Abstract
The Agouti Related Protein (AgRP) is an orexigenic peptide that plays a significant role in energy balance regulation. It is expressed in the hypothalamus, the adrenal glands, and the testis but sequences determining its spatial and temporal expression have not been identified. Using an elaborate in vitro screening approach, we show here that two adjacent enhancers inside the first intron of the neighboring (1.4 kb downstream) ATPase gene (ATP6V0D1) modulate the human AgRP promoter with profound spatiotemporal variation despite their diminutive sizes (221 nt and 231 nt). In transgenic mice, the proximal enhancer displayed specificity for the testis, tail, and ears, and the distal one for the testis, front feet, bone, heart, muscle, brain, spinal cord, and tongue, while dietary fat and overnight fasting had differential effects on enhancer activities. AgRP in the testis was localized to pachytene spermatocytes and in the tongue to epithelial cells. Comparative sequence analysis showed that the AgRP-ATP6V0D1 intergenic region is two times longer in humans than in mice, and that the two enhancers are conserved in the rhesus monkey but not the mouse genome. These data show that spatiotemporal expression of the human AgRP gene is influenced by diversified primate-specific intronic sequences in its neighboring ATP6V0D1 gene.
Keywords: AgRP, ATPase, orexigen, intron, enhancer, promoter
Introduction
AgRP1; 2 is an important regulator of energy balance.3 It binds antagonistically to melanocortin receptors 3 and 4 (MC3&4R) in the hypothalamus while peripherally it is taken up by various organs with preference for the liver, the adrenal glands, and the heart.4 Hypothalamic AgRP is elevated in obese and diabetic mice2 and recent studies have shown that AgRP-expressing neurons are essential for the control of energy homeostasis.5; 6; 7 In humans, functional single nucleotide polymorphisms (SNPs) in the 5′-UTR of AgRP (−3019G>A and −38C>T)8 were associated with reduced fatness in African Americans and absence of diabetes in Sierra Leoneans.9; 10 A SNP in the coding region was associated with resistance to late-onset obesity11 and with preference for carbohydrate consumption over fat12 suggesting that AgRP may also affect macronutrient selection.
The human AgRP gene is a relatively short gene spanning 1.1 kb on chromosome 16q22. It consists of 4 exons (3 coding) and encodes a 132 amino acid protein.2 It is expressed in the arcuate nucleus and a shorter transcript lacking the 5′ non-coding exon is expressed in the adrenal gland, the testis, lung, kidney, spinal cord, and dorsal root ganglia.2; 13 It is unclear whether these transcripts arise from differential splicing or from two independently regulated promoters, but we have shown that the 5′ untranslated exon possesses significant promoter activity. 14 Transgenic mice made with Bacterial Artificial Chromosomes have revealed that the upstream 20 Mb region is required for the expression of the mouse AgRP in the arcuate nucleus. 15; 16 Little is known however, about specific enhancer and/or repressor sequences that determine the spatial and temporal expression of the human AgRP gene.
In this study we report specific regions with silencer and enhancer effects on the promoter of the human AgRP gene. Hypothalamic and periphery-specific cell lines were initially used to narrow down large segments of enhancers. One region in particular, downstream of the human AgRP polyadenylation site and inside the first intron of the neighboring ATPase, H+ transporting lysosomal V0 subunit D1 (ATP6V0D1) gene, was found to contain two adjacent enhancers. These two enhancers were used to make luciferase-expressing transgenic mice to determine their in vivo spatiotemporal effects and evaluate the course of expression of the reporter gene in response to dietary fat and fasting. We found that the proximal enhancer drove expression in the ears, tail, and the testes while the neighboring one had ubiquitous effects with greater specificity for the tongue, front feet, spinal cord, and the testis among other tissues. Using immunocytochemistry we determined that, in the testis, AgRP is expressed in pachytene stage spermatocytes and, in the tongue, in epithelial cells. Dietary fat and overnight fasting affected the activity of the two enhancers that were found to be unique to the human genome while maintaining high conservation with other primates but not rodents. The intergenic region between the human and mouse AgRP and ATP6V0D1 genes was not conserved, with the human locus being twice as long. These studies identify two trans-acting enhancers in the first intron of the ATP6V0D1 gene that influence the spatiotemporal expression of AgRP.
Results
In vitro promoter/enhancer analysis
We examined upstream and downstream regions surrounding the human AgRP locus in an effort to identify regions with enhancer properties. The overall strategy is depicted in Fig. S1a. Four regions were studied: a region upstream of the translation initiation codon (A) that was divided into four 2 kb-long sub-regions, and three regions (B, C, and D) downstream of the polyadenylation signal. All constructs were made in the luciferase pGL3-basic vector (Fig. S1b). Region B spanned the intergenic region between AgRP and ATP6V0D1, and Regions C and D spanned the first exon of ATP6V0D1 and the first intron between exons 1 and 2 of ATP6V0D1.
Because of its length (> 9 kb) Region A was divided into 4 sub-regions as follows: Ai(-9253/-7157)(-18/+312), Aii(-7234/-5117)(-18/+312), Aiii(-5238/-3144)(-18/+373), and Aiv(-3246/+373). Sub-regions Aii and Aiv had significantly and consistently lower activity than the basal promoter construct (-18/+312) (Fig. S2), suggesting that these two regions may contain silencers or binding sites for repressors of activation. These regions were not pursued further because the goal of the present study was to identify neuron-specific enhancers.
Intergenic region B was cloned downstream of luc but using three different segments of the AgRP promoter to examine for potential influences from regions beyond the minimal promoter (-18/+312) (Fig. S3). In the end of the analysis, sub-region Biib(-18/+312)(+1827/+1959) was found to have as much activity as region B (Fig. S3b, lower panel), suggesting that this 137 nt region lying 609 nt downstream of the AgRP polyadenylation site, contained enhancer elements for the AgRP minimal promoter that were specific to the adrenocortical cells. This region was not pursued further in the study described here because we were interested in enhancers that were active specifically in neuronal cells.
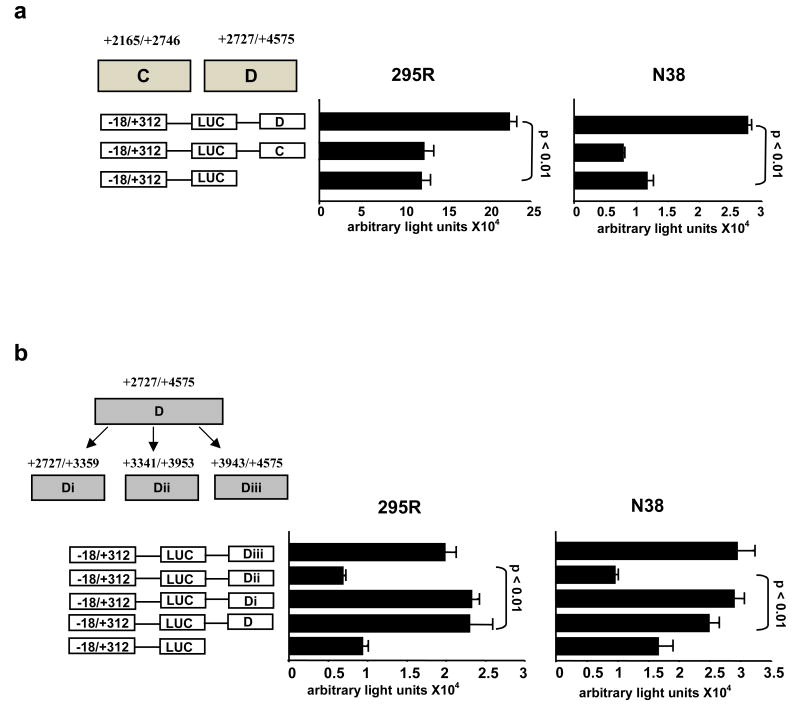
Regions C and D were evaluated in the same fashion. Region D (but not region C) contained enhancer elements in both neuronal and somatic cell types (Fig. 1a). Region D was therefore divided into three overlapping sub-regions: Di(-18/+312)(+2727/+3359), Dii(-18/+312)(+3341/+3941), and Diii(-18/+312)(+3943/+4575). Sub-regions Di and Diii (but not Dii) had enhancer properties for the AgRP basal promoter in both h295R and N38 cell lines (Fig. 1b).
Fig. 1. In vitro determination of enhancer elements.
(a) Region D (but not C) increased activity of the human AgRP promoter, equally in the human adrenocortical NCI-h295R and the mouse clonal hypothalamic N38 cell lines. (b) Region D was divided into 3 sub-regions of which only Di and Diii increased basal promoter activity in both cell lines.
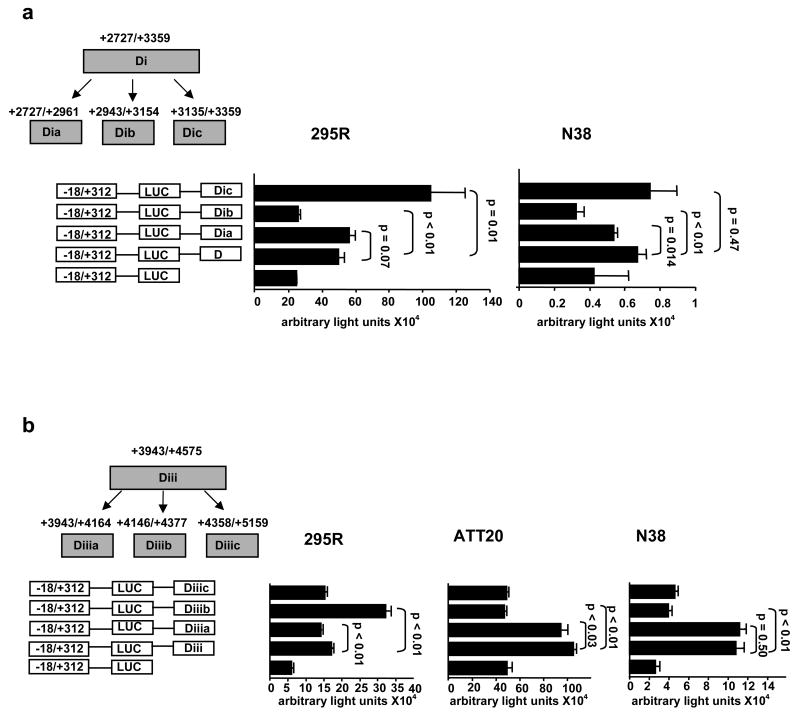
Di and Diii were further engineered into three overlapping sub-regions each (Fig. 2a) and the experiments were performed in the N38 and h295R mouse hypothalamic cells. Sub-region Dib had a lesser effect than the parent construct Di in both cell lines, while, sub-region Dic had higher activity than Di, again in both cell lines albeit not always at statistically significant levels (Fig. 2a). Because these effects were marginal, these sub-regions were not investigated further. Sub-regions of construct Diii however, had more distinct effects. Specifically, Diiib had as much activity as sub-region Diii on its own but only in the h295R cells while sub-region Diiia had the same activity as the parent Diii region but only in the N38 cells (Fig. 2b).
Fig. 2. In vitro elimination of non-enhancer regions.
(a) Sub-region Di was divided into smaller portions; Dia, Dib, Dic. Sub-region Dic had the most significant enhancer effect on AgRP basal promoter in both cell lines (b). Sub-region Diii was also divided into smaller regions. Sub-region Diiia had significant enhancer effect in two neuronal cell lines (N38 and ATT20), while sub-region Diiib had significant enhancer effect specifically in the adrenocortical cells (NCI-h295R).
The latter experiment was repeated, this time including a second neuronal cell line, ATT20, and the results were recapitulated (Fig. 2b). These data suggest that sub-region Diiib may be influencing expression of AgRP in adrenal cells whereas sub-region Diiia may be regulating expression of AgRP in neuronal cells.
We also performed in silico analysis using the Alibaba2 and TESS databases to compare the putative binding sites of transcription factors between enhancers Diiia and Diiib. We found that enhancer Diiia had putative binding sites for Sp1, Ets-1 MyoD, NF-kB, CACCC, and USF, while, enhancer Diiib had a high frequency of OCT-1 binding sites (a total of six OCT-1 sites) and putative binding sites for C/EBPα, SP1, PIT-1a, HNF-1C, and HNF-3.
In vivo enhancer analysis
To examine further the properties of the two candidate enhancers (Diiia and Diiib), luciferase-expressing transgenic mice were made. Schematics of the transgenes used are shown in Fig. 3. Essentially, Diiia and Diiib constructs were digested out of the pGL3-basic vector using the Nhe I and Sal I restriction endonucleases to release the linear inserts. The transgene for Diiib is shown in Fig. 3 in its reverse orientation (3′ to 5′) for the sake of showing the overlap (18 nt) between the two intronic enhancers.
Fig. 3. Schematic of transgenes and bioluminescence of offspring representing the two enhancers.
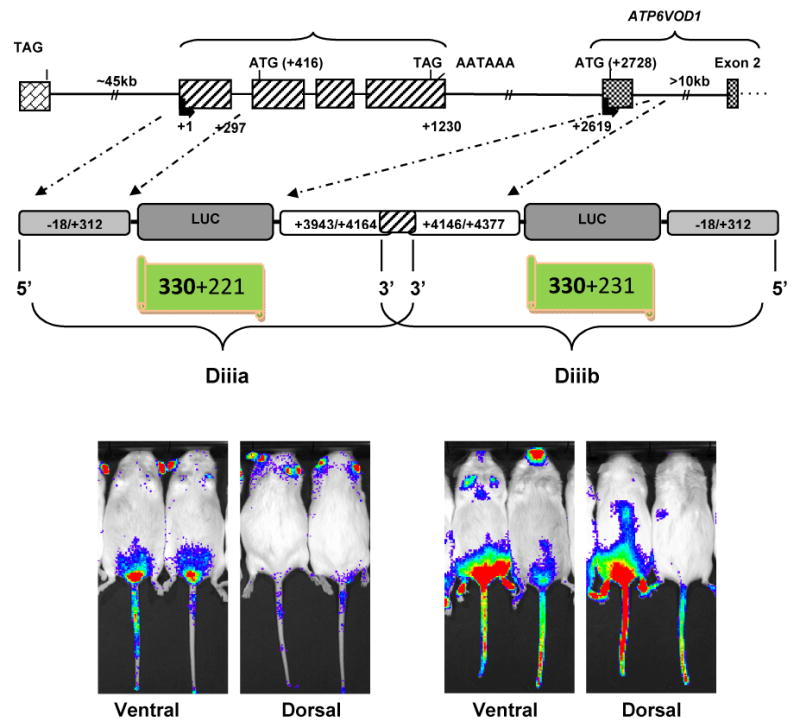
The upper panel shows a schematic of the AgRP-ATP6V0D1 locus. The middle panel shows schematics of the two constructs (Diiia & Diiib) that were used to make the transgenic mice. The transgene for enhancer Diiib is shown in the reverse orientation (3′ to 5′) so that its exact physical position is represented in the middle panel. The 18 nt overlap of the two enhancers is shown by the shaded region. The bottom panel shows bioluminescence images of dorsal and ventral views from second generation transgenic mice. Images of offspring from the same founder are shown for enhancer Diiia (left, bottom). Images from offspring from two different founders are shown for enhancer Diiib (bottom, right).
Following pronuclear injections, three founders per enhancer (transgene) were generated. For enhancer Diiia, only one founder was able to successfully breed and pass the transgene (Fig. 3). For enhancer Diiia, two founders bred successfully and passed the transgene (Fig. 3). Examples of offspring are shown in Fig. 3: two mice from the single founder for Diiib (left-hand-side), and one mouse per founder for Diiib (right-hand-side). Essentially, enhancer Diiia drove expression in the testis, ears, and tail, while enhancer Diiib drove expression more ubiquitously with greater concentration in the testis and the abdominal area, the front limps, and the facial region. Offspring of founder A displayed significantly more intense expression of luciferase than offspring of Founder B.
Localization of AgRP in testis and tongue
Luciferase-expressing transgenic mice were further characterized by luminometric analysis to confirm the presence of the luciferase signal in the suggested tissues/organs visualized by bioluminescence. Mice were thus euthanized and tissues excised and homogenized. In Diiia mice, the highest expression of luciferase was found in the ears, followed by the testis and the tail (Fig. 4a). Real time PCR (qPCR) was then used to examine for the presence of AgRP in the testis which confirmed its abundant expression (Fig. 4b). Using immunocytochemistry, AgRP was found to be in the center of seminiferous tubules, while, as a negative control, it was completely absent in the tubules of AgRP-deficient (AgRP-/-) mice (Fig. 4c). AgRP was further localized in pachytene stage spermatocytes and colocalized with the alpha-synaptonemal complex protein 3 (α-SCP3) (Fig. 4d) that is a meiotic entry-specific protein shown previously to be present in pachytene spermatocytes. 17; 18
Fig. 4. Luminometric analysis of luciferase signal in Diiia mice and localization of AgRP by immunofluorescence.
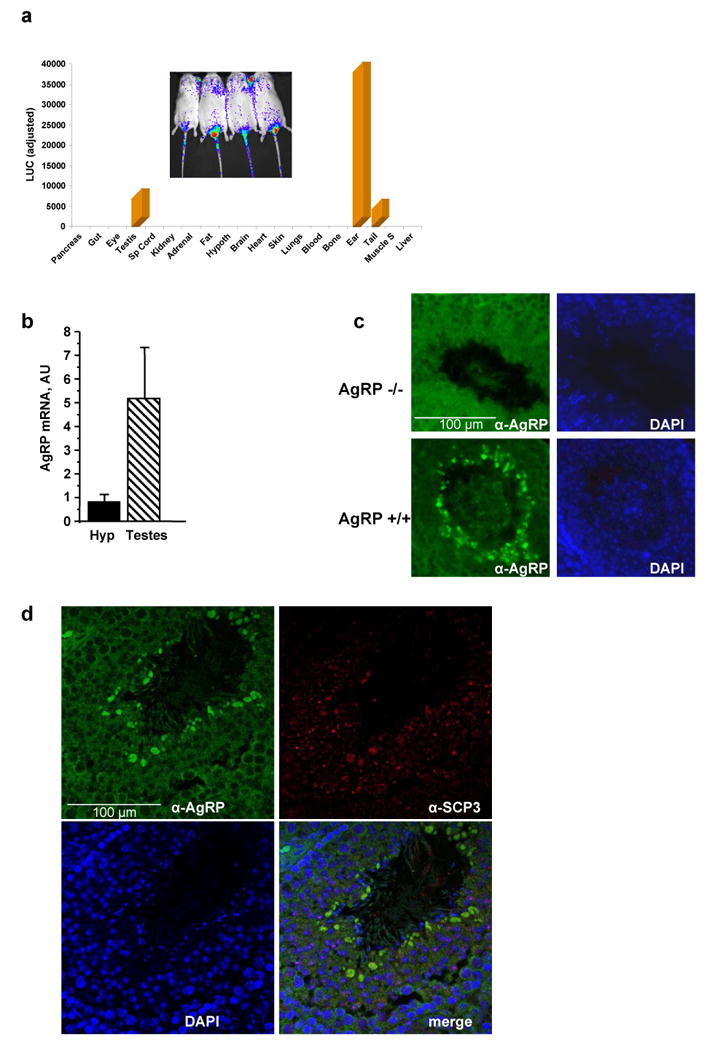
(a) Luminometric analysis (light units) of luciferase expression in various tissues of Diiia mice (inset shows bioluminescence of a ventral view of these mice). Luciferase is predominantly expressed in the ears, testes, and tails. (b) Real time PCR shows robust expression of AgRP in the testis in comparison to AgRP in the hypothalamus. (c) Immunohistochemistry of mouse testis sections localized AgRP in the middle of seminiferous tubules in wild type (AgRP+/+) but not AgRP-deficient (AgRP-/-) mice. (d) Immunohistochemistry colocalized AgRP with the alpha-synaptonemal complex protein 3 (α-SCP3) in pachytene stage spermatocytes.
Enhancer Diiib had more ubiquitous effects on luciferase expression, as mice from the two founders showed (Fig. 5a). There was also significant variation between the two founders (Fig. 5a). Emphasis was placed on the tongue because there was high luciferase expression in this organ. Hence, a Diiib mouse was euthanized and had its skin removed and was re-imaged. The image on the tongue remained detectable even after euthanasia and even after the tongue had been removed from the mouth (Fig. 5b). At higher magnification, the signal was found to be exclusively present in the pharyngeal end of the tongue (Fig. 5b). qPCR confirmed that AgRP was also expressed in the tongue but at significantly lower levels than in the hypothalamus (Fig. 5b). Using immunocytochemistry AgRP was localized in the epithelial cells of wild-type but not AgRP-/- mice (Fig. 5c). This finding was confirmed by the colocalization of AgRP with α-14-cytokeratin that is an epithelial cell-specific protein19; 20 (Fig. 5d).
Fig. 5. Luminometric analysis of luciferase signal in Diiib mice and localization of AgRP by immunofluorescence.
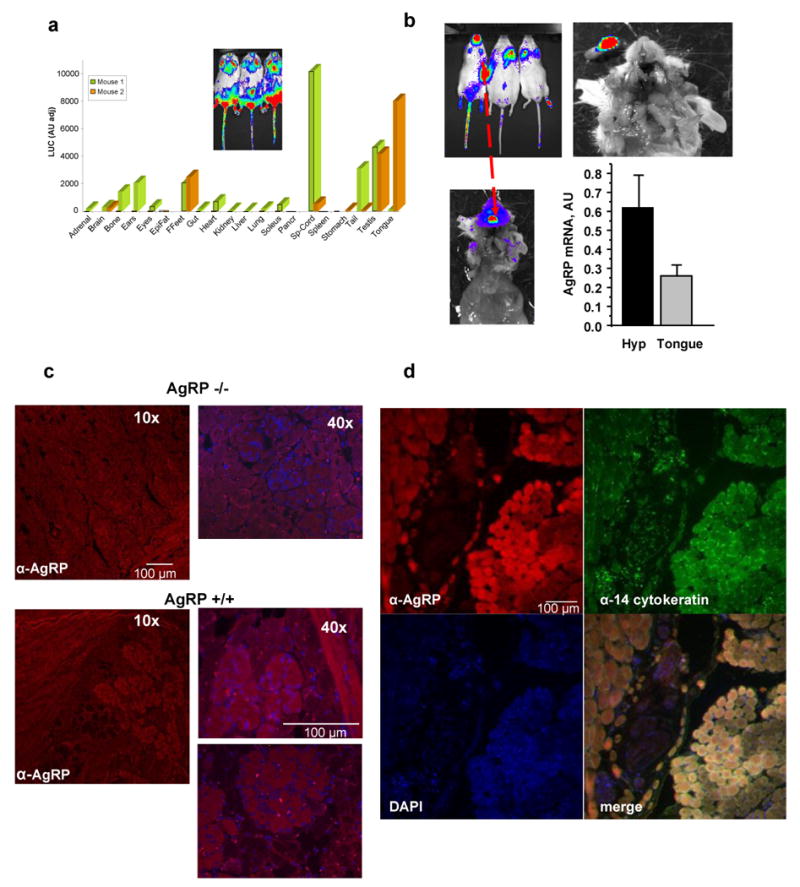
(a) Luminometric analysis (light units) of luciferase expression in various tissues in Diiib mice (inset shows bioluminescence of a ventral view of these mice). Luciferase is ubiquitously expressed with greater preference for the spinal cord, the tongue, the testis, the ears, the bone, the brain, front feet, soleus muscle, among other tissues. There was significant variation between the two founders indicated by different colors of the bars. (b) Bioluminescence of the signal in the tongue is shown it different zoom modes. Real time PCR shows robust expression of AgRP in the tongue but at lower levels than in the hypothalamus. (c) Immunohistochemistry of mouse tongue sections shows AgRP expression in epithelial cells of wild type (AgRP+/+) but not AgRP-deficient (AgRP-/-) mice. (d) Immunohistochemistry colocalized AgRP with the α-14-cytokeratin that is an epithelial cell-specific protein.
Effects of dietary fat on enhancer properties
The effect of dietary fat content on the enhancers was examined by feeding Diiia and Diiib mice chow, low fat diet (LFD), and very high fat diet (VHFD). Four Diiia mice were imaged while consuming chow (26% fat kcal). Mice were then placed on LFD (10% kcal fat) for one week and imaged again. The luciferase signal was reduced by 34% in all the mice (Fig. 6). Mice were then switched to VHFD (60% kcal fat) for one week and imaged again. The luciferase signal recovered back to the chow levels (Fig. 6). Mice were then fed chow diet for two weeks, imaged again, fasted overnight, and then imaged again the following morning. Overnight fasting reduced the luciferase signal to levels similar to those achieved by the LFD (Fig. 6). The same experimental design was repeated with Diiib mice. The results were marginal relative to those observed in the Diiia mice whereby VHFD in general increased luciferase signal mostly in the front legs and chest area (Fig. 7). The effect was more pronounced by the overnight fasting that led to a more significant elevation of the signal by an estimated 49% at all sites of luciferase expression (Fig. 7). Diiib mice from Founder B also displayed a new signal, as a result of the LFD and the overnight fast, on the dorsal left-hand-side, at a position where the left adrenal gland is located. It should be noted that mice from Founder B did exhibit luciferase activity in their adrenal glands, according to luminometric analysis (Fig. 5a).
Fig. 6. In vivo effects of dietary fat and fasting on enhancer Diiia activity.
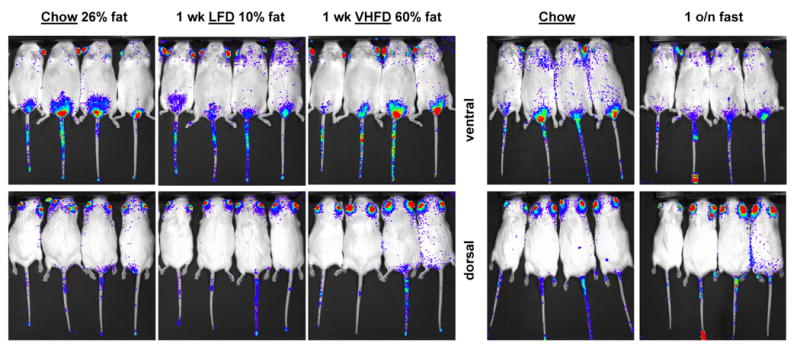
Bioluminescence images of dorsal and ventral views of Diiia offspring while consuming chow, low fat diet (LFD) or very high fat diet (VHFD) for one week each or fasted overnight (o/n) after chow diet. LFD and overnight fasting reduced luciferase expression in a similar fashion.
Fig. 7. In vivo effects of dietary fat and fasting on enhancer Diiib activity.
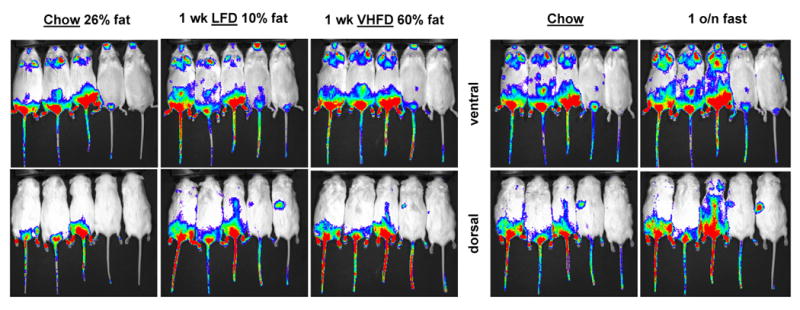
Bioluminescence images of dorsal and ventral views of Diiib offspring from two founders (first three from Founder A and the other two from Founder B) while consuming chow, low fat diet (LFD) or very high fat diet (VHFD) for one week each or fasted overnight (o/n) after chow diet. LFD and overnight fasting enhanced luciferase expression in a similar fashion. Overnight fasting also led to the appearance of a new signal on the left-hand-side dorsal side in offspring from Founder B.
Human/mouse comparative analysis of the AgRP-ATP6V0D1 locus
We compared the human and mouse syntenic AgRP loci to examine for conservation of functional elements. In general, there was high conservation between exonic sequences but introns differ significantly (Fig. 8a). The first coding exon of the ATP6V0D1 gene (that also encompasses a large non-coding region) had the highest identity score (92%) between the two species. There were differences, however, in the regions downstream of the polyadenylation signal of AgRP. In the human, the distance between the AgRP polyadenylation signal and the “ATG” of the ATPase was 1.49 kb whereas in the mouse it was almost half of that, at 0.81 kb (Fig. 8a). Moreover, this region did not contain any conserved sequences between the two species. The regions encompassing enhancers Diiia and Diiib were completely non-homologous suggesting, again, that the two enhancers do not exist in the mouse. We performed further analytical comparisons using the BLAST tool and found that enhancers Diiia and Diiib were extremely well-conserved throughout their entire lengths with the rhesus monkey (Macaca mulatta) equivalent region. The 600 nt region immediately downstream of enhancer Diiib was still highly homologous with the equivalent position in the rhesus monkey genome but not the mouse. The entire AgRP-ATP6V0D1 intergenic region displayed partial conservation with primates (Macaca mulatta, Pan trlodytes, Pongo abelii) but not with rodents or any other species.
Fig. 8. Comparative analysis of human and mouse AgRP-ATP6V0D1 loci.
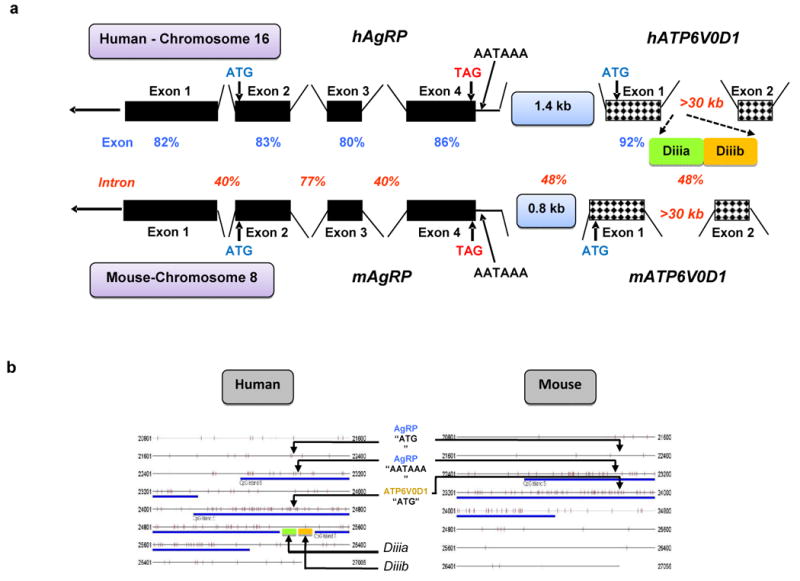
(a) Schematic representation of the human and mouse syntenic AgRP/ATP6V0D1 loci indicating the identity scores (percent) for exons and introns between the two species and presence of enhancers Diiia and Diiib in human only. (b) Algorithmic analysis comparing the CpG islands between the human and mouse syntenic loci encompassing the AgRP-ATP6V0D1 genes. The human AgRP-ATP6V0D1 locus contains three separate CpG islands whereas the mouse locus contains a single continuous CpG island. Enhancers Diiia and Diiib were found to be inside the gap between the second and third CpG islands.
We used in silico analysis21 to compare the presence of CpG islands between the human and mouse syntenic loci encompassing the AgRP-ATP6V0D1 genes. We found that the human AgRP-ATP6V0D1 locus contained three separate CpG islands whereas the mouse locus contained a single continuous CpG island that spanned the fourth exon of AgRP and the first exon of ATP6V0D1 (Fig. 8b). Enhancers Diiia and Diiib were positioned inside the gap between the second and third islands (Fig. 8b).
Discussion
Previous studies have identified upstream regions that direct expression of the mouse AgRP gene to the hypothalamus16 and identify conserved regions between the human and mouse first 760 nucleotides upstream of the translation initiator. 15 Little is known, however, about the functional structure of the human AgRP gene other than information about its minimal promoter that lies in the 5′ non-coding exon of the gene. 10; 14 In the present study we report intronic elements that are unique to the human and primate genomes that determine the spatiotemporal expression of the human AgRP gene.
Using an approach that involved the minimal promoter of the human AgRP gene and concatamers from upstream and downstream regions we identified sequences with silencer and enhancer effects with respect to the activity of the minimal promoter. Upstream of the translation initiator, we identified two regions (Aii and Aiv) that consistently reduced the activity of the minimal promoter (-18/+312), suggesting that they may encompass binding sites for silencers of gene expression. In contrast, in the intergenic region between AgRP and ATP6V0D1 we identified an element (Biib, 137 nt long) that significantly enhanced activity of the AgRP minimal promoter but only in adrenocortical cells.
Because we were interested in enhancers that were active mostly in neuronal cells we looked further downstream in the first exon/intron of the ATP6V0D1 gene. A region termed Region D, fulfilled this criterion and was gradually reduced to smaller fragments. Finally, sub-regions Diiia and Diiib from within the first intron of the ATP6V0D1 gene displayed enhancer effects in either neuronal (Diiia) or adrenocortical (Diiib) cells.
To confirm the tissue specificity of Diiia and Diiib, luciferase-expressing transgenic mice were generated using the exact same constructs that were used for the in vitro studies. Mice for both enhancers displayed robust expression of luciferase suggesting that the two “promoter-luc-enhancer” constructs were also effective in vivo. However, enhancer Diiia drove luciferase expression in the testis, the ears and the tail, instead of in neuronal cells as it had been predicted by the in vitro studies. Similarly, enhancer Diiib that was expected to be adrenocortical cell-specific drove expression more ubiquitously with greater concentration in the testis, the front limps, the spinal cord, and the tongue. We speculate that the difference in the outcome between the in vitro data and the transgenic mice may be due the presence of serum or other media factors that contribute to activation or suppression of the enhancers in cell culture. Two transgenic lines were established for enhancer Diiib from two founders and only one for Diiia from three founders. Two of the three Diiia founders did not produce transgenic colonies because they did not pass the transgene or completely failed to reproduce. Diiib mice in the homozygous state also became infertile which could be due to overexpression of luciferase in the testes. Hence, mice were bred always to wild type FVB, which is the parental strain of these transgenic mice, to maintain the hemizygous state.
The two enhancers displayed a diverse phenotype that could be due to differences in the available binding sites for transcription factors. Hence, we performed in silico analysis to compare the putative binding sites of transcription factors between enhancers Diiia and Diiib. We found that enhancer Diiib had a high abundance of binding sites for OCT-1 which is a ubiquitously expressed transcription factor found in almost every eukaryotic cell. 22 The more ubiquitous expression of luciferase in the Diiib mice could therefore be attributed to the high content of OCT-1 binding sites.
Enhancers Diiia and Diiib were located in the first intron of ATP6V0D1 that is an essential gene for survival. 23 Intronic enhancers have been shown to play significant roles in the spatiotemporal expression of their own genes24; 25 as well as heterologous, neighboring, genes. 26 ATP6V0D1 is a component (subunit D1) of the vacuolar-type H(+)-ATPase (V-ATPase) that is responsible for the acidification of endosomes, lysosomes, and other intracellular organelles. 27 ATP6V0D1 is expressed in the apical pole of narrow and clear cells in the epididymis and vas deferens where it is required for the maintenance of low pH that is essential for the maturation of sperm and its storage in a quiescent state. 28 Given the high intensity of the luciferase signal in the testis of both Diiia and Diiib mice, and the presence of AgRP in this organ as shown here by qPCR and as previously reported, 2 we used immunohistochemistry to identify the exact testicular cell types of AgRP expression. AgRP was co-localized in pachytene spermatocytes with the alpha-synaptonemal complex protein 3 (α-SCP3) that is a meiotic entry-specific protein previously shown to be present in pachytene spermatocytes. 17; 18 There was no luciferase signal in the epididymis and seminal vesicles of Diiia and Diiib mice (as measured by luminometric analysis not shown here) and AgRP was not detected in mature spermatocytes by real time PCR, which suggests that its expression is restricted to pachytene spermatocytes. The significance of AgRP's expression in pachytene spermatocytes warrants further investigation as earlier studies have shown that AgRP stimulates the hypothalamo-pituitary-gonadal axis thus potentially acting as a link between the control of appetite and reproductive function. 29
Because there was significant expression of luciferase in the tongue of Diiib mice, we examined whether AgRP is expressed in this organ. After all, AgRP has been proposed to play a role in food selection in rats and humans. 12; 30 qPCR confirmed that AgRP was expressed in the tongue but at much lower levels than in the hypothalamus. Using immunocytochemistry AgRP was colocalized in the epithelial cells with α-14-cytokeratin that is an epithelial cell-specific protein. 19; 20 The exact epithelial cell-type has not been determined but, based on the features of the structures, we speculate that AgRP in the tongue may be expressed in the Von Ebner salivary glands that secrete lingual lipase which begins the process of lipid hydrolysis in the mouth. 31 As in the case of its expression in the testis, AgRP's expression in the tongue warrants further investigation as the possibility that it may act as a link between energy balance, reproduction, appetite, and food selection appears plausible given its pattern of expression in these important organs.
A landmark feature of AgRP is its upregulation by fasting. 3 Hence we tested the hypothesis that enhancers Diiia and Diiib are involved in the regulation of AgRP by dietary fat and by fasting. In Diiia mice, LFD attenuated luciferase expression and subsequent feeding with VHFD reversed this effect. Overnight fasting also significantly attenuated the luciferase signal. These findings suggest that enhancer Diiia is not likely to play a role in the activation of AgRP by fasting but it can be regulated by dietary fat and/or the complete absence of food. In the Diiib mice, on the other hand, dietary fat content did not have a significant effect on luciferase signal but overnight fasting did enhance the signal. The effect was consistent in mice from both Diiib founders suggesting that it was specific to the enhancer and not due to the site of integration of the transgene in the two founders. We speculate that DNA-binding repressors may be activated (or activators repressed) by dietary and overnight fasting leading to differential regulation of the two enhancers. It has been reported that fasting of transgenic mice carrying large portions of regions upstream of AgRP (but missing the downstream sequences presented here) leads to increased reporter signal in the hypothalamus. 16 However, as discussed below, the human and mouse AgRP-ATP6V0D1 genomic structures are not well conserved suggesting that AgRP may be differentially regulated in the two species.
The conservation level of the AgRP-ATP6V0D1 locus was compared between the human and mouse to determine whether the two enhancers could also play a role in the mouse AgRP regulation. Indeed, conserved intronic enhancers in mammalian genomes play significant roles in gene regulation. 32; 33 We found high conservation levels between exons but the introns were poorly conserved. Furthermore, the human intergenic AgRP-ATP6V0D1 locus was 1.5 kb long whereas the mouse equivalent was only 0.8 kb, without any homologies between the two species. As such, the human Biib region that was found to have significant activity in the human adrenocortical cells (Supplemental data) does not exist in the mouse. Importantly, enhancers Diiia and Diiib were non-homologous with the mouse analogous region (i.e. equidistant from the “ATG” of the ATP6V0D1) but they were well-conserved in the genomes of the rhesus monkey and other primates. Further analysis of the AgRP-ATP6V0D1 locus using the DCODE database (ECR browser) confirmed the absence of the intergenic region in dogs, rodents, chicken, fish, and dogs, and re-affirmed the high homology of the two enhancers with the rhesus monkey genome. This finding presents a case of evolutionary complexity among mammalian species that has risen from diversification of the functional role of primate non-coding sequences. 34
The overall structure of the AgRP-ATP6V0D1 locus was also compare between the human and mouse using algorithmic analysis of the size and frequency of CpG islands. CpG islands are GC-rich regions that are commonly found around transcription start sites35 and are associated with gene silencing due to methylation. 36 We found that the human AgRP-ATP6V0D1 locus contained three separate CpG islands whereas the mouse equivalent locus contained only one. Of particular interest was the finding that human enhancers Diiia and Diiib were positioned inside the gap of the second and third CpG islands. This could explain their properties as enhancers as they are likely to escape methylation and silencing at native conditions. The lack of homology between the human and mouse AgRP-ATP6V0D1 orthologous regions and differences in their CpG island landscapes underlines significant differences of genomic structure between the two species at this locus.
The studies presented here describe two trans-acting enhancers that significantly affect promoter activity of the human AgRP gene. They were found inside the first intron of the neighboring ATP6V0D1 gene and were unique to primates. The two enhancers were constitutively active in multiple organs and were predicted to escape silencing due to their unique genomic locale in-between two CpG islands. Both enhancers responded to overnight fasting suggesting that they may be regulated by energy-dependent homeostatic factors.
Materials and Methods
Luciferase expression constructs
Luciferase expression constructs to evaluate enhancer or silencer effects of the genomic regions surrounding the AgRP locus, were made as previously described10 Specifically, the minimal promoter of the human AgRP gene14 covering the 5′ non-coding exon (-18/+312) was cloned into the pGL3-basic vector (Promega, Madison, WI) in its polylinker at the Nhe I and Sma I sites. Additional genomic regions upstream of the 5′ non-coding exon were subcloned immediately upstream of the -18/+312 portion also at the polylinker site but using different restriction enzyme sites (Kpn I and Nhe I). Downstream regions were cloned into the Bam HI and Sal I sites of the pGL3-basic vector that is downstream of in the natural 5′ to 3′ orientation. The primers used for the amplification and sub-cloning of the various regions into the pGL3-Basic vector are summarized in Table S1. All amplicons were directionally cloned and confirmation of integrity of the DNA was done by bidirectional sequencing with the manufacturers' (Promega, Madison, WI) recommended primers.
The various constructs are referred to by specific names that are also descriptive in terms of the length of the insert sequence. A common characteristic of all constructs is that they were compound: i.e. containing the minimal promoter -18/+312 to account for basal activity plus the additional sequences upstream or downstream of the minimal promoter. The only exception was construct Aiv(-3246/+373) that was one continuous segment of DNA. The numbering of DNA sequences for the description of the upstream constructs is based on the human AgRP gene sequence with Accession number AF314194 in combination with the genomic DNA clones from human chromosome 16q22 with Accession numbers AC027682 and AC009061. Nucleotide number “1” was the same as that shown in the publication by Brown and colleagues. 14
Cell culture
Cell culture was carried out under standard conditions in a humidified incubator at 37 °C and 5% CO2. NCI-h295R cells (human adrenal cortex cancer cell line) were grown in DMEM/F12 50/50 mix medium 500 ml (Cellgro) and in the presence of BD ITS+CULTURE SUPPLEMENT 5ml (BD Biosciences, Palo Alto, CA) and Nu-serum 13 ml (BD Biosciences, Palo Alto, CA), 0.75M HEPES 10.5 ml and penicillin-streptomycin to 1%. The N38 cells (mouse hypothalamic neuronal cell line) were grown in DMEM medium (Cellgro: Cat No: 10-017-cv), FBS to 10%, and penicillin-streptomycin to 1%. The ATT-20/D16V-F2 cells were grown in DMEM medium (ATCC) containing 10% FBS and 1% penicillin-streptomycin.
All cell lines were authenticated and routinely examined for the presence of mycoplasma. As a general rule, cell lines were not passed more than seven times.
Cells were transfected in triplicates for each construct and each (triplicate) experiment was performed at least twice in each cell line. Each (triplicate) experiment was averaged and the standard deviation was calculated. Transient cotransfections were also carried out with Renilla Luciferase plasmids as a control and for adjustment of the luciferase signal (Gene Therapy Systems, San Diego, CA). Cells were harvested using 1× Passive Lysis buffer and the lysates were assayed for luciferase and Renilla Luciferase activities, as prescribed by the assay manufacturer (Promega, Madison, WI) in a luminometer (Zylux Corporation, Pforzheim, Germany). All luciferase activity measurements were normalized to Renilla Luciferase values.
Double-labeling immunofluorescence
Double-labeling fluorescent immunohistochemistry was performed to examine whether AgRP colocalized with SCP3 in the testes. In brief, testis paraffin sections were deparaffinized in xylene, hydrated in decreasing concentrations of ethanol, washed in water and PBS, treated with antigen retrieval buffer (0.1 M Tris, 5% urea, pH 9.5) at 95 °C for 15 min, permeabilized in 0.1 % Triton X-100 for 15 min, pretreated with 10% normal goat serum in PBS, and then incubated in the following mixture: rat monoclonal antibody against AgRP 1:800 (Alpha Diagnostic) and rabbit anti-SCP3 1:1500 (Santa Cruz). Secondary Alexa Fluor 488 goat anti-rat and Alexa Fluor 594 goat anti-rabbit were diluted 1:200 in PBS and applied for 1 h at room temperature. Sections were mounted in Prolong Gold Antifade Reagent with DAPI.
For localization of AgRP in the tongue, immunohistochemistry was performed on longitudinal cryo sections. Sections were fixed in 4% paraformaldehyde, washed in PBS, treated with antigen retrieval buffer, permeabilized in 0.1 % Triton X-100, pretreated with 10% normal goat serum in PBS, and then incubated in the following mixture: rabbit polyclonal antibody against AgRP 1:1500 (Phoenix Pharmaceuticals) and mouse anti-cytokeratin-14 1:200 (Gene Tex. Inc.). Secondary Alexa Fluor 488 goat anti-mouse and Alexa Fluor 594 goat anti-rabbit were diluted 1:200 in PBS and applied for 1 h at room temperature. Sections were mounted in Prolong Gold Antifade Reagent with DAPI.
Double-labeled preparations in testes were analyzed using a confocal microscope (Zeiss Axiovert 200M + 510 META Multiphoton Confocal). For illustrations, the images were taken with ×40 oil immersion objectives. Localization of AgRP in tongue and testis and colocalization of AgRP and Cytokeratin-14 in tongue was examined using the 3I Everest Imaging System. Color images were captured with the Photometrics CoolSNAPHQ Monochrome camera under 10×, 20× or 40× oil magnification.
Transgenic and AgRP-/- mice
The study protocols for all mouse studies were approved by the Institutional Animal Care and Use Committee of the Pennington Biomedical Research Center. Mice were maintained on a 12h:12h day:light cycle in a pathogen-free animal facility. The Diiia and Diiib constructs were linearized by digestion with the Nhe I and Sal I restriction endonucleases. Gel-purified Diiia and Diiib inserts were used for pronuclear injections of FVB donor eggs and subsequent transplantation of embryos into FVB pseudopregnant females. Founders and subsequent generations were screened for presence of the transgenes by PCR. The same forward primer corresponding to the luc gene was used for both transgenics: 5′-TGTTTGTGGACGAAGTACCG-3′. Different reverse primers specific for each enhancer were used: Diiia, 5′-ATTACGAGGCTATCAGCACC-3′ and Diiib, 5′-ACTGGTCCAGTTTGCAAGGC-3′.
AgRP knockout (AgRP-/-) mice were the kind gift of Dr. Qian (Merck Laboratories, Rahway, NJ) 37. Mice used as controls for the studies presented here were interbred in our laboratory.
In vivo bioluminescence
In vivo bioluminescence imaging was conducted on a cryogenically cooled IVIS 100 system (Xenogen Corp., CA, USA) using LivingImaging acquisition and analysis software (Xenogen). Mice were anesthetized with isoflurane and received an i.p. injection of the substrate D-luciferin (100 mg/kg). An integration time of 3 min with a binning of 100 pixels was used for luminescence image acquisition. Signal intensity was quantified as the sum of all detected photon counts within the region of interest after subtraction of background luminescence. Mice were fed chow diet ad libitum and imaged the day before the start of the feeding experiment with PMI5011 and imaged again one week later.
Transcription factor prediction
The prediction of putative binding sites for transcription factors inside enhancers Diiia and Diiib was conducted by using the open access Alibaba2 (http://www.gene-regulation.com/pub/programs/alibaba2/index.html?) and TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess) databases.
Statistical and sequence analyses
Statistical significance was evaluated using one-way analysis of variance (ANOVA) and the students T-test were used for pairwise comparisons. The data were expressed as means ± S.D. P < 0.05 was considered as statistically significant.
The conservation status of enhancers Diiia and Diiib, as well as the surrounding regions, was examined by performing nucleotide BLAST comparisons (http://blast.ncbi.nlm.nih.gov/Blast.cgi). CpG island searches were performed using the web-based tool of Takai and colleagues (http://www.cpgislands.com/). 21
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Diabetes and Digestive and Kidney Diseases (Grant number: DK62156). The authors wish to thank Dr. Denise Belsham of the University of Toronto, (Ontario, Canada) for the kind gift of the N38 neuronal cells. We thank Dr. Adrian Stutz for assistance with the transgene plasmids and Dr. Michael Salbaum of the Pennington Biomedical Research Center for helpful discussions on the manuscript. We also thank the Transgenics Core, the Bioimaging Core (CBBC), and veterinary staff of Comparative Biology at the Pennington Biomedical Research Center.
Footnotes
Supplemental data: A file of supplemental data is provided.
Accession numbers: The AgRP-ATP6V0D1 intergenic and downstream nucleotide sequence data reported are available in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases under the Genbank accession number TPA: BK006637 It should be noted that the precise location of each enhancer is according to this GenBank accession number. The numbers used for the various enhancers are for reference and descriptive of the lengths of the inserts (subtract the first number from the second one to determine the length).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997;278:135–8. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- 2.Shutter JR, Graham M, Kinsey AC, Scully S, Luthy R, Stark KL. Hypothalamic expression of ART, a novel gene related to agouti, is up- regulated in obese and diabetic mutant mice. Genes Dev. 1997;11:593–602. doi: 10.1101/gad.11.5.593. [DOI] [PubMed] [Google Scholar]
- 3.Ilnytska O, Argyropoulos G. The Role of the Agouti-Related Protein in Energy Balance Regulation. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan W, Kastin AJ, Yu Y, Cain CM, Fairburn T, Stutz AM, Morrison C, Argyropoulos G. Selective tissue uptake of agouti-related protein(82-131) and its modulation by fasting. Endocrinology. 2005;146:5533–9. doi: 10.1210/en.2005-0578. [DOI] [PubMed] [Google Scholar]
- 5.Bewick GA, Gardiner JV, Dhillo WS, Kent AS, White NE, Webster Z, Ghatei MA, Bloom SR. Post-embryonic ablation of AgRP neurons in mice leads to a lean, hypophagic phenotype. Faseb J. 2005;19:1680–2. doi: 10.1096/fj.04-3434fje. [DOI] [PubMed] [Google Scholar]
- 6.Gropp E, Shanabrough M, Borok E, Xu AW, Janoschek R, Buch T, Plum L, Balthasar N, Hampel B, Waisman A, Barsh GS, Horvath TL, Bruning JC. Agouti-related peptide-expressing neurons are mandatory for feeding. Nat Neurosci. 2005;8:1289–91. doi: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- 7.Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–5. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 8.Bai F, Rankinen T, Charbonneau C, Belsham DD, Rao DC, Bouchard C, Argyropoulos G. Functional dimorphism of two hAgRP promoter SNPs in linkage disequilibrium. J Med Genet. 2004;41:350–3. doi: 10.1136/jmg.2003.014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Argyropoulos G, Rankinen T, Bai F, Rice T, Province M, Leon A, Skinner J, Wilmore J, Rao D, Bouchard B. The agouti related protein and body fatness in humans. International Journal of Obesity. 2003;27:276–280. doi: 10.1038/sj.ijo.802201. [DOI] [PubMed] [Google Scholar]
- 10.Mayfield DK, Brown AM, Page GP, Garvey WT, Shriver MD, Argyropoulos G. A role for the agouti related protein promoter in obesity and type 2 diabetes. Biochem Biophys Res Commun. 2001;287:568–573. doi: 10.1006/bbrc.2001.5600. [DOI] [PubMed] [Google Scholar]
- 11.Argyropoulos G, Rankinen T, Neufeld DR, Rice T, Province MA, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. A polymorphism in the human agouti-related protein is associated with late-onset obesity. J Clin Endocrinol Metab. 2002;87:4198–202. doi: 10.1210/jc.2002-011834. [DOI] [PubMed] [Google Scholar]
- 12.Loos RJ, Rankinen T, Rice T, Rao DC, Leon AS, Skinner JS, Bouchard C, Argyropoulos G. Two ethnic-specific polymorphisms in the human Agouti-related protein gene are associated with macronutrient intake. Am J Clin Nutr. 2005;82:1097–101. doi: 10.1093/ajcn/82.5.1097. [DOI] [PubMed] [Google Scholar]
- 13.Beltramo M, Campanella M, Tarozzo G, Fredduzzi S, Corradini L, Forlani A, Bertorelli R, Reggiani A. Gene expression profiling of melanocortin system in neuropathic rats supports a role in nociception. Brain Res Mol Brain Res. 2003;118:111–8. doi: 10.1016/j.molbrainres.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Brown AM, Mayfield DK, Volaufova J, Argyropoulos G. The gene structure and minimal promoter of the human agouti related protein. Gene. 2001;277:231–8. doi: 10.1016/s0378-1119(01)00705-3. [DOI] [PubMed] [Google Scholar]
- 15.Kaelin CB, Cooper GM, Sidow A, Barsh GS. Mammalian comparative sequence analysis of the agrp locus. PLoS ONE. 2007;2:e702. doi: 10.1371/journal.pone.0000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaelin CB, Xu AW, Lu XY, Barsh GS. Transcriptional regulation of agouti-related protein (Agrp) in transgenic mice. Endocrinology. 2004;145:5798–806. doi: 10.1210/en.2004-0956. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata M, Nishimori K. Mice deficient in Dmrt7 show infertility with spermatogenic arrest at pachytene stage. FEBS Lett. 2006;580:6442–6. doi: 10.1016/j.febslet.2006.10.066. [DOI] [PubMed] [Google Scholar]
- 18.Wong EC, Ferguson KA, Chow V, Ma S. Sperm aneuploidy and meiotic sex chromosome configurations in an infertile XYY male. Hum Reprod. 2008;23:374–8. doi: 10.1093/humrep/dem377. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa Y, Toyosawa S, Ishida T, Ijuhin N. Keratin 14 immunoreactive cells in pleomorphic adenomas and adenoid cystic carcinomas of salivary glands. Virchows Arch. 2000;437:58–68. doi: 10.1007/s004280000186. [DOI] [PubMed] [Google Scholar]
- 20.Shinozaki A, Nagao T, Endo H, Kato N, Hirokawa M, Mizobuchi K, Komatsu M, Igarashi T, Yokoyama M, Masuda S, Sano K, Izumi M, Fukayama M, Mukai K. Sebaceous Epithelial-Myoepithelial Carcinoma of the Salivary Gland: Clinicopathologic and Immunohistochemical Analysis of 6 Cases of a New Histologic Variant. Am J Surg Pathol. 2008 doi: 10.1097/PAS.0b013e318160852a. [DOI] [PubMed] [Google Scholar]
- 21.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–40. [PubMed] [Google Scholar]
- 22.Zhenilo S, Deyev I, Serov S, Polanovsky O. Regulation of oct-1 gene transcription is different in lymphoid and non-lymphoid cells. Biochimie. 2003;85:715–8. doi: 10.1016/s0300-9084(03)00121-4. [DOI] [PubMed] [Google Scholar]
- 23.Miura GI, Froelick GJ, Marsh DJ, Stark KL, Palmiter RD. The d subunit of the vacuolar ATPase (Atp6d) is essential for embryonic development. Transgenic Res. 2003;12:131–3. doi: 10.1023/a:1022118627058. [DOI] [PubMed] [Google Scholar]
- 24.Feng W, Huang J, Zhang J, Williams T. Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme. Mol Cell Biol. 2008;28:315–25. doi: 10.1128/MCB.01168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones I, Ng L, Liu H, Forrest D. An intron control region differentially regulates expression of thyroid hormone receptor beta2 in the cochlea, pituitary, and cone photoreceptors. Mol Endocrinol. 2007;21:1108–19. doi: 10.1210/me.2007-0037. [DOI] [PubMed] [Google Scholar]
- 26.Faurholm B, Cochrane S, Millar RR, Katz AA. Gene structure and promoter functional analysis of the marmoset type II GnRH receptor. J Mol Endocrinol. 2007;39:91–104. doi: 10.1677/JME-06-0064. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal AK, White PC. Structure of the VPATPD gene encoding subunit D of the human vacuolar proton ATPase. Biochem Biophys Res Commun. 2000;279:543–7. doi: 10.1006/bbrc.2000.4003. [DOI] [PubMed] [Google Scholar]
- 28.Pietrement C, Sun-Wada GH, Silva ND, McKee M, Marshansky V, Brown D, Futai M, Breton S. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod. 2006;74:185–94. doi: 10.1095/biolreprod.105.043752. [DOI] [PubMed] [Google Scholar]
- 29.Stanley SA, Small CJ, Kim MS, Heath MM, Seal LJ, Russell SH, Ghatei MA, Bloom SR. Agouti related peptide (Agrp) stimulates the hypothalamo pituitary gonadal axis in vivo & in vitro in male rats. Endocrinology. 1999;140:5459–62. doi: 10.1210/endo.140.11.7248. [DOI] [PubMed] [Google Scholar]
- 30.Hagan MM, Rushing PA, Benoit SC, Woods SC, Seeley RJ. Opioid receptor involvement in the effect of AgRP- (83-132) on food intake and food selection. Am J Physiol Regul Integr Comp Physiol. 2001;280:R814–21. doi: 10.1152/ajpregu.2001.280.3.R814. [DOI] [PubMed] [Google Scholar]
- 31.Fukami H, Bradley RM. Biophysical and morphological properties of parasympathetic neurons controlling the parotid and von Ebner salivary glands in rats. J Neurophysiol. 2005;93:678–86. doi: 10.1152/jn.00277.2004. [DOI] [PubMed] [Google Scholar]
- 32.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 33.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 34.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 35.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–5. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 36.Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: Causes and implications. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Qian S, Chen H, Weingarth D, Trumbauer ME, Novi DE, Guan X, Yu H, Shen Z, Feng Y, Frazier E, Chen A, Camacho RE, Shearman LP, Gopal-Truter S, MacNeil DJ, Van Der Ploeg LH, Marsh DJ. Neither Agouti-Related Protein nor Neuropeptide Y Is Critically Required for the Regulation of Energy Homeostasis in Mice. Mol Cell Biol. 2002;22:5027–35. doi: 10.1128/MCB.22.14.5027-5035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.