Abstract
DNA Polymerase β is a multifunctional enzyme involved in base excision repair of nuclear DNA in vertebrate cells. It has been extensively studied as a model for mechanistic studies of the nucleotidyl transferase reaction, DNA synthesis fidelity, and protein-DNA interactions. Previous studies of 13C-methyl-methionine labeled Rat pol β revealed extensive dynamics in response to various DNA repair substrates (Bose-Basu et. al, 2004). We present here the first assignments of the full-length protein (335 residues, 39 kDa) in the presence of a double gap - double hairpin DNA, which we have utilized as a model DNA substrate for repair (22 nucleotides, 7 kDa) (Kirby et al., 2005). Out of a predicted 319 HNCO cross-peaks, only 282 were observed, but 95% of these could be confidently assigned. Most missing assignments were in the catalytic sub-domain, in the vicinity of predicted protein-DNA interactions, indicating significant conformational exchange in this region. Assignments for the 1HN, 13Cα, 13Cβ, 13CO and 15N shifts were deposited in the BMRB accession # 7319.
Keywords: Polymeraseβ, Base Excision Repair
Biological Context
DNA damage occurs continuously in normal cells due to a variety of endogenous and exogenous sources. Sites of damage that are not repaired can lead to a variety of potential outcomes, ultimately including necrosis, apoptosis, or transformation, i.e. cancer. Polymerase β (pol β), is part of the base excision repair pathway. The role of the 31 kDa polymerase domain of pol β is to catalyze the insertion of the correct nucleotide opposite the site of damage, after the damaged base has been excised. The lyase domain of pol β(8 kDa) subsequently cleaves the remaining 5’deoxyribose phosphate flap. Crystal structures of various repair intermediates reveal extensive side chain, domain, and sub-domain motions during catalysis. Previous NMR studies of [methyl-13C]-methionine-labeled pol β indicate characteristic conformational changes and conformational exchange dynamics upon formation of the activated, abortive enzyme-DNA-dNTP ternary complex (Bose-Basu et al., 2004, Kirby et al., 2005). In order to gain more insight into the conformational and dynamic response of the enzyme to various substrates, we undertook the assignment the backbone resonances of the enzyme-DNA complex.
Herein we report the assignment of the full-length rat DNA polymeraseβ. The lyase domain (1–87) and the “alm-thumb” (149–335) of the polymerase domain have been studied previously by NMR in isolation (Gryk et al., 2002, Liu et al., 1994). This study represents the first study of the full length protein (1–335) in complex with a double-gapped DNA, a model substrate for pol β (Kirby et al., 2005).
Methods and Experiments
The sample was expressed in E. coli and isolated using previously published protocols. Isotopic labeling utilized media containing 2H2O, U-[13C-2H]-glucose, 15N-amonium sulfate, and 2H-13C-15N -Bio-express (Cambridge Isotopes) used as a supplement at 1% (v/v). The sample required over a week of preparation time in 1H2O, which was found to be sufficient to “ack-exchange”the amide sites with 1H, as assessed by mass spectrometry. The double-gap double-hairpin DNA sequence was: 5’ CAGCGAAGCTGGTCGCGAAGCG-3’ The DNA was examined by 1H NMR to confirm proper Watson-Crick base pairing. In addition to 0.5 mM pol β-DNA complex (46 kDa), the NMR sample contained 150 mM KCl, 5mM NaN3, 10% D2O, and 1:1000 dilution of Calbiochem Protease Inhibitor cocktail III. Under these conditions, the triple resonance assignment experiments performed at 600 MHz provided superior signal/noise relative to studies at 800 MHz, presumably due to a combination of salt and relaxation time (T1) effects. The following 3D triple resonance experiments (TROSY versions) were used in the sequential assignment: HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, HN(CA)CB, HN(COCA)CB, and CBCA(CO)NH (Grzesiek and Bax, 1992, Ikura et al., 1990, Matsuo et al., 1996, Wittekind and Mueller, 1996, Pervushin et al., 1997). Additionally, a 3D 15N separated NOESY was acquired at 800 MHz (Marion et al., 1989). The data was analyzed with NvAssign (Kirby et al., 2004). Various attempts to find suitable conditions to measure residual dipolar couplings failed due to precipitation of the protein.
The input data for assignment included the aforementioned experiments, the crystal structure 1BPX, and the previous assignments of the lyase and palm-thumb domains. All data were input into the program MONTE and the relative weightings of the various datasets were optimized empirically until satisfactory solutions were found (Hitchens et al., 2003). MONTE is one of the few programs we found that could incorporate unassigned NOE spectra in backbone assignments. The connectivity suggested by MONTE was visually inspected for verification. The program PACES was used primarily for verification and bookkeeping (Coggins and Zhou, 2003).
Extent of Assignments and Data Deposition
Out of a predicted 319 HNCO main-chain peaks, only 282 were observed but 95% of these were confidently assigned. The extent of assignments was very different than what might have been expected from the input data from the previous assignments. The DNA binding domain (“fingers”domain, residues 90–145) had not previously been assigned, but, with the exception of two short sequences, was fully assigned in the pol β-DNA complex described here. This result probably indicates stabilization of the DNA-binding domain by the DNA. Assignment of the catalytic domain (“palm”) was the most problematic. Specifically, the assignments of the resonances of the β-sheets were discontinuous, particularly in the regions expected to interact with the DNA. Overall, the average intensity of the HNCO peaks for the nascent base pair binding domain (alternatively referred to as the "thumb") were lower than for the lyase and DNA binding domains, suggesting concerted motion/conformational exchange. Crystal structures predict conformational changes in the thumb domain upon recognition of the correct incoming nucleotide, which then positions key residues for catalysis.
There were numerous subtle chemical shift changes relative to the previously studies of the isolated lyase domain and the isolated palm-thumb construct. Of note, most of the changes in Cα and Cβ were in the lyase domain. For example, there were 47 Cα or Cβ shifts that changed by more than 1 ppm in the whole protein, but 34 of those were distributed throughout the lyase domain indicating significant conformational changes due to interactions with the DNA and with the catalytic domain.
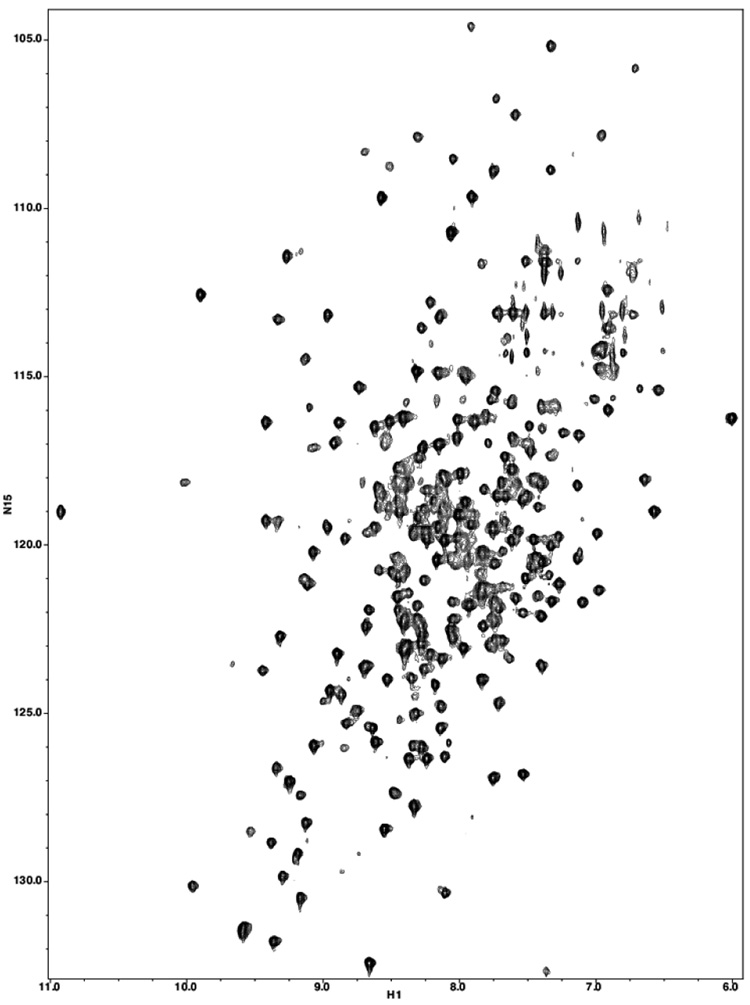
Figure 1.
TROSY-HSQC of Rat Polymerase β.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
All the assignment data has been submitted to the BMRB, accession number 7319.
References
- 1.Bose-Basu B, DeRose EF, Kirby TW, Mueller GA, Beard WA, Wilson SH, London RE. Dynamic characterization of a DNA repair enzyme: NMR studies of [methyl-13C]methionine-labeled DNA polymerase beta. Biochemistry. 2004;43:8911–8922. doi: 10.1021/bi049641n. [DOI] [PubMed] [Google Scholar]
- 2.Coggins BE, Zhou P. PACES: Protein sequential assignment by computer-assisted exhaustive search. J Biomol NMR. 2003;26:93–111. doi: 10.1023/a:1023589029301. [DOI] [PubMed] [Google Scholar]
- 3.Gryk MR, Maciejewski MW, Robertson A, Mullen MA, Wilson SH, Mullen GP. 1H, 13C and 15N resonance assignments for the perdeuterated 22 kD palm-thumb domain of DNA polymerase beta. J Biomol NMR. 2002;22:197–198. doi: 10.1023/a:1014237724868. [DOI] [PubMed] [Google Scholar]
- 4.Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Reson. 1992;99:201–207. [Google Scholar]
- 5.Hitchens TK, Lukin JA, Zhan Y, Mccallum SA, Rule GS. MONTE: An automated Monte Carlo based approach to nuclear magnetic resonance assignment of proteins. J Biomol NMR. 2003;25:1–9. doi: 10.1023/a:1021975923026. [DOI] [PubMed] [Google Scholar]
- 6.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance threedimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 7.Kirby NI, DeRose EF, London RE, Mueller GA. NvAssign: protein NMR spectral assignment with NMRView. Bioinformatics. 2004;20:1201–1203. doi: 10.1093/bioinformatics/bth064. [DOI] [PubMed] [Google Scholar]
- 8.Kirby TW, DeRose EF, Beard WA, Wilson SH, London RE. A thymine isostere in the templating position disrupts assembly of the closed DNA polymerase beta ternary complex. Biochemistry. 2005;44:15230–15237. doi: 10.1021/bi0511742. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, DeRose EF, Prasad R, Wilson SH, Mullen GP. Assignments of 1H, 15N, and 13C resonances for the backbone and side chains of the N-terminal domain of DNA polymerase beta. Determination of the secondary structure and tertiary contacts. Biochemistry. 1994;33:9537–9545. doi: 10.1021/bi00198a020. [DOI] [PubMed] [Google Scholar]
- 10.Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H- 15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo H, Kupce E, Li H, Wagner G. Increased sensitivity in HNCA and HN(CO)CA experiments by selective C beta decoupling. J Magn Reson B. 1996;113:91–96. doi: 10.1006/jmrb.1996.0161. [DOI] [PubMed] [Google Scholar]
- 12.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. B. 1996;101:201–205. [Google Scholar]