Abstract
Haloferax volcanii, an extreme halophile originally isolated from the Dead Sea, is used worldwide as a model organism for furthering our understanding of archaeal cell physiology. In this study, a combination of approaches was used to identify a total of 1,296 proteins, representing 32% of the theoretical proteome of this haloarchaeon. This included separation of (phospho)proteins/peptides by 2-dimensional gel electrophoresis (2-D), immobilized metal affinity chromatography (IMAC), metal oxide affinity chromatography (MOAC), and Multidimensional Protein Identification Technology (MudPIT) including strong cation exchange (SCX) chromatography coupled with reversed phase (RP) HPLC. Proteins were identified by tandem mass spectrometry (MS/MS) using nano-electrospray ionization hybrid quadrupole time-of-flight (QSTAR XL Hybrid LC/MS/MS System) and quadrupole ion trap (Thermo LCQ Deca). Results indicate that a SCX RP HPLC fractionation coupled with MS/MS provides the best high-throughput workflow for overall protein identification.
Keywords: Proteome, Archaea, haloarchaea, halophile
Introduction
Haloferax volcanii is a halophilic archaeon (of the family Halobacteriaceae) commonly isolated from hypersaline environments such as the Dead Sea.1 Like other extreme halophiles, Hfx. volcanii maintains homeostasis by accumulating high concentrations of intracellular cations or counterions (e.g., K+).2 This contrasts with the typical biological strategy in which cells accumulate organic solutes and pump salt ions out. As a result, most haloarchaeal proteins require salt for activity and possess a highly acidic surface that serves as a hydration shell, enabling catalysis to occur under high salt conditions and preventing protein aggregation.2 The unusual ability of archaea, such as Hfx. volcanii, to thrive in environments of low water activity (e.g., high solvent) has made these organisms ideal candidates for advancements in biotechnology.3 Their ranking among some of the earliest life-forms and ability to tolerate high doses of UV and radiation have also made them of interest in the field of astrobiology including discussions on the origins of life4 and survival on Mars.5
Hfx. volcanii has become one of the international models of choice for studies of archaeal cell physiology based on its ease of culturing in the laboratory, growth on minimal medium, rapid methods of transformation and gene knockout, reporter proteins detected in whole cells, stable genome, and advanced biochemical tools including the ability to express and rapidly purify affinity-tagged multisubunit complexes directly from these cells.6, 7 With the genome of Hfx. volcanii now complete (http://archaea.ucsc.edu/), global surveys of transcript and protein levels are possible in efforts to expand our understanding of archaeal systems and how these compare to systems from the other two domains of life. Detailed proteomic and transcriptomic analyses can now be employed to answer lingering questions about Hfx. volcanii from a functional point of view.
In this communication, we describe the first large-scale proteome map of Hfx. volcanii. The overall aims were to optimize methods for high-throughput analysis and achieve a baseline-coverage of the proteome of this extreme halophile. Two-dimensional gel electrophoresis and multi-dimensional liquid separation and fractionation were paired with MS/MS technology for protein identification.
Methods and Materials
Materials
Biochemicals were purchased from Sigma-Aldrich (St. Louis, MO). Other organic and inorganic analytical grade chemicals were from Fisher Scientific (Atlanta, GA) and Bio-Rad (Hercules, CA).
Cell Growth and Protein Extraction
Haloferax volcanii DS70 and its derivates8, 9 were grown in ATCC 974 and defined high-salt liquid media10 at 42°C with orbital shaking at 200 rpm. Growth was monitored by measuring optical density (OD) at a wavelength of 600 nm using a 50-2000 μl, 220-1600 nm Uvette (Eppendorf) and SmartSpec 3000 spectrophotometer (BioRad). Cells at OD (600 nm) of 0.7 to 1.0 were harvested by centrifugation at 10,000 × g for 15 min at 4°C. Cell pellets were thawed on ice and extracted for analysis using Trizol11 and standard methods.12 Resulting protein pellets were dried and stored at -80°C.
Protein Reduction, Alkylation and Tryptic Digestion
Protein (300 μg) from Trizol extractions was resuspended in 100 μl of 50 mM NH4HCO3 (pH 7.5). Samples were reduced by the addition of 5 μl of 200 mM dithiothreitol (DTT solution) (1 h, room temperature or 21°C). Samples were alkylated by the addition of 4 μl of 1M iodoacetamide (1 h, 21°C). Alkylation was stopped by the addition of 20 μl of DTT solution (1 h, 21°C). Samples were digested with a 1:20 mg ratio of trypsin to protein for 18-24 h at 37°C. Digested peptides were purified using 300 μl C18 spin columns and dried under vacuum centrifugation. In-gel proteins were reduced, alkylated, and digested with trypsin using an automated platform for protein digestion (ProGest, Genomics Solutions, Ann Arbor, MI).
Peptide Methyl Esterification
A methanolic acid solution of 2 M was generated by adding 60 μl of 99% (v/v) acetyl chloride dropwise to 300 μl of 100% anhydrous methanol in a glass tube. The mixture was sealed and incubated (10-15 min, 21°C). Dried tryptic peptides were mixed with 30 μl of the methanolic acid reagent and incubated in a sealed jar containing desiccate material for 90 min at 21°C. Methyl esterified samples were dried under vacuum centrifugation and reconstituted in mobile phase B [0.1% (v/v) acetic acid, 0.01% (v/v) trifluoroacetic acid and 95% (v/v) acetonitrile] for analysis by MS/MS.
Immobilized Metal Affinity Chromatography
Phosphoprotein enrichment by immobilized metal affinity chromatography (IMAC) was performed using a Phosphopurification System according to supplier's instructions (Qiagen) with the following modifications: protein (2.5 mg) extracted by either the Trizol or standard method (see above) was resuspended in 25 ml of supplied lysis buffer to a final concentration of 0.1 mg protein per ml. Six 500 μl fractions per sample were collected for analysis.
Titanium Dioxide Phosphopeptide Enrichment
Metal oxide affinity chromatography (MOAC) of Hfx. volcanii tryptic peptides was performed using the Phos-Trap Phosphopeptide Enrichment Kit (Perkin Elmer, cat. no. PRT301001KT) with the following modifications: samples were agitated in the presence of 40 μl of elution buffer as opposed to the recommended 20 μl. Samples were incubated with the TiO2 resin for 10 min instead of 5 min and were incubated with elution buffer for 15 min instead of 10 min. Resulting samples were dried under vacuum centrifugation at 42°C for 30 min and reconstituted in mobile phase B for MS/MS analysis.
In-gel proteome analysis
Isoelectric focusing (IEF) was performed using 11-cm immobilized pH gradient (IPG) strips (Bio-Rad) with a pI range of 3.9–5.1. The strips were loaded with 150 μg of protein in rehydration buffer (7M urea, 2M thiourea, 4% [w/v] CHAPS, 2mM TBP, 0.2% [v/v] Biolyte mixture, 0.001% [w/v] bromophenol blue) at 20 °C for 18-24 h. Rehydrated IPG strips were placed in an 11-cm focusing tray (Bio-Rad) and covered with 2-3 ml of mineral oil. The proteins were focused at a maximum of 8,000 V for 35,000 volt-hours (V-h) at 20°C. Once complete, the strips were removed from the mineral oil, rinsed with proteomics grade water, and equilibrated for 10 min in 2 ml of equilibration buffer A (375 mM Tris–HCl, pH 8.8 with 6 M urea, 2% [w/v] SDS, 20% [v/v] glycerol, and 2% [w/v] dithiothreitol [DTT]) and again for 10 min in 2 ml equilibration buffer B (375 mM Tris–HCl, pH 8.8 with 6 M urea, 2% [w/v] SDS, 20% [v/v] glycerol, and 2.5% [w/v] iodoacetamide). The equilibrated IPG strips were placed in the upper well of an 11-cm Criterion precast gel (Bio-Rad) and set in place with a 0.5% (w/v) agarose overlay (Bio-Rad). The second dimension was run at 200 V for 55 min at 16°C. The completed gel was removed from the cassette and stained overnight in 150 ml of SYPRO Ruby fluorescent protein stain (Bio-Rad) or ProQ Diamond phosphoprotien stain (Molecular Probes) and destained according to the supplier's instructions. The gels were imaged with a Molecular Imager FX Scanner (Bio-Rad) with a 532-nm excitation laser and a 555-nm LP emissions filter. Acquired images were analyzed with PDQuest (version 7.0.1) software (Bio-Rad).
SCX/Reversed Phase HPLC Coupled with Nano-ESI-QTOF (QSTAR) MS/MS
P Tryptic peptides were separated by strong cation exchange (SCX) chromatography using a polysulfoethanyl A column (100 × 2.1 mm, 5 μm i.d.) (PolyLC, Columbia, MD) and a linear gradient of 10% to 100% 20 mM KCl for 55 min at 200 nl/min. Tryptic peptides (desalted with a PepMap C18 cartridge) were further separated by capillary RP HPLC using a PepMap C18 column (15 cm × 75 μm i.d.) and Ultimate Capillary HPLC System (LC Packings, San Francisco, CA). A linear gradient of 5% to 40% (v/v) acetonitrile for 25 min at 200 nl/min was used for separation. MS/MS analysis was performed online using a hybrid quadrupole time-of-flight instrument (QSTAR XL hybrid LC/MS/MS) equipped with a nanoelectrospray source (Applied Biosystems, Foster City, CA) and operated with the Analyst QS v1.1 data acquisition software. Information dependent acquisition (IDA) was employed in which each cycle consisted of a full scan from m/z 400-1500 (1 sec) followed by MS/MS (3 sec) of the two ions that exhibited the highest signal intensity. In the full scan acquisition mode, ions were focused through the first quadrupole by focusing and declustering potentials of 275 V and 55 V, respectively, and guided to the TOF region via two quadrupole filters operated in rf-only mode. Ions were orthogonally extracted, accelerated through the flight tube (plate, grid, and offset voltages were 340, 380, and -15 V, respectively), and refocused to a 4-anode microchannel plate detector via an ion mirror held at 990 V. The same parameters were utilized with MS/MS mode of operation; however, the second quadrupole was employed to filter a specific ion of interest while the third quadrupole operated as a collision cell. Nitrogen was used as the collision gas and collision energy values were optimized automatically using the rolling collision energy function based on m/z and the charge state of the peptide ion.
Three-Dimensional LCQ Deca Ion Trap MS
A portion of the IMAC-enriched samples were separated by 1D SDS PAGE and were analyzed by quadrupole ion trap MS (Thermo LCQ Deca) in line with a 5 cm × 75 μm inner diameter Pepmaptm C18 5 μm/ 300 Å capillary column (LC Packings). The RP HPLC C18 column operating upstream of the MS system was run with a 60-min gradient from 5% to 50% mobile phase B with a flow rate of 12 μl·min-1. MS parent ion scans were followed by four data-dependent MS/MS scans.
MS Data and Protein Identity Analyses
Spectra from all experiments were converted to DTA files and merged to facilitate database searching using the Mascot search algorithm v2.1 (Matrix Science, Boston, MA) against the deduced Hfx. volcanii proteome (http://archaea.ucsc.edu/, April 2007 version). Search parameters included trypsin as the cleavage enzyme. Carbamidomethylation was defined as the only fixed modification in the search while methionine oxidation, pyro-glu from glutamine or glutamic acid, acetylation, and phosphorylation of serine, threonine and tyrosine residues were set as variable modifications. Modifications were not considered if they could be a result of sample processing including: deamidation of asparagine and glutamine residues, oxidation of methionine residues and methyl-esterification of C-terminal, aspartate and glutamate residues. Mass tolerances for all LCQ analyses were 2 Da for MS and 1 Da for MS/MS. Mass tolerances for all QSTAR analyses were 0.3 Da for both MS and MS/MS. Protein identifications for which a probability-based MOWSE score average of 30 or above was not assigned were excluded. Criteria for positive modification of N-terminal peptides included: peptide ion scores ≥ 10 with a rank ≤ 2, a logical assignment, multiple identifications of the same peptide, and peptides with less than 3 modifications. Transmembrane spanning helices were predicted using TMHMM v2.0.13 Phosphosites were predicted using NetPhos v2.0.14 Proteins were categorized into Clusters of Orthologous Groups (COGS) using COGNITOR.15 Basic local alignment search tool (BLAST) was used locally at www.ncbi.nlm.nih.gov.
Results and Discussion
Statistical Analysis of the Hfx. volcanii Proteome Map
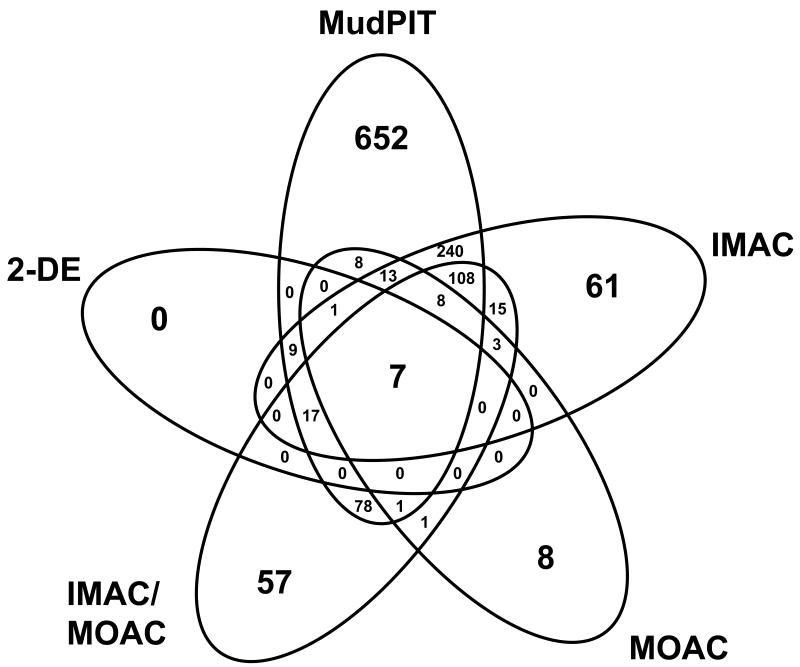
Aggregate data from five independent proteomic experiments resulted in a pool of protein identifications of considerable size and scope (Fig. 1)(Suppl. Tables S1 and S2). In total, 1,296 proteins of the Hfx. volcanii proteome were mapped, constituting nearly 32% of the theoretical coding capacity of this organism. The proteins were identified through 14,553 statistically significant top-ranking peptide matches (hits) with an average of 5.56 matching peptides per protein identification. Of those proteins identified, only 111 (8.5%) were single-hit identifications, leaving 1,185 proteins (91.5%) identified through multiple hits. These data primarily represent cytosolic components and may be expanded in future analyses where consideration is given to proteome subfractions such as secreted or membrane-bound proteins. However, this level of genome-wide coverage is consistent studies of other archaea, including Halobacterium salinarum 16, 17 and Natronomonas pharaonis 18 in which 802 to 929 proteins were mapped to 29 to 33% of the deduced proteome.
Fig. 1.
Venn diagram depicting the proportion of each experiment and overlap between experiments used to generate the current Hfx. volcanii proteome map.
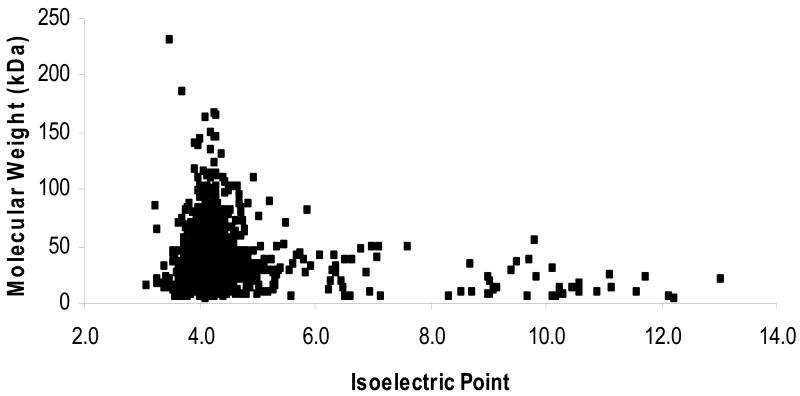
In this study of the Hfx. volcanii proteome, an average probability-based MOWSE score of 80.9 was assigned overall with a range of 35 to 1,028 for the entire dataset. Moreover, these identifications were made over a broad range of masses and isoelectric point (pI) values, likely resulting from the use of multiple complementary protein separation and detection methods for the generation of a unified proteome map. Proteins were identified with a calculated mass range of 4.8 kDa to 232 kDa and an average of 39.9 kDa. These same proteins represented a pI range of 3.10 to 13.04 with an average of 4.39 (Fig. 2). These value ranges are representative of the theoretical proteome which ranges in pI from 2.7 to 13.04 with an average of 4.52 and has a calculated molecular weight range of less than 10 Da to more than 235 kDa. Of the proteins identified by MS/MS, only 13.7% were out of the 97.4 kDa to 14.4 kDa range of resolution for standard 12% SDS-PAGE gels. Likewise, the vast majority of proteins identified (83.4%) were within the narrow pI range of 3.9 to 5.1 determined to be optimal for separation of Hfx. volcanii proteins by a single 2-DE gel run12. Of the proteins identified, only 6.2% (81) were below a pI of 3.9 and 10.5% (135) were above a pI of 5.1. This narrow range in 2-DE gel parameters for the majority of proteins detected in the Hfx. volcanii expressed proteome is likely to account for the protein crowding observed our previous 2-DE analytical studies.11, 12, 19
Fig. 2.
Predicted molecular weights and pI values for Hfx. volcanii proteins identified by MS/MS.
All of the proteins identified in this study were detected by 1D and 2D liquid chromatography (LC/MS/MS approaches) and covered regions of the proteome inaccessible by our gel-based (2-DE/MS) proteomic methods alone. 2-DE/MS approaches were not extensively pursued since these methods were rapidly found to be limited in the number and rate of protein identification. Only 53 proteins were identified by 2-DE/MS and none of these were exclusive to the gel-based separation methods.
In addition to high-throughput, contributions made by the LC/MS/MS approaches included identification of membrane-associated proteins containing trans-membrane helix (TMH) domains. In total, 61 proteins predicted to possess TMH domains were identified by LC/MS/MS, constituting 4.7% of the proteins in this mapping dataset and 6.4% of the 949 proteins in the total proteome predicted to possess at least one TMH domain (Suppl. Table S1). Expectedly, of these 61 TMH domain-containing proteins, none were identified through 2D in-gel separation; a consequence of sample preparation methods ideal for enhanced yield of total protein but not necessarily optimal for hydrophobic protein recovery. The number of TMH domains possessed by individual proteins identified in this data set ranged from 1 to 15 with an average of 3.8.
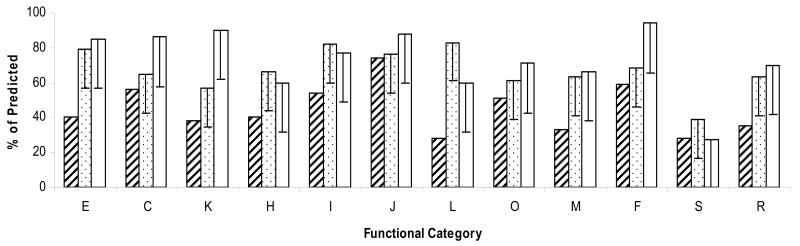
The majority of proteins in the mapped proteome (79.2%) are coded for by genes residing on the largest 2,848-kb chromosome while the remaining 20.8% are distributed over the remaining three replicons in proportion to their respective sizes (Table 1). None were mapped to the pHV2 plasmid, which is cured from the Hfx. volcanii strains used for this analysis (DS70 and derivatives). Proteins identified were categorized into COGs (clusters of orthologous groups) based on proposed function and are summarized in Fig. 3 and Suppl. Table S1. All protein identifications were categorized into one (or as many as 3) of 20 general functional categories. Those functional groups that were best represented included translation, ribosomal structure and biogenesis (74%), nucleotide transport and metabolism (59%) and energy production and conversion (56%). Those categories with the fewest representative protein identifications were intracellular trafficking and secretion (20%), inorganic ion transport and metabolism (20%) and cell motility (27%). Deficiencies in these latter categories are likely attributed to the fact that most of the components in these functional categories are extracytoplasmic and/or membrane-associated and are therefore unaccounted for due to common physical properties rather than skewed levels of expression of these particular proteins.
Table 1.
Protein identifications categorized by contig of origin.
| Contig1 | Size (kb) | No. Identified | No. Deduced | % Identified | % Deduced |
|---|---|---|---|---|---|
| Chromosome | 2848 | 1027 | 2960 | 79.2 | 34.7 |
| pHV4 | 636 | 147 | 638 | 11.3 | 23.0 |
| pHV3 | 438 | 99 | 438 | 7.6 | 25.9 |
| pHV1 | 85 | 23 | 85 | 1.8 | 25.8 |
| pHV2 | 6 | -- | 6 | -- | -- |
Plasmid pHV2 does not contribute to the current dataset due to the use of parent strain DS70, which is cured of this particular contig.
Fig. 3.
Comparison of the proteome maps of three archaea (Hfx. volcanii, S. solfataricus and M. jannaschii). Comparison was based on the 12 major functional categories used in common for protein classification of the S. solfataricus20 (spotted bars), M. jannaschii21 (white bars) and Hfx. volcanii (this study)(diagonal striped bars) proteomes. Y-error bars represent the average difference in identified proteins as a percentage of deduced proteins clustering to each of the 12 COGs for Hfx. volcanii compared to S. solfataricus and M. jannaschii. COGs included amino acid transport and metabolism (E), energy production and conversion (C), transcription (K), coenzyme metabolism (H), lipid metabolism (I), translation, ribosome structure and biogenesis (J), DNA replication, recombination and repair (L), post-translational modification, protein turnover and chaperones (O), cell envelope biogenesis and outer membrane (M), nucleic acid transport and metabolism (F), general function prediction only (R) and proteins of unknown function (S).
Comparative COG Analysis of the Hfx. volcanii Proteome Map
The number of Hfx. volcanii proteins detected by MS/MS were compared those of other archaea, including Methanocaldococcus jannaschii20 and Sulfolobus sulfataricus.21 Twelve of the COGs listed in Fig. 3 and Suppl. Table S1 were common to all three studies and, thus, were used for comparison. Overall, the proteome maps of M. jannaschii20 and S. solfataricus21 consisted of 963 and 1,399 proteins constituting 54% and 47% of the deduced proteome, respectively. Thus, while a comparable number of proteins (1,296) were identified for the Hfx. volcanii proteome map, this translated into a lower percent coverage of the deduced proteome (32%). This difference can be explained by the large genome size of Hfx. volcanii (4.01 Mb) relative to that of M. jannaschii (1.76 Mb) and S. solfataricus (3 Mb) coupled with the limitations of current MS-technology in detecting large numbers of individual proteins in complex mixtures. Thus, when comparing the identified proteins as a percentage of deduced proteins clustering to each of the 12 major functional categories or COGs, it is not surprising that the averages are 22 and 28 % lower for Hfx. volcanii than for S. solfataricus and M. jannaschii (Fig. 3). While these figures are roughly on par with that predicted by differences among the total mapping percentages, there were a few COGs that exhibited a greater difference than the average. The largest difference was in the number of proteins associated with DNA replication, recombination and repair (L) which was much higher in the S. solfataricus proteome than either Hfx. volcanii or M. jannaschii proteomes (Fig. 3). Likewise, proteins involved in transcription (K) were relatively fewer in the Hfx. volcanii and S. solfataricus than the M. jannaschii proteome. Proteins clustering to functional groups associated with translation, ribosome structure and biogenesis (J) as well as those of unknown function (S) were proportionally more highly represented in Hfx. volcanii than the other two archaeal proteomes. Despite these obvious differences, many of the other categories were relatively similar when percent coverage of genome was considered.
N-termini of the Hfx. volcanii proteome
The Hfx. volcanii proteomic dataset was systematically searched for N-terminal peptides and their derivatives including those modified co- and/or post-translational. Overall, 297 unique peptides were identified that mapped to the N-termini of 236 proteins, representing 18 % of the MS/MS-detected proteome (Suppl. Table S3). None of the MS/MS-detected peptides were formylated, consistent with previous findings that archaea like eukaryotes initiate translation with methionine, in contrast to bacteria which use formyl-methioinine.22 Instead, the majority of N-terminal peptides detected in the Hfx. volcanii proteome had patterns consistent with modification by methionine aminopeptidase and/or N-terminal aminotransferase. In fact, over 70% of the proteins with MS/MS-detectable N-termini included peptides in which the initiator methionine was removed (N2) and/or the initiator methionine and/or penultimate (second) residue of the deduced polypeptide was Nα-acetylated (Ac1 and Ac2, respectively) (Suppl. Table S3). The remaining ∼30% of the proteins detected by MS/MS had retained their initiator methionine and were not modified (N1). Multiple N-terminal peptide variants were detected for 22% of the 236 total proteins. Most of these variants were apparent processing intermediates (i.e., mixtures of N1 and N2, N1 and Ac1, N1 and Ac2, N2 and Ac2 or N1, N2 and Ac2). Only a small percentage (5.5%) of the 236 total proteins with MS/MS-detected N-termini appeared to be protein isoforms including mixtures of N2 and Ac1 as well as Ac1 and Ac2 (Suppl. Table S3).
Although Nα-acetylation is relatively rare in Bacteria with only a few protein examples 23-26, it is becoming apparent that this type of modification is common among Archaea (at least haloarchaea) 27, 28 as well as Eukarya.29 Over one-fourth (29%) of the proteins with MS/MS-detectable N-termini were Nα-acetylated in the Hfx. volcanii proteome (Suppl. Table S3). Most of these Nα-acetylated proteins were singly modified with a relatively equal ratio (30:33) of Ac1 to Ac2 forms. Interestingly, of the five proteins with both Ac1 and Ac2 isoforms, three were previously found to accumulate at high levels when Hfx. volcanii is grown in the presence of the irreversible proteasome inhibitor clasto-lactacystin-β-lactone.19 These included Hvo0860, Hvo1545 and Hvo2784 annotated as a SufB-like FeS assembly protein, dihydroxyacetone kinase L subunit, and rpsM ribosomal protein S13p/S18e, respectively (Suppl. Table S3). Although the N-end rule (in which the half-life of a protein is determined by its N-terminal residue) is conserved in Bacteria and Eukarya;30 its function including the relationship of protein stability to Nα-acetylation is not known in Archaea. Thus, the relationship of these Ac1 and Ac2 isoforms to protein stability and susceptibility to proteasome-mediated degradation is highly speculative.
The relative abundance of Hfx. volcanii proteins with small residues (i.e., Gly, Ala, Pro, Val, Ser, Thr) in the penultimate position of their deduced primary sequence was significantly greater for N-terminal MS/MS-detected peptides modified by methionine aminopeptidase and/or N-terminal aminotransferase (87%) compared to those that were unmodified (N1 alone) (32%) (Suppl. Table S3). In particular, the penultimate residue that was dominant for the modified proteins was serine (at 40% of the total proteins modified) compared to its limited presence in this same position of unmodified proteins (16% of total proteins unmodified). This bias, of small residues at the penultimate position, contrasts with the theoretical Hfx. volcanii proteome in which about 60% of the deduced proteins have small residues and 21% have serine residues at this position.
Based on analysis of the MS/MS-detected N-terminal peptides, initiator methionine removal occurred nearly exclusively when the penultimate residue of the protein was small, consistent with the substrate preferences of methionine aminopeptidases characterized from Bacteria, Archaea and Eukarya 31-34 and similar to other archaeal proteomes.27 Of the 16 proteins detected in the Hfx. volcanii proteome that were exceptional to this rule (i.e., in which the initiator methionine was removed from a bulkier residue), most resulted in exposure of an acidic N-terminal residue (i.e., Asp or Glu) (Suppl. Table S3) and, thus, may be a reflection of the increased number of acidic residues used for hydration of halophilic proteins in high salt.2 For the Nα-acetylated proteins, the majority (nearly 80%) of those in an Ac2 form appeared to products of a NatA-like acetyltransferase, which in yeast preferentially transfers acetyl groups to the α amino group of small N-terminal residues after the initiator methionine has been removed.29 In addition, a number of the Ac1 modified proteins detected in the Hfx. volcanii proteome appeared to be a result of a yeast-like NatB acetyltransferase activity in which the initiator methionine residue preceding a penultimate Asp, Glu, Asn or Met was Nα-acetylated.29, 33 However, none of Nα-acetylated peptides detected in the Hfx. volcanii proteome were related in N-terminal sequence to products of the yeast NatC acetyltransferase (i.e., Ac1 forms with penultimate Ile, Leu, Trp or Phe residues).29 Instead, the majority of Ac1 modified proteins (over 80%) had small penultimate residues suggesting an additional acetyltransferase activity distinct from the yeast-like NatA, NatB and NatC is present.
Identification of Paralogs in Hfx. volcanii
Within this current mapping dataset for the Hfx. volcanii proteome, multiple proteins were identified which possess one or two other paralogs. Less common, however, was the MS/MS detection of 4 or more closely related protein paralogs. Nine separate proteins which possessed 4 or more paralogs were detected in this mapping dataset. These paralogs included proteins annotated as FtsZ of which 6 were detected out of a predicted total of 8. Six of the 16 cell division control protein 6/origin recognition complex proteins of the deduced proteome were also detected. Other paralogs included ArcR transcriptional regulators (6 of 16), bacterio-opsin activator-like proteins (7 of 14), non-descript oxidoreductases (6 of 8) and transposases of the IS4 family (5 of 31). Four of each of the following were also identified by MS/MS: cell division control protein 48 (out of 5 total), MutS DNA mismatch repair protein (all accounted for in this mapping dataset) and metallo-β-lactamase superfamily domain protein (out of 9 total). The extent of functional overlap between many of these paralogs is unclear. A few, such as those involved in cell division or transcriptional regulation, may operate in a specialized way or may be co-functional, forming multi-subunit complexes with related paralogs. Those paralogs not identified may have missed detection by our methods or are conditionally expressed. Exploration of a greater variety of growth conditions may assist to fully identify these “missing” proteins. Future characterization of the identified paralogous proteins by the haloarchaeal community may facilitate the assignment of a more specific function and reduce apparent redundancy within the currently annotated proteome.
High-Scoring Hypotheticals
Of the 371 proteins that were assigned to a COG of unknown/general function, 175 were designated as hypothetical or conserved hypothetical proteins. In order to assign putative function to a larger portion of the annotated genome, these proteins were prioritized based on MOWSE score and ratios of MOWSE score to assigned peptide hits. Those that exceeded the arbitrary MOWSE score limit of 60 and MOWSE/peptide ratio of 10 were subjected to protein basic local alignment sequence tool (BLAST) searches. Of the 37 individual proteins analyzed, 4 were related to proteins with assigned function within the non-redundant protein NCBI database. These latter proteins included two putative transcriptional regulators (Hvo_0184 and Hvo_2718) with respective similarity to Natromonas pharaonis NP1352A (E value of 2e-18) and Halorubrum lacusprofundi ZP_02016688 (E value of 2e-45). Hvo_1704 was also identified and found to be related to proteins which undergo reversible glycosylation (e.g., Solanum tuberosum ABA81861; E value of 7e-16), and the identified Hvo_2187 protein is a likely Fe-S oxidoreductases with close relationship to Halorubrum lacusprofundi elongator protein 3/MiaB/NifB-like protein (ZP_02016357; E value of 1e-158). Hvo_2187 also possessed some overlap with the MiaB protein from organisms such as Thermotoga, which has been shown to catalyze the posttranscriptional methylation and thiolation of N-6-isopentenyladenosine in tRNAs.35 Of the remaining 37 proteins not assigned to any putative function, 28 were conserved with hypothetical proteins of other haloarchaea with E values in excess of 1e-80. The remaining 5 proteins were unique to Hfx. volcanii yet had probability-based MOWSE scores ranging from 36 to 111 with MOWSE/peptide ratios of 1.1 to 57.0.
Conclusion
This work reports the identification of approximately one-third of the total proteome of Hfx. volcanii with coverage of both molecular weight and isoelectric point ranges related to those of the deduced proteome. This project is a significant advance toward furthering our understanding of Hfx. volcanii and improving the current annotation of its proteome through careful analysis of high-scoring hypothetical proteins. The mapped regions of the proteome and future expansion of this current map through proteomic analyses of changing growth conditions and various stress challenges will serve as a useful resource for the Hfx. volcanii research community. This information will be particularly useful in coordination with forthcoming transcriptome data and the release of the first published Hfx. volcanii genome. Collectively, this work will also serve those studying other members of the Archaea domain.
Supplementary Material
Supporting information is available free of charge via the internet at http://pubs.acs.org.
Acknowledgments
Thanks to Stan Stevens Jr. and Scott McClung for their assistance with MS analysis at the ICBR Proteomics Core (University of Florida, Gainesville, FL). This research was funded in part by grants from the NIH R01 GM057498 and DOE DE-FG02-05ER15650 to JMF and NSF MCB 0620005 and DOE DE-FG02-91ER20041 to CJD.
Abbreviations
- IPG
immobilized pH gradient
- LC/MS/MS
liquid chromatography tandem-mass spectrometry
- IEF
isoelectric focusing
- IDA
information dependent acquisition
- MOAC
metal oxide affinity chromatography
- IMAC
immobilized affinity chromatography
- MudPIT
Multidimensional Protein Identification Technology
- SCX
strong cation exchange chromatography
- RP
reversed phase
- 2-D
2-dimensional polyacrylamide gel electrophoresis
- COG
Clusters of Orthologous Groups
- TMH
transmembrane spanning helix
- N1
protein retaining the initiator methionine
- Ac1
protein with an Nα-acetyl group on the initiator methionine
- N2
protein with initiator methionine removed
- Ac2
protein with the initiator methionine removed and an Nα-acetyl group on the exposed penultimate residue
References
- 1.Mullakhanbhai MS, Larsen H. Halobacterium volcanii spec. nov., a Dead Sea Halobacterium with a moderate salt requirement. Arch Microbiol. 1975;104:207–214. doi: 10.1007/BF00447326. [DOI] [PubMed] [Google Scholar]
- 2.Mevarech M, Frolow F, Gloss LM. Halophilic enzymes: proteins with a grain of salt. Biophys Chem. 2000;86:155–164. doi: 10.1016/s0301-4622(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 3.Oren A. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol. 2002;28:56–63. doi: 10.1038/sj/jim/7000176. [DOI] [PubMed] [Google Scholar]
- 4.Stan-Lotter H, Sulzner M, Egelseer E, Norton CF, Hochstein LI. Comparison of membrane ATPases from extreme halophiles isolated from ancient salt deposits. Orig Life Evol Biosph. 1993;23:53–64. doi: 10.1007/BF01581990. [DOI] [PubMed] [Google Scholar]
- 5.Litchfield CD. Survival strategies for microorganisms in hypersaline environments and their relevance to life on early Mars. Meteorit Planet Sci. 1998;33:813–819. doi: 10.1111/j.1945-5100.1998.tb01688.x. [DOI] [PubMed] [Google Scholar]
- 6.Allers T, Mevarech M. Archaeal genetics - the third way. Nat Rev Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 7.Soppa J. From genomes to function: haloarchaea as model organisms. Microbiology. 2006;152:585–590. doi: 10.1099/mic.0.28504-0. [DOI] [PubMed] [Google Scholar]
- 8.Wendoloski D, Ferrer C, Dyall-Smith ML. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology. 2001;147:959–964. doi: 10.1099/00221287-147-4-959. [DOI] [PubMed] [Google Scholar]
- 9.Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkland PA, Busby J, Stevens S, Jr, Maupin-Furlow JA. Trizol-based method for sample preparation and isoelectric focusing of halophilic proteins. Anal Biochem. 2006;351:254–259. doi: 10.1016/j.ab.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Karadzic IM, Maupin-Furlow JA. Improvement of two-dimensional gel electrophoresis proteome maps of the haloarchaeon Haloferax volcanii. Proteomics. 2005;5:354–359. doi: 10.1002/pmic.200400950. [DOI] [PubMed] [Google Scholar]
- 13.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 14.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 15.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein C, Garcia-Rizo C, Bisle B, Scheffer B, Zischka H, Pfeiffer F, Siedler F, Oesterhelt D. The membrane proteome of Halobacterium salinarum. Proteomics. 2005;5:180–197. doi: 10.1002/pmic.200400943. [DOI] [PubMed] [Google Scholar]
- 17.Tebbe A, Klein C, Bisle B, Siedler F, Scheffer B, Garcia-Rizo C, Wolfertz J, Hickmann V, Pfeiffer F, Oesterhelt D. Analysis of the cytosolic proteome of Halobacterium salinarum and its implication for genome annotation. Proteomics. 2005;5:168–179. doi: 10.1002/pmic.200400910. [DOI] [PubMed] [Google Scholar]
- 18.Konstantinidis K, Tebbe A, Klein C, Scheffer B, Aivaliotis M, Bisle B, Falb M, Pfeiffer F, Siedler F, Oesterhelt D. Genome-wide proteomics of Natronomonas pharaonis. J Proteome Res. 2007;6:185–193. doi: 10.1021/pr060352q. [DOI] [PubMed] [Google Scholar]
- 19.Kirkland PA, Reuter CJ, Maupin-Furlow JA. Effect of proteasome inhibitor clasto-lactacystin-β-lactone on the proteome of the haloarchaeon Haloferax volcanii. Microbiology. 2007;153:2271–2280. doi: 10.1099/mic.0.2007/005769-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhu W, Reich CI, Olsen GJ, Giometti CS, Yates JR., III Shotgun proteomics of Methanococcus jannaschii and insights into methanogenesis. J Proteome Res. 2004;3:538–548. doi: 10.1021/pr034109s. [DOI] [PubMed] [Google Scholar]
- 21.Chong PK, Wright PC. Identification and characterization of the Sulfolobus solfataricus P2 proteome. J Proteome Res. 2005;4:1789–1798. doi: 10.1021/pr0501214. [DOI] [PubMed] [Google Scholar]
- 22.Ramesh V, RajBhandary UL. Importance of the anticodon sequence in the aminoacylation of tRNAs by methionyl-tRNA synthetase and by valyl-tRNA synthetase in an Archaebacterium. J Biol Chem. 2001;276:3660–3665. doi: 10.1074/jbc.M008206200. [DOI] [PubMed] [Google Scholar]
- 23.Cumberlidge AG, Isono K. Ribosomal protein modification in Escherichia coli. I. A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J Mol Biol. 1979;131:169–189. doi: 10.1016/0022-2836(79)90072-x. [DOI] [PubMed] [Google Scholar]
- 24.Isono K, Isono S. Ribosomal protein modification in Escherichia coli. II. Studies of a mutant lacking the N-terminal acetylation of protein S18. Mol Gen Genet. 1980;177:645–651. doi: 10.1007/BF00272675. [DOI] [PubMed] [Google Scholar]
- 25.Isono S, Isono K. Ribosomal protein modification in Escherichia coli. III. Studies of mutants lacking an acetylase activity specific for protein L12. Mol Gen Genet. 1981;183:473–477. doi: 10.1007/BF00268767. [DOI] [PubMed] [Google Scholar]
- 26.Arai K, Clark BF, Duffy L, Jones MD, Kaziro Y, Laursen RA, L'Italien J, Miller DL, Nagarkatti S, Nakamura S, Nielsen KM, Petersen TE, Takahashi K, Wade M. Primary structure of elongation factor Tu from Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falb M, Aivaliotis M, Garcia-Rizo C, Bisle B, Tebbe A, Klein C, Konstantinidis K, Siedler F, Pfeiffer F, Oesterhelt D. Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. J Mol Biol. 2006;362:915–924. doi: 10.1016/j.jmb.2006.07.086. [DOI] [PubMed] [Google Scholar]
- 28.Aivaliotis M, Gevaert K, Falb M, Tebbe A, Konstantinidis K, Bisle B, Klein C, Martens L, Staes A, Timmerman E, Van Damme J, Siedler F, Pfeiffer F, Vandekerckhove J, Oesterhelt D. Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J Proteome Res. 2007;6:2195–2204. doi: 10.1021/pr0700347. [DOI] [PubMed] [Google Scholar]
- 29.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 30.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Bassat A, Bauer K, Chang SY, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsunasawa S, Izu Y, Miyagi M, Kato I. Methionine aminopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus: molecular cloning and overexpression in Escherichia coli of the gene, and characteristics of the enzyme. J Biochem. 1997;122:843–850. doi: 10.1093/oxfordjournals.jbchem.a021831. [DOI] [PubMed] [Google Scholar]
- 33.Huang S, Elliott RC, Liu PS, Koduri RK, Weickmann JL, Lee JH, Blair LC, Ghosh-Dastidar P, Bradshaw RA, Bryan KM. Specificity of cotranslational amino-terminal processing of proteins in yeast. Biochemistry. 1987;26:8242–8246. doi: 10.1021/bi00399a033. [DOI] [PubMed] [Google Scholar]
- 34.Miller CG, Strauch KL, Kukral AM, Miller JL, Wingfield PT, Mazzei GJ, Werlen RC, Graber P, Movva NR. N-terminal methionine-specific peptidase in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1987;84:2718–2722. doi: 10.1073/pnas.84.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez HL, Pierrel F, Elleingand E, Garcia-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information is available free of charge via the internet at http://pubs.acs.org.