Abstract
Recent studies indicate that lipid droplets isolated from a variety of different cells are rich in proteins known to regulate membrane traffic. Among these proteins are multiple Rab GTPases. Rabs are GTP switches that regulate intracellular membrane traffic through an ability to control membrane-membrane docking as well as vesicle motility. Here we present evidence that the multiple Rabs associated with droplets have a function in regulating membrane traffic. Droplet Rabs are removed by Rab GDP-dissociation inhibitor (RabGDI) in a GDP-dependent reaction, and are recruited to Rab-depleted droplets from cytosol in a GTP-dependent reaction. Rabs also control the recruitment of the early endosome (EE) marker EEA1 from cytosol. We use an in vitro reconstitution assay to show that transferrin receptor positive EEs bind to the droplet in a GTP/Rab-dependent reaction that appears not to lead to membrane fusion. This docking reaction is insensitive to ATPγs but is blocked by ATP. Finally, we show that when GTP bound active or GDP bound inactive Rab5 is targeted to the droplet, the active form recruits EEA1. We conclude that the Rabs associated with droplets may be capable of regulating the transient interaction of specific membrane systems, probably to transport lipids between membrane compartments.
Introduction
Scientists have variously designated sites of lipid accumulation in cells as lipid droplets, lipid bodies, lipid inclusions, oil droplets and oil bodies, which reflects the view that these are simple lipid storage compartments (1, 2). Recent proteomic, lipidomic and genetic evidence suggests, however, that these sites are metabolically active organelles with essential roles in cell signaling, membrane traffic and lipid homeostasis. Nearly all prokaryotic and eukaryotic cells are able to store lipids. Moreover, sites of lipid accumulation are associated with specific membrane systems. In eukaryotic cells, lipid accumulation appears to be associated with endoplasmic reticulum (ER) membranes (3) while in prokaryotic cells special regions of the plasma membrane are involved (2). Therefore, cells appear to have special compartments that contain the molecules necessary to synthesize and degrade specific classes of lipids as well as manage the traffic of these lipids among membrane systems. For this reason, we have proposed the name adiposome to designate a cellular compartment specialized for packaging and distributing various lipids (4). The lipid droplet in this nomenclature is a lipid filled adiposome.
The adiposome hypothesis predicts that in eukaryotic cells portions of the ER are specialized for both accumulating lipids and distributing them among various membrane systems. They may also be involved in distributing specific classes of proteins (5). Although lipid droplets seem an unlikely target for membrane traffic because they lack an intact lipid bilayer, the proteins they contain suggest otherwise. These include: TIP47 (6), a protein involved in mannose-6-phosphate receptor traffic (7); p22, a calcium binding protein implicated in exocytosis (8); PKD2, a kinase that regulates Golgi-plasma membrane traffic (9); the caveolar membrane protein caveolin-1, which may transport newly synthesized cholesterol to caveolae (10) as well as lysosomal cholesterol to the adiposome (11); and multiple small GTPases that regulate membrane traffic (4). While the function of these proteins is unclear, it is possible they are linked to the traffic of essential lipids needed for proper membrane function. For example, cholesterol regulates the traffic of the mannose-6-phosphate receptor between the lysosome and the Golgi apparatus (12) through the activity of NADH sterol dehydrogenase-like protein (NSDHL), which is localized to lipid droplets (6) but can traffic to the Golgi apparatus (13). Sterols are also critical for the traffic of the yeast tryptophan transporter Tat2 to the plasma membrane (14). The availability of stored or newly synthesized fatty acids generated by acetyl Co-A carboxylase, another class of lipid droplet-associated enzymes (4), is linked to the acylation of proteins involved in yeast vacuolar traffic (15) as well as the maintenance of the nuclear envelope (16). Thus, feedback loops appear to exist between the lipids synthesized and stored in droplets and specific membrane systems that carry cargo around in the cell.
An unexpected family of proteins in the adiposome proteome is the Rab GTPases (4). We detected 9 different Rabs by mass spectrometry and four of these were confirmed by immunoblotting. The presence of these Rabs suggests lipid droplets play a central role in membrane traffic. Indeed, overexpression of Rab11 impairs the movement of endosomal cholesterol to sites of cholesterol esterification (17), which appears to take place in droplets (18). In addition, microinjected Rab GDP-dissociation inhibitor (19) blocks mobilization of endosomal cholesterol. Finally, Rab18 appears to regulate the interaction of ER with lipid droplets (20) as well as the level of neutral lipid in the cell (21). Here we present evidence that Rabs regulate the interaction of early endosomes with lipid filled adiposomes.
Materials and Methods
Materials
pAb α-caveolin-1, mAb α-EEA1, mAb α-Rab4, mAb α-Ral B and mAb α-Rab5 were from BD Biosciences (San Jose, CA). pAb α-Rab11 and mAb α-transferrin receptor were from Zymed Laboratories Inc. (San Francisco, CA). mAb α-ADRP was from Research Diagnostics Inc. (Flanders, NJ). pAb α-Rab18, pAb α-Rab5, and DNAse I-protease inhibitor cocktail set III were from Calbiochem (La Jolla, CA). pAb α-Myc from Upstate (Charlottesville, VA). Alexa labeled second antibodies were from Molecular Probes (Eugene, OR). Bovine liver Rab GDP-dissociation inhibitor (lot #090K4116), PMSF and all nucleotides were from Sigma (St. Louis, MO). Calf serum, fetal bovine serum and DMEM were from Hyclone (Logon, UT).
Cell culture
CHO K2 cells were cultured in 150 mm dishes in high glucose DMEM containing 10% calf serum, 40 μg/ml proline, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were either grown until confluent for droplet isolation or to 60% confluent for use in isolating endosomes. HeLa cells were grown overnight in high glucose DMEM with 10% fetal bovine serum and antibiotics were cultured in 12-well plates (seeded at 50,000 cells/well) containing a coverslip and grown over night before transfection.
Lipid droplet purification
Lipid filled adiposomes (designated droplets) were purified as previously described (4) with several modifications designed to increase the purity. Briefly, 10, 150 mm dishes of cells were collected by scraping in ice-cold PBS, re-suspending in buffer A (25 mM Tricine, pH 7.6, 250 mM sucrose), and homogenizing by N2 cavitation (450 psi for 15 min on ice). A post-nuclear supernatant fraction was obtained by centrifugation at 1,000 × g and loaded into a SW41 tube. This sample was centrifuged at 274,000 × g for 1 hr at 4°C. The white band (droplet fraction) at the top of gradient (∼0.5 ml) was collected and resuspended in 6 ml of buffer A in a SW41 tube. The droplet fraction was overlaid with 4 ml buffer B (20 mM HEPES, pH 7.5, 100 mM KCl, 2 mM MgCl2) and centrifuged at 274,000 × g for 1 hr at 4°C. The droplet fraction from this spin (∼0.5 ml) was collected, transferred to an Eppendorf tube and centrifuged at 20,000 × g for 4 min. The clear buffer underlying the white band was removed, the droplets resuspended in 200 μl of buffer B and centrifuged again. This step was repeated a total of 4 times. The droplet fraction was resuspended in 1 ml of buffer B, vortexed to resuspend the droplets, subjected to a second round of washes and finally centrifuged at 265,000 × g in a TLA 100.3 tube for 5 min to remove any contaminating membranes as previously described (4).
Endosome purification
CHO K2 cells at ∼60% confluency were washed twice with ice-cold PBS and detached gently from the dish using a cell scraper. Cells from 6, 100 mm dishes were pooled into one 15 ml tube and spun for 5 min at 500 × g at 4°C. After removing the supernatant, cells were resuspended in 600 μl buffer C (250 mM sucrose, 3 mM imidazole, pH 7.4 containing 1/1,000 dilution of protease inhibitor cocktail set III) and incubated on ice for 20 min. Cells were then broken using a 22.5 gauge needle. Breaking efficiency was determined by phase contrast microscopy. Unbroken cells and nuclei were removed by spinning for 10 min at 1,000 × g at 4°C. The pooled post-nuclear supernatant fraction (PNS) from 30, 100 mm dishes was transferred to the bottom of a centrifuge tube (volume: 38 ml) and adjusted to a sucrose concentration of 40.6% using a stock solution of 62% sucrose, 3 mM imidazole, pH 7.4. The sucrose concentration was determined with a refractometer. The solution was then carefully overlaid with 12 ml of 35% sucrose, 3 mM imidazole, pH 7.4, followed by 8 ml of 25% sucrose, 3 mM imidazole, pH 7.4 and filled to the top with buffer C. Gradients were centrifuged at 108,000 × g in a swinging bucket rotor for 3 h at 4°C. A visible 25-35% interphase enriched for endosomes (early and recycling endosomes) was collected (22, 23). Fractions were used directly or snap-frozen in liquid nitrogen and stored for up to 2 months at -80°C.
Preparation of Rab GDP-dissociation inhibitor
In some experiments we used commercial RabGDI and in others we used recombinant protein purified in the laboratory. For the latter, we used E. coli strain BL21 (DE3) expressing a His-tagged, bovine RabGDI in a pRESET plasmid, which was kindly provided by Dr. Oliver Ullrich, Hamburg, Germany. Bacteria were grown under standard conditions and processed to lyse the bacteria by freeze-thawing in buffer D (50 mM Na2HPO4, pH 8.0, 0.3 M NaCl, 10 mM 2-mercaptoethanol) containing 1 mg/ml lysozyme and 5 μg/ml of DNase I-protease inhibitor cocktail III. The suspension was incubated on ice for 30 min and then sonicated on ice 15 times for 30 sec each at 50 Joule Watt-sec before centrifuging for 1 hr at 60,000 × g at 4°C to remove the cell debris. To purify the His-tagged protein, 45 ml of the supernatant fraction was mixed with 1.5 ml of Ni-NTA beads (Qiagen, Valencia, CA) that had been pre-equilibrated in buffer D containing 10 mM imidazole. The suspension was rotated on a wheel at 4°C for 1 h. The beads were spun down at 1,000 × g for 2 min and washed (resuspend and pellet again) three times with 40 ml of buffer D containing 10 mM imidazole, pH 8.0; three times with 40 ml of buffer D containing 10 mM imidazole, 0.3% Triton X-100, pH 6.0; and two times in 40 ml of buffer D containing 10 mM imidazole, pH 8.0. Beads were loaded onto a 10 ml Poly-Prep chromatography column (Bio-Rad, Richmond, CA), washed with 10 bed volumes buffer D containing 10 mM imidazole, pH 8.0, and finally eluted with 20 ml buffer D containing 200 mM imidazole, pH 8.0. Fractions (1.5 ml) were collected and the fractions with the most protein pooled and dialyzed overnight at 4°C with 3 times buffer change against buffer E (20 mM Hepes, KOH, pH 7.2, 10 mM DTT) in Pierce Dialysis cassettes (Rockford, Il) that had a molecular weight exclusion <10,000, and stored at -80°C. Pooled samples were separated by gel electrophoresis and the purity checked by Coomassie blue staining.
RabGDI treatment of isolated endosomes and droplets
To remove Rabs from purified droplets or endosomes, each was incubated in the presence of 2 mM GDP and 10 μM RabGDI in buffer B at 37°C for 1 hr. At the end of the incubation, the droplets were separated from the incubation mixture by flotation and the endosomes by sedimentation, all at 20,000 × g for 5 min. Both the droplets and the endosomes were washed three times with buffer B and either analyzed directly or used in the various experiments.
Recruitment assay
The basic recruitment assay used droplets, endosomes and cytosol all purified from CHO K2 cells. To measure recruitment of molecules from the cytosol, 50 μl (∼20 μg of protein) of droplets or liposomes were mixed with 100 μl of cytosol (∼80 μg protein) in buffer B containing the indicated nucleotide for 1 hr at 37°C. At the end of the reaction, the droplets or liposomes were separated from the cytosol, washed and processed for immunoblotting to detect the indicated proteins. The same general protocol was used to study membrane recruitment. Droplets (50 μl) were mixed with 50 μl endosomes (100 μg/ml) in buffer B and incubated at 37°C for 1 hr in the presence of the indicated nucleotide and cytosol concentration. In some experiments, the droplets were pre-incubated in the presence of GTPγs. At the end of the incubation, the droplets were separated from the cytosol and endosomes by centrifugation at 10,000 × g. To remove non-specifically bound endosomes, the droplets were subsequently washed three times by repeated resuspension of the droplet fraction in buffer B and centrifuging again at 10,000 × g. The lower g force was necessary to retain endosomes on the droplets during the separation and wash steps.
Electron microscopic immunogold labeling
Isolated droplets suspended in 200 μl of buffer B was mixed with 200 μl of 6% paraformaldehyde in buffer F and incubated for 1 hr at room temperature. The fixed sample was washed by flotation (20,000 × g for 3 min) with buffer B three times, 5 min each at room temperature, resuspended in 50 mM NH4Cl in buffer B for 10 min, and washed again with buffer B two times, 5 min each. The sample was mixed with buffer B containing 0.15% crystallized BSA (buffer F) and incubated for 30 min at room temperature. Buffer F was removed and the sample of droplets mixed with either pAb α-Rab5, pAb α-Rab11, or pAb α-Rab18, diluted 1:25 with buffer F, or 10 μg/ml rabbit IgG in buffer F and incubated further for 15 hrs at 4°C. The droplets were washed by flotation with buffer F and mixed with goat α-rabbit IgG conjugated to 10 nm gold diluted 1:30 in buffer F for 2 hrs at room temperature. Droplets were washed again three times, 5 min at 4°C. Finally, samples were washed with buffer G (0.1 M sodium phosphate, 3 mM KCl and 3 mM MgCl2 pH 7.6), fixed with 1% GTA and post-fixed in 1% OsO4, all in buffer G. Samples were embedding in Epon and viewed with a JEOL 1200 electron microscope.
Targeting Rab5 to droplets
cDNAs were constructed in pcDNA3.1Myc/His that code for Q79L (GTP-bound) or S34N (GDP-bound) Rab5 lacking the C-terminal CAAX box and fused at the N-terminus to the first 73 amino acids of AAM-B (GI 89145417). The N-terminal fusion targets proteins to the droplet (24). HeLa cells cultured on coverslips were transfected with the cDNA for either the Rab5 GTP or Rab5 GDP chimera protein using Fugene 6 according to the manufacturer's protocol, and then cultured for 4-6 hrs before changing to media containing 100 μM oleate and incubating an additional 15 hours to cause lipid accumulation in adiposomes. The cells were then washed, fixed in 3% paraformaldehyde in phosphate buffered saline, permeabilized with 0.1% Triton X-100 and processed for indirect immunofluorescence to localize the indicated protein using pAb α-Myc (1:300) for Rab5 and mAb α-EEA1 (1:200). Cells were viewed using a Zeiss Axioplan 2E fluorescence microscope fitted with a Hamamatsu monochromatic digital camera.
Preparation of liposomes
Liposomes were made using the method of Grüner et al. (25). Briefly, lipids were extracted by the method of Bligh and Dyer (26) from 1/4 of the total membrane pellet obtained during the standard preparation of droplets. The solvent was removed from the lipids using a rotary evaporator followed by 20 minutes under low torr vacuum. Diethyl ether (5 ml) and buffer B (300 μl) were added to the lipid film. The ether was driven off under a stream of nitrogen with bath sonication. The resulting concentrated liposomes were diluted with 1.7 ml of buffer B and drawn through a 22 gauge needle three times.
Immunoblotting
Droplets were processed to detect the indicated protein by precipitating proteins with 1 ml of 100% acetone, dissolving the sample in SDS PAGE sample buffer, separating them by gel electrophoresis (8% gels for EEA1 and 12% for other proteins) and transferring to PVDF membranes for immunoblotting. Immunoblotting was carried out using ECL reagents as previously described (4).
Results
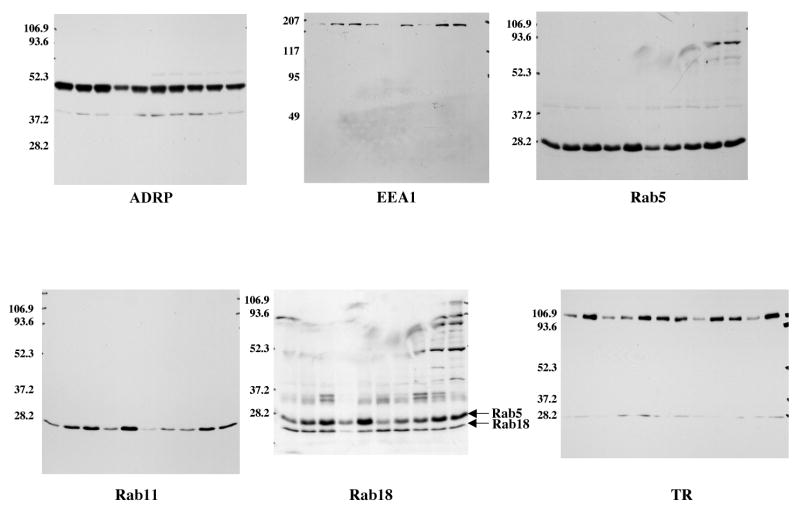
Previously we identified nine different Rab GTPases in the partial proteome of lipid filled adiposomes isolated from CHO cells (4). Other proteomic studies have also found Rabs in droplets isolated from several different cell types (27, 28). We focused our attention on Rab5 and 11 because they have clear roles in regulating membrane traffic and 18 because it has been implicated in droplet interaction with the ER. Commercial antibodies against these three proteins each immunoblot a single band of the correct molecular weight (Rab5, ∼28 kDa; Rab11, ∼28 kDa; Rab18, ∼23 kDa) in fractions of purified droplets (Figure 1B-D, IB). Immunogold labeling with these antibodies show gold particles primarily on the surface of both large and small droplets (arrows, Fig 1B-D, Immunogold). Membrane contaminants were rarely seen in these fractions. Gold labeling, however, is heterogeneous. For example, pAb against Rab5 (B) appear to label the smaller droplets (*) more heavily than the larger ones, which suggests they contain a higher concentration of the protein. No labeling is seen with non-immune antibody (A). We conclude that these three Rabs are associated with the surface of purified droplets.
Fig. 1.
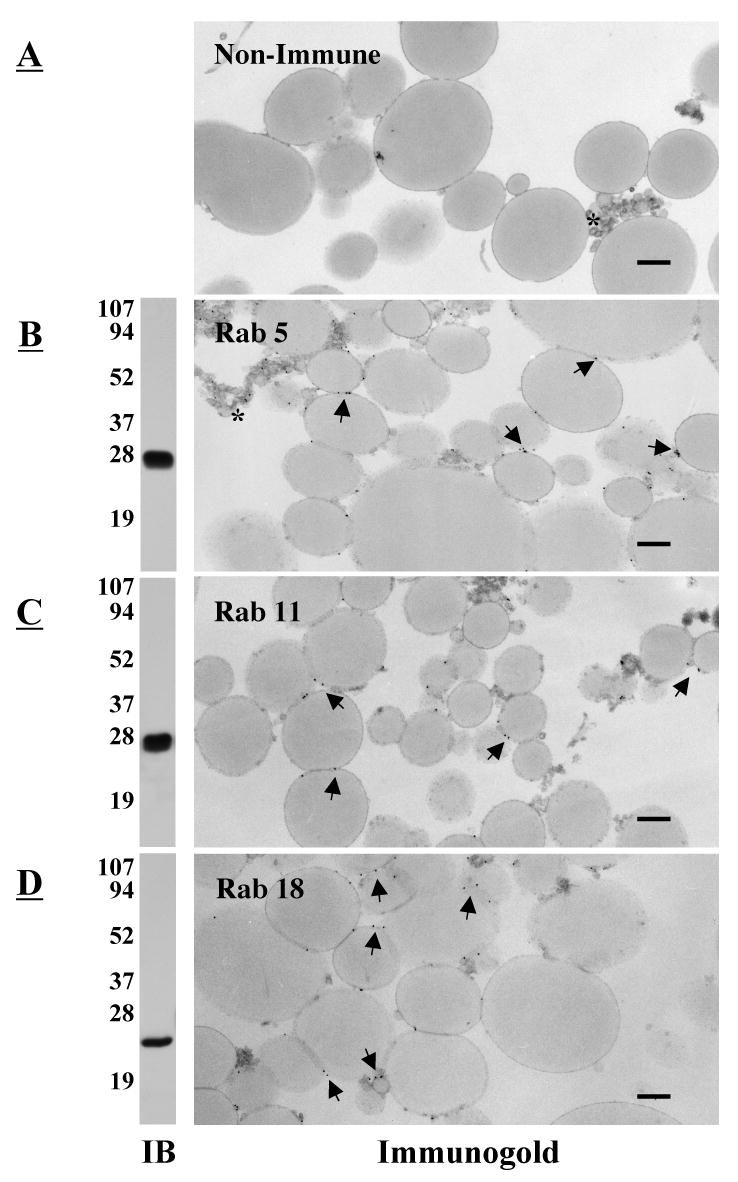
Multiple Rab proteins are on the surface of lipid filled adiposomes. Purified droplets were processed either for immunoblotting (IB) or immunogold localization of the indicated Rab protein (Immunogold). For immunogold, samples were fixed and incubated in the presence of rabbit IgG (A), pAb α-Rab5 (B), pAb α-Rab11 (C), or pAb α-Rab18 (D). The samples were then incubated with the α-rabbit IgG conjugated to 10 nm gold. Gold particles (arrows) are distributed on the surface of the droplet. An asterisk indicates clusters of small droplets. Immunoblotting shows that each antibody recognizes a single band. Bar, 0.5 μm.
Removal and recruitment of Rabs from purified droplets
Rabs are prenylated proteins that dynamically associate with various membrane systems. RabGDI (Rab GDP dissociation inhibitor) is a cytoplasmic protein that releases membrane-associated GDP bound Rabs, by binding the hydrophobic geranylgeranyl group, and effectively solubilizes the protein in a cytosolic complex (29). We tested the ability of purified RabGDI to remove Rabs from purified CHO cell droplets (Figure 2A). We purified droplets, incubated them in the presence (lane 2) or absence (lane 1) of purified RabGDI before separating the droplets from the incubation mixture by flotation and processing each for immunoblotting. After RabGDI treatment, nearly all of the Rab5 and Rab11 (lane 2) were in the incubation solution (lane 4), indicating they had been extracted. Most of the ADRP and all of the caveolin-1, by contrast, remained with the droplets. Loss of ADRP was independent of RabGDI (compare lane 3 with 4). To see if RabGDI preferentially removed GDP bound Rabs (Fig 2B), we looked at the effects of non-exchangeable analogs of GTP and GDP on extraction. RabGDI failed to extract Rab5 from droplets that had been pre-incubated with GTPγS to load the Rabs with non-hydrolyzable GTP (compare lanes 2 and 3). By contrast, Rab5 was efficiently removed if the GTPγS was replaced with GDPβS (lane 4). ATP also did not block RabGDI removal of Rab5 (lane 5). RabGDI did not remove prenylated Ral B under any conditions, indicating that RabGDI specifically works on Rabs. These in vitro results suggest that Rabs may be able to cycle on and off droplets in the cell, and that their interaction with the droplet is not due to a non-specific binding of the prenyl group. Moreover, the ability of Rab GDP-dissociation inhibitor to release these Rabs indicates they are prenylated.
Fig. 2.
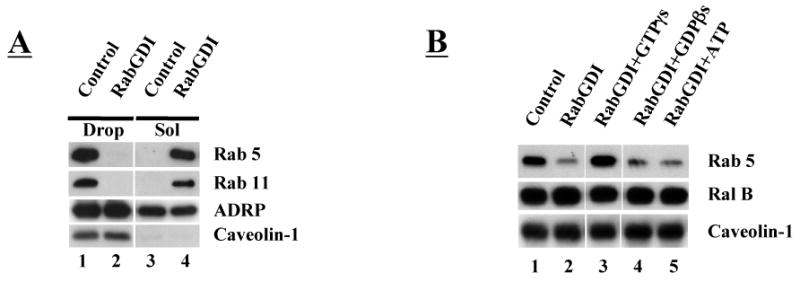
GDP-dependent removal of Rab from droplets by RabGDI. Purified droplets (50 μl, ∼20 μg protein/reaction) were incubated in the presence or absence of 10 μM RabGDI plus 2 mM of the indicated non-exchangeable nucleotide at 37°C for one hour. At the end of the reaction, the droplets were separated from reaction solution by flotation, washed 3 times, the proteins precipitated by acetone and processed for immunoblotting. A) RabGDI transfers Rab5 and 11 from droplets (Drop) to the reaction solution (Sol). The loss of ADRP occurs in buffer alone. Caveolin-1 is not removed. B) RabGDI cannot remove Rab5 when GTPγs is in the reaction mixture. Ral B, another prenylated protein, and caveolin-1 are not removed under any condition.
The soluble Rab/RabGDI complex is recruited to membranes in a multistep process that involves: a) targeting to a specific membrane system by unidentified targeting factors (29); b) insertion of the Rab prenyl group into the membrane; c) dissociation of the RabGDI (30) and d) exchange of GDP for GTP mediated by an exchange factor. Recruitment is dependent on temperature and GTP. Having established that RabGDI removes GDP bound Rabs, we set out to determine if cytosolic Rabs can be recruited back to purified droplets under these physiological conditions (Fig 3). We first looked at the nucleotide requirement (Fig 3A). We purified droplets and processed them to remove Rabs with RabGDI as described above. We then mixed them with cytosol plus or minus GTPγs (lanes 2 and 1, respectively) and incubated for 1 hr at 37°C before separating the lipid droplets from the cytosol by flotation, washing and processing them for immunoblotting to detect Rabs 5, 11 and 18. The presence of GTPγs stimulated the appearance of both Rabs 5 and 11 in the droplet fraction (compare lane 2 with 1). The amount of Rab18 in the fractions remained unchanged under all experimental conditions we tested. If we pretreated the droplet fraction with GTPγs and then incubated with cytosol in the absence of nucleotide, neither Rab5 nor 11 was recruited to the droplet (lane 3). GTPγs had to be present during the incubation (lane 4) for recruitment to occur. We also found that the Rab5 effector EEA1 (early endosome antigen 1) is recruited to isolated lipid droplets and that recruitment is dependent on GTPγs (Fig 3A). Rab recruitment is also temperature dependent (Fig 3B). When Rab-depleted droplets are incubated in the presence of different concentrations of cytosol at 37°C (lanes 4-6), Rab5 and 11 (as well as Rab4) are robustly recruited to droplets. By contrast, very little recruitment occurs at 4°C (lanes 1-3). Recruitment of Rabs 5 and 11 appears to be dependent on the concentration of cytosol in the incubation mixture (Fig 3C). Above ∼1 mg/ml of cytosol, the concentration of these two Rabs in the droplet fraction appeared to plateau, suggesting that there may be a limited number of Rab binding sites on the droplet. In addition to Rab5 and 11, association of EEA1 with the droplet also appears to be dependent on cytosol concentration. ADRP, caveolin-1 and Rab18 were unchanged under all conditions. Finally, liposomes prepared from lipids that we extracted from a CHO cell total membrane pellet could not substitute for the droplets in the Rab recruitment assay (Fig 3D). These results suggest that adiposomes contain the molecular machinery required to recruit Rabs from the cytosol.
Fig. 3.
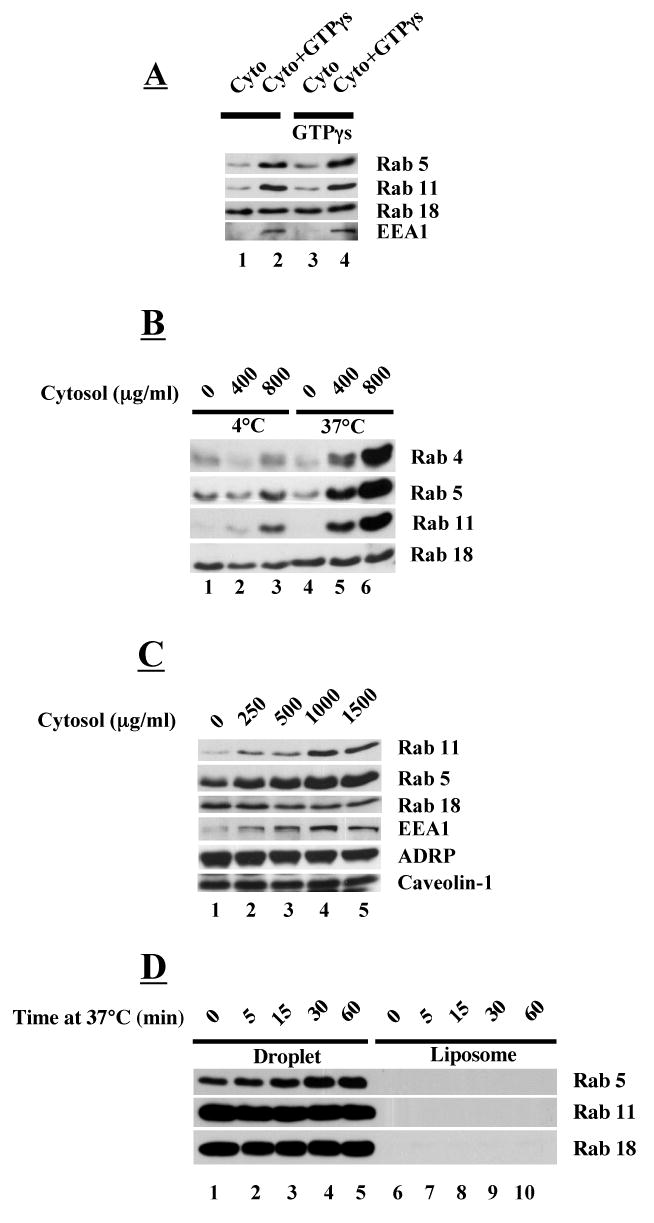
GTP-dependent recruitment of Rabs from cytosol to droplets (A-C) but not to liposomes (D). Rab proteins were released from droplets (A-C) with RabGDI as described in Figure 2. Droplets (50 μl, ∼20 μg protein/reaction) were mixed with either 80 μg of cytosol (A) or varying amounts of cytosol (B-C) in the presence or absence of 2 mM GTPγs. After incubation, droplets were washed three times and the proteins precipitated by acetone and processed for immunoblotting with an antibody against the indicated protein. A) GTPγs pre-bound to droplet is not sufficient to recruit Rabs and EEA1 to droplets. Droplets were either not treated (1,2) or pretreated with GTPγs and mixed with cytosol in the presence or absence of GTPγs at 37°C for 1 hr. B) Rab recruitment is dependent on temperature and cytosol concentration. Incubations were carried out at 4°C and 37°C with either 0, 400 μg/ml or 800 μg/ml of cytosol (volume 100 μl). C) Recruitment of Rabs and EEA1 appears to be saturable. Incubation was carried out at 37°C with the indicated concentration of cytosol (volume 100 μl). D) Rabs are not recruited to liposomes. Liposomes were prepared using lipids extracted from total CHO K2 cell membranes and mixed with 80 μg/ml of cytosol and incubated at 37°C for the indicated time. A companion set of droplets that had not been treated with RabGDI were processed the exact same way.
Rab-dependent interaction of early endosomes (EE) with droplets
The flotation properties of the purified droplets offer the opportunity to explore the possibility that Rabs regulate the interaction of adiposomes with a specific membrane system. We chose early endosomes as the membrane system to study for three reasons; we detected Rab-dependent binding of EEA1 to droplets (Fig 3), there are several membrane markers specific for this compartment and methods are available to partially purify endosomal membranes from tissue culture cells (22). We mixed purified droplets either with buffer alone (Fig 4A lane 1), 5 μg of EE (lane 2), EE plus 2 mM GTPγs (lane 3), EE plus 100 μl of cytosol (lane 4) or EE plus cytosol and 2 mM GTPγs (lane 5) and incubated for 60 min at 37°C. The droplets were re-isolated and washed by flotation and processed for immunoblotting. Isolated droplets alone contained Rab5 and 11 but no EEA1 or transferrin receptor (lane 1). Droplets mixed with EE alone contained a small amount of transferrin receptor. The addition of GTPγs, by contrast, caused a marked increase in the amount of both EEA1 and transferrin receptor. We estimate that under these conditions ∼25% of the transferrin positive membranes in the mixture become associated with the droplets. The addition of cytosol to the mixture had little effect on the amount of EEA1 and transferrin receptor that bound to droplets. In addition, the amount of EE binding to the droplets appeared to be saturable (B), which suggests there are a limited number of binding sites on the droplet. We conclude that EE can interact with the droplet in a GTP-dependent process that does not require cytosolic factors.
Fig. 4.
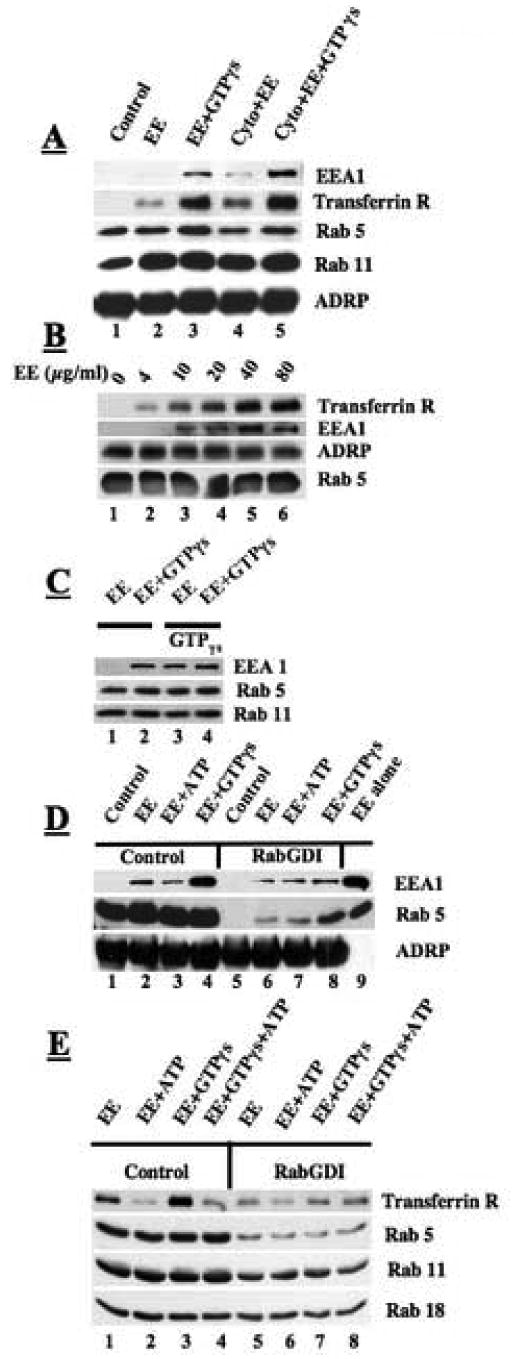
GTP-dependent recruitment of early endosomes (EE) to droplets in vitro. Purified droplets (50 μl, ∼20 μg of protein/reaction) were incubated with EE (50 μl, 5 μg of protein/reaction) at 37°C for 1 hr in the presence or absence of 2 mM GTPγs, cytosol or ATP. At the end of the reaction, free EE was separated from bound EE by flotation of droplets, and the droplet fraction subsequently washed three times with buffer B by repeated centrifugation at 10,000 × g. Samples were processed for immunoblotting as described. A) GTPγs–dependent recruitment of EE to droplets does not require the presence of cytosol. B) EE recruitment to droplets is dependent on concentration of EE (50 μl of droplets, plus 2 mM GTPγs and the indicated amount of EE). All incubations were at 37°C for 1 hr. C) Pretreatment of droplets with GTPγs supports EE recruitment. D) The Rabs on the EE are sufficient to recruit EE to droplets. Droplets were treated with 10 μM RabGDI for 1 hr to remove Rabs. These droplets were then mixed with EE and incubated in the presence of the indicated nucleotide for 1 hr at 37°C. Lane 9 is a sample of EE (1.25 μg). E) Rabs are necessary for recruitment of EE. Both droplets and EE were treated to remove Rabs with RabGDI. These fractions were then mixed in the presence of the indicated nucleotide (2 mM) and incubated for 1 hr at 37°C.
Since the interaction of EE with the droplets does not require the addition of cytosol, we were interested in determining if droplets pretreated with GTPγs will interact with isolated EE in the absence of GTPγs in the incubation mixture (Fig 4C). We pre-incubated one set of isolated droplets with buffer alone (lanes 1, 2) and another with buffer plus GTPγs (lanes 3, 4) at 37°C. We washed the droplets, and mixed each set with isolated EE in the presence or absence of additional GTPγs before re-purifying the droplets and processing them for immunoblotting to detect EEA1. Droplets pre-incubated with GTPγs supported EE binding in the absence of GTPγs in the reaction mixture (compare lane 1 with 3). Moreover, adding the GTPγs to the mixture did not cause further EE recruitment. These results suggest that the GTP bound Rabs on isolated droplet are fully capable of recruiting EE.
To see if Rabs on the EE could substitute for those on the droplet, we removed Rabs from the droplet with RabGDI and then assayed for the recruitment of the EE marker EEA1 (Fig 4D). Untreated droplets recruited some early endosomes in the absence of GTPγs but the amount was markedly increased when GTPγs was present (compare lane 2 with 4). The presence of ATP in the mixture actually caused a decrease in EE recruitment (lane 3). Droplets that lacked detectable Rabs (lane 5) were still able to bind both EEA1 and Rab5 when mixed with isolated early endosomes plus GTPγs, but recruitment was blunted compared to the control (compare lane 8 with lane 4). While Rabs on the EE may be able to control EE interaction with droplets, these interactions work more efficiently when Rabs are on the droplets,. Lane 9 shows that isolated early endosomes contain EEA1 and Rab5.
These experiments do not rule out the possibility that a GTP binding protein other than the Rabs is required for EE recruitment to droplets. Therefore, we prepared droplets and EE and pretreated them both with RabGDI to remove Rabs (Fig 4E). We then used a cytosol-free mixture to compare the ability of Rab depleted droplets and EE to interact (lanes 5-8) relative to untreated samples (lanes 1-4). With the untreated samples, EE binding to droplets increased with the addition of GTPγs (compare lane 1 with 3) but not ATP (lane 2). By contrast, virtually no increase in binding occurred in Rab-depleted samples incubated in the presence of EE and GTPγs. We noted in this and other experiments that ATP caused the dissociation of EE from the droplets (lane 2). Indeed, the addition of ATP and GTPγs together blocked the recruitment of EE to the droplet (lane 4). Non-hydrolyzable ATP would not substitute for ATP (data not shown), indicating that ATP hydrolysis was required for this effect. We also found that ATP stimulated the dissociation of EE that had pre-bound to the droplet (data not shown). Finally, the binding of EE to droplets was not inhibited by N-ethyl-maleimide (NEM) or EDTA (data not shown).
Rab5 will recruit EEA1 to droplets in vivo
The in vitro experiments suggest that Rab GTPases can regulate the interaction between early endosomes and lipid filled adiposomes. To more firmly establish that the in vitro interaction we characterized reflects a potential in vivo activity of the droplets, we carried out experiments to determine if Rab5 can regulate the interaction of early endosomal markers with droplets in the cell. We reasoned from the in vitro experiments that the interactions between droplets and endosomes would be regulated and, therefore, difficult to detect without experimental manipulation. Therefore, to determine if droplets are able to engage endosomal markers in situ, we constructed two cDNAs, one coding for activated Rab5Q79L and the other for inactivate Rab5S34N (Fig 5). The two Rab5 constructs lacked a C-terminal CAAX box but contained an N-terminal 73 amino acid extension that we have established targets and anchors proteins specifically on the droplet (23). HeLa cells were transiently transfected with the cDNA for these proteins and processed for immunofluorescence staining of the Myc-tag (Rab5) and endogenous EEA1. In non-transfected cells, α-EEA1 IgG gave a characteristic punctate staining pattern indicative of endosomal staining (data not shown). The Rab5Q79L and Rab5S34N expressed in cells were primarily located on the periphery of droplets (Fig 5A, D, G). EEA1 in cells expressing 5S34N had a normal punctate staining pattern that was dispersed in the cell and not associated with the droplets (G-I). In Rab5Q79L expressing cells, by contrast, EEA1 had a punctate staining pattern but the puncta were almost exclusively associated with droplets (A-F). We saw very little endosomal EEA1 elsewhere in these cells, which suggests that under these conditions most of the EEA1 in the cells is bound to the Rab5Q79L on the droplet. We conclude that the monolayer surrounding each droplet supports the Rab5-dependent, GTP-regulated interaction of endosomal maker proteins.
Fig 5.
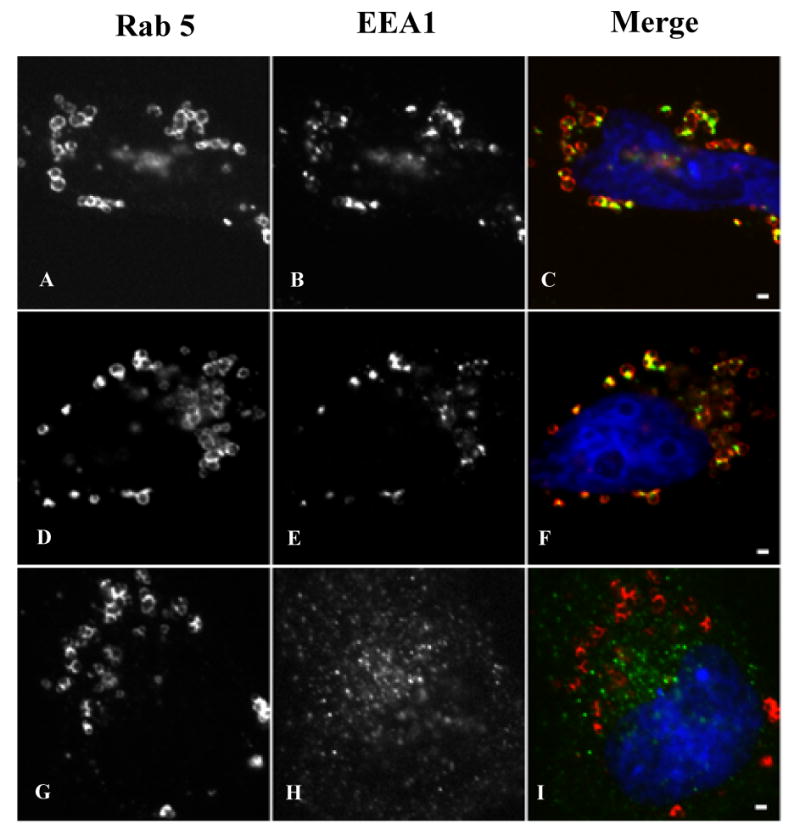
Recruitment of EEA1 to droplets in vivo. We transfected HeLa cells with cDNAs coding for either the Myc-tagged Rab5Q79L (A-F) or Myc-tagged Rab5S34N (G-I) chimeric proteins. The cells were incubated for 15 hours in media containing 100 μM oleic acid to induce lipid droplets and then fixed and processed for indirect immunofluorescence localization of Myc. Both chimeric proteins efficiently targeted droplets (A, D, G). Rab5Q79L (A, D) caused the recruitment of EEA1 (B, E) to distinct sites on the droplet (C, F). EEA1 was largely depleted from other sites within these cells. By contrast, in cells expressing Rab5S34N (G) EEA1 had a normal punctate distribution (H) that was not associated with the droplet (I). The scale bar, 1 μm.
Discussion
Rabs are generally thought to be involved in regulating the movement and interactions of specific membrane systems. For example, Rab5 regulates the traffic and interaction of early endosomes (31) by a mechanism that depends on the presence of Rab5 in this membrane compartment (29). Therefore, the appearance of multiple Rab species in isolated lipid droplets raises important questions about the function of these regulatory proteins in lipid homeostasis. Is the adiposome a promiscuous organelle that normally interacts with multiple membrane systems or do Rabs have novel functions in this compartment that do not fit standard paradigms? The results of these experiments indicate that one function of Rabs on droplets may be to regulate membrane traffic to this organelle.
Rabs on adiposomes are potentially functional
Immunogold EM showed that three Rab species (Rab5, 11, 18) detected by immunoblotting in the droplet fractions are located on the surface of the organelle and not on contaminating membranes. Our first concern was that the phospholipid monolayer surrounding the droplet might non-specifically bind prenylated proteins during the isolation procedure. We carried out multiple tests to rule out this possibility. Rab GDP-dissociation inhibitor (RabGDI) removed multiple Rabs from the droplet, which was inhibited by GTPγs. RabGDI did not remove Ral B, another prenylated protein in the droplets. Unexpectedly, RabGDI also did not remove Rab18, suggesting this Rab is anchored to the droplet by a different mechanism than simply prenylation. In addition to RabGDI removing specific Rab species in a GDP-dependent step, multiple Rabs (but not Rab18) rebound to Rab-depleted droplets incubated in the presence of cytosol plus GTPγs. Binding of each Rab species seemed to be independent of the other, suggesting each Rab species binds to a specific site. Finally, coordinate with the recruitment of Rabs from the cytosol to the droplet, the Rab5 effector EEA1 also appeared on the droplet. These results suggest that the Rabs in purified droplets may be functional within the cell.
To directly test for the functionality of the Rabs in the isolated droplets, we used an in vitro assay. We prepared fractions of droplets and early endosomes and allowed them to interact under defined conditions. The droplets were then re-isolated and assayed for the presence of the EE markers EEA1 and transferrin receptor. Interaction between the two organelles had several unique characteristics; binding of EEs was dependent on 37°C, the presence of Rabs, GTPγs but not cytosol. When we increased the concentration of EE protein in the mixture, there was an increase in EE binding to the droplet that plateaued at ∼40 μg/ml of EE. This suggests there are specific binding sites on the droplet for EE. EE binding to the droplets was not abolished by removing Rabs selectively from droplets, which indicates Rabs may only need to be present on one or the other of the two interacting compartments for interaction to occur. Finally, ATP, but not ATPγs, blocked EE binding and caused the dissociation of pre-bound EE, which is additional evidence that EE interaction with adiposomes involves specific molecular machinery. The effect of ATP may mean a kinase on either the droplet or the early endosome regulates the interaction between the two compartments.
Both the recruitment of EEA1 from the cytosol and the regulated interaction with EE suggests that the Rabs on the surface of the droplet are functional. In support of this conclusion, we found that Rab5-GTP is capable of recruiting EE markers to droplets in living cells. Rather than simply overexpressing dominant inactive and active Rab5 and risk disruption of membrane traffic, we decided to target Rab5 constructs to the droplet and see if any known Rab5-interacting components now bind. Droplets containing Rab5Q79L robustly recruited EEA1, while those coated with Rab5S34N did not. EEA1 staining on the droplet had a punctate appearance, similar in size and morphology to EE in non-transfected cells. Nevertheless, transferrin and transferrin receptor were not detected with the EEA1 on the droplet. This is consistent with our finding that the interaction of transferrin receptor positive EE with the droplets in vitro is tightly regulated by ATP, which may make it difficult to detect transferrin-positive EE traffic to the droplet-associated population of EEA1. Alternatively, the massive relocation of EEA1 to the droplet may have disrupted the membrane traffic of endosomes containing transferrin receptors. Nevertheless, the results of these experiments verify that the phospholipid monolayer surrounding the neutral lipid core of each droplet is able to engage EEA1 in a Rab-GTP dependent reaction. This is significant because Rab-GTP mediated binding of EEA1 to endosomes depends on an interaction between the FYVE motif in EEA1 and PI3P (32), which implies that, like endosomes, droplets are able to recruit the PI3K necessary for PI3P synthesis. Indeed, Yu et al have reported that isolated droplets contain a PI3K (33).
Endosome-adiposome interaction during lipid traffic
The interaction between endosomes and lipid filled adiposomes appears to be different from what occurs during endosome-endosome fusion in vitro (23), but, nevertheless, has the hallmarks of a membrane kiss and run event. Rather than fusion and fission, however, the interaction we detected appears to be transient and involve a regulated docking step without subsequent membrane fusion. Based on what is known about endosome-endosome fusion, docking of EE on adiposomes may use EEA1 as a tethering factor (34). EEA1 mediated endosome-endosome docking is not inhibited by ATPγs (34). We also found that endosome-adiposome docking was not sensitive to ATPγs. During endosome-endosome fusion, the presence of ATP leads to fusion of endosomes in an NSF dependent step (34). By contrast, the presence of ATP caused EE to dissociate from adiposomes and docking was insensitive to NEM (data not shown). EE docking may occur without fusion because droplets are covered by a phospholipid monolayer that is expected to be a poor substrate for membrane-membrane fusion. Nevertheless, we cannot rule out the possibility that under some conditions hemifusion occurs between the adiposome monolayer and the outer leaflet of the endosome or that adiposomes do not contain the molecular machinery required for fusion.
We speculate that each Rab species on the adiposome regulates interaction with a specific membrane system. For example, Rab5 controls interaction with EE and Rab18 with ER (20). Mitochondria (27, 35) and peroxisomes (36) are also known to interact with droplets so there may be Rab species specific for these two compartments too. In support of this speculation, recent proteomic studies have determined that like adiposomes both purified synaptic vesicles (37) and purified secretory membranes (38) contain multiple Rab species. The interaction of adiposomes with different membrane systems may be part of a network of transient inter-compartmental contact sites (TICCS) that are critical for transferring lipids, small molecules and ions without membrane fusion [reviewed in (39)]. There may be both common and specific machinery for bringing together each set of membrane compartments. For example, TICCS may be involved in the interaction of ER subdomains with mitochondria and the transfer of phosphatidylserine (PtdSer) for conversion to phosphatidylethanolamine (PtdEtn) (40). A similar interaction occurs between ER and plasma membrane, which also appears to be important for lipid transfer (41, 42). TICCS may also mediate interactions between adiposomes and peroxisomes (36). Recent studies suggest how TICCS might work (Fig 6). Sterols can move from the cell surface through endosomal membranes to lipid droplets and back by an ATP independent pathway (43). The binding of EE to droplets also does not require ATP, which raises the possibility that a series of TICCS, which can connect together multiple endosomal compartments, allows the passage of sterols from the cell surface to the droplet without expending energy. The physiologic significance of connecting together internal membranes in this way is to allow rapid, bi-directional, regulated movement of cholesterol between HDL (and other lipoproteins) bound to SR-B1 at the cell surface and internal compartments. (44). Identifying the molecular basis of TICCS function is an important future goal that is very approachable using endosome-adiposome interaction as a model system.
Fig 6.
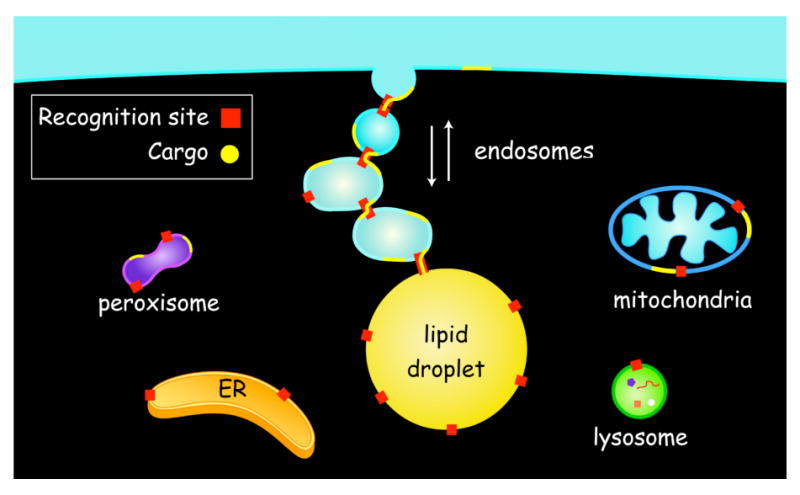
Transient Inter-Compartmental Contact Sites (TICCS) mediate lipid traffic between membrane compartments and lipid filled adiposomes. Here depicted is the movement of a lipid cargo such as membrane sterol (yellow) between the cell surface and lipid droplets using endosomal intermediates. Migration of the sterol is bidirectional and depends on the formation endosome-endosome as well as endosome-droplet TICCS. Recognition sites (red) for each compartment contain the molecular machinery necessary for the formation of TICCS. Compartmental specificity of TICCS formation is regulated by Rab-GTP.
Acknowledgments
We would like to thank Tracy Diaz and Charles Hall for their valuable technical assistance, Brenda Pallares for administrative assistance and Angela Diehl for the artwork. We also would like to thank Dr. Oliver Ullrich for the RabGDI cDNA vector. This work was supported by grants from the National Institutes of Health, HL 20948, GM 52016, the Perot Family Foundation and the Cecil H. Green Distinguished Chair in Cellular and Molecular Biology to RGWA and NIH GM 70117 to JKZ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murphy DJ. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 2.Waltermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stoveken T, von Landenberg P, Steinbuchel A. Mol Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. [DOI] [PubMed] [Google Scholar]
- 3.van Meer G. J Cell Biol. 2001;152:F29–34. doi: 10.1083/jcb.152.5.f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 5.Cermelli S, Guo Y, Gross SP, Welte MA. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 6.Ohashi M, Mizushima N, Kabeya Y, Yoshimori T. J Biol Chem. 2003;278:36819–36829. doi: 10.1074/jbc.M301408200. [DOI] [PubMed] [Google Scholar]
- 7.Diaz E, Pfeffer SR. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- 8.Barroso MR, Bernd KK, DeWitt ND, Chang A, Mills K, Sztul ES. J Biol Chem. 1996;271:10183–10187. doi: 10.1074/jbc.271.17.10183. [DOI] [PubMed] [Google Scholar]
- 9.Yeaman C, Ayala MI, Wright JR, Bard F, Bossard C, Ang A, Maeda Y, Seufferlein T, Mellman I, Nelson WJ, Malhotra V. Nat Cell Biol. 2004;6:106–112. doi: 10.1038/ncb1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smart EJ, Ying Y, Donzell WC, Anderson RG. J Biol Chem. 1996;271:29427–29435. doi: 10.1074/jbc.271.46.29427. [DOI] [PubMed] [Google Scholar]
- 11.Pol A, Martin S, Fernandez MA, Ferguson C, Carozzi A, Luetterforst R, Enrich C, Parton RG. Mol Biol Cell. 2004;15:99–110. doi: 10.1091/mbc.E03-06-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miwako I, Yamamoto A, Kitamura T, Nagayama K, Ohashi M. J Cell Sci. 2001;114:1765–1776. doi: 10.1242/jcs.114.9.1765. [DOI] [PubMed] [Google Scholar]
- 13.Caldas H, Herman GE. Hum Mol Genet. 2003;12:2981–2991. doi: 10.1093/hmg/ddg321. [DOI] [PubMed] [Google Scholar]
- 14.Umebayashi K, Nakano A. J Cell Biol. 2003;161:1117–1131. doi: 10.1083/jcb.200303088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneiter R, Guerra CE, Lampl M, Tatzer V, Zellnig G, Klein HL, Kohlwein SD. Mol Cell Biol. 2000;20:2984–2995. doi: 10.1128/mcb.20.9.2984-2995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtta-Vuori M, Tanhuanpaa K, Mobius W, Somerharju P, Ikonen E. Mol Biol Cell. 2002;13:3107–3122. doi: 10.1091/mbc.E02-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGookey DJ, Anderson RG. J Cell Biol. 1983;97:1156–1168. doi: 10.1083/jcb.97.4.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtta-Vuori M, Maatta J, Ullrich O, Kuismanen E, Ikonen E. Curr Biol. 2000;10:95–98. [PubMed] [Google Scholar]
- 20.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. J Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 21.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. J Biol Chem. 2005;280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 22.Bartz R, Benzing C, Ullrich O. Methods Enzymol. 2005;404:480–490. doi: 10.1016/S0076-6879(05)04042-5. [DOI] [PubMed] [Google Scholar]
- 23.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 24.Zehmer JK, Bartz R, Liu P, Anderson RGW. J Biol Chem. submitted. [Google Scholar]
- 25.Gruner SM, Lenk RP, Janoff AS, Ostro MJ. Biochemistry. 1985;24:2833–2842. doi: 10.1021/bi00333a004. [DOI] [PubMed] [Google Scholar]
- 26.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 27.Brasaemle DL, Dolios G, Shapiro L, Wang R. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 28.Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. J Biol Chem. 2004;279:23699–23709. doi: 10.1074/jbc.M310546200. [DOI] [PubMed] [Google Scholar]
- 29.Seabra MC, Wasmeier C. Curr Opin Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Pfeffer S, Aivazian D. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 31.Zerial M, McBride H. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 32.Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Cassara J, Weller PF. Blood. 2000;95:1078–1085. [PubMed] [Google Scholar]
- 34.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 35.Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. J Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- 36.Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J, Wieland F, Jahn R. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 39.Levine T, Loewen C. Curr Opin Cell Biol. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Voelker DR. Biochem Soc Trans. 2005;33:1141–1145. doi: 10.1042/BST20051141. [DOI] [PubMed] [Google Scholar]
- 41.Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Zellnig G, Perktold A, Daum G. Eur J Biochem. 2001;268:2351–2361. doi: 10.1046/j.1432-1327.2001.02116.x. [DOI] [PubMed] [Google Scholar]
- 42.Schnabl M, Daum G, Pichler H. Biochim Biophys Acta. 2005;1687:130–140. doi: 10.1016/j.bbalip.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Wustner D, Mondal M, Tabas I, Maxfield FR. Traffic. 2005;6:396–412. doi: 10.1111/j.1600-0854.2005.00285.x. [DOI] [PubMed] [Google Scholar]
- 44.Nieland TJ, Penman M, Dori L, Krieger M, Kirchhausen T. Proc Natl Acad Sci U S A. 2002;99:15422–15427. doi: 10.1073/pnas.222421399. [DOI] [PMC free article] [PubMed] [Google Scholar]