Summary
Thiopeptides, with potent activity against various drug-resistant pathogens, contain a characteristic macrocyclic core consisting of multiple thiazoles, dehydroamino acids, and a 6-membered nitrogen heterocycle. Their biosynthetic pathways remain elusive in spite of great efforts by in vivo feeding experiments. Here, cloning, sequencing and characterization of the thiostrepton and siomycin A gene clusters unveiled a new biosynthetic paradigm for the thiopeptide specific core formation, featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. The paradigm generality for thiopeptide biosynthesis was supported by genome mining and ultimate confirmation of the thiocillin I production in Bacillus cereus ATCC 14579, a strain that was previously unknown as a thiopeptide producer. These findings set the stage to accelerate the discovery of novel thiopeptides by prediction at the genetic level and to generate structural diversity by applying combinatorial biosynthesis methods.
Introduction
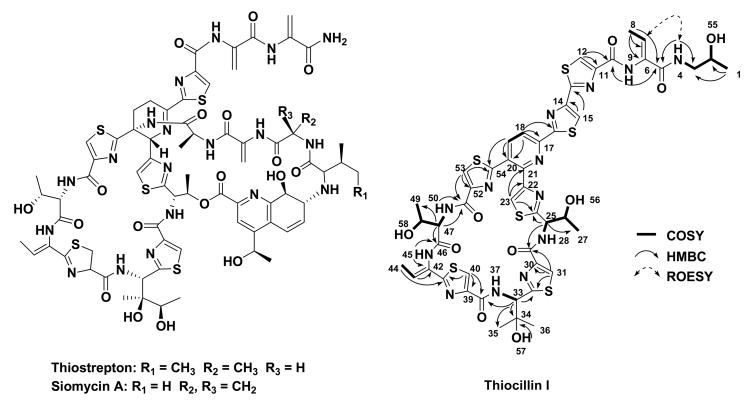
Thiopeptides are a class of polythiazolyl antibiotics (Bagley et al., 2005). The clinical interest in this family was recently renewed since many members show potent activity against various drug-resistant pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), penicillin-resistant Streptococcus pneumoniae (PRSP), and vancomycin-resistant enterococci (VRE). Thiopeptides share a characteristic macrocyclic core, consisting of multiple thiazoles, dehydroamino acids, and a 6-membered, tri- or tetra-substituted nitrogen heterocycle, with side chain(s) appending additional structural diversity (Figure 1 and S1). The complex architectures pose a tremendous challenge to chemical synthesis (Nicolaou et al., 2005; Hughes and Moody, 2007). Although previous isotope-labeled experiments, which aimed at the elucidation of the biosynthetic origins of a few members, established that all moieties exclusively derive from proteinogenic amino acids (Frenzel et al., 1990; Mocek et al., 1993; Smith et al., 1993; Priestley et al., 1996), the biosynthetic pathways of thiopeptides remain elusive. Here, we set out to investigate their biosynthesis by exploiting the genetic basis. Cloning, sequencing and characterization of the thiostrepton and siomycin A gene clusters unveiled a new biosynthetic paradigm for the thiopeptide specific core formation, featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Genome mining and ultimate confirmation of the thiocillin I production in Bacillus cereus ATCC 14579, a strain that was previously unknown as a thiopeptide producer, validated that this paradigm is common in thiopeptide biosynthesis.
Figure 1.
Structures of thiostrepton, siomycin A and thiocillin I (whose 1H-1H COSY, HMBC, and selected ROESY correlations in this study were labeled).
Results and Discussion
Cloning, sequencing and characterization of the thiostrepton biosynthetic gene cluster
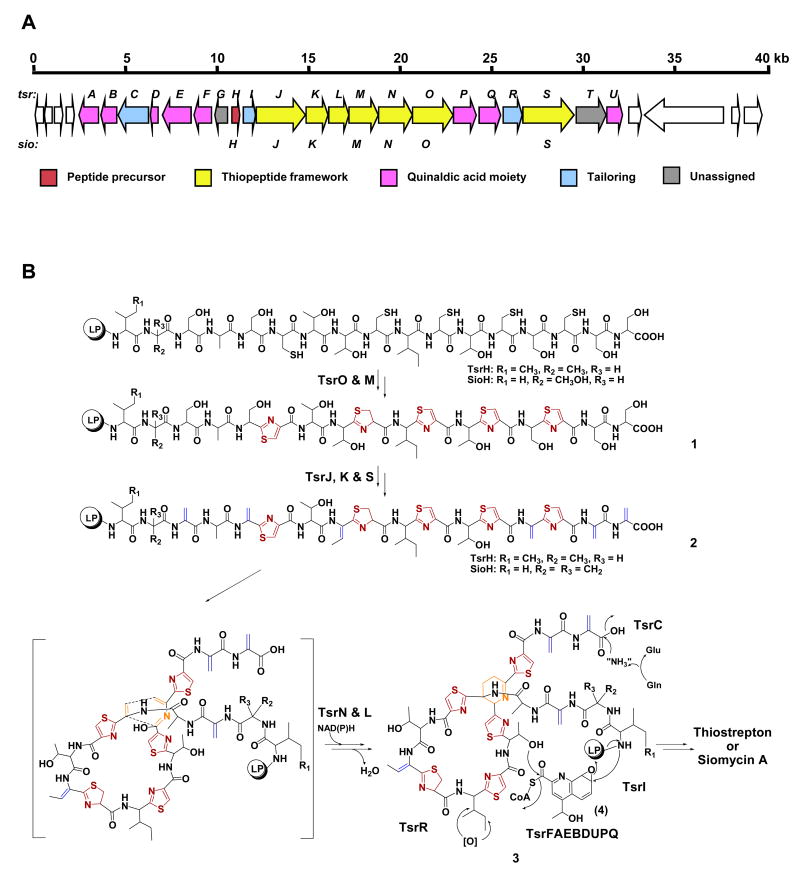
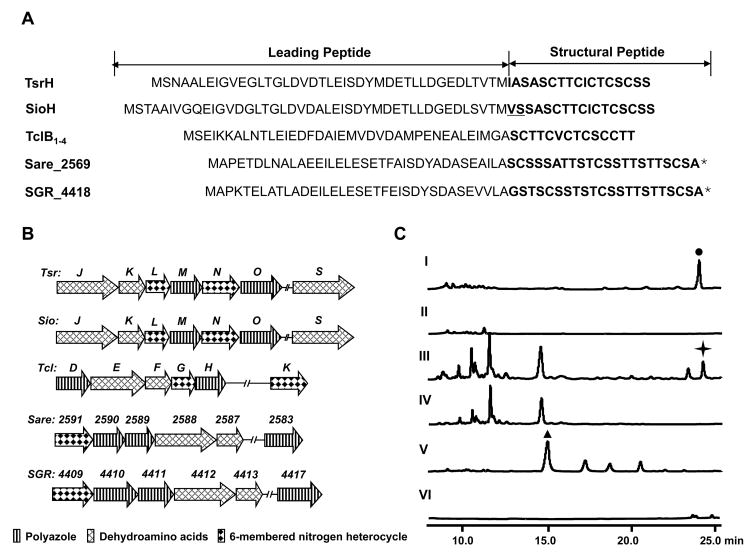
Thiostrepton (Donovick et al., 1955; Dutcher and Vandeputte, 1955; Jambor et al., 1955) (Figure 1), often referred to as the parent compound in this family, was chosen to be the model molecule for accessing the genetic basis of thiopeptide biosynthesis. Cloning and sequencing of the tsr gene cluster (deposited into GenBank under the accession number FJ436358) from Streptomyces laurentii ATCC 31255 revealed 21 open reading frames (orfs), whose deduced gene products supported a new paradigm for thiopeptide biosynthesis featuring a ribosomally synthesized precursor pepetide and conserved posttranslational modifications (Figure 2A and Table 1). The 58-aa precursor peptide TsrH contains a 41-aa leading peptide (LP) and a 17-aa structural peptide (SP). The SP sequence IASASCTTCICTCSCSS is in perfect agreement with the amino acids constituting the thiostrepton peptide backbone, unveiling for the first time the ribosomal origin of thiostrepton (Figure 3A). Central to the tsr gene cluster are the seven orfs, tsrJKLMNOS, the deduced products of which presumably act on the precursor peptide TsrH to afford the characteristic thiostrepton macrocyclic core structure (Figure 2B and 3B). TsrO, with sequence similarity to the cyclodehydratase PatD (38% similarity and 22% identity) in the patellamide biosynthesis (Schmidt et al., 2005), might be functionally associated with the putative dehydrogenase TsrM and able to catalyze the nucleophilic attack of each Cys side chain onto the proceeding carbonyl group followed by dehydration and optional dehydrogenation to afford the thiazoline and thiazole moieties characteristic to 1. TsrJ and TsrK, homologous to the N- (25% similarity and 11% identity) and C-terminal (30% similarity and 15% identity) sequences of SpaB in the subtilin biosynthesis (Xie et al., 2002), respectively, presumably are responsible for multiple dehydrations of Ser or Thr residues, yielding the dehydroamino acids featured in 2 as those in lantibiotics. The existence of the additional dehydratase TsrS (also homologous to the N-terminal SpaB, 24% similarity and 11% identity) suggests a putative residue or region-dependent dehydration pattern in the thiostrepton biosynthesis. The resultant linear intermediate 2 might then be forced into a conformation that can readily undergo an intramolecular [4 + 2] cycloaddition reaction to afford the 6-membered heterocyclic ring and complete the biosynthesis of the 26-membered macrocyclic system, giving the intermediate 3 for further modifications. TsrN and TsrL, showing no significant sequence homology to any proteins of known functions, could serve as candidates to catalyze this process. To provide experimental evidence supporting the above bioinformatics-based proposal, selected genes were inactivated to validate their indispensability. As exemplified by the in-frame deletion of tsrJ, the resulting mutant strain completely lost the ability to produce thiostrepton (Figure 3C, II), clearly confirming its involvement in the thiostrepton biosynthesis.
Figure 2.
Gene cluster and proposed biosynthetic pathway. (A) Organization of the tsr biosynthetic genes, the deduced functions of which are labeled in color and summarized in Table 1. (B) Biosynthetic hypothesis for the thiostrepton (or siomycin A) framework and modifications. Color coding indicates the thiazole/thiazoline (red), dehydroamino acids (blue), and 6-membered nitrogen heterocycle (orange).
Table 1.
Deduced functions of orfs in the thiostrepton biosynthetic gene cluster
| Gene | Sizea | Protein Homologb and Origin | Identity/Simiarity, % | Proposed Function | sio Homolog | tcl Homolog |
|---|---|---|---|---|---|---|
| orf1c | 193 | Upp (NP_628223), from Streptomyces coelicolor A3(2) | 91/97 | uracil phosphoribosyltransferase | ||
| orf2c | 208 | SGR_3903 (YP_001825415), from S. griseus subsp. griseus NBRC 13350 | 41/53 | hypothetical protein | ||
| orf3c | 177 | SSCG_02932 (YP_002192918), from S. clavuligerus ATCC 27064 | 68/84 | hypothetical protein | ||
| orf4c | 142 | MesJ (NP_696675), from Bifidobacterium longum NCC2705 | 57/68 | cytidine and deoxycytidylate deaminase | ||
| tsrA | 362 | HisC2 (YP_001853585), from Mycobacterium marinum M | 44/60 | histidinol-phosphate aminotransferase | ||
| tsrB | 276 | SSEG_03719 (YP_002206063), from S. sviceus ATCC 29083 | 50/65 | predicted hydrolase of the alpha/beta superfamily | ||
| tsrC | 618 | OxyD (AAZ78328), from S. rimosus | 54/65 | amidotransferase | ||
| tsrD | 135 | SnoaL (AAF01813), from S. nogalater | 32/54 | polyketide clyclase-like enzyme | ||
| tsrE | 428 | STH2357 (YP_076186), from Symbiobacterium thermophilum IAM 14863 | 35/51 | acyl-CoA dehydrogenase-like protein | ||
| tsrF | 311 | Sare_0495 (YP_001535415), from Salinispora arenicola CNS-205 | 41/57 | putative methyltransferase | ||
| tsrG | 213 | Mext_1123 (YP_001638598), from Methylobacterium extorquens PA1 | 28/46 | hypothetical protein | ||
| tsrH | 58 | ---------- | --/-- | thiostrepton precursor peptide | sioH | tclB1–4 |
| tsrI | 197 | Bm1_20485 (XP_001895551), from Brugia malayi | 29/42 | hydrolase/esterase | ||
| tsrJ | 933 | SpaB (AAL15564), from Bacillus subtilis | 11/24 | dehydratase | sioJ | tclE |
| tsrK | 346 | SpaB (AAL15564), from B. subtilis | 12/29 | dehydratase | sioK | tclF |
| tsrL | 368 | BC5082 (NP_834750), from B. cereus ATCC 14579 | 21/41 | hypothetical protein | sioL | tclG |
| tsrM | 589 | BAT_3535 (ZP_03054696), from B. pumilus ATCC 7061 | 24/40 | dehydrogenase | sioM | tclH |
| tsrN | 612 | BL02837 (YP_078248), from B. licheniformis ATCC 14580 | 25/39 | hypothetical protein | sioN | tclK |
| tsrO | 681 | PatD (AAY21153), from Prochloron didemni | 22/35 | cyclodehydratase | sioO | tclD |
| tsrP | 474 | PimD (CAC20932), from S. natalasis | 26/42 | P450 epoxidase | ||
| tsrQ | 424 | PstD (CAM56771), from Actinoplanes friuliensis | 29/39 | acyl-CoA synthetase | ||
| tsrR | 396 | RubU (AAM97370), from S. collinu | 30/44 | cytochrome P450 monoxygenase | ||
| tsrS | 936 | SpaB (AAL15564), from B. subtilis | 11/23 | dehydratase | sioS | |
| tsrT | 599 | Gura_3608 (YP_001232335), from Geobacter uraniireducens Rf4 | 27/39 | radical SAM domain-containing protein | ||
| tsrU | 272 | FabG (YP_177255), from B. clausii KSM-K16 | 36/51 | dehydrogenase/reductase | ||
| orf5c | 247 | orf_L7 (CAN89623), from S. collinus | 39/60 | hypothetical protein | ||
| orf6c | 1447 | SsviA_010100017073 (ZP_03195299), from S. sviceus ATCC 29083 | 38/51 | WD-40 repeat protein | ||
| orf7c | 99 | SCO4036 (NP_628218), from S. coelicolor A3(2) | 82/91 | hypothetical protein | ||
| orf8c | 336 | SclaA_010100013725 (ZP_03183088), from S. clavuligerus ATCC 27064 | 79/81 | RNA polymerase sigma factor, sigma F |
Numbers are in amino acids.
NCBI accession numbers are given in parentheses.
orfs beyond the tsr gene cluster
Figure 3.
Comparison and validation of the thiopeptide framework formation. (A) Peptide precursors for thiostrepton (TsrH), siomycin A (SioH), thiocillin I (TclB1–4), and two putative thiopeptides (Sare_2569 and SGR_4418). The SP sequences are labeled in bold (asterisk indicates the proposed SPs). The SP amino acids difference of SioH from TsrH is underlined. (B) Organization of the thiopeptide framework-forming genes identified from the producers of thiostrepton (tsr), siomycin A (sio), and thiocillin I (tcl), and two uncharacterized producers Salinispora arenicola CNS-205 (sare) and S. griseus subsp. Griseus NBRC 13350 (sgr). (C) HPLC analysis of the thiopeptide production in the wild type strain of the thiostrepton (solid dot) producer S. laurentii ATCC 31255 (I), tsrJ mutant S. laurentii strain SL1001 (II), wild type strain of the siomycin A (solid star) producer S. sioyaensis ATCC 13989 (III), sioO mutant S. sioyaensis strain SL2001 (IV), wild type strain of the thiocillin I (solid triangle) producer B. cereus ATCC 14579 (V), and tclE mutant B. cereus strain SL3001 (VI).
Functional assignment of the remaining orfs within the tsr cluster allowed the proposal for tailoring intermediate 3 into thiostrepton. Nine genes, tsrFAEBDUPQI, serve as candidates encoding enzymes responsible for the quinaldic acid formation, affording the 27-membered side ring moiety (Figure 2B and S2). They encode a methyltransferase (TsrF), a pyridoxal phosphate (PLP)-dependent aminotransferase (TsrA), an acyl-CoA α/β dehydrogenase-like protein (TsrE), a hydrolase (TsrB), a polyketide cyclase-like enzyme (TsrD), a dehydrogenase/reductase (TsrU), a P450 epoxidase (TsrP), an acyl-CoA synthetase (TsrQ), and a hydrolase/esterase (TsrI). Together, these enzyme functions are consistent with the previously proposed pathway for the quinaldic acid moiety (4) formation from Trp (Mocek et al., 1993; Smith et al., 1993; Priestley et al., 1996): the biosynthetic process may require methylation, desamination and oxidation, followed by imine ring opening and recyclization, epoxidation and carboxyl group activation. The attachment of 4 onto Ile42 (via amination) and Thr53 (via esterification) may lead to the closure of the side macrocyclic ring with the concomitant release of the 41-aa LP. Finally, tsrR and tsrC, encode a putative P450 hydroxylase (TsrR) and an asparagine synthase-like protein (TsrC), respectively, serving as candidates for the oxidations of Ile51 and C-terminal amide bond formation to furnish thiostrepton (Figure 2B). While the precise timing for many steps remains to be determined, the available tsr gene cluster and proposed pathway for thiostrepton biosynthesis have now set the stage for experimental validation.
Cloning, sequencing and characterization of the siomycin A biosynthetic gene cluster
To examine if the newly unveiled thiostrepton pathway is common for the thiopeptide biosynthesis in general, we next cloned and sequenced the biosynthetic gene cluster of siomycin A (deposited into GenBank under the accession number FJ436355), a naturally occurring analog of thiostrepton (Ebata et al., 1969; Tori et al., 1979) (Figure 1), from S. sioyaensis ATCC 13989 for comparative analysis. Remarkably, within the sio gene cluster, sioH encodes a 61-aa precursor peptide SioH containing a 17-aa SP that is nearly identical to that of TsrH (Figure 3A). The only exception is the N-terminal Val45-Ser46 residues, which is consistent with their structural difference in a Val-dehydroalanine unit of siomycin A in place of an Ile-Ala unit of thiostrepton. The seven genes, sioJKLMNOS, highly conserved relatives to tsrJKLMNOS in both sequence and organization (Figure 3B), were proposed to encode the comparable enzymatic activities for the similar thiopeptide core formation as that in thiostrepton biosynthesis (Figure 2 and 3). The inactivation of the putative cyclodehydratase gene sioO completely abolished the production (Figure 3C, IV), confirming its indispensability to the siomycin A biosynthesis. These findings support a common paradigm for the thiopeptide biosynthesis. Thus, a bacterial thiopeptide biosynthetic machinery should minimally be characterized by: 1) a ribosomally synthesized precursor peptide whose SP is Cys and Ser/Thr-rich, matching the aa sequence of the resultant thiopeptide backbone (Figure 3A); 2) a cyclodehydratase/dehydrogenase complex (i.g. TsrOM and SioOM) to catalyze the formation of the multiple thiazole and thiazoline rings; 3) a dehydratase pair (i.g. TsrJK and SioJK) to generate the dehydroamino acid residues; and 4) at least a TsrN or SioN-like protein to furnish the 6-membered nitrogen heterocycle via putative [4 + 2] cycloaddition. These enzymes are unique to thiopeptide biosynthesis. As exemplified by phylogenetic analysis of the thiopeptide cyclodehydratases, they are distinct from the polythiazole synthetases in both bacteriocin and cyanobactin biosynthesis (Li et al., 1996; Donia et al., 2006; Lee et al., 2008; Walsh and Nolan, 2008) (Figure S3).
Genome mining and confirmation of the thiocillin I production in a previously unknown producer
To demonstrate the generality of the newly emerged thiopeptide biosynthetic paradigm, we further carried out a comprehensive survey of published sequence data of microbial origins, focusing on mining the genomes for the Cys and Ser/Thr-rich precursor peptide and the conserved posttranslational modification enzymes. The characteristic genetic loci were indeed identified from several bacterial genomes, as exemplified by Bacillus cereus ATCC 14579 (NC_004722) (Ivanova et al., 2003), Salinispora arenicola CNS-205 (NC_009953), and S. griseus subsp. griseus NBRC 13350 (NC_010572), none of which, to our knowledge, was previously known as a thiopeptide producer. In the case of B. cereus ATCC 14579, there are four tandem genes, namely, tclB1, B2, B3 and B4, which encode an identical 52-aa precursor peptide (Figure 3A). Intriguingly, the 14-aa SP sequence completely matches the aa sequence predicted for micrococcins or thiocillins (Shoji et al., 1981; Bagley et al., 2005) (Figure 1 and S1), a group of thiopeptides that have previously isolated from Bacillus strains. For the remaining two bacterial strains, the deduced peptide precursors (Sare_2569 from S. arenicola CNS-205, and SGR_4418 from S. griseus subsp. griseus NBRC 13350, Figure 3A) share high similarity (74% identity) to each other and likely serve as the substrates for the conserved posttranslational modification enzymes to synthesize structurally related but new thiopeptides, since their C-terminal SP sequences do not fit any peptide backbone of structurally characterized thiopeptides.
Inspired by the genome mining findings, we fermented B. cereus ATCC 14579 and isolated the product to validate thiopeptide production. HPLC-ESI-MS analysis of the crude extract from 12 L of fermentation revealed one major product (Figure 3C, V), which was further purified for structural elucidation (Supplemental Data). HR-ESI-MS analysis showed its positive [M + Na]+ ion at m/z 1182.19996, establishing the molecular formula as C48H49N13O10S6. Taking the ultraviolet absorptions at λmax 225 nm, 280 nm, and 350 nm into account, this compound was deduced to be a highly unsaturated molecule rich in sulfur, amide bonds and aromatic rings. The 1H, 13C and 2D NMR analyses (including 1H-1H COSY, HSQC, HMBC and ROESY spectra) eventually confirmed its identity as thiocillin I (Figure 1 and Table S2 & Figure S4), which has been previously isolated from B. cereus G-15 (Shoji et al., 1976). Finally, the dehydratase gene tclE was inactivated in B. cereus ATCC 14579, and, as expected, the resulting mutant strain completely lost its ability to produce thiocillin (Figure 3B, VI), unambiguously establishing the predicted thiopeptide genetic locus as responsible for thiocillin production.
Supplementary Material
Acknowledgments
We thank Prof. H.G. Floss, Department of Chemistry, University of Washington, USA, for providing of S. laurentii ATCC 31255 and his pioneer works on thiopeptide biosynthesis. This work was supported in part by grants from NIH of U.S (CA094426 to B.S.), the National Natural Science Foundation of China (30525001, 90713012 and 20832009), the Ministry of Science and Technology of China (2006AA022185), and the Chinese Academy of Science (KJCX2-YW-H08) (all to W.L.).
Footnotes
Significance
In this study, we have uncovered a common paradigm for thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. We discovered this new pathway by first cloning, sequencing, and characterizing the thiostrepton biosynthetic gene cluster from S. laurentii ATCC 31255, subsequently validating its generality by cloning, sequencing, and characterizing the siomycin A biosynthetic gene cluster from S. sioyaensis ATCC 13989, and finally demonstrating its applicability as a new paradigm for thiopeptide biosynthesis by genome mining and ultimate confirmation of thiocillin I production in B. cereus ATCC 14579, a strain that was previously unknown as a thiopeptide producer. The newly discovered pathway is remarkably concise and efficient, in contrast to the heroic efforts of chemical synthesis of these natural products. Sequence permutations to the precursor peptides followed by the diverse tailoring modifications on the resulting thiopeptide scaffold could be a very attractive strategy to access thiopeptide structural diversity. The findings reported here now set the stage to accelerate the discovery of novel thiopeptides by genome mining and to generate structural diversity by applying combinatorial biosynthesis methods.
Supplemental data include bacterial strains and plasmids, gene cluster cloning and analysis, gene inactivation, thiopeptide production and HPLC-MS analysis, and structural elucidation and can be found with this communication online at http//www.xxxx
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- Donovick R, Pagano JF, Stout HA, Weinstein MJ. Thiostrepton, a new antibiotic. I In vitro studies. Antibiot Annu. 1955;3:554–559. [PubMed] [Google Scholar]
- Dutcher JD, Vandeputte J. Thiostrepton, a new antibiotic. II Isolation and chemical characterization. Antibiot Annu. 1955;3:560–561. [PubMed] [Google Scholar]
- Ebata M, Miyazaki K, Otsuka H. Studies on siomycin. I Physicochemical properties of siomycins A, B and C. J Antibiot (Tokyo) 1969;22:364–368. [PubMed] [Google Scholar]
- Frenzel T, Zhou P, Floss HG. Formation of 2-methyltryptophan in the biosynthesis of thiostrepton: isolation of S-adenosylmethionine:tryptophan 2-methyltransferase. Arch Biochem Biophys. 1990;278:35–40. doi: 10.1016/0003-9861(90)90227-p. [DOI] [PubMed] [Google Scholar]
- Hughes RA, Moody CJ. From amino acids to heteroaromatics--thiopeptide antibiotics, nature’s heterocyclic peptides. Angew Chem Int Ed Engl. 2007;46:7930–7954. doi: 10.1002/anie.200700728. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- Jambor WP, Steinberg BA, Suydam LO. Thiostrepton, a new antibiotic. III In vivo studies. Antibiot Annu. 1955;3:562–565. [PubMed] [Google Scholar]
- Lee SW, Mitchell DA, Markley AL, Hensler ME, Gonzalez D, Wohlrab A, Dorrestein PC, Nizet V, Dixon JE. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- Mocek U, Zeng Z, O’Hagan D, Zhou P, Fan LDG, Beale JM, Floss HG. Biosynthesis of the modified peptide antibiotic thiostrepton in Streptomyces azureus and Streptomyces laurentii. J Am Chem Soc. 1993;115:7992–8001. [Google Scholar]
- Nicolaou KC, Safina BS, Zak M, Lee SH, Nevalainen M, Bella M, Estrada AA, Funke C, Zecri FJ, Bulat S. Total synthesis of thiostrepton. Retrosynthetic analysis and construction of key building blocks. J Am Chem Soc. 2005;127:11159–11175. doi: 10.1021/ja0529337. [DOI] [PubMed] [Google Scholar]
- Nicolaou KC, Zak M, Safina BS, Estrada AA, Lee SH, Nevalainen M. Total synthesis of thiostrepton. Assembly of key building blocks and completion of the synthesis. J Am Chem Soc. 2005;127:11176–11183. doi: 10.1021/ja052934z. [DOI] [PubMed] [Google Scholar]
- Priestley ND, Smith TM, Shipley PR, Floss HG. Studies on the biosynthesis of thiostrepton: 4-(1-hydroxyethyl)quinoline-2-carboxylate as a free intermediate on the pathway to the quinaldic acid moiety. Bioorg Med Chem. 1996;4:1135–1147. doi: 10.1016/0968-0896(96)00126-5. [DOI] [PubMed] [Google Scholar]
- Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji J, Hinoo H, Wakisaka Y, Koizumi K, Mayama M. Isolation of three new antibiotics, thiocillins I, II and III, related to micrococcin P. Studies on antibiotics from the genus Bacillus. VIII. 1976;29:366–374. doi: 10.7164/antibiotics.29.366. [DOI] [PubMed] [Google Scholar]
- Shoji J, Kato T, Yoshimura Y, Tori K. Structural studies on thiocillins I, II and III (studies on antibiotics from the genus Bacillus XXIX) J Antibiot (Tokyo) 1981;34:1126–1136. doi: 10.7164/antibiotics.34.1126. [DOI] [PubMed] [Google Scholar]
- Smith TM, Priestley ND, Knaggs AR, Nguyen T, Floss HG. 3,4-Dimethylindole-2-carboxylate and 4-(1-hydroxyethyl)quinoline-2-carboxylate activating enzymes from the nosiheptide and thiostrepton producersStreptomyces actuosus and Streptomyces laurentii. J Chem Soc, Chem Commun. 1993:1612–1614. [Google Scholar]
- Tori K, Tokura K, Yoshimura Y, Okabe K, Otsuka H, Inagski F, Miyazawa T. 1H NMR spectral evidence for the structure and conformation of peptide antibiotic siomycin-A. J Antibiot (Tokyo) 1979;32:1072–1077. doi: 10.7164/antibiotics.32.1072. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Nolan EM. Morphing peptide backbones into heterocycles. Prod Natl Acad Sci USA. 2008;105:5655–5656. doi: 10.1073/pnas.0802300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Chatterjee C, Balsara R, Okeley NM, van der Donk WA. Heterologous expression and purification of SpaB involved in subtilin biosynthesis. Biochem Biophys Res Commun. 2002;295:952–957. doi: 10.1016/s0006-291x(02)00783-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.