Abstract
The CG3252 gene product, DmNAT1, represents the first Nutrient Amino acid Transporter cloned from Drosophila. It absorbs a broader set of neutral amino acids versus earlier characterized insect NATs and mammalian NATs-B0 system transporters from the Sodium Neurotransmitter symporter Family (SNF, a.k.a. solute carrier family 6, SLC6). In addition to B0-specific L-substrates, DmNAT1 equally or more effectively transports D-amino acids with sub millimolar affinities and 1:1 sodium:amino acid transport stoichiometry. DmNAT1 is strongly transcribed in the absorptive and secretory regions of the larval alimentary canal and larval brain, revealing its roles in the primary absorption and redistribution of large neutral L-amino acids as well as corresponding D-isomers. The absorption of D-amino acids via DmNAT1 may benefit the acquisition of fermented and symbiotic products, and may support the unique capacity of fruit fly larvae to utilize a diet with substitution of essential amino acids by D-isomers. It also suggests a remarkable adaptive plasticity of NAT-SLC6 mechanisms via alterations of a few identifiable sites in the substrate-binding pocket. The strong transcription in the brain suggests roles for DmNAT1 in neuronal nutrition and clearance of L-neutral amino acids from the fly brain. In addition, neuronal DmNAT1 may absorb synaptic D-serine and modulate NMDA receptor-coupled signal transduction. The characterization of the first invertebrate B0-like transporter extends the biological roles of the SLC6 family, revealing adaptations for the absorption of D-isomers of the essential amino acids. These findings suggest that some members of the NAT-SLC6 subfamily evolving specific properties which contribute to nutrient symbiotic relationships and neuronal functions.
Keywords: insect NAT, SLC6, symbiosis, NMDA receptor, essential amino acids, D-serine, B0 system, aromatic substrate, synapse
Introduction
Neutral amino acids, which largely represent essential substrates in animals, are absorbed via a B0 transport system (the Broad neutral amino acid spectrum system (Stevens et al., 1982)). This system was initially characterized as a high-capacity sodium-dependent transport in isolated brush border membrane vesicles and cells of mammals and insects (reviewed in (Boudko et al., 2005a; Castagna et al., 1997; Neal, 1996; Palacin et al., 1998)). Recent cloning and heterologous expression of several mammalian transporters unraveled the molecular identity of the B0 system (Bauer et al., 2005; Bohmer et al., 2005; Broer et al., 2006; Kowalczuk et al., 2005; Takanaga et al., 2005a; Takanaga et al., 2005b). These transporters form a specific phylogenetically segregated group of the Sodium Neurotransmitter symporter Family, (SNF, a.k.a. SLC6 solute carrier family 6, by the HUGO nomenclature) in mammals. B0 system transporters were linked to metabolic disorders in humans (Broer et al., 2004; Broer et al., 2005; Seow et al., 2004); however, little is known about similar transport mechanisms in the rest of the Animal Kingdom. Molecular and phylogenetic analysis of insect SLC6 members revealed a Nutrient Amino acid Transporters (NATs) cluster that is phylogenetically close to mammalian B0 transporters and similarly includes transporters for neutral amino acids (Boudko et al., 2005a). It was proposed that the phylogenetic neighbors B0 and insect NATs may be functional counterparts, which together with other lineage-specific NAT groups form the largest subfamily of the SLC6 with putative synergistic functions in the absorption of essential amino acids (Boudko et al., 2005a; Boudko et al., 2005b; Boudko et al., 2005c; Meleshkevitch et al., 2006). However, the insect NATs characterized thus far absorb only a portion of classical B0 substrates. In addition, they act with 2 Na+ per 1 amino acid symport stoichiometry versus the 1:1 stoichiometry of known mammalian B0 transporters. The partitioning of the B0 substrate spectra among characterized insect NATs and obvious lineage-specific duplications of NAT members in insect genomes suggests that B0 system substrates can be distributed between multiple NATs, which together deploy an active core of the transport network absorbing essential amino acids in organisms.
NATs, including mammalian B0 and insect NATs, were identified in the absorptive epithelia of the alimentary canal, suggesting that epithelial NATs mediate the primary absorption of nutrient amino acids. NATs were also localized in the mammalian brain (Broer et al., 2005; Takanaga et al., 2005a) and specific neuronal populations of the insect brain and sensory afferents (Boudko et al., 2005a; Meleshkevitch et al., 2006), suggesting roles for the neuronal NATs as substrate providers in the synthesis of major non-aromatic neurotransmitters e.g. GABA and glutamate (Broer et al., 2005; Takanaga et al., 2005a) and aromatic neurotransmitters e.g. catechol- and indole- amines (Boudko et al., 2005a; Meleshkevitch et al., 2006). So far, a small fraction of identified NATs have been cloned and characterized. This includes the mammalian SLC6A19 (Bohmer et al., 2005), A20 (Takanaga et al., 2005b, Kowalczuk et al., 2005), A15 (Broer et al., 2005; Takanaga et al., 2005a) (a.k.a. B01, IMINO and B02, respectively, listed in chronological order); Manduca sexta msKAAT1 (Castagna et al., 1998) and msCAATCH1 (Feldman et al., 2000); yellow fever mosquito, Aedes aegypti, aeAAT1 (Boudko et al., 2005a); and malaria mosquito, An. gambiae, AgNAT8 (Meleshkevitch et al., 2006) and AgNAT6 (Boudko et al., 2005b). Future identification of new NATs with the rational expansion of experimental model organisms promises major breakthroughs in understanding the evolution, integration and disorders of essential amino acid absorption and metabolism. Based on genomic information (Adams et al., 2000) we have cloned six NATs from Drosophila melanogaster. Here we report the functional characteristics and spatial transcription of the CG3252 gene product, the first functionally expressed NAT-SLC6 member from the fruit fly. Unexpectedly, we identified an unusual broad substrate spectrum transporter, which is expressed in the gut and brain, and displays an extraordinary capacity to absorb the D-isomers of essential amino acids. This discovery substantially extends the physiological, adaptive and evolutionary significance of the SLC6 family members. It generates a concept that in addition to nutrient amino acid absorption and redistribution of essential amino acids, NATs may have evolved properties supporting symbiotic relationships and neuronal functions in metazoan organisms.
Materials and Methods
Experimental organism
Drosophila melanogaster w118 strain was grown at ambient temperature and light conditions on an instant fly food/fresh yeast medium (Carolina Biological Supply Co., Burlington, NC, USA).
Bioinformatics
Sequence analysis and assembly were performed using SeqMan II (DNASTAR, Madison, WI, USA). Topology was predicted using TopPred II (Claros and von_Heijne, 1994) at bioweb.pasteur.fr. Putative glycosylation and phosphorylation sites were identified using cbs.dtu.dk/services/NetNGlyc and cbs.dtu.dk/services/NetPhosservices. Protein sequences of Drosophila SLC6 were retrieved by NCBI BLASTp, aligned with ClustalX (Thompson et al., 1997) and analyzed with MrBayes (Huelsenbeck and Ronquist, 2001) by inferring a mixture of models with a uniform gamma shape substitution for 106 generations. A consensus tree was visualized with TreeView (Page, 1996).
Molecular cloning
Forward AAAATGGAATTGAAAGGAGTTCA and reverse TTGTTTACTTGTTGCTGCCACAGA primers were used. The open reading frame of dmNAT1 was amplified from a Drosophila cDNA collection and cloned into pCR4-TOPO (Invitrogen, Carlsbad, CA, USA). The DmNAT1 ORF was re-amplified from a pCR4-TOPO clone using primers with EcoRI and HindIII restriction sites added. EcoRI/HindIII-digested PCR products were subcloned into the similarly-digested expression vector pXOON (Jespersen et al., 2002).
Heterologous expression
cRNA for oocyte injections was obtained by transcription of XbaI-linearized dmNAT1/pXOON using an mMessage mMachine® kit (Ambion, Austin, TX, USA). Surgically-isolated and collagenase-treated stage V-VI Xenopus oocytes were injected with ∼ 40 ng of dmNAT1 cRNA and incubated for 3-4 days at 16° C in sterile oocyte medium (Boudko et al., 2005a).
Electrophysiological characterization
Experimental oocytes were recorded in a 50 μl constant-flow chamber. OC-725C Oocyte Clamp amplifier (Warner Instruments, Hamden, CT, USA) and 2M KCl-loaded glass microelectrodes with 0.5-1 MΩ resistance were used for recordings. Current/Voltage signals were acquired and analyzed using DigiData 1322A ADC and pCLAMP software (Molecular Devices, Sunnyvale, CA, USA). SigmaPlot 9.0 software (Systat Software, Inc., San Jose, CA, USA) was used for statistical analysis. The composition of solutions used for ion substitution assays has been described earlier (Boudko et al., 2005a). Kinetic profiles and constants were derived by curve fitting of normalized data sets with a three parameter sigmoidal Hill function: y = a*xb/(cb + xb) upon 200 iterative steps, where: a = max(y) = derived normalized mean maximum current; b = η = order of the transport process; c= K0.5, (x, y), substrate concentration at half maximal current.
Isotope uptake assay
Groups of 5-6 cRNA- or water- injected oocytes were incubated in serum-free oocyte medium (Boudko et al., 2005a) for 30 min at room temperature. Competition experiments were then performed, in which uptake of 100 μM L- [methyl - 3H] Methionine (specific activity 3.11*109 Bq Mol l-1; Amersham Biosciences, Piscataway, NJ, USA) was challenged with 1 mM unlabeled amino acid (Sigma-Aldrich, St. Louis, MO, USA). Uptake solution was removed by washing four times with an equimolar ice-cold choline chloride saline. Individual oocytes were transferred to scintillation tubes containing 2% SDS (w/v) to lyse the membranes. Tubes were then filled with 5 mL of scintillation fluid and the radioactivity was determined by liquid scintillation counting in a Beckman-Coulter LS 6500 scintillation counter (Beckman, Fullerton, CA, USA).
Whole-mount in situ hybridization
A purified dmNAT1/pCR4-TOPO plasmid was linearized with NotI or SpeI restriction enzymes to obtain full length run-off transcripts using T3 and T7 promoters for anti-sense and sense probes respectively. Partial probes which included the less-conserved one third 3′ part of the dmNAT1 open reading frame (ORF) were also generated and used in several experiments. DIG-labeled probes were transcribed in vitro using a DIG RNA labeling kit (Roche, Mannheim, Germany). Third instar Drosophila melanogaster larvae were immobilized in ice-cold 0.1 M phosphate buffered saline (PBS), pulled apart by pinching the neck and posterior end, and fixed in 4% paraformaldehyde/PBS overnight. Tissues were treated and hybridized with DIG-labeled probes using a previously described protocol (Meleshkevitch et al., 2006). Labeled preparations were photographed using an inverted DIAPHOT 300 microscope (Nikon Inc., Melville, NY, USA) equipped with Hoffman contrast optics and a Fuji 2S Pro digital SLR camera. Images were assembled using the Corel Draw X3 package (Corel Co., Ottawa, Ontario).
Results
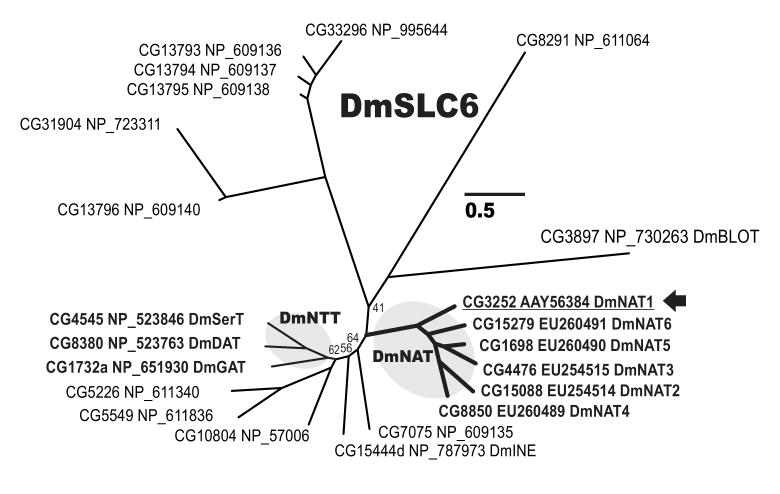
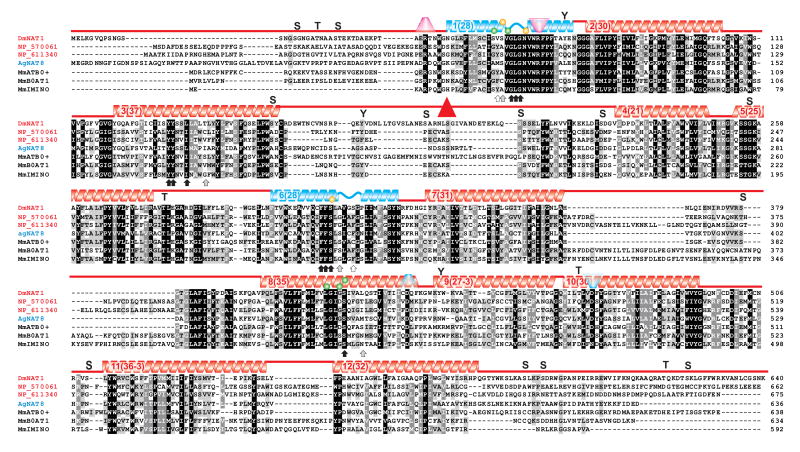
We amplified the dmNAT1 transcript from D. melanogaster third instar larvae cDNA (NCBI accession number DQ073009, protein ID AAY56384). The dmNAT1 transcript is a splice product of the D. melanogaster FlyBase gene CG3252, which is located on the negative strand of chromosome X. The dmNAT1 cDNA contains a 1926 bp open reading frame that gives rise to a protein of 641 amino acid residues with an estimated molecular mass of 71,727 Daltons. The DmNAT1 protein belongs to the insect-specific nutrient amino acid transporters (NATs) which represent the largest subfamily in the solute carrier family 6 (SLC6), comprising Na+(K+)- and Cl- - dependent symporters for amino acids (Fig. 1). Hydropathy analysis revealed 12 transmembrane domains with cytosolic amino and carboxyl termini, which is in agreement with the general structure of transporters in the SLC6 family (Boudko et al., 2005c) (Fig. 2). There was one non-synonymous SNP between the coding sequence of dmNAT1 obtained by us and the predicted coding sequence of NM_131991 in the NCBI database, resulting in an amino acid difference at position 636 (F/L). In addition, 7 synonymous SNPs were identified, in which the sequence of the predicted transcript differed from 3 cloned dmNAT1 transcripts. A high-probability N-glycosylation site was identified at amino acid positions 190-193 (NLSG), which resides in the extracellular loop between the 3rd and 4th transmembrane domains (Fig. 2, red triangle). Based on the sequence alignment with selected characterized NATs and a crystallized bacterial NAT (AaLeuT, sequence not included) it was possible to identify substrate-binding moieties of DmNAT1 (Fig. 2 arrows and spheres, for organic and sodium substrates respectively). Moreover, by screening for unique substitutions in the set of substrate binding residues it was possible to predict the most important residues coordinating D-amino acids in the substrate binding pocket of DmNAT1 (Fig 2. empty and grey arrows).
Figure 1. Evolutionary relationships of fruit fly SLC6 members.
The Bayesian tree of 22 Drosophila SLC6 members. Leaves are Drosophila gene names (CG) followed by predicted (NP) or cloned protein (AAY or EU) accession numbers. The arbitrary nomenclature is shown for NATs and certain neurotransmitter transporters. Posterior probabilities equal to 100% are omitted and < 100% are reported near specific nodes. The bar scale reflects evolutionary distance in substitutions per site.
Figure 2. Alignment of DmNAT1 with selected insect and mammalian SLC6 members.
The amino acid sequence of DmNAT1 (AAY56384) is shown aligned with those of NP_570061 (product of CG10804-PB, phylogenetically proximal to mammalian B0,+), NP_611340 (product of CG5226-PA, phylogenetically close to mammalian B0), An. gambiae AgNAT8 (AAN40409, selected as example of a narrow substrate spectrum transporter), MmATB0,+ (Mus musculus B0,+ transporter AAD49320, a.k.a. SLC6A14, example of D-amino acid absorbing mammalian transporter), MmB0AT1 (Mus musculus B0 transporter Q9D687 a.k.a. SLC6A19, example of mammalian NAT with DmNAT1 similar spectrum, and stoichiometry), and MmIMINO (NP_631881 a.k.a. SLC6A20, example of mammalian NAT with different spectrum). Transmembrane domains (TMDs) are depicted by color gradient helices above the sequences with the TMD # and its length in parentheses. Blue helices represent key TMDs for substrate coordination and translocation. Upright and upside down trapezoids represent putative cationic gates at extra- and intracellular interfaces, respectively. Arrows indicate predicted substrate binding sites (black, conserved; gray, phenotype-specific; and white, presumably the most important for D-amino acid coordination in the substrate binding pocket); Yellow and green spheres indicate sites that may interact with the first and second sodium ion, respectively, in other SLC6 members. A putative N-glycosylation site is indicated by a red triangle. Letters S, T, and Y show sites with high probabilities for serine, threonine, and tyrosine phosphorylation, respectively.
In situ hybridization with antisense probes revealed strong expression of dmNAT1 in the salivary glands, brain, imaginal discs, and posterior midgut (Fig. 3). Similar results were obtained by using a full-length or partial ORF probes. Negative controls with RFP antisense probes produced no detectible hybridization signal (Fig. 3 L). Negative controls with sense dmNAT1 probes generally were not labeled (Fig. 3K); however some weak signals, partially mimicking an antisense labeling pattern, were detected in about 20% of control preparations (this may correspond to transient expression of regulatory antisense transcripts, data not shown).
Figure 3. In situ hybridization of dmNAT1 in Drosophila tissues.
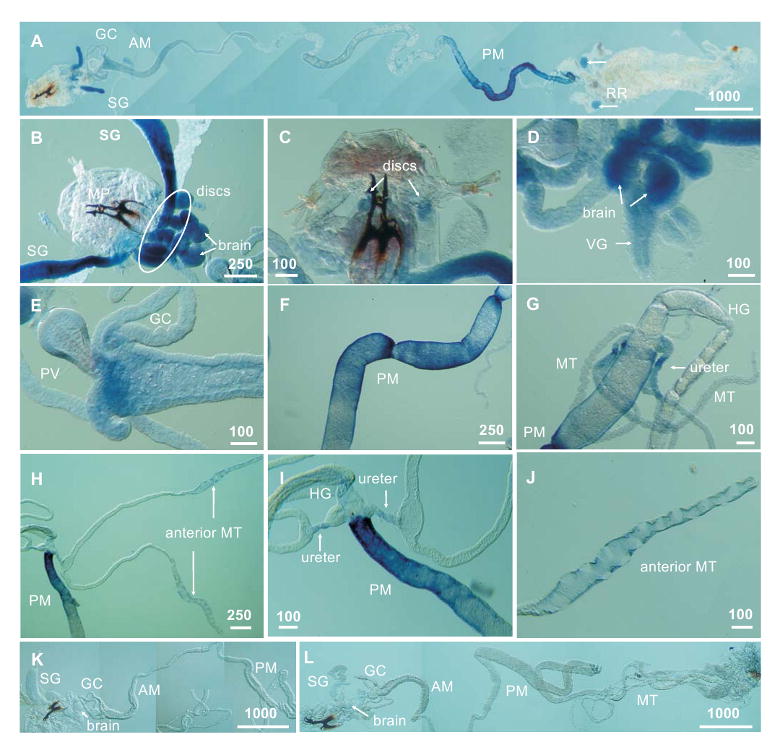
A panoramic reconstruction of dmNAT1 transcription patterns in the larval alimentary canal (A). Magnified images of the salivary glands, brain, and imaginal discs of the thorax (B). Weak hybridization signals in the imaginal discs of the head (C). Labeling in the brain hemispheres together with the ventral ganglion (D); labeling in the anterior midgut (E); in the posterior midgut (F); Strong labeling in the posterior midgut (H and I) vs. relatively weak labeling in the common ureters (G, H and I) and distal part of the anterior pair of Malpighian tubules (H and J). Negative control preparation showing labeling with sense DmNAT1 and antisense Red Fluorescent Protein (RFP) probes (K and L respectively). Tissue and organ abbreviations are: AM, anterior midgut; GC, gastric caeca; MP, mouth parts; MT, Malpighian tubules; RR, reproductive rudiments; SG, salivary gland; VG, ventral ganglion; PV, proventriculus; PM, posterior midgut; HG, hindgut. Scale bars are shown in μm.
The open whole-mount larval preparations exhibited a detailed pattern of spatial expression of dmNAT1 transcripts. Specific but weak labeling was also observed in the anterior midgut just proximal to the gastric caeca, (Fig 3E) reproductive rudiments (Fig 3A), common ureters of the Malpighian tubules (Fig. 3G, H, I), and distal swollen portion of the anterior pair of Malpighian tubules (Fig. 3H and J). Our results are consistent with data from the late embryonic in situ hybridization of CG3252 in the anterior and posterior parts of the embryonic alimentary canal, which has been shown on the web page of the Berkeley Drosophila Genome Project (BDGP; http://www.fruitfly.org/…/insitu26794.jpe, (Tomancak et al., 2002)). The BDGP also detected CG3252 expression in the late embryonic hindgut; however, the larval hindgut did not demonstrate detectable dmNAT1 expression (Fig. 3G, I). In addition, earlier embryonic stages 1-12 showed no labeling reflective of transient expression of CG3252 upon fly development. A comparative study of SLC6 members in adult D. melanogaster detected CG3252 transcripts in tissue sections of the cardia (proventriculus) of the adult alimentary canal (Thimgan et al., 2006). Labeling extended from the long columnar cells of the cardia into the posterior ventricular cells of the midgut, consistent with our observations of staining in the most anterior portion of the larval midgut (Fig. 3E).
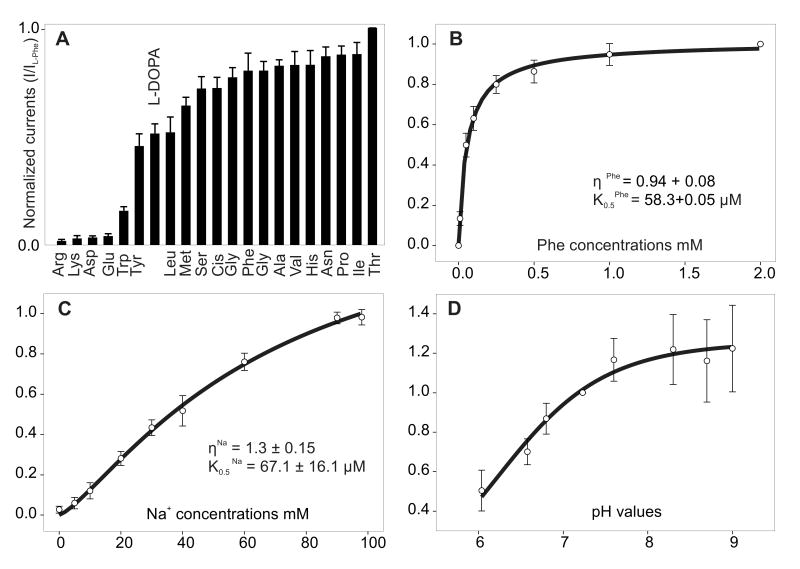
Steady-state heterologous expression of DmNAT1 in Xenopus laevis oocytes was confirmed by both radiolabeled substrate accumulation and electrophysiological measurements of amino acid-induced currents (Figs. 4-7). All neutral amino acids induced inward currents in DmNAT1-expressing oocytes (Fig. 4A). Histidine and the phenylalanine metabolite L-DOPA also induced significant currents. Anionic and cationic amino acids induced small or no currents similar to that in the water-injected control oocytes. The aromatic amino acid tryptophan elicited small but significant currents (Fig. 4A). Neurotransmitters (serotonin, acetylcholine, octopamine, histamine, GABA, and taurine) did not elicit detectable currents (data not shown).
Figure 4. Substrate spectrum and kinetic properties of DmNAT1.
A. L-substrate evoked a current profile of DmNAT1. 1 mM final substrate concentrations were applied at a -50 mV holding potential. Maximum induced current values were normalized to the current evoked by L-threonine. Data are means ± SEM; n ≥ 3 recordings for at least three different oocytes for each data point. B. Phenylalanine-evoked currents as a function of substrate concentration. C. Phenylalanine-evoked currents as a function of cotransported ion concentration. Substrate saturation of kinetic constants determined upon application of different concentrations of Phe and Na+ (substitution with NMDG) at the -50 mV holding potential are shown. D. pH-dependent kinetics of phenylalanine-induced currents. Data are currents measured at increasing values of pH ranging from 6.0 to 8.7 and are means ± SEM for at least 3 oocytes per experiment per data point. Data from different oocytes were normalized in each group relative to values at pH 7.23.
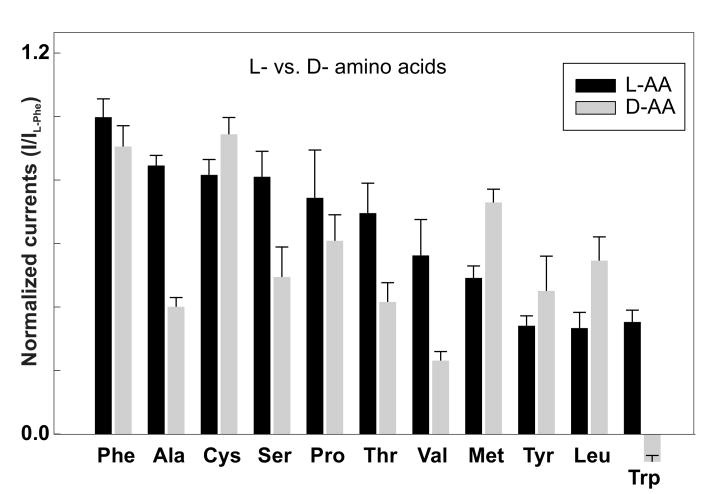
Figure 7. Stereoselectivity of DmNAT1.
The ability of selected D-amino acids to evoke inward currents via DmNAT1 was compared to their L-enantiomers. Amino acid-induced currents (1 mM) at -50 mV were normalized to the current evoked by L-phenylalanine. Data are means ± SEM; n ≥ 3 recordings for at least three different oocytes for each data point.
Substrate-evoked currents were saturable and followed Michaelis-Menten type kinetics (e.g. Phe in Fig. 4B and other substrates, data not shown). The K0.5 values measured for the amino acids and their derivatives that generated inward sodium currents were between 40 μM and 300 μM, with L-enantiomers of methionine and phenylalanine exhibiting the lowest values (Table 1). The Hill coefficients for organic substrates were approximately 1 at a 96 mM concentration of sodium ion. The Hill coefficient for Na+ tested at 1 mM concentrations of transporter-specific organic substrates also was approximately 1 (e.g. Phe, Fig. 4C) suggesting 1:1 stoichiometry of the transport mechanism at the specified conditions. DmNAT1 transport activity was pH-dependent; for example, phenylalanine-induced currents were maximal at around pH 7.6-8.7, but were notably reduced at acidic pH (Fig. 4D).
Table 1.
Apparent kinetic constants for substrate-induced currents in DmNAT1-expressing oocytes
| Substrate | K0.5L-form (μM) ± SEM | K0.5D-form (μM) ± SEM | K0.5L/ K0.5D |
|---|---|---|---|
| Methionine | 39.8 ± 12.7 | 24.7 ± 9.9 | 1.6 |
| Phenylalanine | 58.3 ± 5.3 | 55.2 ± 10.3 | 1.1 |
| Valine | 96.8 ± 20.2 | 170.2± 38.3 | 0.6 |
| Tyrosine | 96.8 ± 17.9 | 64.3 ± 15.3 | 1.5 |
| Serine | 119.7 ± 18.6 | 128.1 ± 21.3 | 0.9 |
| Leucine | 126.2 ± 39.8 | 92.2 ± 34.8 | 1.4 |
| Proline | 127.6 ± 44.9 | 142.4 ± 38.5 | 0.9 |
| Isoleucine | 130.1 ± 37.5 | NA | |
| Histidine | 188.5 ± 37.9 | NA | |
| Cysteine | 199.1 ± 18.8 | 141.0 ± 28.6 | 1.4 |
| DOPA | 242.3 ± 57.9 | NA | |
| Alanine | 255.6 ± 76.8 | NA | |
| Asparagine | 264.2 ± 85.1 | NA | |
| Glycine | 278.6 ± 71.1 | NA | |
| Glutamine | 289.5 ± 85.3 | NA | |
| Threonine | 297.4 ± 140.3 | NA |
Estimated Hill coefficients (η) for all substrates were ≈ 1; n > 3 for all values; NA, analysis was not performed.
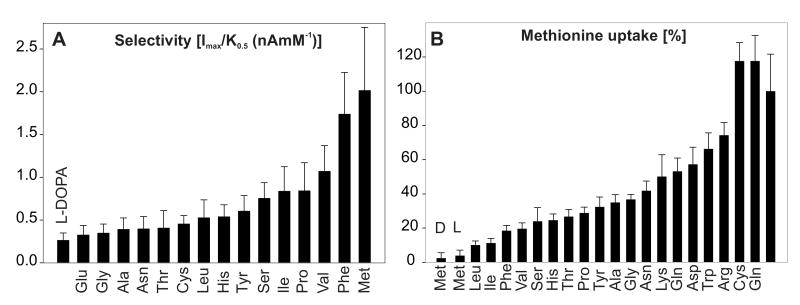
The order of apparent K0.5 values for DmNAT1 substrates (Table I) did not strictly correlate with the observed transport velocities (Imax). We employed the ratio of Imax/K0.5 as an additional index of substrate selectivity, or transport efficiency (Fig. 5A). This analysis confirmed that methionine and phenylalanine are the preferred substrates for DmNAT1, followed by other neutral amino acids. DmNAT1-mediated active transport was confirmed by the uptake of isotope-labeled phenylalanine and methionine, which represent DmNAT1-preferred substrates (Fig. 5B and data not shown). The substrate specificity of DmNAT1 was also tested by challenging the uptake of 100 μM [3H] methionine with a 10- fold excess of unlabeled L-amino acids (Fig. 5B). Methionine uptake was inhibited by all neutral amino acids. In addition, 4 out of 5 amino acids that induced small or insignificant inward currents (Arg, Lys, Asp, Trp) in DmNAT1-expressing oocytes were able to inhibit methionine uptake to some extent, between 25% and 50%. However, we should consider that some amino acids may bind to DmNAT1 and trigger an ion channel state without being transported. Surprisingly, cysteine did not block the transport of methionine, although it appears to be a substrate of DmNAT1, according to inward currents induced by that amino acid. In summary, it appears that neutral amino acids are the preferred substrates for DmNAT1.
Figure 5. Substrate selectivity.
A. Profile of Imax/K0.5 as a relative index of substrate selectivity. B. Inhibition of 100 μM L- [methyl - 3H] methionine uptake by the 20 standard L-amino acids (1 mM) and D- methionine in oocytes expressing DmNAT1, measured over 10 min in 100 mM NaCl medium (pH 7.2). Data are means ± SEM for 5-6 oocytes per group. The transport activity of control oocytes was subtracted in each case.
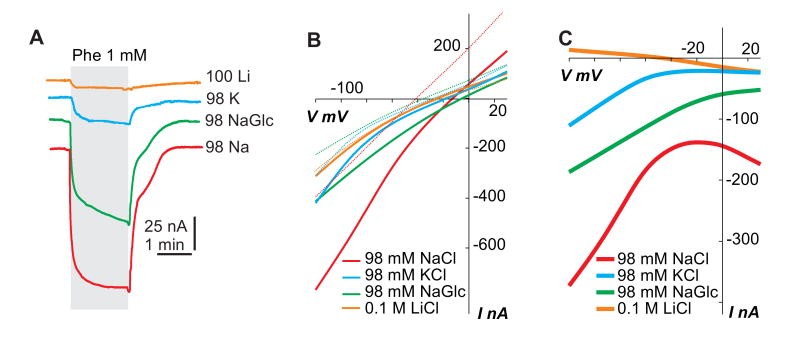
Superfusion of DmNAT1-expressing oocytes with phenylalanine at different holding potentials increased inward currents, whereas outward currents associated with the transporter (i.e. unrelated leak conductance in the oocyte membrane) remained unaltered (data not shown and Fig. 6A). A coupling of substrate transport to Na+ translocation was supported by the voltage-dependence of phenylalanine-induced currents, which remained inwardly directed at all the holding potentials tested (Fig. 6B). The DmNAT1-mediated phenylalanine-evoked current was all but abolished when Na+ was replaced with Li+ (Fig. 6A). Replacement of Na+ with N-methyl-D-glucamine (NMDG) also eliminated substrate-induced currents (data not shown). However, K+ supported a smaller but significant phenylalanine-induced current (Fig. 6A). The ability of DmNAT1 to utilize an inward K+ electrochemical gradient is supported by I/V plots (Fig. 6B and C). Substitution of Cl- with gluconate resulted in a moderate decay of the currents (average of 52% of NaCl), revealing that DmNAT1 exhibits a partial dependence on Cl- (Fig. 6A). This observation is also supported by I/V plots in Fig. 6B and C.
Figure 6. Ion dependence and I/V profile of DmNAT1.
A. Phenylalanine-induced currents associated with ion charge transport are shown. A DmNAT1 expressing oocyte was tested with solutions containing 100 mM LiCl, 98 mM KCl, 98 mM Na gluconate (NaGlc), or 98 mM NaCl, respectively. B. Amino acid-dependent current-voltage (I/V) relations as a function of co-transported ion and organic substrate (Phe). All recordings were made from a representative oocyte expressing DmNAT1. I/V plots show the currents evoked by 1 mM phenylalanine upon Na+ substitution in the bathing medium. Line colors represent different ion profiles of oocyte-bathing solution (see graph inset). Solid and dotted lines of the same color represent I/Vs with or without Phe, respectively. C. I/V profiles in which “background” currents (in the absence of organic substrates) were subtracted from currents obtained in the presence of 1 mM substrate concentrations. All presented characteristics were acquired at holding potential = -50 mV. Decremented square pluses were used for IV reconstruction; first pulse = +30 mV, last pulse = -90 mV; base = -10 mV.
To determine the stereoselectivity of DmNAT1, we measured the currents evoked by D-enantiomers of selected substrates (Fig. 7). Unexpectedly, DmNAT1 exhibited remarkably little stereospecificity and in some cases even preferred the D- vs. L-isomers according with apparent transport turnover (Fig. 7) and affinity (Table 1). Where the L-isomer was preferred, the D-enantiomer still induced comparable currents, except in the case of D-tryptophan, which failed to induce any inward current and apparently jams the transport mechanism. This assumption is based on apparent negative currents, which we tentatively explain as a reduction of transporter associated sodium leak current (Fig. 7, Trp). The results of these experiments strongly suggest that DmNAT1 transports both L- and D-amino acids with little stereoselectivity, a unique property among the currently characterized members of the SLC6 family.
Discussion
DmNAT1 is the first of six NATs present in the D. melanogaster genome (Fig. 1) and the fourth NATs functionally expressed from dipteran model organisms with annotated genomes, the current set of which includes Ae. aegypti transporter AeAAT1 (Boudko et al., 2005a), and two An. gambiae transporters AgNAT8 (Meleshkevitch et al., 2006) and AgNAT6 (Boudko et al., 2005b). DmNAT1 shares the highest sequence and structural similarity with AeAAT1 (pairwise similarity 52.5%, identical sites 359, 51.7%); however, it exhibits a broader substrate spectrum. Two known Anopheles transporters, AgNAT8 and AgNAT6, are more distant from DmNAT1 (pairwise similarity 45.7%, identical sites 229, 33.1%) and have very narrow selectivity for aromatic substrates. AeAAT1 and DmNAT1 represent basal branches of the NAT subfamily with the shortest evolutionary distances from a Last Universal Common Ancestor (LUCA) with SLC6 and NATs. These observations suggest that insect-specific NAT populations evolved from some ancestral broad substrate spectra phenotype and acquired substrate selectivity phenotypes upon successive gene duplications. It also suggests that broad spectra transporters are long-existing members of the NAT population and play important physiological roles in organisms.
Broad selectivity transporter such as DmNAT1 may perform high-throughput absorption of digested free amino acids. Hence, the elevated dmNAT1 transcription in the posterior midgut (PMG) likely corresponds with the primary apical absorption of amino acids from the alimentary canal lumen. Similar expression patterns were reported for characterized NATs in the PMG mosquito larvae, where the apical location of the transporters was also confirmed with AgNAT6- and AgNAT8-specific antibodies (Okech et. al. 2008). In contrast to the straight larval gut of mosquito larvae, the fruit fly larval alimentary canal is folded (Fig. 3). This morphology increases length of the PMG and an absorption area vs. digested volume ratio. This may lead to better and more complete clearing of essential nutrients from the midgut lumen. A similar phenomenon is observed in the case of herbivorous adaptations in mammals which typically possess a longer intestine vs. carnivorous mammals and usually have some specific structure for symbiotic fermentation of the vegetarian diet. A long intestine and balanced amino acid profile are favorable conditions for the rapid absorption of broad spectra of substrate via DmNAT1 mechanisms. However, the DmNAT1 function may be insufficient for comprehensive absorption of dietary-underrepresented amino acids. Five other D. melanogaster NATs are differentially expressed in the larval gut (based on the in situ hybridization of these transporters, BDGP and personal observation, data not shown). They may assist DmNAT1 by mediating selective absorption of amino acids which are present in low concentrations; however, exact functions of these paralogous NATs are currently unknown. The pattern of axial spatial segregation of paralogous transporters, with inferior affinity transporters occupying proximal positions and higher affinity transporters expressed in more distal positions of alimentary structures, was also observed in the mammalian kidney with pairs of Na+-glucose (SGLT1 and SGLT2) and H+-peptide (PEPT1 and PEPT2) transporters (You et al., 1995, Shen et al., 1999).
DmNAT1 can absorb D-amino acids and prefers some D- vs. L-enantiomers. To date, only one other SLC6 member, ATB0,+ (SLC6A14, a.k.a. B0,+ - neutral and cationic amino acid system transporter), has been demonstrated to transport D-amino acids in detectable levels (Hatanaka et al., 2002). Apparently, DmNAT1 has a comparable apparent affinity and transport efficiency for serine enantiomers (∼150 μM). It also can absorb D-enantiomers of alanine, methionine, leucine and tryptophan but not other neutral amino acids. In contrast to DmNAT1, ATB0,+ appears to have a higher apparent transport efficiency for L-enantiomers (Sloan and Mager, 1999; Ugawa et al., 2001). Another striking difference is the failure of DmNAT1 to transport cationic amino acids, which appears to be one of the most important physiological functions of the mammalian transporter. ATB0,+ performs absorption of L-arginine, a principal limiting substrate in the NOS-mediated generation of nitric oxide (reviewed (Boudko, 2007)). Phylogenetically, mammalian SLC6A14 members form a cluster with a pair of uncharacterized insect transporters (e.g. CG3349 and CG7075 in the fruit fly (Meleshkevitch et al., 2006)). Hence, some similarity between DmNAT1 and ATB0,+ functions may be a result of convergence rather than inherited conservation. Residues involved in the substrate binding mechanism of NATs are readily identifiable based on the sequence alignment with the crystallized bacterial homolog of NAT, AaLeuT (Yamashita et al., 2005), homology modeling and in silico substrate docking approaches (Fig. 2 and data not shown). Among 16 sites interacting with organic substrates (Fig. 2: arrows), 10 sites are highly conserved in dipteran NATs (Fig. 2: black arrows). 6 phenotype-specific sites determine the unique shape of substrate binding pockets and substrate specificity (Fig. 2: gray and empty arrows). The most unique substitutions in the substrate-binding pocket of DmNAT1 vs. other SLC6 members are V54, S319 and L421 (Fig. 2: empty arrows). It is reasonable to propose that these substitutions are responsible for the acquisition of D-enantiomers. However, the mammalian D-amino acid transporter has different amino acids in the corresponding positions (Fig. 2: MmATB0,+ and DmNAT1 vs. empty arrows). Once again, this may suggest independent origins and convergence of D-amino acid selectivity among SLC6 members. It also may suggest a different structural basis of D-amino acid binding in the MmATB0,+ and DmNAT1.
ATB0,+ expressed in colonocytes with possible luminal membrane associations was postulated as uniquely suitable for the absorption of bacteria-derived D-serine (Hatanaka et al., 2002). Insect symbiosis with bacteria is the most universal adaptation representing the key to the evolutionary success of this class of arthropods and is usually critical for survival. D-amino acids are common components in the bacterial cell wall and metabolic pathways (Bauer et al., 2005). Insects are reported to accumulate significant levels of D-amino acids from symbiotic sources (Corrigan and Srinivasan, 1966). In particular, D. melanogaster larvae have a remarkable capacity to use several D-forms of essential amino acids for growth in place of their L-isomers (Geer, 1966). Our findings regarding DmNAT1function and distribution explain the capacity of fruit fly larvae to internalize D-forms of neutral amino acids. Therefore, DmNAT1 can substantially benefit larval nutrition and development by acquiring bacterial and yeast fermentation products.
CG3252 is transcribed in the larval brain and adult head (this work and (Thimgan et al., 2006)) where DmNAT1 may support plasma membrane absorption of neutral L-amino acid and provide essential nutrition for neuronal nd glial cells. In addition, by desorbing free amino acids DmNAT1 can mediate neutral amino acids clearances from the Drosophila brain neuropile and neuronal synapses and support an anticipated haemolimph-brain barrier function. The L-amino acid uptake by DmNAT1 correlates with certain neuronal functions; however, little is known regarding specific D-amino acids in the invertebrate brain. In contrast, D-serine is a well-known modulator of mammalian long-term potentiation and visual signal transduction (reviewed in (Bauer et al., 2005; Broer and Brookes, 2001; Miller, 2004)). It mediates glutamatergic neurotransmission by acting as an agonist at the glycine-binding site of NMDA receptors (NMDARs). Drosophila NMDAR has been cloned and characterized (Xia et al., 2005). It shares physiological properties with vertebrate NMDARs and is modulated by glycine, although to a lesser extent. The D-serine modulation of invertebrate NMDAR has been shown in hydra (Schell, 2004). With the supposition of an earlier origin (Schell, 2004), we may expect conserved roles for D-serine in Drosophila and the mammalian brain. DmNAT1 expression in the larval brain is favorable for the absorption of endogenously produced or exogenously provided D-serine. Thus, in addition to universal L-amino acid clearance and essential neuronal nutrition, DmNAT1 may carry on D-serine absorption and modulation of the NMDAR-coupled neuronal signaling. In summary, the present analysis unravels the first invertebrate B0-like transporter and the first transport mechanism with adaptive specialization in the absorption of D-amino acids. These findings considerably broaden the biological significance of the NAT-SLC6 subfamily; showing that its members have evolved unique properties which may support the acquisition of symbiotic metabolites and contribute to basic neuronal functions in metazoan organisms.
Acknowledgments
This work was supported by the NIH-NIAID research grant R01 AI030464 (DB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Bauer D, Hamacher K, Broer S, Pauleit D, Palm C, Zilles K, Coenen HH, Langen KJ. Preferred stereoselective brain uptake of D-serine - a modulator of glutamatergic neurotransmission. Nuclear Medicine and Biol. 2005;32:793–797. doi: 10.1016/j.nucmedbio.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Bohmer C, Broer A, Munzinger M, Kowalczuk S, Rasko JEJ, Lang F, Broer S. Characterization of mouse amino acid transporter B(0)AT1 (slc6a19) Biochem J. 2005;389:745–751. doi: 10.1042/BJ20050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko DY. Bioanalytical profile of the L-arginine/nitric oxide pathway and its evaluation by capillary electrophoresis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:186–210. doi: 10.1016/j.jchromb.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko DY, Kohn AB, Meleshkevitch EA, Dasher MK, Seron TJ, Stevens BR, Harvey WR. Ancestry and progeny of nutrient amino acid transporters. Proc Natl Acad Sci USA. 2005a;102:1360–1365. doi: 10.1073/pnas.0405183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko DY, Meleshkevitch EA, Harvey WR. Novel transport phenotypes in the sodium neurotransmitter symporter family. Faseb J. 2005b;19:A748–A748. [Google Scholar]
- Boudko DY, Stevens BR, Donly BC, Harvey WR. Nutrient Amino acid and Neurotransmitter transporters. In: I K, G SS, Gilbert Lawrence I, editors. Comprehensive Molecular Insect Sci. Vol. 4. Amsterdam: Elsevier; 2005c. pp. 255–309. [Google Scholar]
- Broer A, Klingel K, Kowalczuk S, Rasko JEJ, Cavanaugh J, Broer S. Molecular cloning of mouse amino acid transport system B-0, a neutral amino acid transporter related to Hartnup disorder. J Biol Chem. 2004;279:24467–24476. doi: 10.1074/jbc.M400904200. [DOI] [PubMed] [Google Scholar]
- Broer A, Tietze N, Kowalczuk S, Chubb S, Munzinger M, Bak LK, Broer S. The orphan transporter v7-3 (slc6a15) is a Na+-dependent neutral amino acid transporter (B0AT2) Biochem J. 2006;393:421–30. doi: 10.1042/BJ20051273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer S, Brookes N. Transfer of glutamine between astrocytes and neurons. Journal of Neurochemistry. 2001;77:705–719. doi: 10.1046/j.1471-4159.2001.00322.x. [DOI] [PubMed] [Google Scholar]
- Broer S, Cavanaugh JA, Rasko JEJ. Neutral amino acid transport in epithelial cells and its malfunction in Hartnup disorder. Biochemical Society Transactions. 2005;33:233–236. doi: 10.1042/BST0330233. [DOI] [PubMed] [Google Scholar]
- Castagna M, Shayakul C, Trotti D, Sacchi V, Harvey W, Hediger M. Molecular characteristics of mammalian and insect amino acid transporters: implications for amino acid homeostasis. J Exp Biol. 1997;200:269–286. doi: 10.1242/jeb.200.2.269. [DOI] [PubMed] [Google Scholar]
- Castagna M, Shayakul C, Trotti D, Sacchi VF, Harvey WR, Hediger MA. Cloning and characterization of a potassium-coupled amino acid transporter. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5395–400. doi: 10.1073/pnas.95.9.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, von_Heijne G. TopPred II: an improved software for membrane protein structure predictions. Computer Applications in the Biosciences : Cabios. 1994;10:685–6. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Corrigan J, Srinivasan N. The occurrence of certain D-amino acids in insects. Biochemistry. 1966;5:1185–1190. doi: 10.1021/bi00868a010. [DOI] [PubMed] [Google Scholar]
- Feldman DH, Harvey WR, Stevens BR. A novel electrogenic amino acid transporter is activated by K+ or Na+, is alkaline pH-dependent, and is Cl--independent. J Biol Chem. 2000;275:24518–26. doi: 10.1074/jbc.M907582199. [DOI] [PubMed] [Google Scholar]
- Geer BW. Utilization of D-amino acids for growth by Drosophila melanogaster larvae. J Nutr. 1966;90:31–9. doi: 10.1093/jn/90.1.31. [DOI] [PubMed] [Google Scholar]
- Hatanaka T, Huang W, Nakanishi T, Bridges CC, Smith SB, Prasad PD, Ganapathy ME, Ganapathy V. Transport of D-serine via the amino acid transporter ATB(0,+) expressed in the colon. Biochem Biophys Res Commun. 2002;291:291–5. doi: 10.1006/bbrc.2002.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jespersen T, Grunnet M, Angelo K, Klaerke DA, Olesen SP. Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques. 2002;32:536–8. 540. doi: 10.2144/02323st05. [DOI] [PubMed] [Google Scholar]
- Kowalczuk S, Broer A, Munzinger M, Tietze N, Klingel K, Broer S. Molecular cloning of the mouse IMINO system: an Na+- and Cl--dependent proline transporter. Biochemical J. 2005;386:417–422. doi: 10.1042/BJ20050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshkevitch EA, Assis-Nascimento P, Popova LB, Miller MM, Kohn AB, Phung L, Mandal A, Harvey WR, Boudko DY. Molecular characterization of the first aromatic nutrient transporter from the sodium neurotransmitter symporter family. J Exp Biol. 2006;209:3183–3198. doi: 10.1242/jeb.02374. [DOI] [PubMed] [Google Scholar]
- Miller RF. D-Serine as a glial modulator of nerve cells. Glia. 2004;47:275–83. doi: 10.1002/glia.20073. [DOI] [PubMed] [Google Scholar]
- Neal JJ. Brush border membrane and amino acid transport. Arch Insect Biochem Physiol. 1996;32:55–64. [Google Scholar]
- Okech BA, Meleshkevitch EA, Miller MM, Popova LB, Harvey WR, Boudko DY. Synergy and specificity of two Na+-aromatic amino acid symporters in the model alimentary canal of mosquito larvae. J Exp Biol. 2008;211:1594–602. doi: 10.1242/jeb.017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Palacin M, Estevez R, Bertran J, Zorzano A. Molecular biology of mammalian plasma membrane amino acid transporters. Physiol Rev. 1998;78:969–1054. doi: 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- Schell MJ. The N-methyl D-aspartate receptor glycine site and D-serine metabolism: an evolutionary perspective. Philos Trans R Soc Lond B Biol Sci. 2004;359:943–64. doi: 10.1098/rstb.2003.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow HF, Broer S, Broer A, Bailey CG, Potter SJ, Cavanaugh JA, Rasko JEJ. Hartnup disorder is caused by mutations in the gene encoding the neutral amino acid transporter SLC6A19. Nature Genetics. 2004;36:1003–1007. doi: 10.1038/ng1406. [DOI] [PubMed] [Google Scholar]
- Shen H, Smith DE, Yang T, Huang YG, Schnermann JB, Brosius FC., 3rd Localization of PEPT1 and PEPT2 proton-coupled oligopeptide transporter mRNA and protein in rat kidney. Am J Physiol. 1999;276:F658–65. doi: 10.1152/ajprenal.1999.276.5.F658. [DOI] [PubMed] [Google Scholar]
- Sloan JL, Mager S. Cloning and Functional Expression of a Human Na+ and Cl-- dependent Neutral and Cationic Amino Acid Transporter B0+ J Biol Chem. 1999;274:23740–23745. doi: 10.1074/jbc.274.34.23740. [DOI] [PubMed] [Google Scholar]
- Stevens BR, Wright SH, Hirayama BS, Gunther RD, Ross HJ, Harms V, Nord E, Kippen I, Wright EM. Organic and inorganic solute transport in renal and intestinal membrane vesicles preserved in liquid nitrogen. Membr Biochem. 1982;4:271–82. doi: 10.3109/09687688209065436. [DOI] [PubMed] [Google Scholar]
- Takanaga H, Mackenzie B, Peng JB, Hediger MA. Characterization of a branched-chain amino-acid transporter SBAT1 (SLC6A15) that is expressed in human brain. Biochem Biophys Res Commun. 2005a;337:892–900. doi: 10.1016/j.bbrc.2005.09.128. [DOI] [PubMed] [Google Scholar]
- Takanaga H, Mackenzie B, Suzuki Y, Hediger MA. Identification of mammalian proline transporter SIT1 (SLC6A20) with characteristics of classical system imino. J Biol Chem. 2005b;280:8974–84. doi: 10.1074/jbc.M413027200. [DOI] [PubMed] [Google Scholar]
- Thimgan MS, Berg JS, Stuart AE. Comparative sequence analysis and tissue localization of members of the SLC6 family of transporters in adult Drosophila melanogaster. J Exp Biol. 2006;209:3383–3404. doi: 10.1242/jeb.02328. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugawa S, Sunouchi Y, Ueda T, Takahashi E, Saishin Y, Shimada S. Characterization of a mouse colonic system B0+ amino acid transporter related to amino acid absorption in colon. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2001;281:G365–G370. doi: 10.1152/ajpgi.2001.281.2.G365. [DOI] [PubMed] [Google Scholar]
- Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, Lin IR, Saitoe M, Tully T, Chiang AS. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15:603–15. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–23. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- You G, Lee WS, Barros EJ, Kanai Y, Huo TL, Khawaja S, Wells RG, Nigam SK, Hediger MA. Molecular characteristics of Na(+)-coupled glucose transporters in adult and embryonic rat kidney. J Biol Chem. 1995;270:29365–71. doi: 10.1074/jbc.270.49.29365. [DOI] [PubMed] [Google Scholar]