Abstract
Processing of novel and target stimuli in the auditory target detection or ‘oddball’ task encompasses the chronometry of perception, attention and working memory and is reflected in scalp recorded event-related potentials (ERPs). A variety of ERP components related to target and novelty processing have been described and extensively studied, and linked to deficits of cognitive processing. However, little is known about associations of genotypes with ERP endophenotypes. Here we sought to elucidate the genetic underpinnings of auditory oddball ERP components using a novel data analysis technique. A parallel independent component analysis of the electrophysiology and single nucleotide polymorphism (SNP) data was used to extract relations between patterns of ERP components and SNP associations purely based on an analysis incorporating higher order statistics. The method allows for broader associations of genotypes with phenotypes than traditional hypothesis-driven univariate correlational analyses. We show that target detection and processing of novel stimuli are both associated with a shared cluster of genes linked to the adrenergic and dopaminergic pathways. These results provide evidence of genetic influences on normal patterns of ERP generation during auditory target detection and novelty processing at the SNP association level.
Keywords: event-related potential, single nucleotide polymorphism, auditory oddball, P3 component, norepinephrine, dopamine, parallel independent component analysis
Introduction
The scalp recorded event-related potential (ERP) samples a sequence of cerebral processes leading to discrimination of a target or novel stimulus in an oddball experiment. The waveform comprises a number of ERP components, such as the early negative peak, N1, which peaks around 100ms post-stimulus, N2 at about 200ms and a P3 between 300–500 ms (Polich, 2003; Polich and Kok, 1995). The P3 has been most widely studied to investigate cognitive processes such as attention, memory and decision making processes (Donchin and Coles, 1988; Hansenne, 2000; Kugler et al., 1993; Picton, 1992; Polich, 2007) in healthy controls as well as in a wide range of mental illnesses, such as schizophrenia and Alzheimer disease, where P3 responses are abnormal (Antal et al., 2000; Blackwood, 2000; O’Donnell et al., 2004; Polich and Corey-Bloom, 2005; Turetsky et al., 2000). ERPs elicited by deviant stimuli interspersed with repetitive standard elements demonstrate two subcomponents of P3, a earlier positive peak (~250–270 ms) with frontocentral distribution elicited by task-irrelevant distracters (P3a) or novel stimuli (Novelty P3), indicating an automatic/bottom-up shift of attention, and a later (~300–350 ms) parietal P3b that is elicited by task-relevant stimuli, which is particularly sensitive to probability, stimulus sequence, and target-to-target interval (Croft et al., 2003; Squires et al., 1976; Sutton et al., 1965).
The generating mechanisms of ERPs, in particular those of P3, are of great interest (Polich and Criado, 2006). Generator sites of P3 potentials are widespread across the brain and involve multiple regions in the frontal, temporal and parietal lobes (Eichele et al., 2005; Kiehl et al., 2005). Javitt and colleagues recently reviewed the underlying pathophysiological mechanisms for schizophrenia, and P3 abnormality was associated with abnormal dopaminergic function, as well as GABA-ergic and cholinergic functions (Javitt et al., 2008). To date, the locus coeruleus noradrenergic system and the ventral tegmental area and substantia nigra dopaminergic pathways have been proposed as dominant transmitter systems for P3 responses (Nieuwenhuis et al., 2005a, Polich, 2007). This is (indirectly) supported by fMRI activations in these areas elicited by novelty processing and predictive coding, functions known to drive the P3 (Bunzeck and Duzel, 2006; D’Ardenne et al., 2008).
In twin and family studies, ERP measures show high degrees of heritability (van Beijsterveldt and van Baal, 2002). The P3a and P3b show similar high heritability ranges from 0.6 to 0.8 (Frangou et al., 1997; van Beijsterveldt and Boomsma, 1994). These findings suggest that ERPs can provide reliable endophenotype information for the pursuit of a genetic basis of brain functions and their generation mechanism. However, little is known about the genetic underpinnings of ERPs, the knowledge so far is derived from very limited studies on the P3 (Begleiter et al., 1998; Hammond et al., 1987; Hansenne, 2000; Mulert et al., 2006). Somewhat divergent results show that the P3 is connected with a set of scattered dopamine-related (Mulert et al., 2006), adrenergic related and cholinergic-related genes (loci), and also mapping to additional loci on chromosomes 2 and 6 (Begleiter et al., 1998). In the majority of studies of genetic influences to normal or disordered brain processes performed so far, a small set of genes were preselected based on prior information, and then investigated using a hypothesis-driven approach (Cedazo-Minguez, 2007; Mulert et al., 2006; Vawter et al., 2004b). To avoid problems associated with gene pre-selection biases, here we developed a data-driven method for larger, unbiased, gene pools.
The purpose of the present study was to explore the genetic influences on the ERP using a set of 384 single nucleotide polymorphisms (SNPs) located in 222 genes. This array was initially designed for metabolism studies and includes genes from different physiological systems. We used a three-stimulus auditory oddball discrimination task to elicit target and novelty related responses, yielding the N1, N2b, P3a and P3b component peaks in the ERP waveforms. To identify the underlying relationship between SNP patterns and ERP patterns, where the SNP patterns present associations encoding different physiological processes and ERP patterns reveal independent event-related brain processes, we assumed that multiple associations exist in the SNP pool, and multiple brain functions are represented in the ERP such that the potential connections between them are inherently multivariate. In other words, we proposed that SNP associations may contribute to biological/cognitive functions and that the functions may affect portions of the ERP waveform. This idea is supported by recent findings revealing the associations of multiple SNPs with complex cognitive processes (Roffman et al., 2006; Seshadri et al., 2007). Following this assumption, we adapted a blind source separation method that affords a parallel multivariate decomposition of data from different modalities, based on independent component analysis (ICA). Parallel ICA provides a way to extract the overall strongest connections between two modalities from a multivariate perspective. It has been previously applied to combine functional magnetic resonant imaging and genetic data collected from schizophrenia patients and controls (Liu et al., 2008; Liu et al., 2009), and was able to extract a set of genes linked to schizophrenia.
In this paper, parallel ICA of ERPs and SNPs is employed to extract relationships between patterns of ERP components and SNP associations. We show that novelty and target processing are associated with a common cluster of genes linked to the adrenergic and dopaminergic pathways. These genetic influences on ERP generation during auditory novelty and target processing help to understand the signal generation mechanisms underlying these brain responses, and may ultimately help identify multimodal diagnostic markers of mental illnesses.
Materials and Methods
Subjects
We selected data from 41 healthy Caucasian participants (24 female, age 39±19; 17 male, age 38±14) who were recruited for several related studies at the Olin Neuropsychiatry Research Center at the Institute of Living in Hartford (CT). All participants were free from any history of DSMIV Axis I or Axis II psychopathology as assessed with the SCID (structured clinical interview for diagnosis). All participants provided written, informed, IRB-approved consent at Hartford Hospital.
Experiment design
The auditory oddball discrimination task consisted of responding to an infrequent target sound within a series of frequent standard sounds and infrequently presented task irrelevant novel sounds, as used in our prior studies (Kiehl et al., 2005). The standard stimulus was a 500 Hz tone, the target stimulus a 1000 Hz tone, and the novel stimuli consisted of non-repeating meaningless sounds (e.g., tone sweeps, whistles). The target and novel stimuli occurred with a probability of 0.10 each; the standard stimuli occurred with a probability of 0.80. Each stimulus was presented to participants by a computer-controlled sound system that delivered the auditory stimuli with a 200 ms duration and a 2000 ms stimulus onset asynchrony. Participants were instructed to respond as quickly and accurately as possible with their right index finger every time they heard a target stimulus and not to respond to standards or novels.
Data acquisition
EEG/ERP: Electroencephalograms (EEG) were acquired continuously while participants were performing the auditory oddball task. Data were collected using an SA bioelectric amplifier system capable of amplifying electrical activity from 64 separate single-ended channels. Scalp potentials were recorded from tin electrodes (ElectroCap International) placed over 62 electrode sites according to standard placement guidelines of the International 10–20 System and some additional sites. (EOG) was recorded from electrodes located on the lateral and supra-orbital ridges of the right eye. All electrodes were referenced to the nose. Electrical impedances were maintained below 10 kΩ throughout the experiment. The EEG channels (SA instruments) were amplified (20,000 gain) with a band pass filter of 0.01 to 100 Hz, digitized on-line at a rate of 500 samples per second, and recorded on computer hard disk.
Continuous EEG recordings were filtered by a high pass 0.05 Hz filter, preprocessed using independent component analysis (ICA) to remove eye blink artifacts. The ICA utility built in the EEGLAB software (Delorme and Makeig, 2004) was used to decompose EEG recordings to independent components, and then an in-house template matching algorithm (Jung et al., 2000) was used to identify eye blinks (see supplement Figure 1 for the eye blink template). Then, EEG signals were segmented from 210 ms pre-stimulus to 1050 ms post-stimulus. ERPs were constructed for trials in which participants correctly responded, and those trials were baseline corrected, artifacts rejected with a 100 uV maximum amplitude safeguard, averaged and filtered with a 30Hz low pass filter. The averaged epoch was 1250 ms long with a 200 ms pre-stimulus baseline.
SNP data: A blood or saliva sample was obtained for each subject and DNA was extracted. Genotyping was performed using the Illumina BeadArray™ platform and the GoldenGate™ assay (Fan et al., 2003; Oliphant et al., 2002). The PG Array of Genomas Inc. (Hartford, CT, USA) was used, which contains a SNP array consisting of 384 SNPs from 222 genes from 6 physiological systems: neurobiology, cardiovascular system, inflammation, metabolism, cholesterol biochemistry, and cell proliferation (Liu et al., 2009). The following pathways were represented: neurotransmitter axes (serotonin, dopamine cholinergic, histamine and glutamate), apolipoproteins and receptors, insulin resistance, glucose metabolism, energy homeostasis, adiposity, fatty acid and cholesterol metabolism, lipases, cell signaling and transcriptional regulation, growth factors, drug metabolism, blood pressure, vascular signaling, endothelial dysfunction, coagulation and fibrinolysis, vascular inflammation, cytokines and behavior (satiety). Genotyping analysis software, GenCall, was used to cluster the resultant intensities from the genotyping microarray into three clusters: AA, AB, and BB without assuming dominant or recessive inheritance. SNPs passing quality control were selected, resulting in 326 SNPs. Genotypes are inherently categorical and were represented as discrete numbers, e.g. 1 for one type of homozygous, 0 for heterozygous and −1 for the other type of homozygous. Negative and positive signs are not important in our application, since we look at genotype variation.
Data analysis and statistics
Novel and target ERPs were analyzed separately, in conjunction with the SNP data. The method to analyzing data is parallel ICA, developed in-house and publicly available at (http://icatb.sourceforge.net/fusion/fusion_startup.php). Parallel ICA is a variant of ICA designed for the processing of multimodal data, which maximizes the independence within modality using an entropy-based cost function (Bell and Sejnowski, 1995), while also identifying inter-modality correlations through an additional cost function. The total cost function scales independent information carried by entropy terms and connections between different modalities in a balanced way. Just like ICA, parallel ICA assumes for both modalities that the observation is a linear mixture of independent components, and there is no explicit noise model and particular requirement for input data type. The mixing processes in the two modalities are assumed to be linked in parallel ICA, i.e. components are mixed into observations in a similar or related way. A full description of the parallel ICA algorithm is found in (Liu et al., 2008; Liu et al., 2009). Here, parallel ICA was utilized to extract the independent components in ERPs and SNPs, and to identify the connections between two modalities.
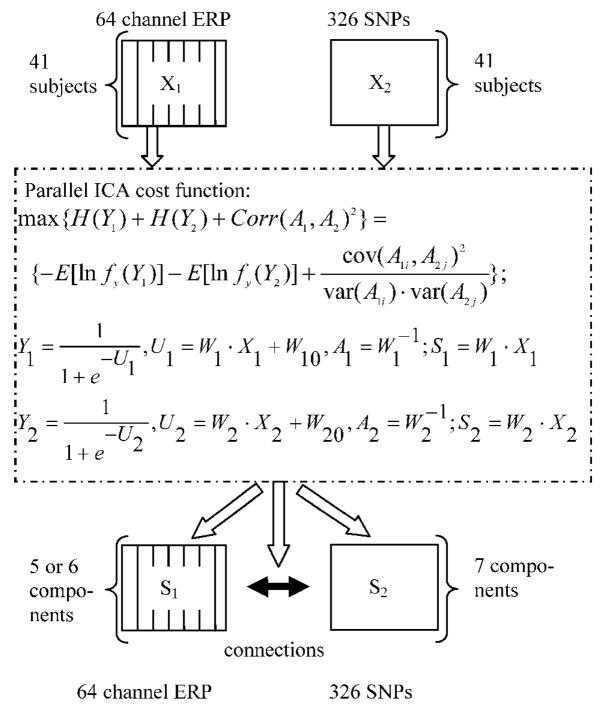
ERP data were constructed as a matrix of subjects-by-ERPs, where ERPs were averaged epoch responses concatenated from 64 channels’, i.e. containing the entire spatio-temporal information available from an averaged dataset. SNP data were organized as a matrix of subjects-by-SNPs. These two matrices were the inputs for the parallel ICA as illustrated in Figure 1. In this work, an ERP component is a weighted set of ERP time points which is maximally temporally independent of the other ERP components, presenting an underlying neuroelectrical signal. A SNP component is a weighted set of SNPs which is maximally independent of the other SNP components. The SNPs with large weights within a given component contribute significantly. We assumed each ERP or SNP component appears to different extents in different subjects. The ERP and SNP components are linked by virtue of their inter-subject variability, i.e. the loading pattern of the SNP and ERP components in 41 subjects. Figure 1 provides a schematic illustration of the parallel ICA implementation in this study. X1 and X2 are the preprocessed ERP and SNP data, respectively. S1 and S2 are the extracted components of ERP and SNP, representing sources. A1 and A2 are the mixing/loading parameter matrices for ERP and SNP components respectively, and they both compose of vectors of 41 elements, with one vector corresponding to one component’s loading pattern in subjects. The correlations of A1 vectors with A2 vectors are the aimed relation between SNP and ERP in the analysis, which enables us to evaluate the likely possibility that multiple SNPs contribute to a given ERP component of interest.
Figure 1.
Illustration of parallel ICA implemented in ERP and SNP. Data from SNP and ERP are entered into an optimization algorithm with a cost function listed in the dash line box, based on entropy terms ( H(Y1) and H(Y2) ) and a correlation term between A1 and A2. Y1 and Y2 are functions of components used to calculate entropy terms. W10 and W20 are bias weights. W1 and W2 are the weights to find components. As results, independent components are extracted from both modalities, as well as their possible correlations.
Estimation of the number of components is still an open and challenging problem. The Akaike information criterion (AIC) is one of validity functions based on information theory to determine data dimensionality (Akaike, 1974). But AIC tends to overestimate component number (De Ridder et al., 2005). To mitigate over-estimation of the dimensionality we used the AIC estimated number as a beginning point and reduced the component number until the resultant components were consistent among different leave-one-out validation runs. The AIC estimated 15 for target ERP data, 13 for novelty ERP data, and 7 for SNP data. Based on component’s consistency, the number of components was finally set to six for the target ERPs, five for the novelty ERPs, respectively, and seven for the SNP data.
We performed the leave-one-out cross evaluation, used in the component number estimation procedure, to validate findings in the study by evaluating the consistency of the SNP/ERP components and their inter-correlations on a random subset of the data with the same parallel ICA specifications. Since the method aims at finding relations between extracted components from both modalities, the derived correlations, therefore, are those between five or six ERP components with seven SNP components. Bonferroni correction for multiple comparisons is performed on the 35 or 42 possible correlation coefficients.
The output of the parallel ICA comprises paired independent components from both modalities. Each ERP independent component (IC) had time courses from 64 channels. The scalp distribution of each IC was identified using the peak value of each channel. Each SNP IC was a SNP association with contribution weights for 326 SNPs. A threshold on the weights (Z score =2.5, set empirically around 1% probability cutoff for an alternation on a normal distribution) was applied to select the dominant/important SNP loci of great effect for each SNP component. The cross-correlations among the selected SNPs were also evaluated to provide an indication of genetic interplay. Possible confounds of the ERPs, i.e. age and gender were also evaluated using linear regression.
Pathway analysis
To have a better understanding of the mechanism by which the set of genes modulate the generation of event related responses, most likely canonical pathways that genes of interest derived from the selected SNPs are involved were indentified using Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA, USA). First, the genes of interest were entered into a pathway entity list. Then, from the Ingenuity’ knowledge base, a repository of molecular interactions, regulatory events, gene to phenotype associations and chemical knowledge, the entities including genes, chemicals, enzymes, complexes and etc. bridging genes of interest with direct and indirect relationship (allowing one entity in between) were added into the list. Finally, well-characterized signaling and metabolic pathways were assigned to the pathway analysis list, when more than 2 entities are included in the pathway.
Results
Target-related independent components
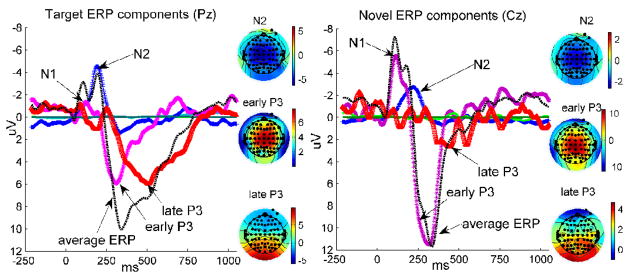
The multi-channel, multi-subject ERP in response to targets was decomposed into six independent components representing independent brain processes (Figure 2, Left), among which one overlapped with the N2 peak (−6.22μV peaking at 196ms and Cz location), another with the early P3 (7.41μV peaking at 304ms at Cz location, and peak width of 233~382 ms measured at half maximum), and a third with a later potion of P3 (5.98μV peaking at 472ms Pz location, peak width of 344~720 ms measured at half maximum). The N1 peak was partially accommodated by the early and late P3 ICs. These three ICs accounted for 97.8% of the variance of the averaged ERPs in all channels. The remaining three ICs had non-distinct/noisy time courses (light and dark green lines in Figure 2, Left), contributing little to the variance of the event-related responses (peak values at Cz location are 0.34±4.33μV, 0.06±3.13μV and 0.05±1.42μV in 41 subjects, respectively), and these were omitted from further analysis. Figure 2, Left shows the averaged ERP time course and IC time courses reconstructed at a parietal midline location (Pz). The black dashed line is the averaged target ERP, the colored lines correspond to the ICs. The scalp distributions of the ICs of interest were also plotted. The N2 IC is distributed in central/frontal region of the scalp. The early P3 IC is frontocentrally located, and the late P3 IC localized to parieto-occipital electrode sites, with polarity inversion over frontal sites.
Figure 2.
Independent components extracted from event related potentials. Left: the averaged ERP waveform responding to targets and six extracted ICs are plotted at location Pz, to have a full strength view of the late P3 IC. The scalp distributions of the N2, early P3 and late P3 ICs are presented with color indication illustrated. The other three ICs are waveforms with relatively smaller amplitude. Right: the averaged ERP waveform responding to novelty and five extracted ICs are plotted at location Cz, to have a better view of the early P3 IC. The scalp distributions of the N2, early P3 and late P3 ICs are also presented with the same color mapping. The other two ICs have waveforms with relatively smaller amplitude.
Novelty-related independent components
Five event-related independent components were extracted from ERPs responding to novel stimuli, including an N2 IC (−2.72μV peaking at 222ms Cz location), an early P3 IC (11.48μV peaking at 318ms and Cz location, peak width of 231~370 ms measured at half maximum) and a small late P3 IC (2.94μV peaking at 450ms and Pz location, peak width of 348~487 ms measured at half maximum), accounting for 99.0% of the variance. The N1 peak was mainly contained in the early P3 IC. Two further ICs (shown in Figure 2, Right, light and dark green lines) were identified as ICs of no interest, since no distinct peak waveform was recognized (peak values at Cz location are 0.35±1.93μV and 0.003±4.72μV in 41 subjects). Figure 2, Right shows the averaged ERP time course and IC time courses at a central midline location Cz. The black dashed line is the averaged novel ERP, and the colored lines are IC time courses, where the late P3 IC is very marginal. The N2 IC is distributed in the central/frontal part of the scalp. The early P3 IC localized to central/frontal areas, and the late P3 IC localized to the parietal areas.
Similarity and difference between target-related and novelty-related ICs
Results from target and novel ERPs were identified across separate decompositions, yet showed strong similarity, besides differences, in the extracted independent components. They both included three important ICs: the N2, early P3 and late P3. These three ICs shared similar pattern of time courses and scalp topographies, and differed mainly in amplitude across conditions. In target responses, the early and late P3 ICs had compatible amplitudes (statistics in 41 subjects: 7.58±.12μV vs. 6.07±4.97μV, no significant difference in two-sample t-test: t=1.02, p =0.31). The novel early P3 IC had relatively larger amplitude compared to the late P3 IC to novel stimuli (statistics in 41 subjects: 11.52±6.35μV vs. 2.982.92μV, significant difference in two-sample t-test t=7.83, p<1E-10). The late P3 components in target ERPs and novelty ERPs show significant difference in terms of amplitude with p <0.001; and the early P3 component also presents differences in amplitude between target and novelty responses with p<0.05. The N1 peak was clustered into P3 ICs (mainly the early P3 IC) under both conditions. From the peak time of time course and scalp distribution map, the early P3 IC is identifiable as the P3a, and the late P3 IC similar to that expected for the P3b. Hereafter, the early P3 IC is termed as P3a IC; the late P3 IC is named as P3b IC.
Connection between target-related ICs and SNPs
The strongest correlation extracted by parallel ICA between the target ERPs and the SNPs was r = 0.51±0.08 (from 50 runs with p<0.0012, Bonferroni corrected) between the P3b IC plotted in Figure 2, Left, and selected SNPs in a SNP component as listed in Table 1-A, including eleven loci. Leave-one-out cross validation of the relation between target ERP and SNP shows that the P3b and the SNP component were correlated with coefficient of 0.43±0.15, and with one sample t-test against zero t=17.55, p<1e-19. No other connection was significant after Bonferroni correction. The loading parameter of the P3b IC in subjects was also related to age with a correlation of r =−0.40 (p<0.01, one correlation test only). No significant relation was found with gender. 17% of the variance of the P3b IC loading pattern in subjects was explained by the SNP association and 2% by age.
Table 1.
The SNP components linked with target and novelty responses
| A | B | ||||
|---|---|---|---|---|---|
| SNPs linked with the target response |
SNPs linked with the novel response |
||||
| SNP loci | Z scores | Genes | SNP loci | Z scores | Genes |
| rs1800545 | 4.00 | ADRA2A | rs1800545 | 4.0105 | ADRA2A |
| rs7412 | 3.73 | APOE | rs6578993 | 3.4649 | TH |
| rs6578993 | 3.22 | TH | rs7412 | 3.3802 | APOE |
| rs1128503 | −2.67 | ABCB1 | rs2278718 | −3.059 | MDH1 |
| rs3842726 | 2.59 | TH | rs429358 | −2.9739 | APOE |
| rs429358 | −2.58 | APOE | rs521674 | −2.7814 | ADRA2A |
| rs2278718 | −2.55 | MDH1 | rs3813065 | 2.6592 | PIK3C3 |
| rs1045642 | −2.51 | ABCB1 | rs4121817 | 2.6592 | PIK3C3 |
| rs521674 | −2.50 | ADRA2A | rs1128503 | −2.5183 | ABCB1 |
| rs3813065 | 2.49 | PIK3C3 | |||
| rs4121817 | 2.49 | PIK3C3 | |||
Connection between novelty-related ICs and SNPs
The strongest correlation between novelty related ICs and SNPs was 0.50±0.08 (from 50 runs with p<0.0014, Bonferroni corrected). The correlated independent components were the P3a IC plotted in Figure 2, Right and the paired SNP component with nine selected SNPs listed in Table 1-B. Leave-one-out cross validation of the relation between novelty ERP and SNP shows that the P3a and the SNP component was correlated with coefficient of 0.47±0.09, one sample t-test t=31.60, p<1e-29. No significant connection between either N2 IC or P3b IC with any SNP association was identified after Bonferroni correction. The loading parameter of the P3a IC in subjects was related to age with a correlation of r =−0.53 (p<0.01, one correlation test only). No significant relation was found with gender. The variance of the P3a IC loading pattern was explained 6% by the SNP association and 14% by age.
Association of specific SNPs linked to event-related ICs
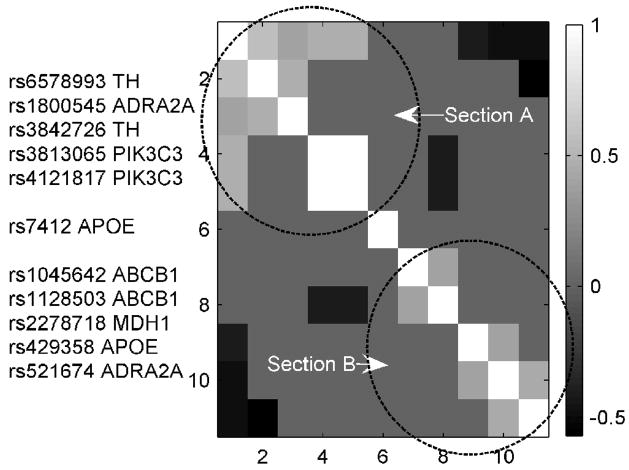
A total of 11 SNPs appeared in the target ERP-SNP and novel ERP-SNP links, which were rs1800545, rs7412, rs1128503, rs3842726, rs6578993, rs1045642, rs2278718, rs521674, rs429358, rs3813065 and rs4121817. These SNPs derive from 6 genes: alpha-2A-adrenergic receptor (ADRA2A), apolipoprotein E (APOE), tyrosine hydroxylase (TH), malate dehydrogenase 1, NAD (MDH1) and ATP-binding cassette, sub-family B, member 1 (ABCB1), and phosphoinositide-3-kinase, class 3 (PIK3C3). Figure 3 shows the cross correlations among the genotypes of 11 SNPs in 41 subjects with absolute value larger than 0.4. Two distinct sections are denoted in the figure: SNPs within section A are correlated positively, as do SNPs within section B. However SNPs in section A are negatively correlated with SNPs in section B. This clustering was also supported by the z-scores indicating SNP contributions to the genetic component (Table 1).
Figure 3.
Cross correlations of 11 SNPs from 6 genes. 11 SNPs are grouped into two sections based on their genotype patterns in 41 subjects. SNPs in section A are positivity related, SNPs in section B are positively related, but SNPs in Sections A and B show negatively correlated patterns.
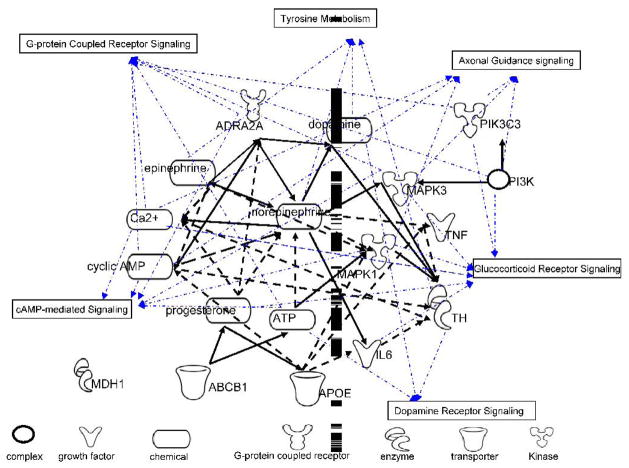
Pathway analyses
The biological significance of a subset of selected genes was further analyzed using biological networks and canonical pathways. The goal of this analysis was to identify possible pathways in which genes of interest may regulate the ERP generation. As shown in Figure 4, six canonical pathways were identified including tyrosine metabolism, dopamine receptor signaling, G-protein receptor signaling, cAMP-medicated signaling, axonal guidance signaling, and glucocorticoid receptor signaling. The nodes are the entities in the analysis list. The node shapes denote complex, enzyme, growth factor, chemical, G-protein coupled receptor, kinase and transporter, respectively. The black edges are biological relationships between nodes (solid line, direct association; dashed line, indirect association). All edges are supported by canonical information stored in the Ingenuity Pathways Knowledge Base. The blue lines indicate involvement in the identified pathways. MDH1, APOE, and ABCB1 are not involved in the selected pathways, and MDH1 does not have any direct and indirect connection with any other gene in the selected set of genes.
Figure 4.
Pathways analysis on the set of genes contained in the SNP component. Nodes are the entities in the analysis list, including genes of interest and entities bridging them. A black edge represents a relationship between two nodes. Blue lines show the node’s involvement with a canonical pathway. Six canonical pathways were indentified to be most likely involved from Ingenuity pathway knowledge base.
Discussion
We designed this study to investigate the genetic underpinning of target detection and novelty processing as indexed by independent components contributing to scalp recorded event-related responses. The P3 and its subcomponents consistently appear as a major response in auditory oddball tasks and since the subcomponents show similar levels of inheritance based on family studies, we hypothesized that their genetic sources likely share a common origin. We also hypothesized that these components would be linked to norepinephrine and/or dopamine related genes (Javitt et al., 2008; Nieuwenhuis et al., 2005a; Nieuwenhuis et al., 2005b; Polich, 2007).
Based on the results derived from target ERPs, the P3b IC was linked to a set of SNPs, whereas in novel ERPs the P3a IC was shown to have a SNP association. The SNP association linked to the P3b in Table 1-A is very similar to that linked to the P3a in Table 1-B (9 out of 11 SNPs are the same), suggesting that the two SNP associations are essentially identical. Our finding that these ICs share a common genetic origin is consistent with phenotypic findings from twin and family studies (van Beijsterveldt and Boomsma, 1994; Frangou et al., 1997).
The genetic source extracted by parallel ICA in our study was a group of SNPs from 6 genes coding for ADRA2A, TH, APOE, MDH1, ABCB1, and PIK3C3. These SNPs were clustered into two sections (Figure 3). These two sections were both centered at ADRA2A, which mediates adrenergic synaptic transmission. TH is involved in the conversion of tyrosine to dopamine, a precursor to norepinephrine and then to epinephrine. Not only does TH play a key role in the tyrosine metabolism pathway, but also, as illustrated in Figure 4, is the rate-limiting enzyme in catecholamine synthesis. Dopamine is converted to norepinephrine by dopamine beta-hydroxylase in some neuronal populations, such as the noradrenergic neurons in the locus ceruleus (LC). Both alpha-1 and the alpha-2 adrenergic receptors are present in LC and likely have an important role in behavioral activation related to novelty (De Sarro et al., 1987). A recent study showed that such LC Alpha-1 receptors are activated not only by norepinephrine, but also by dopamine, which is an endogenous agonist for behaviorally activating LC alpha-1receptors in response to novelty (Lin et al., 2008). Although alpha-1 rather than the alpha-2 adrenergic receptors indicated in our SNP component were identified in this study, the LC has projections to the regional cortical sources of the P3 and plays a key in the P3 generation; determining Norepinephrine/Dopamine interactions in the context of novelty-related activation sheds interesting light on our findings. Altogether, these results support the previously conjectured neurophysiology model of adrenergic and dopaminergic pathways in the state of arousal and attention (Nieuwenhuis et al., 2005a; Polich, 2007).
Phosphoinositide-3-kinases are involved in both receptor-mediated signal transduction and intracellular trafficking. Specifically, PIK3C3 functions in many signaling pathways, and in our study it appears in three pathways: glucocorticoid receptor signaling, axonal guidance signaling and G-protein coupled receptor signaling, which influence brain developmental processes. In particular, PIK3C3 promoter variants have been associated with the development of bipolar disorder and schizophrenia (Lencz et al., 2007; Stopkova et al., 2004). The P3 also presents different levels of associations with bipolar disorder and schizophrenia, respectively (O’Donnell et al., 2004; Turetsky et al., 1998; Turetsky et al., 2000). Given the involvement of PIK3C3 in signal transduction, it is likely that the protein participates in the amplification of the initial dopamine or NE-derived signal during the generation of the P3.
MDH1 catalyzes the reversible oxidation of malate to oxaloacetate. The protein encoded by this gene is localized to the cytoplasm and may play pivotal roles in the malate-aspartate shuttle that operates in the metabolic coordination between cytosol and mitochondria (Musrati et al., 1998). Decreased expression of this gene has been observed in the prefrontal cortex (Middleton et al., 2002; Vawter et al., 2004b) and peripheral lymphocytes of patients with schizophrenia (Vawter et al., 2004a), suggesting that this gene may be involved in the metabolic abnormalities seen in the patients. Event-related response differences between schizophrenia patients and healthy controls might be potential outcome effects of the metabolic abnormalities.
APOE is involved in cholesterol transport, but has been shown to be involved in several neurotransmitter (such as cholinergic, noradrenergic and serotonergic) projection pathways, influencing prefrontal, parietal cortex and the hippocampus in multiple ways (Chapman and Michaelson, 1998). Moreover, APOE has been marked as an important risk gene for Alzheimer’s disease and other neurological diseases (Cedazo-Minguez, 2007; Higgins et al., 1997; Laskowitz and Vitek, 2007). P3 amplitude reduction and latency prolongation have been repeatedly observed in Alzheimer’s disease patients (Polich and Corey-Bloom, 2005) and APOE ε4 allele carriers exhibit reduced P3 amplitude prior to the emergence of clinically relevant cognitive symptoms (Reiman et al., 2001). The membrane-associated protein encoded by ABCB1 is a member of the superfamily of ATP-binding cassette transporters, responsible for decreased drug accumulation in multidrug-resistant cells and also functions as a transporter in the blood-brain barrier. Specific polymorphisms in this gene have been associated with alterations in drug uptake to the central neural system (Basic et al., 2008; Kim et al., 2008). Although we do not know precisely how ABCB1 and APOE interact with dopamine or NE-derived signal transduction pathways, we hypothesize that the above genes may be responsible for signal amplification, thus influencing response magnitude and duration.
We also tested possible confounding effects from age and gender, and the results show that age is a significant factor influencing response magnitude, which has been identified in the literature (Polich, 1997). Based on relational analyses between age and the expression of individual genes, both TH and APOE showed a significant correlation with age (p<0.01). One possible explanation for the age effect could be through its interaction with the APOE and TH alleles. APOE ε4 interacting with aging is a risk factor for the common, late-onset form of Alzheimer’s disease (Serretti et al., 2007), and it is associated with age-associated/dependent symptoms. For example, a decrease in cerebrospinal fluid beta-amyloid 42 concentration was significantly and substantially greater in subjects with the APOE ε4 allele compared with those without the APOE ε4 allele in adults with normal cognition (Peskind et al., 2006). The significant increase in TH mRNA expression in aged animals suggests an enhanced synthetic capability to compensate for LC cell loss (Shores et al., 1999), which directly affects P3 generation.
This report reveals the possible genetic influences on ERP at the SNP association level. Even though the ERP hosts distinct components, from prior research most likely linked to different neurotransmitter systems, our data suggests they appear to be closely linked genetically, as expected from their close neurophysiologic association and similar heritability. In depth studies of the identified genes are by all means necessary in order to fully understand how those genes shape the brain processes. The method used in the study, parallel ICA, is a data driven multivariate association approach. Accurate estimation of component number is required and findings are purely based on data statistics and therefore verifications based on their biological interpretations or other biological-based (such as molecular experiments) approaches are necessary. As we discussed above, some of our findings are highly consisted with ERP generation hypotheses in literature and others are not expected but logically implicated. Overall, the findings are informative and interesting, providing new perspective to understand the ERP generation. Further studies with replication in larger population samples and molecular level investigation are needed for any broader conclusion.
We believe that these results and parallel blind source decomposition methodology have implications for future genomic studies on the understanding of normal brain function and its alternations in mental illnesses. The multivariate method helps to identify complex objective biological measures of psychopathological variables, and such findings may aid in early diagnosis of mental illnesses and lead to development of individual treatment strategies on a genetic variation basis.
Acknowledgments
This research was supported by the National Institutes of Health, under grants R01 EB 000840, R01 EB 005846 (to Dr. Calhoun), and RO1 MH43775, RO1 MH074797 and R01 MH077945 (to Dr. Pearlson), and R01 MH 071896-01, R01 MH 070539-01 and R01 MH 072681-01 (to Dr. Kiehl), and K23 MH 070036-01 (to Dr. Stevens). Dr. Eichele was supported by the L. Meltzer foundation (801616). Particular thanks go to Dr. Michael Stevens who has generously provided us the data for the study. We would also like to thank Genomas Inc. who conducted genotyping for all subjects, and in particular, give thanks to Dr. Gualberto Ruaño and Dr. Andreas Windemuth.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akaike H. A new look at statistical model identification. IEEE Trans Automatic Control. 1974;19:716–726. [Google Scholar]
- Antal A, Dibo G, Keri S, Gabor K, Janka Z, Vecsei L, Benedek G. P300 component of visual event-related potentials distinguishes patients with idiopathic parkinson’s disease from patients with essential tremor. J Neural Transm. 2000;107:787–797. doi: 10.1007/s007020070059. [DOI] [PubMed] [Google Scholar]
- Basic S, Hajnsek S, Bozina N, Filipcic I, Sporis D, Mislov D, Posavec A. The influence of C3435T polymorphism of ABCB1 gene on penetration of phenobarbital across the blood-brain barrier in patients with generalized epilepsy. Seizure. 2008 doi: 10.1016/j.seizure.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, Almasy L, Foroud T, Van Eerdewegh P, Polich J, Rohrbaugh J, Kuperman S, Bauer LO, O’Connor SJ, Chorlian DB, Li TK, Conneally PM, Hesselbrock V, Rice JP, Schuckit MA, Cloninger R, Nurnberger J, Jr, Crowe R, Bloom FE. Quantitative trait loci analysis of human event-related brain potentials: P3 voltage. Electroencephalogr Clin Neurophysiol. 1998;108:244–250. doi: 10.1016/s0168-5597(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Blackwood D. P300, a state and a trait marker in schizophrenia. Lancet. 2000;355:771–772. doi: 10.1016/S0140-6736(99)00261-5. [DOI] [PubMed] [Google Scholar]
- Bunzeck N, Duzel E. Absolute coding of stimulus novelty in the human substantia nigra/VTA. Neuron. 2006;51:369–379. doi: 10.1016/j.neuron.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Cedazo-Minguez A. Apolipoprotein E and Alzheimer’s disease: molecular mechanisms and therapeutic opportunities. J Cell Mol Med. 2007;11:1227–1238. doi: 10.1111/j.1582-4934.2007.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Michaelson DM. Specific neurochemical derangements of brain projecting neurons in apolipoprotein E-deficient mice. J Neurochem. 1998;70:708–714. doi: 10.1046/j.1471-4159.1998.70020708.x. [DOI] [PubMed] [Google Scholar]
- Croft RJ, Gonsalvez CJ, Gabriel C, Barry RJ. Target-to-target interval versus probability effects on P300 in one- and two-tone tasks. Psychophysiology. 2003;40:322–328. doi: 10.1111/1469-8986.00036. [DOI] [PubMed] [Google Scholar]
- D’Ardenne K, McClure SM, Nystrom LE, Cohen JD. BOLD responses reflecting dopaminergic signals in the human ventral tegmental area. Science. 2008;319:1264–1267. doi: 10.1126/science.1150605. [DOI] [PubMed] [Google Scholar]
- De Ridder F, Pintelon R, Schoukens J, Gillikin DP. Modified AIC and MDL model selection criteria for short data records. Instrumentation and Measurement, IEEE Transactions on. 2005;54:144–150. [Google Scholar]
- De Sarro GB, Ascioti C, Froio F, Libri V, Nistico G. Evidence that locus coeruleus is the site where clonidine and drugs acting at alpha 1- and alpha 2-adrenoceptors affect sleep and arousal mechanisms. Br J Pharmacol. 1987;90:675–685. doi: 10.1111/j.1476-5381.1987.tb11220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme A, Makeig S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J Neurosci Methods. 2004;134:9–21. doi: 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Donchin E, Coles MGH. Is the P300 component a manifestation of context updating? Behavioral and Brain Sciences. 1988;11:357–374. [Google Scholar]
- Eichele T, Specht K, Moosmann M, Jongsma ML, Quiroga RQ, Nordby H, Hugdahl K. Assessing the spatiotemporal evolution of neuronal activation with single-trial event-related potentials and functional MRI. Proc Natl Acad Sci U S A. 2005;102:17798–17803. doi: 10.1073/pnas.0505508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Frangou S, Sharma T, Alarcon G, Sigmudsson T, Takei N, Binnie C, Murray RM. The Maudsley Family Study, II: Endogenous event-related potentials in familial schizophrenia. Schizophr Res. 1997;23:45–53. doi: 10.1016/S0920-9964(96)00089-8. [DOI] [PubMed] [Google Scholar]
- Hammond EJ, Meador KJ, Aung-Din R, Wilder BJ. Cholinergic modulation of human P3 event-related potentials. Neurology. 1987;37:346–350. doi: 10.1212/wnl.37.2.346. [DOI] [PubMed] [Google Scholar]
- Hansenne M. The p300 cognitive event-related potential. I. Theoretical and psychobiologic perspectives. Neurophysiol Clin. 2000;30:191–210. doi: 10.1016/s0987-7053(00)00223-9. [DOI] [PubMed] [Google Scholar]
- Higgins GA, Large CH, Rupniak HT, Barnes JC. Apolipoprotein E and Alzheimer’s disease: a review of recent studies. Pharmacol Biochem Behav. 1997;56:675–685. doi: 10.1016/s0091-3057(96)00420-0. [DOI] [PubMed] [Google Scholar]
- Javitt DC, Spencer KM, Thaker GK, Winterer G, Hajos M. Neurophysiological biomarkers for drug development in schizophrenia. Nat Rev Drug Discov. 2008;7:68–83. doi: 10.1038/nrd2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung TP, Makeig S, Westerfield M, Townsend J, Courchesne E, Sejnowski TJ. Removal of eye activity artifacts from visual event-related potentials in normal and clinical subjects. Clin Neurophysiol. 2000;111:1745–1758. doi: 10.1016/s1388-2457(00)00386-2. [DOI] [PubMed] [Google Scholar]
- Kiehl KA, Stevens MC, Laurens KR, Pearlson G, Calhoun VD, Liddle PF. An adaptive reflexive processing model of neurocognitive function: supporting evidence from a large scale (n = 100) fMRI study of an auditory oddball task. Neuroimage. 2005;25:899–915. doi: 10.1016/j.neuroimage.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Kim WS, Weickert CS, Garner B. Role of ATP-binding cassette transporters in brain lipid transport and neurological disease. J Neurochem. 2008;104:1145–1166. doi: 10.1111/j.1471-4159.2007.05099.x. [DOI] [PubMed] [Google Scholar]
- Kugler CF, Taghavy A, Platt D. The event-related P300 potential analysis of cognitive human brain aging: a review. Gerontology. 1993;39:280–303. doi: 10.1159/000213544. [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Vitek MP. Apolipoprotein E and neurological disease: therapeutic potential and pharmacogenomic interactions. Pharmacogenomics. 2007;8:959–969. doi: 10.2217/14622416.8.8.959. [DOI] [PubMed] [Google Scholar]
- Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, Kucherlapati R, Malhotra AK. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Quartermain D, Dunn AJ, Weinshenker D, Stone EA. Possible dopaminergic stimulation of locus coeruleus alpha1-adrenoceptors involved in behavioral activation. Synapse. 2008;62:516–523. doi: 10.1002/syn.20517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Demirci O, Calhoun VD. A Parallel Independent Component Analysis Approach to Investigate Genomic Influence on Brain Function. IEEE Signal Processing Letters. 2008;15:413. doi: 10.1109/LSP.2008.922513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp. 2009 doi: 10.1002/hbm.20508. In press (available online) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulert C, Juckel G, Giegling I, Pogarell O, Leicht G, Karch S, Mavrogiorgou P, Moller HJ, Hegerl U, Rujescu D. A Ser9Gly polymorphism in the dopamine D3 receptor gene (DRD3) and event-related P300 potentials. Neuropsychopharmacology. 2006;31:1335–1344. doi: 10.1038/sj.npp.1300984. [DOI] [PubMed] [Google Scholar]
- Musrati RA, Kollarova M, Mernik N, Mikulasova D. Malate dehydrogenase: distribution, function and properties. Gen Physiol Biophys. 1998;17:193–210. [PubMed] [Google Scholar]
- Nieuwenhuis S, Aston-Jones G, Cohen JD. Decision making, the P3, and the locus coeruleus-norepinephrine system. Psychol Bull. 2005a;131:510–532. doi: 10.1037/0033-2909.131.4.510. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis S, Gilzenrat MS, Holmes BD, Cohen JD. The role of the locus coeruleus in mediating the attentional blink: a neurocomputational theory. J Exp Psychol Gen. 2005b;134:291–307. doi: 10.1037/0096-3445.134.3.291. [DOI] [PubMed] [Google Scholar]
- O’Donnell BF, Vohs JL, Hetrick WP, Carroll CA, Shekhar A. Auditory event-related potential abnormalities in bipolar disorder and schizophrenia. Int J Psychophysiol. 2004;53:45–55. doi: 10.1016/j.ijpsycho.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;(56–58):60–51. [PubMed] [Google Scholar]
- Peskind ER, Li G, Shofer J, Quinn JF, Kaye JA, Clark CM, Farlow MR, DeCarli C, Raskind MA, Schellenberg GD, Lee VM, Galasko DR. Age and apolipoprotein E*4 allele effects on cerebrospinal fluid beta-amyloid 42 in adults with normal cognition. Arch Neurol. 2006;63:936–939. doi: 10.1001/archneur.63.7.936. [DOI] [PubMed] [Google Scholar]
- Picton TW. The P300 wave of the human event-related potential. J Clin Neurophysiol. 1992;9:456–479. doi: 10.1097/00004691-199210000-00002. [DOI] [PubMed] [Google Scholar]
- Polich J. EEG and ERP assessment of normal aging. Electroencephalogr Clin Neurophysiol. 1997;104:244–256. doi: 10.1016/s0168-5597(97)96139-6. [DOI] [PubMed] [Google Scholar]
- Polich J. Detection of Change: Event-Related Potential and fMRI findings. Kluwer Academic Publishers; Norwell, MA: 2003. [Google Scholar]
- Polich J. Updating P300: an integrative theory of P3a and P3b. Clin Neurophysiol. 2007;118:2128–2148. doi: 10.1016/j.clinph.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polich J, Corey-Bloom J. Alzheimer’s disease and P300: review and evaluation of task and modality. Curr Alzheimer Res. 2005;2:515–525. doi: 10.2174/156720505774932214. [DOI] [PubMed] [Google Scholar]
- Polich J, Criado JR. Neuropsychology and neuropharmacology of P3a and P3b. Int J Psychophysiol. 2006;60:172–185. doi: 10.1016/j.ijpsycho.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Polich J, Kok A. Cognitive and biological determinants of P300: an integrative review. Biol Psychol. 1995;41:103–146. doi: 10.1016/0301-0511(95)05130-9. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Chen K, Alexander GE, Bandy D, Frost J. Declining brain activity in cognitively normal apolipoprotein E epsilon 4 heterozygotes: A foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer’s disease. Proc Natl Acad Sci U S A. 2001;98:3334–3339. doi: 10.1073/pnas.061509598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffman JL, Weiss AP, Goff DC, Rauch SL, Weinberger DR. Neuroimaging-genetic paradigms: a new approach to investigate the pathophysiology and treatment of cognitive deficits in schizophrenia. Harv Rev Psychiatry. 2006;14:78–91. doi: 10.1080/10673220600642945. [DOI] [PubMed] [Google Scholar]
- Serretti A, Olgiati P, De Ronchi D. Genetics of Alzheimer’s disease. A rapidly evolving field. J Alzheimers Dis. 2007;12:73–92. doi: 10.3233/jad-2007-12108. [DOI] [PubMed] [Google Scholar]
- Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, Kase CS, D’Agostino RB, Sr, Decarli C, Atwood LD, Wolf PA. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shores MM, White SS, Veith RC, Szot P. Tyrosine hydroxylase mRNA is increased in old age and norepinephrine uptake transporter mRNA is decreased in middle age in locus coeruleus of Brown-Norway rats. Brain Res. 1999;826:143–147. doi: 10.1016/s0006-8993(99)01200-7. [DOI] [PubMed] [Google Scholar]
- Squires KC, Wickens C, Squires NK, Donchin E. The effect of stimulus sequence on the waveform of the cortical event-related potential. Science. 1976;193:1142–1146. doi: 10.1126/science.959831. [DOI] [PubMed] [Google Scholar]
- Stopkova P, Saito T, Papolos DF, Vevera J, Paclt I, Zukov I, Bersson YB, Margolis BA, Strous RD, Lachman HM. Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol Psychiatry. 2004;55:981–988. doi: 10.1016/j.biopsych.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Sutton S, Braren M, Zubin J, John ER. Evoked-potential correlates of stimulus uncertainty. Science. 1965;150:1187–1188. doi: 10.1126/science.150.3700.1187. [DOI] [PubMed] [Google Scholar]
- Turetsky B, Colbath EA, Gur RE. P300 subcomponent abnormalities in schizophrenia: II. Longitudinal stability and relationship to symptom change. Biol Psychiatry. 1998;43:31–39. doi: 10.1016/s0006-3223(97)00261-8. [DOI] [PubMed] [Google Scholar]
- Turetsky BI, Cannon TD, Gur RE. P300 subcomponent abnormalities in schizophrenia: III. Deficits In unaffected siblings of schizophrenic probands. Biol Psychiatry. 2000;47:380–390. doi: 10.1016/s0006-3223(99)00290-5. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt CE, Boomsma DI. Genetics of the human electroencephalogram (EEG) and event-related brain potentials (ERPs): a review. Hum Genet. 1994;94:319–330. doi: 10.1007/BF00201587. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt CE, van Baal GC. Twin and family studies of the human electroencephalogram: a review and a meta-analysis. Biol Psychol. 2002;61:111–138. doi: 10.1016/s0301-0511(02)00055-8. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Ferran E, Galke B, Cooper K, Bunney WE, Byerley W. Microarray screening of lymphocyte gene expression differences in a multiplex schizophrenia pedigree. Schizophr Res. 2004a;67:41–52. doi: 10.1016/s0920-9964(03)00151-8. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Shannon Weickert C, Ferran E, Matsumoto M, Overman K, Hyde TM, Weinberger DR, Bunney WE, Kleinman JE. Gene expression of metabolic enzymes and a protease inhibitor in the prefrontal cortex are decreased in schizophrenia. Neurochem Res. 2004b;29:1245–1255. doi: 10.1023/b:nere.0000023611.99452.47. [DOI] [PubMed] [Google Scholar]