Abstract
The most common form of genetic variation, single nucleotide polymorphisms or SNPs, can affect the way an individual responds to the environment and modify disease risk. Although most of the millions of SNPs have little or no effect on gene regulation and protein activity, there are many circumstances where base changes can have deleterious effects. Non-synonymous SNPs that result in amino acid changes in proteins have been studied because of their obvious impact on protein activity. It is well known that SNPs within regulatory regions of the genome can result in disregulation of gene transcription. However, the impact of SNPs located in putative regulatory regions, or rSNPs, is harder to predict for two primary reasons. First, the mechanistic roles of non-coding genomic sequence remain poorly defined. Second, experimental validation of the functional consequences of rSNPs is often slow and laborious. In this review, we summarize traditional and novel methodologies for candidate rSNPs selection, in particular in silico techniques that aid in candidate rSNP selection. Additionally we will discuss molecular biological techniques that assess the impact of rSNPs on binding of regulatory machinery, as well as functional consequences on transcription. Standard techniques such as EMSA and luciferase reporter constructs are still widely used to assess effects of rSNPs on binding and gene transcription; however, these protocols are often bottlenecks in the discovery process. Therefore, we highlight novel and developing high-throughput protocols that promise to aid in shortening the process of rSNP validation. Given the large amount of genomic information generated from a multitude of re-sequencing and genome-wide SNP array efforts, future focus should be to develop validation techniques that will allow greater understanding of the impact these polymorphisms have on human health and disease.
Keywords: polymorphism, SNPs, gene regulation, functional genomics, microsphere assay
1. Introduction
An individual’s physiologic response to environmental stimuli can be modulated by genetic variation. Millions of genetic alterations (termed polymorphisms) occur at >1% in the human population, and while most have little impact on human health, there is abundant recent evidence that certain variants cause a myriad of phenotypic effects and given a specific context, can strongly impact disease susceptibility [1]. Common polymorphisms include tandem repeated segments (minisatellite, 0.1–20 kb; microsatellite, 2–100 nucleotides), large (copy number variants) and small segmental deletions/insertions/duplications, and single nucleotide polymorphisms (SNPs). SNPs are exceedingly common polymorphisms, accounting for approximately 90% of all known sequence variation [2] and are estimated to occur every 100–300 base pairs. The most recent build of NCBI’s SNP database (dbSNP 127) lists over 11 million identified SNPs in the human population, with over 5 million validated by multiple investigators. By calculation, given evenly spaced SNP incidence throughout the human genome, there are approximately 165,000 SNPs within the 20,000–25,000 estimated genes whose coding regions cover approximately 1.5% of the human genome [3,4].
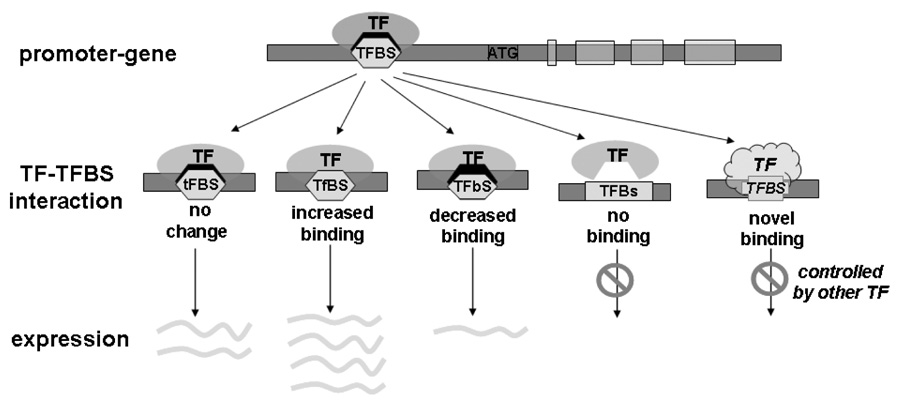
SNPs that affect gene expression occur in all regions of the genome. SNPs located within the coding region of genes have been extensively studied, including those that cause amino acid codon alterations (non-synonymous variants) which can lead to protein misfolding, polarity shift, improper phosphorylation and other functional consequences. Less predictable are variants located within non-coding regions of the genome. While mostly regarded as non-functional, this type of alteration can impact gene regulatory sequences such as promoters, enhancers, and silencers [5]. Termed regulatory SNPs (rSNPs), these variations have become more prevalent in recent literature [6–9]. Transcription factor (TF) binding sites are attractive regions to search for functional rSNPs. A SNP in a TF binding site can have multiple consequences. In most cases, a SNP does not change the TF and binding site interaction nor does it alter expression, since a TF will usually recognize a considerable number of binding sites. In some cases, a SNP may increase or decrease the binding, leading to allele-specific gene expression. In rare cases, a SNP may eliminate the natural binding site or generate a novel binding site. Consequently, the gene can no longer be regulated by the original TF. Thus, functional rSNPs in TF binding sites may predictably lead to differences in gene expression and phenotypes, and ultimately affect susceptibility to environmental exposure (Fig. 1). Indeed, there are numerous examples of rSNPs associated with disease susceptibility, including hypercholesterolemia [10], hyperbilirubinemia [11,12], myocardial infarction [13], acute lung injury [14], and asthma [15].
Figure 1.
The impact of a SNP in a transcription factor binding site (TFBS). In many cases, the SNP will not change TF binding activity or target gene expression because the TF, in general, allows variation in the consensus sequence of the binding site. In some cases, a SNP may increase or decrease the binding, leading to allelic-specific gene expression. In rare cases, a SNP may eliminate the natural binding site or generate a novel binding site, and consequently the gene is no longer controlled by the original TF.
Identification and experimental verification of functional rSNPs are key limiting steps in an efficient functional polymorphism discovery process. Here, we review traditional and novel prioritization methods to generate candidate genes that have polymorphic regulatory regions using sequence analysis, expression and genotype information. Further, we will describe standard and developing molecular biology techniques that quantify the impact of polymorphisms on DNA binding activity and gene regulation (see Fig. 2 for discovery pipeline). Functional analysis using wet lab techniques is a slow and laborious process, yet it remains critical to validation of a phenotypic effect and risk characterization. Additionally, we will highlight developing high-throughput methodologies which promise to revolutionize the field of rSNP discovery and functional genomics.
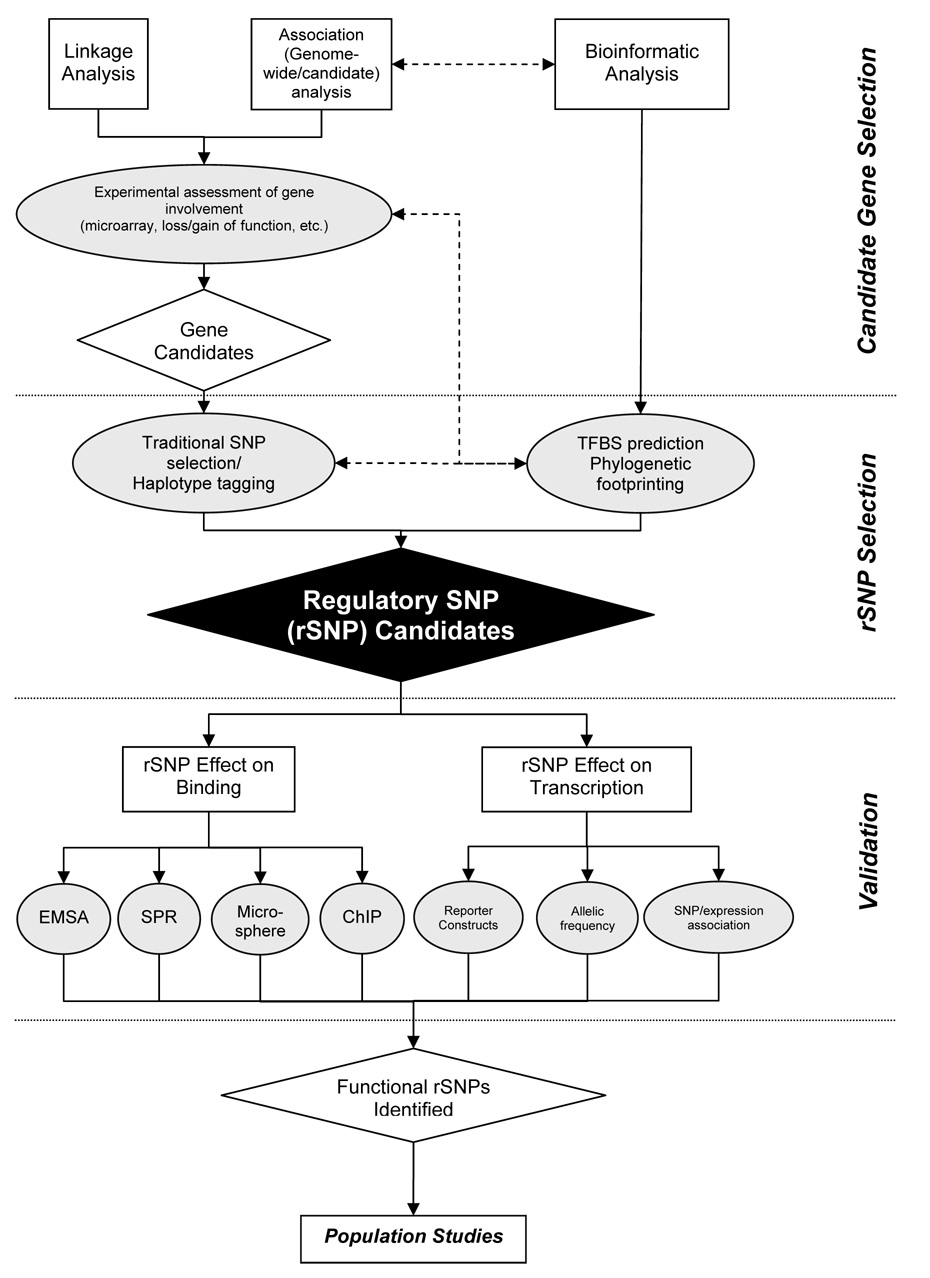
Figure 2.
Flowchart of identification and experimental validation of candidate regulatory SNPs (rSNPs). Briefly, candidate gene selection involves one or more association and predictive processes that incorporate genomic, gene expression, and phenotypic information. An initial candidate gene list can benefit from bioinformatic analysis of this information, and vice versa. Once a list of candidate genes has been generated, polymorphisms in the regulatory region of these genes are selected by informed analysis of the region of interest, available genotyping data, and/or representative SNPs of haplotype regions. Then, experimental validation assessing polymorphic effects on binding affinity and transcriptional regulation can be performed using methodology outlined in this review. Following characterization, rSNPs can then be tested in populations to determine if they are predictive of disease or environmental response.
2. Regulatory SNP discovery
2.1. Identification of candidate gene regions and finding candidate rSNPs
A candidate gene or a gene region becomes of interest in a SNP discovery project through several mechanisms. A knowledge base, such as expertise in DNA repair, may have pointed to a group of genes involved a key cellular process or likely to be involved in a disease and these genes may be sequenced (or resequenced) in a population in order to identify variants. Epidemiological studies draw attention to a gene by detecting the association between disease phenotypes and chromosomal regions, individual genes, haplotypes, or individual SNPs, depending on the methodology and resolution of the markers. A review of these genetic and epidemiological methods is beyond the scope of the present work. However, SNPs or markers used in such studies are often not the causative variants; rather they simply are markers which serve to delimit or discover certain genomic regions that may contain the functional variants.
In recent years, epidemiological studies use SNPs identified through comparisons of genomic sequences obtained from contig or shotgun libraries in the Human Genome Project, or candidate gene-directed resequencing. The vast majority of known SNPs were identified in comparisons of genome sequences from only a few humans (10–20). Resequencing efforts such as the NHLBI Program for Genome Applications and the NIEHS Environmental Genome Project [16] have used larger groups of samples, 24 to 400 or more, with varying degrees of ethnic or racial diversity, but sampling strategies have not been employed to generate a representative population sample. Due to cost considerations, the poorly-defined regulatory regions of genes have not always been sequenced. The result is that the catalog of variants in the existing databases is incomplete, uneven and most lack accurate frequency estimates. A significant number of the SNPs in dbSNP are either sequencing errors or have extremely rare frequencies. Some genomic regions are very poorly characterized, with only few SNPs known for only some ethnic groups while other regions are well-characterize among worldwide groups (e.g. Human Diversity project) [17].
The International HapMap project has improved the situation dramatically by evaluating a large set of SNPs (~4 million distributed across the genome) in samples of African, Asian and European descent (described in more detail later) [18]. This has allowed the identification of haplotypes and haplotype tagging SNPs (tagSNPs) that are common among human populations and those that are specific to some ethnicities [19]. Despite the remarkable coverage achieved by the HapMap, many genome regions still have poor resolution because the SNPs picked for evaluation in the project were found to be monomorphic or very rare. In addition, populations used to define haplotypes may not be representative of other populations of interest [20], resulting in failed associations (both false positives and false negatives) in disease studies [21].
The use of 300,000- 1 million tagSNPs in whole genome association studies has enabled tests of association between common variation and disease; however, SNPs identified as risk factors in these scans are in noncoding regions that frequently have no apparent relationship to a gene’s function. Thus there is still need for sequencing in larger, more diverse populations in order to discover the linked causal variants. The ultimate approach is to test all genetic variation in all individuals of a study cohort. This possibility is near reality as extensive efforts to lower costs of sequencing, i.e. the “$1000 genome” challenge, are currently underway [22–24]. In the near term, as attention turns to noncoding regions, there is a need to develop approaches to identify functional regulatory elements and evaluate how sequence variation alters function.
2.2. A bioinformatics approach
Successful bioinformatics identification of functional rSNPs requires identification of putative regulatory sequences, or motifs, and the co-location of SNPs in these sequences. Prediction of the impact of SNP on TF binding would be useful. Within a given gene network, such as the p53 or NRF2 pathways, members of the pathway typically have cis-regulatory sequences within 10 kb (sometimes larger) of the transcription start site. Computational methods for the identification of cis-regulatory sequences have been successfully applied to simple organisms such as yeast and worm, and while some methods have been plagued by high false positive rates in mammals primarily because of the very large quantity of intergenic sequence present [25], many recent new bioinformatics algorithms have improved prediction [26]. These include examining evolutionarily conserved regulatory sequences in upstream sequences of orthologous genes across species [7,27–29] and identifying statistically over-represented motifs in the upstream regions of genes that are co-regulated in microarray expression profiles [26,30].
With the availability of fully-assembled human and other mammalian genomes along with large-scale genotyping and gene expression databases, our laboratory developed computational tools to systematically evaluate rSNPs in transcription factor binding sites (TFBSs) in the human genome and to predict their impact on the expression of target genes. This approach integrates distinct computational methods and databases to facilitate position weight matrix (PWM) construction, TFBS prediction, phylogenetic footprinting, microarray expression profile analysis and SNP genotype-gene expression association. This system has been used to identify polymorphic antioxidant response elements [7].
Briefly, we built a PWM model based on a set of TFBSs that were discovered by investigators using classical experimental methods to examine DNA-protein interaction, including electrophoretic mobility shift assay, DNase I footprinting, and gene promoter reporter constructs. The PWM is a probabilistic model that assesses the nucleotide preference at each position of a TFBS. Assuming that the highest scoring sequences represent the strongest binding sequences, this model provides an approximation of relative DNA-protein interaction strength [31] and has been used to identify binding sites of p53 [32] and estrogen receptor [33]. Using the PWM model, potential TFBSs were identified for each input SNP sequence from dbSNP in a “sliding window” manner, a quantitative score was assigned, and the impact of a rSNP on a TFBS was estimated by calculating the difference in PWM scores for the allele-pair. Allelic pairs resulting in a larger score difference are predicted to be more likely to influence the binding of transcription factors. The selected rSNPs were then mapped to nearby genes using chromosomal positions and transcription start site information from NCBI dbSNP and Genome databases. To increase the chance of finding truly functional or bone fide binding sites and to identify potential loss of function SNPs, a comparative genomics approach called “phylogenetic footprinting” [34,35] was used. This analysis identifies evolutionary sequence conservation across human, mouse, and rat genomes—presumably, high levels of similarity imply functionality. Phylogenetics is commonly used and accepted for the identification of conserved regulatory elements. However, Horvath et al. [36] demonstrated that many human p53 response elements, particularly those regulating apoptosis genes and DNA repair, while showing strong homology among primate species (chimp, gorilla, rhesus) show very low conservation relative to rodents. Thus phylogenetic methods must be applied with knowledge of the pathway and used with caution. While recent sequencing of rhesus macaque (Macaca mulatta) [37] enables reconstruction of the ancestral state of the primate genome before the divergence of chimpanzees and humans, the extremely high homology among nonhuman primates alignments of primates limits informative comparisons to regions of recent divergence and positive selection [27,38,39]. The draft quality of nonhuman primate genome assemblies has also challenged the ability of current methods to detect insertions, deletions, and copy-number variations between humans, and primates, although this situation is improving rapidly [40].
Identification and annotation of all the DNA regulatory sequences in the human genome is a fundamental challenge in genomics and computational biology. Here we have briefly outlined approaches that examine TF binding motifs and phylogenetics in order to identify variation in sequences most likely to exhibit a phenotypic effect; however, it does not represent the breadth of bioinformatics methods that have been used to identify DNA regulatory sequences.
2.3. Phenotype assessment
An obstacle in the discovery process (e.g. bioinformatic approaches) is that the function of many candidate genes is unknown. An approach for refining gene candidates prior to assessing SNP effects is to determine if the gene in question affects a model phenotypic endpoint of interest. At the cellular level, many molecular biological techniques that curtail loss of function such as dominant negative mutants, antisense oligonucleotides, and RNA interference can be utilized to determine a gene’s effect on phenotypic endpoint. Additionally, the use of transgenic (gene overexpression or knockout) mice or siRNA-induced loss of function allows for in vivo assessment of a gene’s effect on phenotype. Specific reduction or elimination of a gene of interest and subsequent measurement of a quantifiable cellular phenotype can justify further genetic-based studies; however, these processes are relatively low-throughput and not amenable to prioritizing large candidate gene lists. However, high-throughput RNA interference screens have been developed for mammalian cell cultures and, despite current high costs and various technical challenges, these large-scale screens hold future promise as effective tools for gene candidate selection [41].
3. Experimental assessment of regulatory SNPs
3.1. DNA binding assays
This review focuses on approaches for experimentally assessing the relationship between genetic variation and phenotype. rSNP candidates, whether identified through linkage, candidate gene, and/or bioinformatic analysis, can be verified through a number of methods. Some methods test molecular properties such as binding affinity of a protein to TFBSs and the effect of a SNP in that region, while others directly examine the influence of a SNP on gene transcription and, ultimately, phenotypic response. Classically, many of these protocols are slow to process and difficult to optimize, making large scale evaluation of candidate rSNPs impractical. Therefore, many recent techniques have addressed the need to assess polymorphic effects in an accelerated, but quantifiable manner. In this section, we review both current and developing techniques which determine rSNP functionality.
3.1.1. Electrophoretic mobility shift assay
Electrophoretic gel-mobility shift assay, or EMSA, allows in vitro detection and visualization of protein-DNA binding [42]. First used to characterize the interaction of E. coli lac repressor to DNA restriction enzyme fragments [43], EMSA has become a flexible, simple, and relatively sensitive protocol for assessing DNA-protein interaction. Briefly, the assay utilizes a double-stranded oligonucleotide containing a known or putative binding sequence, typically 20–30 nts, which is labeled with either a radioactive or fluorescent marker and added to crude cellular nuclear extract or purified recombinant protein, thereby allowing DNA-protein complexes to form. The DNA probe/protein mixture is loaded onto a polyacrylamide gel and electrophoresed. If proteins are bound to the labeled sequence, the DNA-protein complex will move more slowly through the gel matrix, creating a “shift” relative to unbound oligonucleotide. Typically, a larger complex (i.e., DNA bound protein) results in retarded band migration. Dissociation of the protein from the oligonucleotide is discouraged by the low ionic strength of the gel, as well as the gel matrix itself [43].
Competitive EMSA distinguishes binding affinity of a particular protein for a target binding sequence, i.e. sequence variation caused by a SNP or site-directed mutation, by the use of competing non-labeled sequence in the binding reaction. Different types of unlabeled competing oligonucleotide are added to the binding reaction and attenuation of the signal (shifted band intensity) of the labeled probe is quantified. For example, Pitarque et al. [44] demonstrated the effect of a SNP located 5’ upstream region of the cytochrome P450 2A6 (CYP2A6) gene utilizing competitive EMSA. Furthermore, competitive unlabeled oligonucleotides can define what binding factors create a shift of interest by challenging with a sequence matching the consensus of a binding factor. Additionally, antibodies which recognize epitopes of a bound protein create a less mobile DNA-protein complex, resulting in a “supershift” on the gel (or in some cases, the antibody binds the DNA-binding region of the protein, thereby attenuating intensity of the shifted band), indicating specificity of DNA-protein binding. As an example, Masotti et al. [45] used this approach to determine that the transcription factor YY1 targeted a promoter sequence of the ribosome biogenesis gene TCOF1. Some Treacher-Collins syndrome (TCS) patients were found to harbor a SNP in this region, thereby altering YY1 binding potential and subsequently reducing TCOF1 gene expression – a phenotype suggested to increase TCS susceptibility.
While highly sensitive, EMSA results are often qualitative, as band intensity is difficult to quantify due to issues such as background and band smearing. When assessing SNP effects, in particular those that have exhibit minimal binding differences, more resolute quantification is desired. Additionally, gel electrophoretic-based technologies that assess in vitro DNA-binding potential are time-consuming and often difficult to optimize [46] (also see review [8]).
3.1.2. Fluorescent reporters
Increasingly, there is a demand for higher throughput assays to verify TF binding sites (and the effects of SNPs). One such approach utilizes fluorescence (Förster) resonance energy transfer (FRET). FRET describes the energy transfer of the emission of a donor fluorophore by an accepting fluorophore if the distance between the two is less than ~100 Å [47] resulting in a change in fluorescence (or quenching). FRET has outstanding sensitivity of detection, which can be realized at the level of single biomolecules [48]. A number of recent efforts have utilized FRET to quantify binding of protein to DNA [49–51]. In one scheme, two duplex DNA “half” protein-binding sites are each separately labeled with a donor and acceptor fluorophores [50]. The FRET effect is stabilized by the binding of protein to the full binding site. This procedure, known as the molecular beacon binding assay, allows real-time optical reporting of the binding event. Another approach exploits the use of dual fluorophore labeled duplex DNA with protein binding detected by exonuclease III protection [51]. A linear duplex DNA is designed with two binding sites occupying each end of the dual-labeled probe. Protein binding protects the DNA duplex from exonuclease III digestion, thereby maintaining FRET. In the absence of protein binding, the duplex is digested and the FRET signal extinguished. Both assays demonstrate a quantitative dynamic range in respect to protein concentration, and feasibly could be used to test for SNP effects on TF binding. Additionally, these assays are rapid, and can be multiplexed using different fluorophores, greatly enhancing its utility as a medium-throughput method. Currently, designing molecular beacon assays can be technically challenging and the dual-labeled probes are relatively expensive to manufacture.
3.1.3. Surface plasmon resonance imaging arrays
Surface plasmon resonance (SPR) imaging arrays are a somewhat recent innovation that can achieve determinations of DNA-protein binding kinetics without the use of radioactive or fluorescent nucleic acid base modifications. SPR describes the natural phenomena of photon energy transferring to electrons of a noble metal film (high refraction) present in an air or liquid (low refraction) interface [52]. When the metal film is coupled on a prism, the incident light focused at a certain angle will be absorbed by the metal, thereby attenuating reflected light. However, in the presence of molecules occupying a chemically or biologically modified metal surface, the angle of non-reflectance will change, which can subsequently be measured in real-time by a CCD camera, a process known as SPR imaging. This technique has been utilized to measure many kinds of biomolecular interactions including antibody-epitope recognition (i.e. protein-protein interaction) [53], DNA: DNA or DNA:RNA oligomerization [54–57] and DNA-protein interactions [58–61]. DNA microarray technology has been combined with SPR imaging analysis to create a medium to high-throughput methodology for quantifying DNA-protein interactions [62] in which a recently modified DNA array generation process has enhanced detection of interactions with double-stranded sequences of similar makeup (i.e., SNPs in binding regions) [63]. Indeed, this technique demonstrated quantifiable differences in binding association and dissociation of competing MafG homodimer or MafG:Nrf2 heterodimer transcription factors to systematic mutations of a Maf recognition element (MARE) sequence using single or double base substitutions in its flanking and core binding regions [61]. This method allowed prediction of the binding affinities of native and mutated sequence for the MafG homodimer and MafG:Nrf2 heterodimer which have been shown to repress and enhance downstream antioxidant gene expression, respectively [64].
SPR imaging holds great promise for investigations of protein binding to polymorphic regulatory elements, but at present it is still a challenging technology. Coupling DNA oligonucleotide arrays to SPR imaging technology expands the possibilities, but as yet only one or two recombinant proteins have successfully been quantitatively assayed in combination. The use of experimentally derived cell or nuclear extracts has not been accomplished, although this might be achieved with antibody probing analogous to an EMSA supershift technique [65]. It remains to be seen if cumulative induction-repression dynamics of a regulatory sequence in a treatment/exposure model can be assessed with SPR imaging.
3.1.4. Immobilized oligonucleotide and microbead technologies
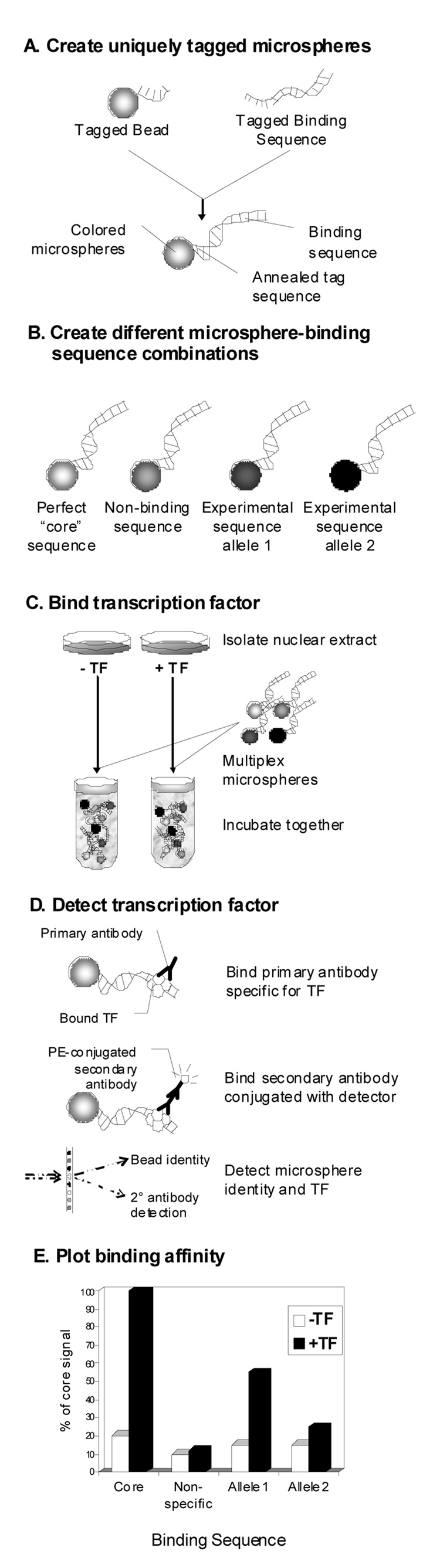
A format in common use is the enzyme-linked immunosorbant assay (ELISA)-like procedure in which immobilized DNA binding regions are incubated with recombinant protein or nuclear extract in a multi-well format, and then probed with an antibody specific for the protein of interest [66,67]. An advantage of this protocol is that any protein of putative DNA-binding potential can be assayed, assuming the antibody is available. Additionally, the assay format has been demonstrated to be more sensitive to binding potential than EMSA, ascribed to the fact that protein-DNA binding equilibrium can be more readily achieved in this format given fixed amounts of protein and oligonucleotide [66]. Assessing differences in binding affinities for polymorphic sequences might be accomplished using this format in the presence of competitive oligonucleotides specific to each allele, similar to competitive EMSA. Unfortunately, much like EMSA, only binding of one protein to one DNA sequence can be assayed per reaction. However, fluidic ELISA-based assays capable of identifying unique bound sequences in a multiplexed oligonucleotide binding reaction have been recently developed. Gorenstein, et al. [68,69] created a polystyrene bead library consisting of unique bead collections synthesized with a single species of nuclease-resistant synthetic phosphodiester-modified oligonucleotides known as thioaptamers. A mixture of differently labeled thioaptamer coated beads was incubated with NF-κB p50 protein, and then the bound protein was detected with a fluorescently labeled anti-p50 antibody. Labeled thioaptamer beads were then detected and sorted using flow-cytometry and the p50-bound thioaptamer sequences were recovered and sequenced for identification [69]. Similarly, Yaoi, et al. multiplexed 24 different double-stranded TF binding sequences, each labeled with biotin, in a reaction with cellular nuclear extract [70]. After purification, denatured double-stranded protein bound biotinylated probes were hybridized to unique fluorescently-labeled beads conjugated with anti-sense sequences specific for each of the 24 TF probes (i.e., one bead, one TF binding sequence). Following phycoerythrin-conjugated streptavidin treatment, each oligonucleotide probe provided a quantitative readout of TF binding to its consensus sequence. Our laboratory has pursued the possibility that an oligo-conjugated fluorescent microsphere-based technique could be utilized to assess differential binding affinities due to sequence polymorphism. We have developed a sensitive, high-throughput microsphere-based assay based on Luminex FlexMap technology to measure both p53 binding to p53-response elements and estrogen receptor α (Erα) binding to ERα response elements. Up to 82 response elements have been tested in a multiplex hybridization. Microspheres are sorted, identified, and bound proteins are detected with phycoerythrin-labeled antibodies (unpublished data, technique outlined in Fig. 3). The approach shows high resolution and utility in both experimental mutagenesis studies of TF binding sites and the functional evaluation of SNPs in TF binding sites.
Figure 3.
Outline of Microsphere Binding Assay. a) First, anneal oligonucleotides containing test sequences to tagged microspheres. b) Microspheres are uniquely dyed to allow identification of tagged oligonucleotides and multiplexing. This example shows four different microspheres and binding sequences. c) This assay allows for binding of pure TF or cellular extract containing activated TF. Cell cultures with and without activated TF are used and nuclear extracts are then mixed with the multiplexed microsphere-oligonucleotides to allow binding of TF. d) After washing, primary antibody specific for the TF of interest is added and detection is achieved with a phycoerythrin-conjugated antibody. A Bioplex flow cytometric device equipped with two excitation lasers and absorption optics is used for detection. The first laser identifies the microsphere (binding sequence) and the second laser identifies the quantitative signal from the TF. e) Signals are averaged for each microsphere type and plotted as signal intensity. Here, signal intensity for each condition is shown as a percentage of the perfect binding sequence. The core binding sequence generated the most signal when TF is added, while non-specific sequence generates only background noise. Our experimental sequence generates signal when TF is added, however the polymorphism modulates signal, and therefore binding potential of the TF.
A recently developed very high-throughput methodology that can evaluate hundreds of thousands of binding sites in a single experiment utilized the incorporation of immobilized oligos into a microarray format. While similar to the previously described ELISA-based formats, microarray binding technology provides both a quantitative and highly scalable platform to assess in vitro protein-DNA binding [71–74]. The first use of this approach on a genome-wide scale was performed by Mukherjee and colleagues using purified GST-tagged yeast transcription factors [75]. This technology, termed ‘Protein Binding Microarrays’ or PBMs, has since been scaled up to represent every possible binding site 8–10 bp in length [76,77]. PBMs will likely significantly enhance our understanding of what individual sequence variants do to alter binding potential in an in vitro setting, allowing for greater predictive capability of a SNP effect in a TFBS.
3.1.5. Chromatin immunoprecipitation
A concern with in vitro binding assays is DNA is presented in an unnatural state, i.e., the sequences are not in a chromatin context, and specific cis- or trans-acting co-factors or enhancer elements are absent, thus creating an artificial protein-DNA binding situation. Chromatin immunoprecipitation (ChIP) is a versatile binding assay that provides insight into gene regulation in an endogenous state. In this technique, DNA binding proteins such as transcription factors or proteins involved in DNA binding complexes are crosslinked to the DNA by formaldehyde and subsequently sonicated in order to fragment the DNA before immunoprecipitation by an antibody specific for the protein of interest [78,79]. Formaldehyde is a powerful and reversible crosslinking agent that essentially acts to “freeze in time” protein-DNA, protein-RNA and protein-protein interactions [80].
Although the crosslinking, sonication, and immunoprecipitation procedures for ChIP have been well-documented in numerous recent studies, there are many techniques for detecting and analyzing immunoprecipitated DNA. Historically, slot blot, Southern blot and cloning and sequencing were used. More recently, immunoprecipitated DNA is PCR amplified and viewed on a gel or with quantitative real-time monitoring [81,82]. Additionally, immunoprecipitated DNA can be measured genome-wide by hybridizing to a genomic tiling microarray, procedurally known as ChIP-on-chip or ChIP-chip [83]. Discovery methods such as ChIP-chip are powerful because they allow genomic scale identification of both known and novel DNA binding sites. Encouragingly, a recent independent and blind test of different ChIP-chip platforms using the same DNA samples with “spiked-in” targets found that ChIP-chip was very reproducible, where variation tended to result from lab, protocol, and computational algorithm differences [84]. However, ChIP-chip detection limit is somewhat sensitive to protein expression levels and may be influenced by protein-protein interactions such as competitive DNA binding of other proteins [83]. In addition, the sequence complexity in the human genome and the relative sequence complexity of the binding motif can affect resolution. However, this approach is likely to become routine as whole genome tiling arrays become more cost-effective. In order to overcome some of these limitations, the DIP-chip (DNA immunoprecipitation with microarray detection) method was developed using yeast Leu3p as a model in which purified protein and naked genomic DNA are mixed in vitro to form protein-DNA complexes. These complexes are then isolated, run on whole genomic microarrays to identify protein bound DNA fragments, and sequenced to identify putative binding sites [85].
Methods to detect protein-DNA interactions using high-throughput DNA sequencing techniques have also been developed using ChIP technology. For example, ChIP-STAGE (Sequence Tag Analysis of Genomic Enrichment) was developed to map regions of protein-DNA interactions in both yeast TATA-box binding protein and human transcription factor E2F4 targets [86]. A similar method, ChIP-PET, combines ChIP with paired-end ditag (PET) sequencing for identification of TF binding sites [87]. Using the sequence specific p53 tumor suppressor binding site as a model, Wei et al. [88] cloned immunoprecipitated DNA into a DNA library from which tags of both the 5’ and 3’ ends of each binding region were created and joined together to create PETs. PETs were then cloned into a final ChIP-PET library, sequenced, and then genomically mapped to determine boundaries of cloned fragments. While nonspecific fragments are distributed randomly, overlapping fragments representing true binding sites will form PET clusters [87].
ChIP methods have also been developed to analyze differential protein-DNA binding among allelic variants of a gene. HaploChIP (haplotype-specific chromatin immunoprecipitation), developed by Knight et al. [89], combined traditional ChIP using Pol II antibody with mass spectrometry to identify differential protein-DNA binding in vivo associated with allelic variants of the imprinted gene SNRPN. Our laboratory has modified this technique to address differential allelic binding. In this method, protein-bound DNA is immunoprecipitated and polymorphic candidate binding regions are then assayed using a probe-based allelic-discrimination genotyping assay to assess differential binding potential. Liu et al. [90] used this technique to demonstrate differential Pol II binding potential in a promoter region of the glutathione-S-transferase gene, GSTM3, caused by a base pair substitution. In a more high-throughput manner, Maynard and colleagues annealed ChIP DNA material to Illumina HumanHap300 GenotypingBeadChip genotyping arrays in order to assess allelic binding differences of Pol II across the genome [91]. ChIP-SNP identified 466 SNPs that altered binding of Pol II, including many known imprinted genes. While this method binding evaluates several thousand polymorphic elements in a single experiment, an important limitation is that the evaluated SNPs in a test sample must be heterozygous, where homozygous locations are uninformative. Evaluation of all polymorphic TFBS of interest would therefore require numerous samples that represent heterozygosity at all putative binding locations, thereby limiting the cost effectiveness of such a study.
3.2. Assessment of SNP effect on gene regulation
While TF-DNA binding is typically necessary for TF transactivation and gene regulation, assessing the effect of SNPs themselves on transcription and phenotype is essential.
3.2.1. Promoter reporter constructs
A putative regulatory sequence can be incorporated into an expression vector construct containing a reporter gene lacking endogenous promoter activity. The plasmid is subjected to regulatory cellular machinery when transfected into a cell and the reporter gene expression is measured (i.e., typically a luciferase gene is downstream of the promoter region of interest). For polymorphisms, separate constructs containing allelic sequences are transfected in parallel and expression is normalized to a reference. For a thorough discussion of the use of gene reporter constructs in the context of assessing gene regulatory regions, see these reviews [8,92,93]. Various factors affect interpretation of data generated from reporter construct assays, including variation in transfection efficiency [94], normalization to co-transfected constitutively expressed constructs, and exclusion/inclusion of enhancer regions in the test reporter construct that cooperate in gene expression of the endogenous gene. While transient expression of reporter gene constructs use the host cell’s transcriptional machinery, high copy number and the lack of any chromatin features and structure make it an imperfect model system. Stable transfection of reporter constructs (i.e., permanent incorporation of the foreign plasmid DNA into the genome) potentially corrects problems of an abnormal chromatin context, but clonal variation can hinder precise quantitative assessment of gene regulation and expression [8,95]. While imperfect, transfection of plasmid constructs in a specific cell system, which may overexpress (or have induced) the TF of interest, gives the researcher a specific and quantitative tool for assessing SNP effects on TF gene transactivation.
3.2.2. Measuring allelic imbalance in mRNA
If SNPs are present in the coding region of genes, then the effect of an rSNP on gene transcription can often be estimated by measuring allelic imbalance in transcript levels. Allelic ratios of a coding region SNP in a heterozygous individual, in which the rSNP of putative functional relevance is in linkage disequilibrium (LD), provides direct evidence for differential transcription of alleles caused by rSNPs. Detection of allele-specific gene expression depends on quantification of mRNA from each allele and assumes that normally each chromosome is equally expressed in a 1:1 ratio [96,97]. Relative allelic expression has been studied using methods such as amplification refractory mutation system (ARMS), PCR followed by RFLP analysis, SNP genotyping arrays, mRNA sequencing and mass spectrometry-based genotyping [98]. PCR followed by primer extension is also common and has been detected using capillary electrophoresis [96,99] or mass spectrometry [100].
Alternatively, probe-based genotyping assays can provide a quantitative way of assessing allelic imbalance [101]. For example, a coding SNP was found to be in LD with a functional rSNP in the promoter of the GSTM3 gene [90]. Using genomic DNA of known genotypes as a standard reference, allelic expression of the coding SNP in GSTM3 transcript was measured using a quantitative probe-based genotyping assay. Using this approach the transcript linked to the strong binding promoter allele was enriched over the transcript linked to the weak allele assessed in three cell lines heterozygous for the functional rSNP (Fig. 4). We are currently applying this method to evaluate the effects of candidate rSNPs in antioxidant response elements [7]. Because measured allele ratios can be impacted by the quantity of input mRNA and the number of amplification cycles, it is important to validate allelic imbalance methods using reference mixtures of DNA of different concentration and composed of known allele ratios. Allelic imbalance evaluation using probe-based assays are amenable to high-throughput assessment from a large set of samples. Indeed, other high-throughput methodologies, including pyrosequencing techniques [102,103], are being utilized to accurately and reliably quantify allelic imbalance.
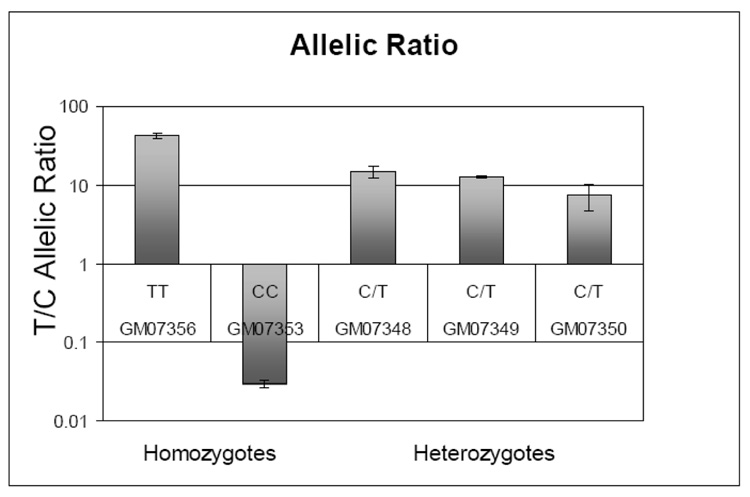
Figure 4.
An example of allelic imbalance measured by the use of quantitative allele discrimination assay for SNPs in coding regions performed on cDNA samples. A 5’ nuclease-based assay for rs1537236 was applied to homozygote and heterozygote cell lines to demonstrate the low level of expression of the C allele in GSTM3 gene [90].
3.2.3. Evaluation of genotype-expression phenotype association in humans
The expression phenotype for many genes is highly variable in humans. Several studies indicate a considerable portion is heritable and is associated with SNPs in both coding and regulatory regions of the genome [104]. The development of the International HapMap project has provided a convenient resource to examine this association. This project so far has genotyped more than 3 million SNPs in four distinct populations (CEU: 90 Utah residents with ancestry from northern and western Europe; CHB: 45 Han Chinese in Beijing; JPT: 45 Japanese in Tokyo; YRI: 90 Yoruba in Ibadan, Nigeria). Several research groups have generated gene expression profiles in lymphoblastoid cell lines of HapMap individuals and analyzed the correlation of SNP genotype to gene expression phenotype. For example, the genome-wide linkage analysis has been performed for expression phenotypes in CEU individuals to map the genetic determinants of variation in human gene expression [104]. In addition, cis-acting rSNPs have been discovered using a regional association approach [105].
On a larger scale, genome-wide association has been performed to seek statistically significant association of SNPs with expression variation in lymphoblastoid cell lines of all 210 unrelated HapMap individuals [106]. Our laboratory has also utilized the HapMap resource to test the association between genotypes of bioinformatically identified antioxidant response element SNPs and gene expression phenotypes in 60 unrelated CEU individuals [7]. There are, however, numerical limitations with these approaches utilizing the HapMap data. First, HapMap genotyping data and expression are currently only available for 3.1 million SNPs and 270 individuals. Second, many SNPs (or haplotypes) have very low allele frequencies. Third, and most importantly, the microarray expression profiles used in our association analysis were from untreated human lymphoblastoid cell lines. Ideally, expression information from additional tissues and stress exposures could be evaluated in order to find association between exposure-inducible gene expression and SNPs that affect transcriptional activation. Because polymorphic variants that are known to affect expression are often found at population frequencies of less than 5% [107–109], issues of sample size become crucial in a survey like this one. Ideally, expression profiles from a larger sample (n > 400) of stress-exposed human cells could be used in this analysis, combined with a higher resolution of HapMap genotyping data, making this bioinformatics-driven approach particularly effective.
4. Conclusions
A key challenge in identifying functional polymorphisms in gene regulatory regions is the shear amount of genomic data and the undefined nature of these regions. Linkage analysis has had success in identifying chromosomal regions relevant to disease susceptibility, and genome-wide SNP association studies will identify many additional important candidate genes. However, it remains a challenge to resolve the role by which specific polymorphisms affect gene expression. Candidate gene approaches can be effective; however, novel genes are often overlooked using this method. By using bioinformatics-based methods that access genomic information and expression databases, one can retain the power of a candidate gene approach yet still uncover novel gene regions. Additionally, resources such as dbSNP and the International HapMap database can provide detailed polymorphism data on gene candidates whose impact can be predicted further using bioinformatics tools such as PWM scoring.
It is still necessary, however, to verify the functional impact of novel gene polymorphism using basic molecular biological techniques. Methods such as EMSA, ChIP, and luciferase reporter constructs can be used to test the effect of a SNP on regulatory element function. These processes tend to be laborious and not well matched for screening large numbers of DNA elements and SNPs. It is, therefore, imperative to develop high-throughput methods to assess regulatory regions in the genome. Indeed, methodologies covered in this review, such as SPR analysis, oligo-conjugated microsphere binding assays, PBMs, and allelic imbalance methods, have contributed greatly to this endeavor. Unfortunately, obstacles exist with these technologies (such as reproducibility, quantification, limit of detection, and affordability), and continued efforts to develop these and new technologies are needed. Methodology advancement to allow effective analysis of genomic data will be necessary to understand the genetic basis of disease susceptibility and effectively predict and prevent disease in individuals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 2.Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.I.H.G.S. Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 5.Ponomarenko JV, Orlova GV, Merkulova TI, Gorshkova EV, Fokin ON, Vasiliev GV, Frolov AS, Ponomarenko MP. rSNP_Guide: an integrated database-tools system for studying SNPs and site-directed mutations in transcription factor binding sites. Hum Mutat. 2002;20:239–248. doi: 10.1002/humu.10116. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Tomso DJ, Liu X, Bell DA. Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes. Toxicol Appl Pharmacol. 2005;207:84–90. doi: 10.1016/j.taap.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight JC. Functional implications of genetic variation in non-coding DNA for disease susceptibility and gene regulation. Clin Sci (Lond) 2003;104:493–501. doi: 10.1042/CS20020304. [DOI] [PubMed] [Google Scholar]
- 9.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ono S, Ezura Y, Emi M, Fujita Y, Takada D, Sato K, Ishigami T, Umemura S, Takahashi K, Kamimura K, Bujo H, Saito Y. A promoter SNP (-1323T>C) in G-substrate gene (GSBS) correlates with hypercholesterolemia. J Hum Genet. 2003;48:447–450. doi: 10.1007/s10038-003-0055-x. [DOI] [PubMed] [Google Scholar]
- 11.Sugatani J, Yamakawa K, Yoshinari K, Machida T, Takagi H, Mori M, Kakizaki S, Sueyoshi T, Negishi M, Miwa M. Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Biophys Res Commun. 2002;292:492–497. doi: 10.1006/bbrc.2002.6683. [DOI] [PubMed] [Google Scholar]
- 12.Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GNJ, Jansen PLM, Elferink RPJO, Chowdhury NR. The Genetic Basis of the Reduced Expression of Bilirubin UDP-Glucuronosyltransferase 1 in Gilbert's Syndrome. N Engl J Med. 1995;333:1171–1175. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura S, Kugiyama K, Sugiyama S, Miyamoto S, Koide S, Fukushima H, Honda O, Yoshimura M, Ogawa H. Polymorphism in the 5'-flanking region of human glutamate-cysteine ligase modifier subunit gene is associated with myocardial infarction. Circulation. 2002;105:2968–2973. doi: 10.1161/01.cir.0000019739.66514.1e. [DOI] [PubMed] [Google Scholar]
- 14.Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, Kleeberger SR. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. Faseb J. 2007;0:0. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- 15.Jinnai N, Sakagami T, Sekigawa T, Kakihara M, Nakajima T, Yoshida K, Goto S, Hasegawa T, Koshino T, Hasegawa Y, Inoue H, Suzuki N, Sano Y, Inoue I. Polymorphisms in the prostaglandin E2 receptor subtype 2 gene confer susceptibility to aspirin-intolerant asthma: a candidate gene approach. Hum Mol Genet. 2004;13:3203–3217. doi: 10.1093/hmg/ddh332. [DOI] [PubMed] [Google Scholar]
- 16.Wilson SH, Olden K. The Environmental Genome Project: phase I and beyond. Mol Interv. 2004;4:147–156. doi: 10.1124/mi.4.3.4. [DOI] [PubMed] [Google Scholar]
- 17.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 18.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Yakub I, Birren BW, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 20.Salisbury BA, Pungliya M, Choi JY, Jiang R, Sun XJ, Stephens JC. SNP and haplotype variation in the human genome. Mutat Res. 2003;526:53–61. doi: 10.1016/s0027-5107(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 21.Clark AG, Boerwinkle E, Hixson J, Sing CF. Determinants of the success of whole-genome association testing. Genome Res. 2005;15:1463–1467. doi: 10.1101/gr.4244005. [DOI] [PubMed] [Google Scholar]
- 22.R.F. Service Gene sequencing. The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 23.Chan EY. Advances in sequencing technology. Mutat Res. 2005;573:13–40. doi: 10.1016/j.mrfmmm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Mardis ER. Anticipating the 1,000 dollar genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang LW, Nagarajan R, Magee JA, Milbrandt J, Stormo GD. A systematic model to predict transcriptional regulatory mechanisms based on overrepresentation of transcription factor binding profiles. Genome Res. 2006;16:405–413. doi: 10.1101/gr.4303406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs' target genes and functions. Nat Methods. 2008;5:347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 28.Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 30.Haverty PM, Hansen U, Weng Z. Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res. 2004;32:179–188. doi: 10.1093/nar/gkh183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benos PV, Bulyk ML, Stormo GD. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci U S A. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bajic VB, Tan SL, Chong A, Tang S, Strom A, Gustafsson JA, Lin CY, Liu Dragon ET. ERE Finder version 2: A tool for accurate detection and analysis of estrogen response elements in vertebrate genomes. Nucleic Acids Res. 2003;31:3605–3607. doi: 10.1093/nar/gkg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J Mol Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 35.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 36.Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O'Brien W E, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 38.Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris RA, Rogers J, Milosavljevic A. Human-specific changes of genome structure detected by genomic triangulation. Science. 2007;316:235–237. doi: 10.1126/science.1139477. [DOI] [PubMed] [Google Scholar]
- 41.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user's guide. Nat Rev Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 43.Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitarque M, von Richter O, Rodriguez-Antona C, Wang J, Oscarson M, Ingelman-Sundberg M. A nicotine C-oxidase gene (CYP2A6) polymorphism important for promoter activity. Hum Mutat. 2004;23:258–266. doi: 10.1002/humu.20002. [DOI] [PubMed] [Google Scholar]
- 45.Masotti C, Armelin-Correa LM, Splendore A, Lin CJ, Barbosa A, Sogayar MC, Passos-Bueno MR. A functional SNP in the promoter region of TCOF1 is associated with reduced gene expression and YY1 DNA-protein interaction. Gene. 2005;359:44–52. doi: 10.1016/j.gene.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forster VT. Zwischenmoleculare Energiewanderung und Fluorescenz. Annals of Physics. 1948;2:57–75. [Google Scholar]
- 48.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 49.Furey WS, Joyce CM, Osborne MA, Klenerman D, Peliska JA, Balasubramanian S. Use of fluorescence resonance energy transfer to investigate the conformation of DNA substrates bound to the Klenow fragment. Biochemistry. 1998;37:2979–2990. doi: 10.1021/bi9719758. [DOI] [PubMed] [Google Scholar]
- 50.Heyduk T, Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Li T, Guo X, Lu Z. Exonuclease III protection assay with FRET probe for detecting DNA-binding proteins. Nucleic Acids Res. 2005;33:e23. doi: 10.1093/nar/gni021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brockman JM, Nelson BP, Corn RM. Surface plasmon resonance imaging measurements of ultrathin organic films. Annu Rev Phys Chem. 2000;51:41–63. doi: 10.1146/annurev.physchem.51.1.41. [DOI] [PubMed] [Google Scholar]
- 53.Wegner GJ, Lee HJ, Corn RM. Characterization and optimization of peptide arrays for the study of epitope-antibody interactions using surface plasmon resonance imaging. Anal Chem. 2002;74:5161–5168. doi: 10.1021/ac025922u. [DOI] [PubMed] [Google Scholar]
- 54.Nelson BP, Grimsrud TE, Liles MR, Goodman RM, Corn RM. Surface plasmon resonance imaging measurements of DNA and RNA hybridization adsorption onto DNA microarrays. Anal Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 55.Lee HJ, Wark AW, Li Y, Corn RM. Fabricating RNA microarrays with RNA-DNA surface ligation chemistry. Anal Chem. 2005;77:7832–7837. doi: 10.1021/ac0516180. [DOI] [PubMed] [Google Scholar]
- 56.Jordan CE, Frutos AG, Thiel AJ, Corn RM. Surface Plasmon Resonance Imaging Measurements of DNA Hybridization Adsorption and Streptavidin/DNA Multilayer Formation at Chemically Modified Gold Surfaces. Anal. Chem. 1997;69:4939–4947. doi: 10.1021/ac961012z. [DOI] [PubMed] [Google Scholar]
- 57.Thiel AJ, Frutos AG, Jordan CE, Corn RM, Smith LM. In Situ Surface Plasmon Resonance Imaging Detection of DNA Hybridization to Oligonucleotide Arrays on Gold Surfaces. Anal. Chem. 1997;69:4948–4956. [Google Scholar]
- 58.Jost JP, Munch O, Andersson T. Study of protein-DNA interactions by surface plasmon resonance (real time kinetics) Nucleic Acids Res. 1991;19:2788. doi: 10.1093/nar/19.10.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Majka J, Speck C. Analysis of protein-DNA interactions using surface plasmon resonance. Adv Biochem Eng Biotechnol. 2007;104:13–36. [PubMed] [Google Scholar]
- 60.Tsoi PY, Yang M. Surface plasmon resonance study of the molecular recognition between polymerase and DNA containing various mismatches and conformational changes of DNA-protein complexes. Biosens Bioelectron. 2004;19:1209–1218. doi: 10.1016/j.bios.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto T, Kyo M, Kamiya T, Tanaka T, Engel JD, Motohashi H, Yamamoto M. Predictive base substitution rules that determine the binding and transcriptional specificity of Maf recognition elements. Genes Cells. 2006;11:575–591. doi: 10.1111/j.1365-2443.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- 62.Brockman JM, Frutos AG, Corn RM. A Multistep Chemical Modification Procedure To Create DNA Arrays on Gold Surfaces for the Study of Protein-DNA Interactions with Surface Plasmon Resonance Imaging. J. Am. Chem. Soc. 1999;121:8044–8051. [Google Scholar]
- 63.Kyo M, Yamamoto T, Motohashi H, Kamiya T, Kuroita T, Tanaka T, Engel JD, Kawakami B, Yamamoto M. Evaluation of MafG interaction with Maf recognition element arrays by surface plasmon resonance imaging technique. Genes Cells. 2004;9:153–164. doi: 10.1111/j.1356-9597.2004.00711.x. [DOI] [PubMed] [Google Scholar]
- 64.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 65.Teh HF, Peh WY, Su X, Thomsen JS. Characterization of protein--DNA interactions using surface plasmon resonance spectroscopy with various assay schemes. Biochemistry. 2007;46:2127–2135. doi: 10.1021/bi061903t. [DOI] [PubMed] [Google Scholar]
- 66.Benotmane AM, Hoylaerts MF, Collen D, Belayew A. Nonisotopic quantitative analysis of protein-DNA interactions at equilibrium. Anal Biochem. 1997;250:181–185. doi: 10.1006/abio.1997.2231. [DOI] [PubMed] [Google Scholar]
- 67.Renard P, Ernest I, Houbion A, Art M, Le Calvez H, Raes M, Remacle J. Development of a sensitive multi-well colorimetric assay for active NFkappaB. Nucleic Acids Res. 2001;29:E21. doi: 10.1093/nar/29.4.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang X, Bassett SE, Li X, Luxon BA, Herzog NK, Shope RE, Aronson J, Prow TW, Leary JF, Kirby R, Ellington AD, Gorenstein DG. Construction and selection of bead-bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002;30:e132. doi: 10.1093/nar/gnf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang X, Li X, Prow TW, Reece LM, Bassett SE, Luxon BA, Herzog NK, Aronson J, Shope RE, Leary JF, Gorenstein DG. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yaoi T, Jiang X, Li X. Development of a fluorescent microsphere-based multiplexed high-throughput assay system for profiling of transcription factor activation. Assay Drug Dev Technol. 2006;4:285–292. doi: 10.1089/adt.2006.4.285. [DOI] [PubMed] [Google Scholar]
- 71.Drobyshev AL, Zasedatelev AS, Yershov GM, Mirzabekov AD. Massive parallel analysis of DNA-Hoechst 33258 binding specificity with a generic oligodeoxyribonucleotide microchip. Nucleic Acids Res. 1999;27:4100–4105. doi: 10.1093/nar/27.20.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bulyk ML, Gentalen E, Lockhart DJ, Church GM. Quantifying DNA-protein interactions by double-stranded DNA arrays. Nat Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 73.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci U S A. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zasedateleva OA, Krylov AS, Prokopenko DV, Skabkin MA, Ovchinnikov LP, Kolchinsky A, Mirzabekov AD. Specificity of mammalian Y-box binding protein p50 in interaction with ss and ds DNA analyzed with generic oligonucleotide microchip. J Mol Biol. 2002;324:73–87. doi: 10.1016/s0022-2836(02)00937-3. [DOI] [PubMed] [Google Scholar]
- 75.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc Natl Acad Sci U S A. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- 80.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 81.Menendez D, Krysiak O, Inga A, Krysiak B, Resnick MA, Schonfelder G. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci U S A. 2006;103:1406–1411. doi: 10.1073/pnas.0508103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci U S A. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hudson ME, Snyder M. High-throughput methods of regulatory element discovery. Biotechniques. 2006;41:673. doi: 10.2144/000112322. 675, 677 passim. [DOI] [PubMed] [Google Scholar]
- 84.Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, Koch CM, Dunham I, Bieda M, Xu X, Farnham PJ, Kapranov P, Nix DA, Gingeras TR, Zhang X, Holster H, Jiang N, Green RD, Song JS, McCuine SA, Anton E, Nguyen L, Trinklein ND, Ye Z, Ching K, Hawkins D, Ren B, Scacheri PC, Rozowsky J, Karpikov A, Euskirchen G, Weissman S, Gerstein M, Snyder M, Yang A, Moqtaderi Z, Hirsch H, Shulha HP, Fu Y, Weng Z, Struhl K, Myers RM, Lieb JD, Liu XS. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu X, Noll DM, Lieb JD, Clarke ND. DIP-chip: rapid and accurate determination of DNA-binding specificity. Genome Res. 2005;15:421–427. doi: 10.1101/gr.3256505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim J, Bhinge AA, Morgan XC, Iyer VR. Mapping DNA-protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nat Methods. 2005;2:47–53. doi: 10.1038/nmeth726. [DOI] [PubMed] [Google Scholar]
- 87.Peters BA, Velculescu VE. Transcriptome PETs: a genome's best friends. Nat Methods. 2005;2:93–94. doi: 10.1038/nmeth0205-93. [DOI] [PubMed] [Google Scholar]
- 88.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 89.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 90.Liu X, Campbell MR, Pittman GS, Faulkner EC, Watson MA, Bell DA. Expression-based discovery of variation in the human glutathione S-transferase M3 promoter and functional analysis in a glioma cell line using allele-specific chromatin immunoprecipitation. Cancer Res. 2005;65:99–104. [PubMed] [Google Scholar]
- 91.Maynard ND, Chen J, Stuart RK, Fan JB, Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat Methods. 2008;5:307–309. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- 92.Alam J, Cook JL. Reporter genes: application to the study of mammalian gene transcription. Anal Biochem. 1990;188:245–254. doi: 10.1016/0003-2697(90)90601-5. [DOI] [PubMed] [Google Scholar]
- 93.Arnone MI, Dmochowski IJ, Gache C. Using reporter genes to study cis-regulatory elements. Methods Cell Biol. 2004;74:621–652. doi: 10.1016/s0091-679x(04)74025-x. [DOI] [PubMed] [Google Scholar]
- 94.Hollon T, Yoshimura FK. Variation in enzymatic transient gene expression assays. Anal Biochem. 1989;182:411–418. doi: 10.1016/0003-2697(89)90616-7. [DOI] [PubMed] [Google Scholar]
- 95.Carey M, Smale S. Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. New York: CSHL Press; 1999. [Google Scholar]
- 96.Buckland PR. Allele-specific gene expression differences in humans. Hum Mol Genet. 2004;13(Spec No 2):R255–R260. doi: 10.1093/hmg/ddh227. [DOI] [PubMed] [Google Scholar]
- 97.Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 98.Knight JC. Analysis of allele-specific gene expression. Methods Mol Biol. 2006;338:153–165. doi: 10.1385/1-59745-097-9:153. [DOI] [PubMed] [Google Scholar]
- 99.Wang D, Sadée W. Searching for polymorphisms that affect gene expression and mRNA processing: Example ABCB1 (MDR1) AAPS Journal. 2006;8:E515–E520. doi: 10.1208/aapsj080361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jurinke C, Denissenko MF, Oeth P, Ehrich M, van den Boom D, Cantor CR. A single nucleotide polymorphism based approach for the identification and characterization of gene expression modulation using MassARRAY. Mutat Res. 2005;573:83–95. doi: 10.1016/j.mrfmmm.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 101.Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang H, Elbein SC. Detection of allelic imbalance in gene expression using pyrosequencing. Methods Mol Biol. 2007;373:157–176. doi: 10.1385/1-59745-377-3:157. [DOI] [PubMed] [Google Scholar]
- 103.Loeuillet C, Weale M, Deutsch S, Rotger M, Soranzo N, Wyniger J, Lettre G, Dupre Y, Thuillard D, Beckmann JS, Antonarakis SE, Goldstein DB, Telenti A. Promoter polymorphisms and allelic imbalance in ABCB1 expression. Pharmacogenet Genomics. 2007;17:951–959. doi: 10.1097/FPC.0b013e3282eff934. [DOI] [PubMed] [Google Scholar]
- 104.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pastinen T, Sladek R, Gurd S, Sammak A, Ge B, Lepage P, Lavergne K, Villeneuve A, Gaudin T, Brandstrom H, Beck A, Verner A, Kingsley J, Harmsen E, Labuda D, Morgan K, Vohl MC, Naumova AK, Sinnett D, Hudson TJ. A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- 108.Koivisto UM, Palvimo JJ, Janne OA, Kontula K. A single-base substitution in the proximal Sp1 site of the human low density lipoprotein receptor promoter as a cause of heterozygous familial hypercholesterolemia. Proc Natl Acad Sci U S A. 1994;91:10526–10530. doi: 10.1073/pnas.91.22.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, Cheng JF, Rubin EM, Wood WG, Bowden D, Higgs DR. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]