Oncol 19: 992–1000 [DOI] [PubMed] [Google Scholar]
- Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K (2001) Transcriptional activities of p73 splicing variants are regulated by inter-variant association. Biochem J 356: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HG, Takayama S, Rapp UR, Reed JC (1996) Bcl-2 interacting protein, BAG-1, binds to and activates the kinase Raf-1. Proc Natl Acad Sci USA 93: 7063–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chernenko G, Hao Y, Ding Z, Pater MM, Pater A, Tang SC (1998) Human BAG-1/RAP46 protein is generated as four isoforms by alternative translation initiation and overexpressed in cancer cells. Oncogene 17: 981–989 [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, Lu H, Kharbanda S, Weichselbaum R, Kufe D (1999) p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 399: 814–817 [DOI] [PubMed] [Google Scholar]