Summary
Spirochetes represent one of the bacterial groups often observed in hydrogen-sulfide-rich layers from coastal microbial mats. However, relatively few spirochetes from these microbial mats have been described and characterized. We have used 16S rDNA phylogenetic analysis to investigate the spirochetal diversity of microbial mats from two different geographic locations in the western Mediterranean (Ebro Delta, Spain, and La Camargue, France). Samples from each delta was monitored in the spring and winter over 1 to 2 year’s duration. In sequence analysis of 332 clones derived from samples of both deltas, 42 novel phylotypes of not-yet-cultivated spirochetes belonging to the genus Spirochaeta were detected. None of the phylotypes were identified as known culturable species of Spirochaeta or previously identified phylotyepes cloned from other hypersaline microbial mat such as Guerrero Negro. Eight of the phylotypes were common to Ebro and Camargue mats, two of them, IF058 and LL066 are present both in spring and winter. Some phylotypes appeared to show seasonal variation, i.e. they were found only in the spring, but not in the winter. On the other hand, Ebro and Camargue phylotypes compared with phylotypes from Guerrero Negro grouped according to the vertical gradient of oxygen and sulfide in the mat. Some phylotypes, such as LH073, IE028, LH042 or LG013 are harbored in low H2S or H2S-O2 interface zone. In contrast, major phylotypes were detected presumably in deeper layers and they are likely to be strict anaerobes and high tolerance to H2S. The presence of spirochetes in different located microbial mats suggests that constitutes a very diverse and stable population involved in a well-integrated “symbiosis” (physiological-cell-cooperatively) with other gild communities in the mats to maintain a coordinated functional community.
Keywords: Spirochaeta, phylogenitic diversity, temporal variation, microbial mats
Introduction
Biofilms are bacterial surface-associated communities attached to solid substrata, growing into and embedded in a polymer matrix produced by the bacteria. Microbial mats can be considered complex biofilms. Microbial mats are stratified microbial ecosystems. Lamination results from a light gradient along the vertical axis due to light extinction with depth, and from physicochemical microgradients due to the metabolism of different bacterial populations These laminae reflect a complex structure organized at the mm and μm spatial scale. This ecosystem contains within it members of all trophic levels. It is characterized by cyclic seasonal fluctuations of flooding and desiccation, and by diel fluctuations in the concentrations of oxygen, sulfide, and other chemical nutrients (Decker et al., 2005). Microorganisms rapidly respond to changes in various physicochemical gradients, and locate themselves according to the most favorable environmental conditions. This behavior is likely to govern the vertical species stratification that results from the active migration of motile cells in response to the shifting gradients of electron donors and/or acceptors observed within microbial mats (Guerrero & Berlanga, 2006). Microbial mats exemplify functionally integrated, self sustaining, laminated microbial consortia systems (Guerrero et al., 2002; Paerl et al., 2000; Des Marais, 2003). Mats exhibit a remarkably high degree of biodiversity compressed into a few millimeters. Molecular analysis of Guerrero Negro microbial mats revealed the most complex bacteria populations described to date in any environment, with 42 phyla including 15 novel candidate phyla (Ley et al., 2006). In comparison, soil can contain more than 20 bacterial divisions (Dumbar et al., 2002), approximately 12 division are represented in Sargasso Sea (Venter et al., 2004) and the lowest identified to date is the adult human gastrointestinal with 8 divisions (Bäckhed et al., 2005) or in healthy human skin also with 8 divisions (Gao et al., 2007), but it appears to be tremendously diverse at the strain and species level.
Free-living spirochetes represent one of the bacterial groups often observed in hydrogen sulfide rich layers from microbial mats habitats (Ley et al., 2006; Abed et al., 2007; Bordenave et al., 2007], and spirochetes may constitute a stable population, in deed, it has been observed the cosmopolitan distribution of Spirosymplokos deltaeiberi, described first in Ebro Delta microbial mats (Guerrero et al., 1993; Margulis et al., 1993], and lately in samples from Sippewissett salt marsh at Woods Hole, Massachusetts, US, and from microbial mats at North Pond, Laguna Figueroa, Baja California Norte, Mexico. The identity of these spirochetes was confirmed by electron microscopy (Margulis et al., 1998). Spirochetes are a group of helical, motile bacteria that are widely distributed in nature. The spirochetes represent a monophyletic phylum characterized phenotipically by a cellular ultrastructure and a movement that are unique among the bacteria. [Paster & Dewhirst, 2000).
The primary purpose of this study was to use culture-independent molecular methods to determine and compare the Spirochaeta diversity from two Mediterranean microbial mats Ebro Delta (Northeastern coastline of Spain) and La Camargue (South coastline of France) and compared with some cultivable species, “fifteen” cultivable species of Spirochaeta are presently known (Leschine et al., 2006) or phylotypes previously reported. The major difference between the two geographical sites is their salinity. Ebro Delta salinity ranges from 40 to 70 ‰ (Mir et al., 2002), whereas La Camargue has a higher salinity, from 60 to 140‰ (Wieldant & Kühl, 2006). It has been described that salinity influence microbial mat composition and it reflected in the diversity of the entire prokaryotic community (Abed et al., 2007; Yannarell & Paerl, 2007), and hence may control also spirochete diversity. The secondary purpose was to determine the seasonal variation of Spirochaeta diversity by monitoring samples taken over the course of 1 to 2 years.
Materials and methods
Sample collection
Samples were collected from laminated, intertidial microbial mats from Alfacs Peninsula, Ebro Delta, Northeastern Spain (0° 35′E, 0° 56′E; 40° 33′N, 40° 47′N), and La Camargue, Rhône Delta, Southern France (04° 11′E, 04° 57′E; 43° 40′N, 44° 40′N). Samples were collected in cores (1 cm × 3 cm) of mats and sliced horizontally in 2-mm increments (from the top to a depth of 6 mm) at 12.00 h. Samples were obtained from the Ebro Delta in May 2001, November 2002, and May 2003, and from La Carmargue in April 2002, November 2002, and April 2003.
DNA extraction
DNA was extracted from 2-6 mm depth slices. Pieces of microbial mat of approximately 1 mm3 from each slice were suspended in 100 μl TE buffer in 2.0 ml vials with a capful of 0.1 mm glass beads. The mixture was then homogenized for 1 min in a Minibeadbeater-8 (Biospec Products, Inc., Bartlesville, OK) and centrifuged at high speed for 2 min. Carefully avoiding the transfer of beads, 50 μl of each sample was pipetted into sterile 0.5 ml Eppendorf tubes. DNA was extracted with a phenol-chloroform mixture and precipitated in the cold with 95% ethanol. Aliquots were resuspended in 20 μl TE pH 7.5 and stored at -20°C to preserve the DNA until needed.
PCR and cloning
A spirochetal selective reverse primer 5′-GTTACGACTTCACCCYCCT-3′ was used with a universal forward primer 5′-GAGTTTGATYMTGGCTCAG-3′ to selectively amplify spirochetal 16S rRNA genes from environmental samples (Dewhirst et al., 2000). PCR amplification was performed in 50 μl final volume of reaction mix (1 μl of the DNA template, 20 pmol of each primer, 40 nmol of dNTPs, 1.5 mM MgCl2 and 1 U of Taq platinum polymerase (Invitrogen Corporation, San Diego, CA). Samples were preheated at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 45 s, annealing at 60°C for 45 s, and elongation at 72°C for 1.5 min, and finally an elongation step at 72°C for 15 min. The results of PCR amplification were examined by electrophoresis in a 1% agarose gel. DNA was stained with ethidium bromide and visualized under short-wavelength UV light. PCR products were purified by a QIAquick Gel extraction kit (Qiagen Inc., Valencia, CA) and cloned using the TOPO TA cloning kit (Invitrogen Corporation, San Diego, CA), according to the manufacturer’s recommendations.
16S rRNA sequencing
Purified PCR products were sequenced using an ABI prism cycle-sequencing kit (BigDye® Terminator Cycle Sequencing kit with AmpliTaq DNA polymerase FS, Perkin-Elmer, Boston, MA). The primers used for 16S rRNA sequencing were as previously described (Paster et al., 2001). Half dye or quarter dye chemistry was used with 3.2 mM primers, 3 μl PCR product in a final volume of 20 μl. Cycle sequencing was performed using an ABI 9700 with 25 cycles of denaturation at 96°C for 10 s, and annealing and extension at 60°C for 4 min. Sequencing reactions were run on an ABI 3100 DNA sequencer.
Data analysis
Over 300 clones with the correct size insert of approximately 1,500 bases were analyzed. About 500 bases were first sequenced to determine approximate phylogenetic position. Then, full sequences were obtained for representatives of novel Spirochaete phylotypes. The 16S rRNA sequences were compared to known sequences in GenBank with the advanced gapped BLAST (basic local alignment search tool) algorithm. Phylogenetic analyses were performed using MEGA version 2.1. The dendrogram was constructed using the neighbor-joining algorithm and the kimura 2-parameter distance estimation method. Two hundred bootstrap trees were generated, and bootstrap confidence levels were determined using the MEGA 2.1 program. Chimeric sequences were identified by using the Chimera check program in the Ribosomal Database Project II (Cole et al., 2003), by treeing analysis, or base signature analysis.
Nucleotide sequence accession numbers
The complete 16S rRNA gene sequences of clones representing novel phylotypes defined in this study and published sequences are available for electronic retrieval from the EMBL, GenBank, and DDBJ nucleotide sequence databases.
Results
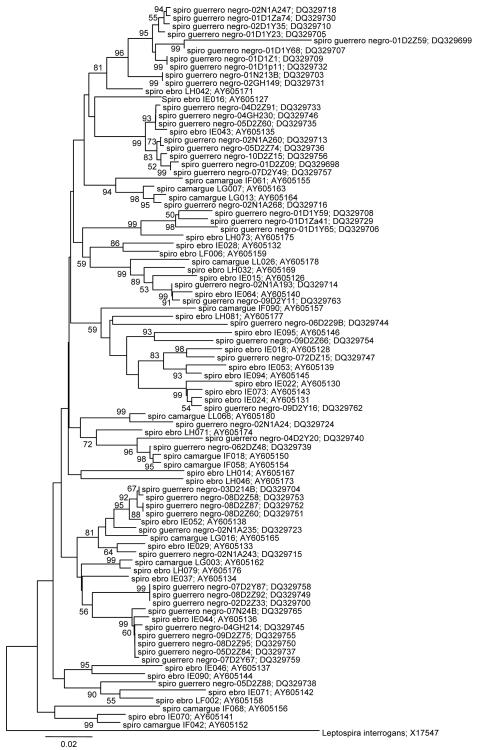
A total of 332 clones were sequenced in order to establish the diversity of spirochetes in two microbial mats located in the Mediterranean (Ebro Delta, and La Camargue); temporal variation of spirochete diversity at each location was also examined. New 42 Spirochaete phylotypes were detected in these microbial mats (Fig. 1). Collectively, there were 33 phylotypes detected in Ebro Delta samples, of which 25 were unique to this location. In La Camargue samples, 17 phylotypes were detected, of which 9 were found only in La Camargue. There were 8 phylotypes common to both samples.
Fig.1.
Spirochetal phylotypes of the genus Spirochaeta detected from microbial mats of the Ebro Delta and La Camargue. The information presented includes spirochetal species or phylotype clone ID and sequence accession number. Novel phylotypes are defined as those taxa that are <98.5 similar in sequence comparisons to its closest relative. Columns to the right indicate presence of phylotype in each delta at different sampling dates. Numbers in boxes represent the number of clones that the phylotype was detected in each library. Boxes with no number indicate that the phylotype was not detected at that time point. Bar = 0.02 difference in nucleotide sequence. On thousand bootstrap trees were generated and bootstrap confidence levels as percentages (only values over 50%) are shown at tree nodes.
None of the phylotypes were identified as known cultivable species of Spirochaeta. Phylotype IE071 grouped with Spirochaeta bajacaliforniensis and S. smaragdinae, both species are obligate anaerobes. S. bajacaliforniensis is a marine spirochete isolated from Laguna Figueroa microbial mats (Baja California, Mexico) (Frasek & Stolz, 1985); and S. smaragdinae was isolated from an oil field (Congo, Africa), it grew optimally with a sodium chloride concentration of 5%, and reduced thiosulfate and sulfur to H2S (Magot et al., 1997). LH073 clusted with S. alkalica, S. africana and S. halophila. The two firsts species are aerotolerant anaerobes, halophilic, and isolated from sediments of alkaline lakes; S. halophila is a facultative anaerobe isolated from a high-salinity pond. These five spirochaeta species may share physiological characteristics with our phylotypes because of the environment conditions may be similar. Whereas, there are no close relationship with S. asiatia, S. thermophila, S. zuelzerae, S. caldaria, S. stenostrepta, and S. aurantia (see the review about the genera Spirochaeta, Leshine et al., 2006). In deed, the species S. caldaria, S. stenostrepta and S. zuelzerae although free living spirochetes grouped within the genus Treponema on the basis of 16S rRNA sequence (Paster et al., 2000].
Temporal variation was observed in the spirochetal phylotypes IE029, IE044, IE090, IE070 that were detected only in springtime samplings, but seem to “disappear” in the cold season. This fluctuation, seasonal variation, in population distribution of 16S rRNA phylotypes has been observed in a hot spring microbial mat (Ferris et al., 1997; Bhaya et al., 2007), diazotrophic bacteria in Chesapeake Bay (Short et al., 2004) or in temperate stream habitats (Hullar et al., 2006) that have distinct nonramdom spatial and seasonal distribution that are either specific physical processes or adapted to different environmental niches. Phylotypes IF058 and LL066 ere present both in spring and winter and in both mats, Ebro and Camargue.
None of the Ebro-Camargue-spirochetes phylotypes were identified as previously phylotypes reported from other hypersaline microbial mat such as Guerrero Negro (Fig. 2). Ley et al. (2006) described microbial diversity within the Guerrero Negro mat vertically on millimeter scale. Samples were collected in June and October at 4 am (night) and 1 pm (day). The majority of spring Mediterranean spirochetes phylotypes grouped with October samples from Guerrero Negro, in which the temperature and irradiance during the autumn and the spring could be similar. Although we have not studied spirochete diversity through depth in Ebro and Camargue mats, when we compare them with spirochetes from Guerrero Negro we can observed two major groups: spirochetes that were located in the oxic zone, and spirochetes that were placed in the anoxic zone. The phylotypes LH073, LH042 and IE016 cluster with phylotypes detected at 0-2 mm depth in Guerrero Negro mats picked up during the day, that correspond on aerobics, facultative anaerobes or aerotolerant spirochetes. These phylotypes also grouped with aerotolerant or facultative anaerobe cultivable Spirochaeta (see Fig. 1). Mediterranean spirochetes that cluster with Guerrero Negro phylotypes observed at > 2 mm depth collected both during the day or night may consider obligate anaerobes.
Fig.2.
Spirochaeta phylotypes detected from Ebro and Camargue microbial mats vs. Spirochaeta phylotypes from Guerrero Negro mats. Phylotypes from Guerrero Negro were named according to the layer they originated from (01 to 10), the time of collection (D for day, N for night), and the month of collection (1, June; 2, October).
Discussion
Although spirochetes population in microbial mats seems to represent only 1% to 4% of the total population, spirochetal population at specie-taxonomical level is remarkable higher in microbial mats of the Ebro Delta and La Camargue as observed previously in Guerrero Negro mat (Ley et al., 2006). It has been suggested that diversity of competing or related phylotypes coexistence may be associated with both spatial and temporal segregation in spatially heterogeneous environments that challenges the metabolic and regulatory repertoire of the indigenous populations (Kerr et al., 2002; Hansen et al., 2004; Ward et al., 2006). Phylotypes may complement each other in space, so spatial gradients, allows occupation with more diverse species. Diversity may acts as insurance for ecosystem functioning against temporal environmental change to functional compensations among phylotypes as showed in both seasonally and diel fluctuations in microbial mats (Ley et al., 2006; Bordenave et al., 2007; Villanueva et al., 2007).
The temperature change dramatically between summer and winter season. During winter time ambient temperatures are lower and daily temperature variations (day-night) less pronounced than in summer, so less pronounced daily temperatures variation in winter could have resulted in an adaptation of the microbial population to lower temperatures, that could explain the temporal season distribution of several phylotypes, such as the phylotypes IE029, IE044, IE090 and IE070. Adaptation of populations to different temperatures may help to provide homeostasis within the mat community, i.e temperature-adapted populations could stabilize community functions against flow-related or seasonal variation in temperature (Wieland & Kühl, 2006; Ward et al., 2006, Bhaya et al., 2007).
Diel fluctuations in the concentrations of oxygen and sulfide shape the chemical environment and provide daily contrasting microenvironments that are separated on a scale of a few millimeters. During the day the redoxcline may be ca. 1-2 mm depth. Oxygen concentration within the mat increased during the day until the afternoon and then decreased with decreasing irradiance. When oxygenic photosynthesis ceases at night, the upper layers of the mat become highly reduced and sulfidic (Mir et al., 2002; Ley et al., 2006; Wieland & Kühl, 2006). Microorganisms orient along microscale chemical (i.e. O2, pH, Eh) gradients to meet and optimize the biogeochemical processes (C, N, S cycling) essential for survival, growth and maintenance of genetic diversity, needed to sustain functional systems. Phylotypes LH073, LH042 and IE016 may be aerobe, aerotolerant or facultative anaerobe, that may be present in the upper layers and they may be important population in the sharp transition between the oxic and the sulfidic zone that is characterized by a narrow zone of overlapping O2 and sulfide gradients.
The main link in microbial community is the trophic interaction. The functional grouping within the community depends on the quality of substrates and products. This type of interdependence is exampled by an anaerobic community where a network operates from hydrolytic to fermenting primary anaerobes, then to syntrophic bacteria and to homoacetocetic, methanogenic, or sulfidogenic secondary anaerobes. In diverse anoxic environments, spirochetes are the trophic intermediate between the hydrolytic bacteria and these secondary anaerobes, because of the main compounds produced by spirochete are acetate, H2, and CO2, which are normally consumed by sulfate-reducing bacteria and methanogens (Fernández et al., 1999; Blazejak et al., 2005). Spirochetes may constitute a ubiquitous component of microbial mats that may play an important functional role in the community supplying carbon sources and electron donors to the other populations. Spirochaeta phylotypes persist in anoxic-microoxic sediments and compete effectively with other heterotrophic organisms
This study is a good “first-step” estimate of the diversity of spirochetes in microbial mats of the Ebro Delta and La Camargue. Additional 16S rDNA sequencing needs to be done to establish the spirochetal diversity through depth in the mat. Once achieved, 16S rRNA-based checkerboard DNA-DNA hybridization assays or microarrays can be used to more accurately determine the bacterial populations’ distribution and function within microbial mats.
Acknowledgements
This work was supported by grant CGL2005-04990/BOS (Spanish Ministry of Science and Technology) to RG, and by NIH grants DE-11443 and DE-10374 from the National Institute of Dental & Craniofacial Research to BJP
References
- Abed RMM, Kohls K, de Beer D. Effect of salinity changes on the bacterial diversity, photosynthesis and oxygen consumption of cyanobacterial mats from an intertidal flat of the Arabian Gulf. Environ Microbiol. 2007;9:1384–1392. doi: 10.1111/j.1462-2920.2007.01254.x. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Grossman AR, Steunou A-S, Khuri N, Cohan FM, Hamamura N, Melendrez MC, Bateson MM, Ward DM, Heidelberg JF. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. The ISME J. 2007;1:703–713. doi: 10.1038/ismej.2007.46. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Blazejak A, Erséus C, Amann R, Dubilier N. Coexistence of bacterial sulfide oxidizers, sulfate reducers, and spirochetes in a gutless worm (Oligochaeta) from the Peru margin. Appl Environ Microbiol. 2005;71:1553–1561. doi: 10.1128/AEM.71.3.1553-1561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenave S, Goñi-Urriza MS, Caumette P, Duran R. Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat. Appl Environ Microbiol. 2007;73:6089–6097. doi: 10.1128/AEM.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker KLM, Potter CS, Bebout BM, Des Marais DJ, Carpenter S, Discipulo M, Hoehler TM, Miller SR, Thamdrup B, Turk KA, Visscher PT. Mathematical simulation of the diel O, S, and C Biogeochemistry of a hypersaline microbial mat. FEMS Microbiol Ecol. 2005;52:377–395. doi: 10.1016/j.femsec.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Des Marais DJ. Biogeochemistry of hypersaline microbial mats illustrates the dynamics of modern microbial ecosystems and the early evolution of the biosphere. Biol Bull. 2003;204:160–167. doi: 10.2307/1543552. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Tamer MA, Ericson RE, Lau CN, Levanos VA, Boches SK, Galvin JL, Paster BJ. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol Immunol. 2000;15:196–202. doi: 10.1034/j.1399-302x.2000.150308.x. [DOI] [PubMed] [Google Scholar]
- Dumbar J, Barns SM, Ticknor LO, Kuskei CR. Empirical and theoretical bacterial diversity in four Arizona soils. Appl Environ Microbiol. 2002;68:3035–3045. doi: 10.1128/AEM.68.6.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández A, Huang S, Seston S, Xing J, Hickey R, Criddle C, Tiedje J. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697–3704. doi: 10.1128/aem.65.8.3697-3704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MJ, Ward DM. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracek SP, Jr, Stolz JF. Spirochaeta bajacaliforniensis sp. n. from a microbial mat community at Laguna Figueroa, Baja California Norte, Mexico. Arch Microbiol. 1985;142:317–325. doi: 10.1007/BF00491897. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng C-H, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero R, Ashen J, Solé M, Margulis L. Spirosymplokos deltaeiberi nov. gen., nov. sp.: variable-diameter composite spirochete from microbial mats. Arch Microbiol. 1993;160:461–470. doi: 10.1007/BF00245307. [DOI] [PubMed] [Google Scholar]
- Guerrero R, Piqueras M, Berlanga M. Microbial mats and the search for minimal ecosystems. Int Microbiol. 2002;5:177–188. doi: 10.1007/s10123-002-0094-8. [DOI] [PubMed] [Google Scholar]
- Guerrero R, Berlanga M. Life’s unity and flexibility: the ecological link. Int Microbiol. 2006;9:225–235. [PubMed] [Google Scholar]
- Hansen SH, Molin S. Temporal segregation: succession in biofilms. In: Miller RV, Day MJ, editors. Microbial Evolution. Gene establishment, survival, and exchange. ASM Press; Washington DC: 2004. pp. 192–213. [Google Scholar]
- Hullar MAJ, Kaplan LA, Stahl DA. Recurring seasonal dynamics of microbial communities in stream habitas. Appl Environm Microbiol. 2006;72:713–722. doi: 10.1128/AEM.72.1.713-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in real-life gane of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Leschine S, Paster BJ, Canale-Parola E. Free-living saccharolytic spirochetes: the genus Spirochaeta. The Prokaryotes; Springer, NY: 2006. DOI: 10.1007/0-387-30747-8. [Google Scholar]
- Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM, Maresca JA, Bryant DA, Sogin ML, Pace NR. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mats. Appl Environ Microbiol. 2006;72:3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magot M, Fardeau ML, Arnauld O, Lanau C, Olliver B, Thomas P, Patel BK. Spirochaeta smaragdinae sp. nov., a new mesophilic strictly anaerobic spirochete from an oil field. FEMS Microbiol Lett. 1997;155:185–191. doi: 10.1111/j.1574-6968.1997.tb13876.x. [DOI] [PubMed] [Google Scholar]
- Margulis L, Ashen JB, Solé M, Guerrero R. Composite, large spirochetes from microbial mats: spirochete structure review. Proc Natl Acad Sci USA. 1993;90:6966–6970. doi: 10.1073/pnas.90.15.6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Navarrete A, Solé M. Cosmopolitan distribution of the large composite microbial mat spirochete, Spirosymplokos deltaeiberi. Int Microbiol. 1998;1:27–34. [PubMed] [Google Scholar]
- Mir J, Martínez-Alonso M, Caumette P, Guerrero R, Esteve I. Sulfide fluxes in a microbial mat from the Ebro Delta, Spain. Int Microbiol. 2002;5:133–138. doi: 10.1007/s10123-002-0076-x. [DOI] [PubMed] [Google Scholar]
- Paerl HW, Pinckney JL, Steppe TF. Cyanobacterial-bacterial mat consortia: examining the functional unit of microbial survival and growth in extreme environments. Environ Microbiol. 2000;2:11–26. doi: 10.1046/j.1462-2920.2000.00071.x. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE. Phylogenetic foundation of spirochetes. J Mol Microbiol Biotechnol. 2000;2:341–344. [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, Jenkins BD, Zehr JP. Spatial and temporal distribution of two diazotrophic bacteria in the Chesapeake Bay. Appl Environ Microbiol. 2004;70:2186–2192. doi: 10.1128/AEM.70.4.2186-2192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargassoo Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Villanueva L, Navarrete A, Urmeneta J, White DC, Guerrero R. Analysis of diurnal and vertical microbial diversity of a hypersaline microbial mat. Arch microbiol. 2007;188:137–146. doi: 10.1007/s00203-007-0229-6. [DOI] [PubMed] [Google Scholar]
- Ward DM, Bateson MM, Ferris MJ, Kühl M, Wieland A, Koeppel A, Cohan FM. Cyanobacteria ecotypes in the microbial mat community of mushroom spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition structure and function. Phil Trans R Soc B. 2006 doi: 10.1098/rstb.2006.1919. DOI:10.1098/rstb.2006.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland A, Kühl M. Regulation of photosynthesis and oxygen consumption in a hypersaline cyanobacterial mat (Camargue, France) by irradiance, temperature and salinity. FEMS Microb Ecol. 2006;55:195–210. doi: 10.1111/j.1574-6941.2005.00031.x. [DOI] [PubMed] [Google Scholar]
- Yannarell AC, Paerl HW. Effects of salinity and light on organic carbon and nitrogen uptake in a hypersaline microbial mat. FEMS Microbiol Ecol. 2007;62:345–353. doi: 10.1111/j.1574-6941.2007.00384.x. [DOI] [PubMed] [Google Scholar]