Summary
Suppression of viral infection by RNA in a nucleotide sequence homology-dependent manner was first reported in plants in early 1990s. Studies in the past 15 years have established a completely new RNA-based immune system against viruses that is mechanistically Riverside, CA, USA. related to RNA silencing or RNA interference (RNAi). This viral immunity begins with recognition of viral double-stranded or structured RNA by the Dicer nuclease family of host immune receptors. In fungi, plants and invertebrates, the viral RNA trigger is processed into small interfering RNAs (siRNAs) to direct specific silencing of the homologous viral genomic and/or messenger RNAs by an RNaseH-like Argonaute protein. Deep sequencing of virus-derived siRNAs indicates that the immunity against viruses with a positive-strand RNA genome is induced by Dicer recognition of dsRNA formed during the initiation of viral progeny (+)RNA synthesis. The RNA-based immune pathway in these organisms overlaps the canonical dsRNA-siRNA pathway of RNAi and may require amplification of viral siRNAs by host RNA-dependent RNA polymerase in plants and nematodes. Production of virus-derived small RNAs is undetectable in mammalian cells infected with RNA viruses. However, infection of mammals with several nucleus-replicating DNA viruses induces production of virus-derived microRNAs capable of silencing host and viral mRNAs as found for viral siRNAs. Remarkably, recent studies indicate that prokaryotes also produce virus-derived small RNAs known as CRISPR RNAs to guide antiviral defense in a manner that has yet to be defined. In this article, we review the recent progress on the identification and mechanism of the key components including viral sensors, viral triggers, effectors, and amplifiers, of the small RNA-directed viral immunity. We also highlight some of the many unresolved questions.
Keywords: viral, pattern recognition receptors, RNA silencing
Introduction
Innate immunity represents an ancient defense mechanism conserved in diverse multicellular organisms that responds immediately upon pathogen attack. The discovery of host innate immune receptors played a key role in understanding both the importance and mechanism of innate immunity in the defense against pathogens (1, 2). These receptors are collectively referred to as pattern recognition receptors (PRRs), because they recognize conserved molecular patterns associated with microbes. The transmembrane Toll-like receptors (TLRs) and the cytosolic NOD-like receptors (NLRs) and RIG-I-like DExD/H box RNA helicases (RLRs) are among the best characterized families of PRRs. PRRs typically contain two key functional domains. One domain interacts directly with microbial signatures shared by major classes of microbes, whereas the second protein—protein interaction domain activates the downstream signaling events, leading to transcription of immunity effector genes with broad-spectrum anti-microbial activities (2).
The dsRNA-specific Dicer nucleases represent a distinct family of PRRs. Dicer also recognizes a viral RNA trigger like some PRRs to initiate protective immunity against RNA and DNA viruses; however, Dicer further processes the viral RNA trigger into virus-derived small RNAs, which are assembled into effector complexes to guide specific antiviral defense via the RNA silencing pathway (3). RNA silencing refers to related gene silencing mechanisms guided by three broadly defined classes of small RNAs (4). Both small interfering RNAs (siRNAs) and microRNAs (miRNAs) are Dicer products processed from perfect-base paired double-stranded RNA (dsRNA) and hairpin dsRNA regions of single-stranded RNA precursors, respectively. By contrast, PIWI-interacting RNAs (piRNAs) are Dicer independent. All three classes of small RNAs are found in effector complexes, such as RNA-induced silencing complex (RISC), that contain an Argonaute protein (AGO) as an essential component, and guide AGO-mediated, specific gene silencing by base pairing between small RNAs and target genes. Upon viral infection, fungi, plants, and invertebrate animals produce virus-derived siRNAs (viRNAs) to direct antiviral immunity (3). By contrast, infection of mammals with certain nucleus-replicating DNA viruses induces production of virus-derived miRNAs capable of silencing mRNAs of the cognate viruses (5). In this article, we review the recent progress on understanding the mechanism of the key components of this Dicer-initiated viral immunity (DVI) and highlight some of the many unresolved questions.
The Dicer family of type III nucleases
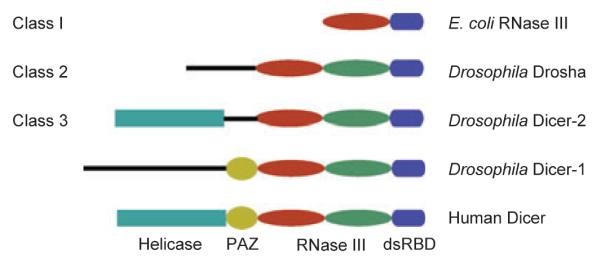
Dicer proteins are type III endoribonucleases, which specifically cleave dsRNA or hairpin dsRNA regions of single-stranded RNAs. All RNase III enzymes encode a homologous ribonuclease domain known as the RNase III domain, and dsRNA cleavages by RNase III produce duplex fragments with a characteristic terminal structure consisting of a 5′-phosphate group and a two-nucleotide overhang at the 3′-end. Type III RNases are divided into three classes (6) (Fig. 1). Class 1 RNases contain a single RNase III domain joined to a dsRNA-binding domain (dsRBD). Members of Class 1 RNases are found in bacteria and yeast, and they play an important role in processing cellular and viral RNA targets. Class 1 RNase III acts as a tight homodimer in which the RNase III domain from two molecules combines to form a single processing center, with each domain contributing to the hydrolysis of one RNA strand of the dsRNA substrate. In vitro cleavage of perfect-base paired dsRNA by bacterial RNase III occurs with little regard for sequence. However, a recent study (7) suggests that the dsRNA sequence extending 10 bp from the cleavage site can affect RNase III activity by influencing substrate affinity or catalysis.
Fig. 1.
Domain structures of representatives from the three classes of RNase III.
Class 2 and class 3 RNases from fungi, plants, invertebrates, and vertebrates are essential for the biogenesis of miRNAs and/ or siRNAs. Known as Drosha and Dicer, respectively, both classes of RNases contain two tandem RNase III domains. In addition, most of the Dicer RNases from class 3 encode an N-terminal RNA helicase domain closely related to RLRs and a PAZ (Piwi/Argonaute/Zwille) domain shared between Dicer and AGOs. Drosophila melanogaster encodes two functionally distinct Dicers: Dicer-2 (DCR2) produces siRNAs from dsRNA precursors, whereas DCR1 recognizes stem-loop structures present in single-stranded miRNA precursors (8, 9). Arabidopsis thaliana encodes four Dicer-like proteins (DCLs), all of which recognize dsRNA, although the primary role of DCL1 is in the biogenesis of miRNAs (8, 9). However, mammals and the nematode Caenorhabditis elegans encode a single Dicer required for the biogenesis of both miRNAs and endogenous siRNAs (endo-siRNAs). Dicer can cleave any dsRNA with a simple preference toward the terminus of dsRNA and produce small duplex fragments of discrete sizes progressively from the terminus.
The presence of the helicase/adenosine triphosphatase (ATPase) domain is consistent with the observation that generation of siRNAs from dsRNA in vitro by D. melanogaster DCR2 and C. elegans DCR1 is stimulated by the addition of adenosine triphosphate (ATP). Genetic studies also supported a role for the helicase domain of D. melanogaster DCR2 in dsRNA processing but not in the subsequent loading of siRNAs into RISC. However, ATP has no effect on the in vitro dsRNA processing activity of human Dicer. Moreover, a functional helicase domain is not found in DCR1 of D. melanogaster, either of the two Dicer-related proteins of amoeba Dictyostelium which supports RNAi, or the protozoan Giardia intestinalis Dicer that contains a PAZ domain and tandem RNase III domains and can process dsRNA into 25–27 nucleotide (nt) small RNAs.
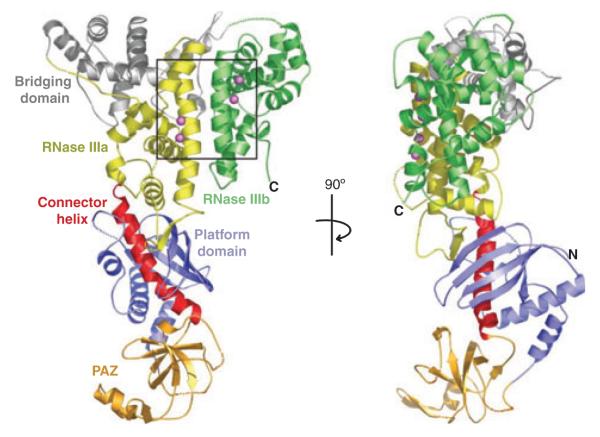
The PAZ domain acts as a novel RNA-binding module that specifically recognizes the 2-nt 3′-overhang of the siRNA duplex. However, a PAZ domain is not recognized in either DCR2 of D. melanogaster or the fission yeast Dicer, which is required for the biogenesis of heterochromatic siRNAs. Complementary biochemical and structural studies have illustrated that the two RNase III domains of Dicer form an intramolecular dimer that resembles the homodimer of class 1 RNases (10). The crystal structure of Giardia Dicer further revealed Dicer as an elongated molecule with the RNase III dimer and PAZ domain connected by a long helix run (11) (Fig. 2). Based on these findings, a model has been proposed to explain why distinct Dicer RNases generates dsRNA fragments of discrete sizes. For example, DCL4, DCL2, and DCL3 of A. thaliana produce endo-siRNAs of 21, 22, and 24 nt, respectively (12–14). Similarly, miRNAs produced by DCR1 in D. melanogaster are predominantly 22 nt in length in contrast to endo-siRNAs produced by DCR2 which show preference for a length of 21 nt (15). In this model, the PAZ domain of Dicer is responsible for anchoring the end of a dsRNA so that the connector helix between the RNase III dimer and PAZ domain measures the distance from the dsRNA end to the cleavage site (16).
Fig. 2. Crystal structure of Giardia Dicer.
Depicted are the PAZ (Piwi/Argonaute/Zwille) domain (orange), the connector helix (red), the RNase IIIa domain (yellow), and the RNase IIIb domain (green). Reprinted from (11).
Class 2 RNase Drosha is essential for the biogenesis of miRNAs in both invertebrates and vertebrates. Drosha cleaves the hairpin regions of primary miRNA transcripts to yield pre-miRNAs in the nucleus which are then processed into mature miRNAs in the cytoplasm by Dicer. Drosha alone is not active, and specific pri-miRNA processing by Drosha requires its binding partner Pasha (DGCR8), a protein with tandem dsRBDs. Dicer also has a binding partner with tandem dsRBDs. Such a dsRBD protein, including Loquacious (Loq) in D. melanogaster, hyponastic leaves 1 (HYL1) in A. thaliana, and TAR RNA-binding protein (TRBP) in humans, is ubiquitously required for the Dicer-dependent processing of pre-miRNAs into mature miRNAs. By contrast, the dsRBD protein R2D2 of D. melanogaster, which forms a heterodimer with DCR2, functions in siRNA loading but is dispensable for the biogenesis of siRNAs from exogenous dsRNA. Thus, it appears that a dsRNA-binding protein is required for the production of miRNAs from hairpin RNAs but not of siRNAs from perfect dsRNA substrates by class 2 and class 3 RNase III enzymes. However, the dsRNA-binding protein Rde-4 is essential for the biogenesis of both exo- and endo-siRNAs in C. elegans, and recent studies (17) also revealed a role of Loq in a DCR2 complex for the production of endo-siRNAs in D. melanogaster.
Viral sensors
Detection of virus-specific small RNAs of both polarities in plant and fruit fly cells upon viral infection had predicted a role for Dicer in the initiation of the RNA silencing-based antiviral immunity (18, 19). However, for a number of reasons detailed below, identification of a specific Dicer(s) responsible for the production of viral siRNAs and the initiation of the small RNA-directed viral immunity in D. melanogaster and A. thaliana was only reported in 2006.
D. melanogaster
RNA silencing in D. melanogaster includes three well-defined small RNA pathways. DCR2 and DCR1 initiate the canonical siRNA pathway and the miRNA pathway, respectively, whereas the piRNA pathway is Dicer independent. Use of a well-characterized Flock house virus (FHV) (20) facilitated the genetic analysis of the small RNA-directed viral immunity. FHV is a natural insect pathogen and contains a bipartite positive-strand RNA [(+)RNA] genome (Fig. 3) that replicates via dsRNA intermediates on the outer mitochondria membrane. RNA1 of FHV is 3.1 kb long and serves as both a genomic RNA and an mRNA for the translation of the viral RNA-dependent RNA polymerase (RdRP). FHV RNA2 (1.2 kb) encodes the coat protein precursor that is required for packaging the genomic RNAs 1 and 2 into virions. Thus, FHV RNA1 can self-replicate independent of RNA2. Protein B2 is also encoded by RNA1 but is translated from RNA3, a non-packaged subgenomic RNA produced after the replication of RNA1. B2 can suppress RNA silencing in both plant and fruit fly cells (19) and contains a novel dsRBD that binds to long dsRNA and siRNA as a homodimer (21, 22). B2 inhibits both the in vitro DCR2-dependent processing of dsRNA into siRNA and RNA interference (RNAi) induced by synthetic siRNAs in a dsRNA binding-dependent manner, suggesting a dual mode of RNAi suppression by the B2 protein (21, 23, 24).
Fig. 3.
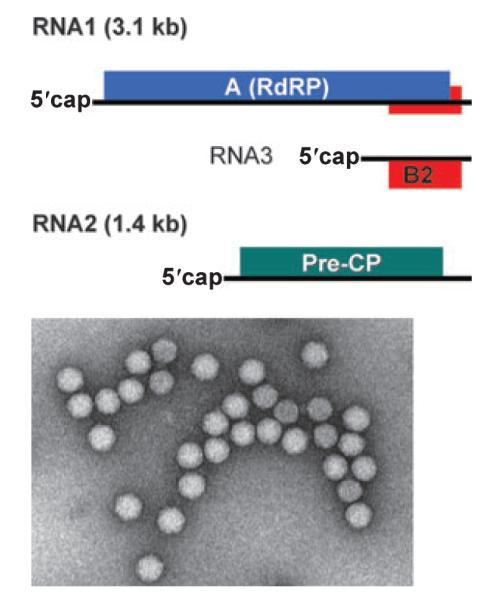
Structures of the bipartite (+)RNA genome and virions of Flock house virus.
Infection of S2 cells by FHV results in the production of 21-nt viral siRNAs that can be detected in Northern blots by probes hybridizing to any regions of either the (+) or (-) genomic RNAs (19). Self-replication of wildtype FHV RNA1 in S2 cells led to abundant accumulation of both RNA1 and RNA3. By contrast, self-replication of a B2-deficient FHV RNA1 mutant (FR1-ΔB2) resulted in readily detectable accumulation of RNA1 and RNA3 in S2 cells only after treatment with dsRNA targeting AGO2 mRNA (19). Intriguingly, treatment of cultured fruit fly S2 cells with dsRNA targeting DCR1, DCR2, or both was unable to rescue the accumulation of FR1-ΔB2, even though it is known that dsRNA treatment efficiently depletes DCR1 and DCR2 proteins and suppresses exo-RNAi as effectively as Ago-2 depletion (19, 25, 26). As Ago-2 was known to act in the exo-RNAi pathway at the time (26), these findings implicated but did not identify DCR2 as the Dicer to initiate the small RNA-directed viral immunity in D. melanogaster.
Identification of D. melanogaster mutants carrying genetic lesions in DCR2 including a null allele dcr-2L811fsX (27), made it possible to re-examine the role of DCR2 in antiviral silencing (28–31). Containing a premature stop codon before the RNase III domains, dcr-2L811fsX flies are defective for exo-RNAi but display no obvious developmental defect, because DCR2 is dispensable for miRNA function. Independent studies from three groups showed that the DCR2 mutant flies exhibit enhanced disease susceptibility to all of the four (+)RNA viruses examined (28–30). These include FHV, Sindbis virus and two polio-like insect viruses from the Dicistroviride, Drosophila C virus (DCV) and cricket paralysis virus (CrPV). These viruses accumulated to much higher levels and caused earlier and more complete lethality in the DCR2 mutant flies than in wildtype flies, indicating that DCR2 provides protection against diverse (+)RNA viruses in D. melanogaster (28–30).
Fruit fly embryos support robust self-replication of FHV RNA1 following microinjection of FHV RNA1 synthesized in transcription reaction in vitro (29). However, self-replication of FR1-ΔB2 resulted in readily detectable accumulation of RNA1 and RNA3 in mutant embryos carrying a homozygous null allele of ago-2 (ago-2414) but not in wildtype embryos. Thus, FHV RNA replication in fly embryos induces Ago-2-dependent antiviral silencing which effectively inhibits FHV replication without suppression of RNAi by B2 (29), which is similar to that found in fly S2 cells (19). Notably, replication of FR1-ΔB2 was rescued in dcr-2L811fsX embryos as effectively as in ago-2414 embryos (19), and replication of FR1-ΔB2 encoded in a transgene was also restored in dcr-2L811fsX flies but not in wildtype flies (28). These findings establish an essential and specific role for DCR2 in the small RNA-directed viral immunity in D. melanogaster. Furthermore, antiviral silencing induced by FR1-ΔB2 is inhibited by the null allele of dcr-2 (28, 29) but not by the incomplete depletion of DCR2 in S2 cells treated with dcr-2 dsRNA (19), suggesting that the viral immunity can be initiated by DCR2 at significantly reduced expression levels.
Ultimately, identification of a host Dicer protein as the viral sensor of the small RNA-directed viral immunity requires demonstration of a specific role of the Dicer in the biogenesis of viral siRNAs (3). FHV infection in both fruit fly cell culture and adult animals induces production of viral siRNAs (19, 28, 29). Whereas significantly enhanced accumulation of FHV was detected in both dcr-2L811fsX and r2d2S165fsX mutant flies, abundant viral siRNAs were produced by r2d2S165fsX flies but not by dcr-2L811fsX flies (19). Production of viral siRNAs was also examined in wildtype, dcr-2L811fsX, and ago-2414 embryos following self-replication of FR1-ΔB2 in a recent study (32). Removal of B2 is necessary to probe the capacity of the fly immune system in the recognition of viral triggers and in the production of viral siRNAs because B2 expression inhibited the biogenesis of viral siRNAs. Under these conditions, production of viral siRNAs was demonstrated in wildtype embryos following replication of FR1-ΔB2, even though the induced small RNA immunity potently inhibits FR1-ΔB2 replication, thereby restricting the accumulation of viral triggers for dicing. Rescue of FR1-ΔB2 replication in ago-2414 embryos resulted in a dramatic increase in the production of viral siRNA. However, viral siRNAs were undetectable in dcr-2L811fsX embryos in which FR1-ΔB2 replication was efficiently rescued as in ago-2414 embryos and the miRNA-producing dcr-1 was wildtype (32). Taken together, these data indicate that DCR1 may not be involved in the biogenesis of viral siRNAs and that DCR2 acts as the viral sensor in the initiation of the small RNA-directed viral immunity in D. melanogaster.
Drosophila X virus (DXV) contains a bi-segmented dsRNA genome and flies infected with DXV become sensitive to CO2. Enhanced susceptibility to DXV was observed in several RNAi mutants, including ago-2 and r2d2 mutants in the siRNA pathway and piwi and aubergine (aub) in the piRNA pathway but, intriguingly, not in either dcr-2L811fsX flies or a heterozygous dcr-1 mutant (31). This suggests that the biogenesis and activity of viral siRNAs from dsRNA viruses, which remains to be experimentally verified, may follow a genetic pathway distinct from the viral siRNAs of (+)RNA viruses.
A. thaliana
The four Dicers of A. thaliana have specific roles in the biogenesis of distinct classes of endogenous small RNAs (33). DCL3 produces 24-nt repeat-associated siRNAs (rasiRNAs) that target transposons, retro-element loci, and repetitive DNA. The DCL4-depependent trans-acting siRNAs are 21 nt long and guide RNA silencing of endogenous targets. miRNAs are predominantly made by DCL1. DCL1 and DCL2 are also required for the production of natural anti-sense transcript-derived siRNA induced by biotic and abiotic stresses (34). Unlike DCR1 and DCR2 of D. melanogaster, there is functional redundancy among the DCLs of A. thaliana; for example, DCL1, DCL2, and DCL4 can produce 21- and 22-nt rasiRNAs in the absence of DCL3 (33). Functional redundancy was a major reason why early studies using single dcl mutants of A. thaliana were unable to convincingly link viral susceptibility to Dicer (3). A number of (+)RNA viruses (Fig. 4) have been used to investigate the immune responses of A. thaliana, including cucumber mosaic virus (CMV), turnip mosaic virus (TuMV), turnip crinkle virus (TCV), tobacco rattle virus (TRV), and oilseed rape mosaic virus (ORMV). The titers, disease symptoms, and viral siRNAs of both TuMV and CMV in single dcl mutants were indistinguishable from those in their respective wildtype parents (35). A reduction in the accumulation of viral siRNAs was observed in dcl2 mutant plants 7 days postinfection with TCV, but this effect was transient and did not result in a predicted increase in the accumulation of TCV in the infected plants (35).
Fig. 4.
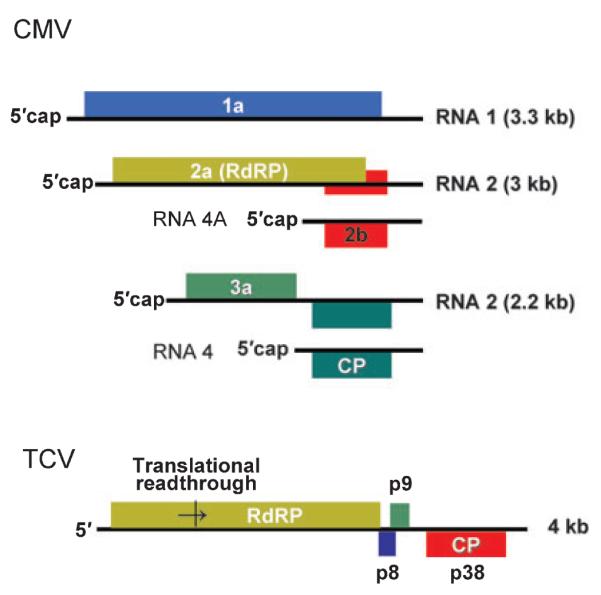
Structures of the (+)RNA genomes of cucumber mosaic virus (CMV) and turnip crinkle virus (TCV).
Recent studies have shown that DCL4 and DCL2 act redundantly and either is sufficient to initiate the small RNA-directed viral immunity in A. thaliana against diverse (+)RNA viruses (36–39). DCL4 is the predominant viral sensor as most of the viral siRNAs detected in the infected plants are 21 nt long. An exception was in the TCV-infected plants where 22-nt siRNA species is dominant due to viral suppression of DCL4 (37). Genetic inactivation of DCL4 enhances production of 22-nt viral siRNAs by DCL2, which can initiate effective antiviral silencing in the absence of DCL4. Thus, inactivation of both DCL4 and DCL2 is required to eliminate the production of 21- and 22-nt viral siRNAs and to dramatically increase both viral titers and symptom severity in A. thaliana infected with CMV, TCV, and TRV. Lack of 21- and 22-nt viral siRNAs and enhanced viral titers were also observed in dcl2 dcl4 double mutant plants infected with ORMV, although, in this case, the infected dcl2 dcl4 plants did not exhibit more severe disease symptoms than wildtype and other mutant plants (40). By contrast, abundant production of 24-nt viral siRNAs by DCL3 in dcl2 dcl4 plants infected with (+)RNA viruses failed to either degrade homologous host mRNAs or inhibit infection of (+)RNA viruses. Thus, DCL3 alone is unable to initiate antiviral silencing in contrast to DCL4 and DCL2.
Deletion of viral silencing suppressor such as 2b of CMV and P38 of TCV produces mutant viruses with multiple defects in plant infection (41). However, these defects associated with TCV-ΔP38 and CMV-Δ2b were efficiently rescued in mutant plants in which both DCL4 and DCL2 were inactivated, but not in single or other double dcl mutants such as dcl2 dcl3 and dcl3 dcl4 plants (37, 38). These findings provided further support that either DCL4 or DCL2, but not DCL3, can act as the viral sensor to trigger antiviral silencing so that both pathways must be suppressed to achieve successful infection.
Cauliflower mosaic virus (CaMV) and geminivirus CaLCuV, containing a circular dsDNA and ssDNA genome, respectively, are also targeted for RNA silencing in A. thaliana (40, 42). However, antiviral silencing against DNA viruses exhibit several distinct features when compared with antiviral silencing against (+)RNA viruses. First, the 24-nt viral siRNAs produced by DCL3 were more abundant than 21- and 22-nt siRNAs in plants infected with either virus. Second, DCL1 plays a more prominent role in the biogenesis of viral siRNAs from DNA viruses. For example, DCL1-dependent production of 21-nt viral siRNAs was readily detectable in dcl2 dcl3 dcl4 triple mutant plants infected with either CaMV or CaLCuV, which may explain why simultaneous inactivation of DCL2, DCL3, and DCL4 does not increase the severity of symptoms caused by either CaMV or CaLCuV when compared with wildtype plants (40, 42). The titers of CaLCuV were also similar in dcl2 dcl3 dcl4 and wildtype plants. An increased accumulation of CaMV in dcl2 dcl3 dcl4 plants was observed in one study, which, however, was not reproduced in an independent study. Thus, DCL1 may be essential for the small RNA-directed viral immunity against DNA viruses and use of a quadruple dcl mutant plants (36) will be necessary to determine the specific role of DCL1.
Fungi, C. elegans, and mammals
Although fission yeasts contain a single Dicer that acts in the nucleus (43), many fungi, such as Neurospora crassa and the chestnut blight fungus Cryphonectria parasitica, encode two Dicers that differ by the presence of a dsRBD at the C-terminus in DCR2 but not in DCR1 (44, 45). DCR1 and DCR2 of N. crassa act redundantly in transgene-induced RNA silencing (45). However, DCR2, but not DCR1, of C. parasitica is required for viral siRNA production and antiviral silencing against CHV1-EP713, a (+)RNA virus member of the Hypoviridae (44, 46). Infection of cultured C. elegans cells by VSV, a (-)RNA virus, induces production of viral siRNAs (47). It is likely that the single C. elegans Dicer acts as the viral sensor. DCR1 of C. elegans is required for the RNAi-based immunity against VSV (48) and several genes essential for the canonical dsRNA—siRNA pathway downstream of DCR1 participate in antiviral silencing against both VSV and FHV (23, 47, 48). These include RDE1, RDE4, and RRF1 that are dispensable for the biogenesis of piRNAs that are DCR1 independent (49).
Infection of mammalian cells with nucleus-replicating DNA viruses induces production of virus-derived miRNAs. The first viral miRNAs were identified from the γ-herpersvirus Epstein—Barr virus (EBV) and subsequent studies showed that 10 viruses from the herpesvirus family, two simian polyomaviruses and human adenovirus produce miRNAs ranging from 1 in SV40 to 23 in EBV (5, 50). These findings predict that viral nuclear transcripts are recognized by the biogenesis pathway in mammals, which includes processing of primary miRNAs into pre-miRNAs by Drosha/DGCR8 in the nucleus and final maturation of miRNAs by Dicer/TRBP in the cytoplasm. A comprehensive understanding of the function of these mammalian viral miRNAs is currently lacking. Viral miRNAs may function to target and regulate the expression of host genes in mammals (5), as has been shown for siRNAs derived from CaMV, a plant dsDNA virus (42). However, several viral miRNAs, such as miR-S1 of SV40, miR-BART2 of EBV, and miR-H2-3p of herpes simplex virus 1, are transcribed anti-sense to and shown to guide either cleavages or translational repression of, key mRNAs of the respective viruses (5, 50–52). Thus, at least some of the known mammalian viral miRNAs exhibit features of viral siRNAs produced by plant and invertebrate hosts, suggesting that they may act to silence viral sequences as part of mammalian immune responses to infection. This hypothesis is supported by the observation that most of the herpesviral miRNAs are not conserved among related viruses (5). Thus, it is unlikely that viral miRNAs have evolved to inhibit conserved functions of hosts.
Viral triggers
What is recognized by the host as the substrate of Dicer to trigger the small RNA-directed viral immunity had been under debate. In principle, three different forms of viral RNA may serve as the precursor of viral small RNAs. Either the double-stranded viral replicative intermediate RNAs (vRI-dsRNA) of viruses with an RNA genome or highly structured hairpin regions in single-stranded viral genomic RNA and mRNA of RNA and DNA viruses may directly be processed into viral small RNAs. In addition, any single-stranded viral RNA may be targeted and converted first to dsRNA by a cellular RdRP before recognition by Dicer. Detection of small RNAs corresponding to both the positive and negative strands of (+)RNA viruses in the infected plant and insect cells implicated vRI-dsRNA as the viral trigger (18, 19, 53). However, cloning and sequencing of small RNAs in plants infected with Cymbidium ringspot tombusvirus (CymRSV), a (+)RNA virus, revealed that 80% of the sequenced viral small RNAs were derived from the positive-strand viral RNA and 85% of them were mapped to several clusters (54). Far more abundant (+) viral small RNAs were also demonstrated in the TCV-infected plants by small RNA sequencing and in plants infected with potato virus X (PVX) and tobacco mosaic virus (TMV) by Northern blot hybridizations (54, 55), both of which contain a (+)RNA genome. These findings led to a hypothesis that imperfect duplexes originating from highly base paired structures from the single-stranded (+) and (-) genomic RNAs act as the substrates of a virus-specific Dicer, similar to the recognition of pre-miRNAs by miRNA-producing Dicers (54, 55).
This hypothesis is not supported by the genetic characterization of the host antiviral silencing pathway. The viral sensors identified in both D. melanogaster and A. thaliana for diverse (+)RNA viruses are the known siRNA-producing Dicers and the miRNA-producing DCR1 and DCL1 play no detectable role in the biogenesis of (+)RNA virus-derived small RNAs. In addition, antiviral silencing requires host proteins from the canonical dsRNA—siRNA pathway(s) that are dispensable for miRNA function, which include SGS2, SDE-3, SDE5, and RDR6 of A. thaliana (56–58), Ago-2 and R2D2 of D. melanogaster (19, 30), and RDE-1, RDE-4, and RRF-1 of C. elegans (23, 47, 48). Moreover, approximately equal ratios of (+) and (-) strand viral small RNAs were found in plant, fungal, and fruit fly cells infected with diverse (+)RNA viruses (32, 46, 55, 59). These findings therefore support vRI-dsRNA of (+)RNA viruses as the viral trigger of DVI, although they do not rule out a prior recognition of viral ssRNA targets by RDR in organisms that encode RDR.
The replication cycle of an RNA genome is predicted to yield dsRNA, which is at least 40 bp long, because it was detectable by a dsRNA-specific monoclonal antibody during infection of a number of plant and animal (+)RNA viruses (60, 61). vRI-dsRNA has been proposed to trigger recognition by Dicer in plants and invertebrates and by PRRs in mammals including TLRs and RLHs (2). However, RNA replication occurs inside the intracellular membrane structures for (+)RNA viruses or the virions for (-)RNA and dsRNA viruses, so that vRI-dsRNA may be embedded and protected from immune recognition. In addition, (+)RNA viruses may produce dsRNA during the synthesis of (i) the (-)RNA template from the incoming (+)RNA genome, (ii) the viral progeny (+)RNA from the (-)RNA template, or (iii) subgenomic RNAs from the (-)RNA template of some (+)RNA viruses. Thus, it is unknown if vRI-dsRNA synthesized from a particular step of viral RNA replication is involved in the induction of any of these dsRNA-specific innate immune responses.
The population of viral small RNAs in Drosophila cells infected abortively by a B2-deficient mutant of FHV (FHV-B2) was recently examined by pyrosequencing and gel blot hybridizations. The results showed that 57% and 43% of the sequenced viral small RNAs were mapped, respectively, to the (+) and (-) strands of the FHV genome, most (89%) were 20–22 nt long with a major peak at 21 nt (60.3%), and more than 60% of FHV RNA1-specific small RNAs were clustered in the 5′-terminal region of about 400 nt long (32). These findings indicate that replication of the FHV (+)RNA genome produces an approximately 400-bp dsRNA at the 5′-terminal region that is recognized by Dicer-2 as the major precursor of viral siRNAs. Notably, B2 interacts with both viral dsRNA and RNA replicase and potently inhibits production of the 5′-terminal viral siRNAs (32). These observations therefore provide a cell biology model in which DVI is induced during the initiation of viral progeny (+)RNA synthesis but is suppressed by B2 inside the viral RNA replication complex (Fig. 5).
Fig. 5. Model for the induction and suppression of the Dicer-initiated viral immunity in Drosophila against Flock house virus.
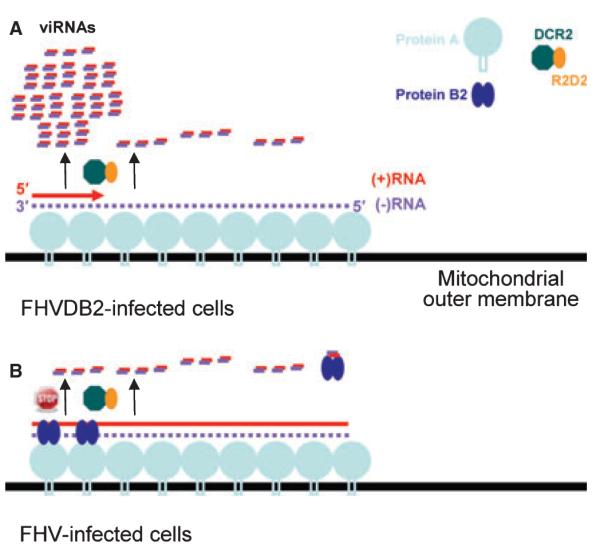
Asymmetric RNA synthesis in the replication of (+)RNA viruses involves multiple initiation of the progeny (+)RNA synthesis on the low abundant (-)RNA template complexed with the viral RdRP and other host factors. The resulting dsRNA of approximate 400 nt in length formed between the 5′-terminal nascent progeny (+)RNA1 and the (-)RNA1 template in FHV-infected cells, termed the initiating vRI-dsRNA, serves as substrates of DCR2. This results in the predominant production of 5′-terminal viRNAs, thereby triggering the RNAi-mediated viral immunity and abortive infection by FHV-ΔB2. In addition to binding to viRNAs, B2 is part of the viral RNA replication complex by direct interactions with viral RdRP (protein A) and vRI-dsRNA and inhibits DCR2-dependent production of viRNAs, thus ensuring successful infection by FHV. We propose that sequestering the initiating vRI-dsRNA and inhibiting their processing into the 5′-terminal viRNAs by B2 play a particularly important role in the suppression of the viral immunity. Reprinted from (32). RdRP, RNA-dependent RNA polymerase; FHV, Flock house virus; DCR, Dicer.
Effectors
All types of small RNAs guide specific gene silencing in an effector complex such as RISC, of which AGO is an essential component. Initially described as developmental regulators in plants and first identified as an RNAi component in C. elegans (RDE-1), AGO proteins are conserved in eukaryotes and are divided into three subfamilies based on their sequence and functional similarities (62, 63). Members of the Argonaute and PIWI subfamilies interact, respectively, with siRNAs/miRNAs and piRNAs, whereas the third subfamily contains an expanded group of C. elegans proteins. In addition to the PAZ domain found also in Dicer, Ago/Piwi proteins contain the MID domain responsible for binding the 5′-phosphate of the loaded small RNA and the C-terminal RNase H-like PIWI domain (64). In RNAi, asymmetry of siRNAs and the heterodimer of DCR2 and R2D2 play a key role in siRNA loading and the selection of the guide strand siRNA in RISC (65). Recent studies further revealed the importance of the 5′-terminal nucleotide of the small RNA in the sorting process in A. thaliana (66–68).
Small RNA-loaded AGOs find their specific targets by base pairing between the small RNA and its target ssRNA, and the subsequent gene silencing may be achieved by three distinct mechanisms. When there is extensive base pairing between small RNA and its target RNA with minimal mismatches, the target ssRNA may be cleaved or sliced at the central position opposite guide RNA by the RNase H-like activity of the AGO in the effector complex (17). D. melanogaster encodes two (AGO1 and AGO2) and three members in the Argonaute and PIWI subfamily, respectively, all of which exhibit slicer activity, although AGO1 is a weak slicer when compared with AGO2 (17). A. thaliana encodes 10 AGOs, all of which belong to Argonaute subfamily, and the slicer activity has been established for AGO1, AGO4, and AGO7 (63). However, many AGOs including three of the four human AGO subfamily members do not exhibit slicer activity. Animal miRNAs are partially complementary in the 5′-seed region to their target mRNAs and inhibit translation without slicing by sequestering mRNA away from the translational machinery into cytoplasmic foci termed P-bodies. The third effector mechanism is transcriptional gene silencing (TGS) in which the AGO complex may be directed to the nascent transcripts to induce modification of either DNA or histone.
An antiviral role of AGOs was first reported in 2002 for AGO1 of A. thaliana and AGO2 of D. melanogaster (19, 69). In C. elegans, at least two members from the third AGO subfamily, RDE1 and C04F12.1, participate in antiviral RNAi against RNA viruses (23, 47, 48). A. thaliana AGO1 has an essential role in the miRNA pathway; however, hypomorphic ago1 mutants of A. thaliana are viable and exhibit hypersensitivity to CMV infection (69). Genetic inactivation of AGO2 in either cultured cells or embryos of D. melanogaster or of RDE1 in C. elegans is sufficient to rescue the accumulation of the B2-deficient mutant of FHV, which is defective in the suppression of antiviral silencing (19, 23, 29). In D. melanogaster cells infected with FHV, viral siRNAs are loaded in AGO2 (Fig. 6), and AGO2-loaded viral siRNAs are methylated at their 3′-ends (32), similar to the endogenous siRNAs of D. melanogaster (17). Viral siRNAs in the input prior to co-immunoprecipitation by a monoclonal antibody to AGO2 was partially sensitive to periodate oxidation and β elimination treatments. Thus, a portion of viral siRNAs in the infected cells contain unmethylated 3′-ends, possibly because of saturation of AGO2 loading by the highly abundant viral siRNA. However, viral siRNAs accumulated to high levels in ago-2414 embryos in which antiviral silencing is defective. Thus, AGO2 is essential for the antiviral activity but dispensable for the biogenesis, of viral siRNAs and unloaded free-floating viral siRNAs may be stable (17). Interestingly, although the B2-deficient mutant of FHV replicated to high levels in both ago-2414 and DCR2L811fsX embryos (29), efficient rescue of the FHV mutant occurred in D. melanogaster cells after dsRNA-mediated depletion of AGO2 but not of DCR2 (19). These findings indicate that the role of AGO2 in DVI is dosage sensitive, in contrast to DCR2.
Fig. 6. A framework of the Dicer-initiated viral immunity.
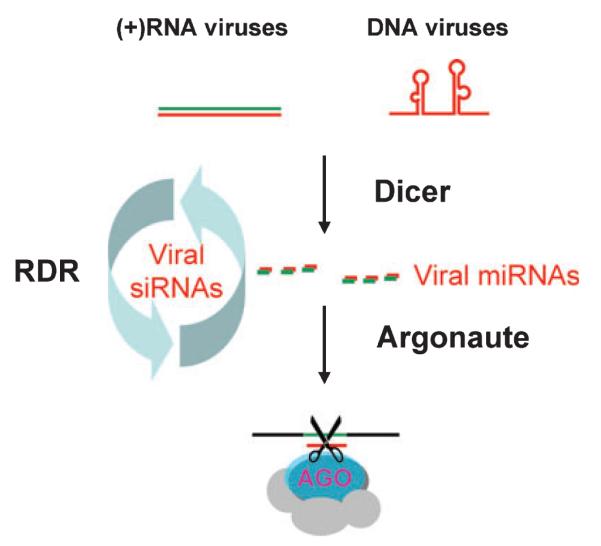
The immunity begins with the recognition of the viral triggering RNA molecules by the viral sensor (Dicer) of the host. The products of Dicer, the virus-derived small RNAs, are loaded in Argonaute protein-containing effector complexes to guide cleavage or translational arrest of viral RNAs. In plants and Caenorhabditis elegans, virus-derived siRNAs may be further amplified by a host RNA-dependent RNA polymerase (RdRP).
Enhanced dicing of viral RNA into 24-nt siRNAs by DCL3 in dcl2 dcl4 mutant A. thaliana plants is unable to silence either TCV-ΔP38 or CMV-Δ2b, both of which are defective in silencing suppression. As the 24-nt siRNAs guide TGS in the nucleus and do not guide RNA degradation in the cytoplasm, these findings suggest a key role for slicing in antiviral silencing in plants and that dicing alone is insufficient (37, 38). Infection of Nicotiana benthamiana with p19-deficient tombusviral mutants induced assembly of a discrete, high molecular weight RISC-like complex, which contains siRNAs derived from the infecting viruses and exhibits specific slicer activity (70, 71). Sensor mRNAs containing either a sense or anti-sense viral insert are sliced in the infected plants, and those sensor mRNAs complementary to (+)viRNAs are cleaved more efficiently than (-)viRNA targets (70), which appears to be consistent with the earlier observation that (+)viRNAs are more abundant than (-)viRNAs in tombusvirus-infected plants (54). There are slicing hotspots within the 190-nt viral insert of both sense and anti-sense RNA sensors, but these hotspots or lack of them do not correlate with the relative abundance of viRNAs generated in the infected plants (70). Mapping of cleavage sites suggest that the positive-strand genomic RNA was indeed cleaved in infected plants and the cleavage hotspots were mapped to the same region revealed by the non-replicating sensor RNAs. However, no cleavage of the viral negative-strand RNA at any position was detected in spite of abundant effector complexes loaded with (+)viRNAs. Thus, viral templates for replication may be inaccessible to slicing (70), similar to the targeting of RNA viruses by designed siRNAs in mammalian cells (72–74). The non-overlapping nature of the hot spots for dicing and slicing also suggests that the biogenesis of viral siRNA by dicing may be independent of slicing, which has been proposed for the production of secondary siRNAs in C. elegans, transacting-siRNAs in plants and nuclear siRNAs of fission yeasts.
These studies in N. benthamiana suggested but did not identify an AGO (or AGOs) in the slicing or the assembly of an RISC-like effector complex in the tombusvirus-infected cells (70, 71). Whether or not A. thaliana AGOs bind to viral siRNAs in the infected cells have been investigated by co-immunoprecipitation of epitope-tagged AGOs expressed from stably integrated transgenes. Both AGO2 and AGO5 bind to viral siRNAs in CMV-infected A. thaliana plants (68) (Fig. 6). Binding of AGO1 to siRNAs derived from three (+)RNA viruses was undetectable in one study (75), but physical association of AGO1 with viral siRNAs was demonstrated in a subsequent study using the same AGO1 transgenic plants infected with CMV (76). However, CMV-Δ2b infection was as defective in any of the three ago1 mutants (ago1-11, -12, and -27) tested as in wildtype plants (X. Wang and S.W. Ding, unpublished data), suggesting that AGO1 is either not required or redundantly required for antiviral silencing. Future work is necessary to resolve this discrepancy and to determine if viRNA-loaded AGOs have antiviral activities and if genetic inactivation of specific AGO genes is able to inhibit silencing of virus mutants defective in silencing suppression.
Amplifiers
The canonical dsRNA—siRNA pathway of RNA silencing is amplified in fungi, C. elegans, and plants by an RDR-dependent pathway (77). Eukaryotic RDRs share little sequence homology with the RdRPs encoded by RNA viruses and only a few studies on their biochemical activities have been reported (77, 78). However, genetic studies have clearly established an essential role for RDR in RNA silencing and the production of secondary siRNAs and various types of endo-siRNAs in fungi, C. elegans, and plants. For example, A. thaliana encodes six RDRs grouped into four clusters, RDR1, RDR2, RDR3/4/5, and RDR6. RDR2 is required for the biogenesis of the 24-nt siRNAs that mediate DNA and histone modifications in the nucleus, whereas ta-siRNAs and nat-siRNAs are RDR6 dependent. Notably, RDR6-dependent production of transgene secondary siRNAs plays a key role in mediating the cell-to-cell spread of RNA silencing in A. thaliana (79).
A role for the RDR activity in an RNA silencing-based antiviral response was first proposed in 1993, before its molecular cloning in tomato and its identification as an RNAi component in N. crassa (77, 80–82). A. thaliana rdr6 mutant plants contain increased virus titers and develop more severe symptoms than wildtype plants infected with CMV (56). Degradation of viral mRNAs of the DNA virus CaLCuV was also inhibited in rdr6 plants, which, however, was not accompanied with an increase in virus titers or symptom severity (83). In addition, the loss-of-function mutations in RDR6 had no effect on the infection by many (+)RNA viruses including TRV and TuMV (57, 84). Similarly, tobacco plants with reduced expression of RDR1 or N. benthamiana plants with reduced expression of RDR6 exhibit enhanced susceptibility to some (+)RNA viruses but not to others including CMV (85, 86). N. benthamiana plants contained a defective RDR1, and over-expression of a functional RDR1 increased resistance to several tobamoviruses but not to PVX or CMV (87). The somatically active RDR of C. elegans, rrf-1, also contributes to antiviral RNAi against VSV (48). These studies support an antiviral role for cellular RDRs (Fig. 6).
It is unclear if and how host RDRs mediate antiviral response via an RNA silencing pathway. None of the above studies has convincingly established a link between antiviral silencing and RDR-dependent amplification of viral siRNAs. Use of CMV-Δ2b for infection revealed a dramatic reduction in the biogenesis of viral siRNAs in A. thaliana rdr1 mutant plants when compared with wildtype plants (38). A similar reduction in the accumulation of viral siRNAs was recently observed in rdr1 rdr2 rdr6 triple mutant plants infected with TRV (88). Thus, these studies provided the first evidence for RDR-dependent production of secondary viral siRNAs in plants. However, both studies failed to detect any effect of secondary viral siRNAs on either viral accumulation or symptom severity in the infected plants. It remains to be determined if this is due to either functional redundancy of the uncharacterized RDR3/4/5 or masking of RDR’s effect by the encoded viral suppressor of RNA silencing.
Conclusions and future directions
In addition to producing distinct classes of endogenous small RNAs, the Dicer nuclease acts as a PRR in diverse eukaryotes to initiate the small RNA-directed viral immunity. Available data show that recognition of (+)RNA viruses is mediated by a specific Dicer(s) in organisms that encode more than one Dicer protein. For example, detection of four (+)RNA viruses in D. melanogaster is mediated specifically by DCR2, and similarly, only one of the two fungal Dicers appears to participate in antiviral silencing. In A. thaliana, two of the four Dicers, DCL4 and DCL2, act redundantly to mediate antiviral silencing against diverse (+)RNA viruses, whereas neither DCL1 nor DCL3 is essential or sufficient. By contrast, all four DCLs of A. thaliana participate in the defense against DNA viruses. Many questions on the detection of viruses by the Dicer family of host immune receptors remain unresolved. How is the observed specificity in virus sensing by Dicer determined? All of the Dicer proteins that have been implicated in antiviral silencing contain a dsRBD at the C-terminus, although whether or not the C-terminal dsRBD is involved in a direct binding of Dicer to the viral RNA trigger remains to be investigated. Is any of the antiviral Dicers induced transcriptionally upon virus challenge? Is the single Dicer of C. elegans and human indeed involved in the production of siRNAs from RNA viruses and miRNAs from DNA viruses, respectively? In contrast to robust Dicer-dependent immune responses to (+)RNA viruses in plants and invertebrates, available data suggest that these viruses are not detected at all by the human Dicer, for which the molecular basis is unknown.
Viral dsRNA has been proposed as the molecular signature of RNA viruses to trigger recognition of TLR3 in the endosome and of RIG-I and MDA5 in the cytosol, in addition to Dicer. The replication cycle of RNA viruses predicts production of dsRNA during the synthesis of either (+)RNA or (-)RNA as well as during transcription of subgenomic mRNAs. However, RNA replication occurs in intracellular membrane structures and viral dsRNA produced may be transient in time, short in length, and inaccessible to host immune receptors because of containment within membrane structures. Thus, it remains to be verified in vivo if viral RNA replication produces a viral dsRNA of sufficient length and accessible to binding by TLR3, RIG-I, and MDA5 to trigger innate immune responses and if the triggering viral dsRNA is synthesized from a particular step of viral RNA replication. Unlike TLR3, RIG-I, and MDA5, recognition of a viral RNA trigger by Dicer is accompanied with the processing of the triggering molecule into small RNAs, which can be sequenced and mapped to specific positions along the genomic RNA. This strategy led to the demonstration that a ∼400-bp dsRNA from the 5′-terminus of the genomic RNA, which is produced during the initiation of the progeny (+)RNA synthesis from the 3′-end of the (-)RNA template, serves as the viral RNA trigger in Drosophila. This finding therefore identifies the step of the progeny (+)RNA synthesis as critical in the induction of DVI. It will be necessary to determine if this represents a shared mechanism for responding to infection of (+)RNA viruses in insects and other organisms. Interpretation of the distribution patterns of viral siRNAs along the viral genomic RNA in plants and C. elegans may not be as straightforward as in D. melanogaster. In plants and C. elegans, production of secondary viral siRNAs must be considered unless the cellular RDR (or RDRs) that amplifies viral siRNAs is genetically inactivated.
The innate immunity mediated by TLRs and RLHs has broad spectrum antiviral activity, almost inevitably leads to destruction of the infected cell, and may involve multiple effectors. By contrast, DVI has a well-defined effector molecule, Argonaute, with a specific antiviral activity determined by the small guide RNAs derived from the challenge virus. In this regard, the RNA-based immunity has strong parallels in adaptive immunity in which short peptide epitopes are processed from a viral protein and used as specificity determinants of the immunity. In both plants and fruit flies, the viral siRNAs are methylated at their 3′-ends. AGOs exhibit structural similarity to RNase H and mediate cleavage of the target viral RNAs as the major effector mechanism. However, many AGOs are not active RNase and instead repress translation without cleaving the target RNA. Argonaute loaded with a guide small RNA acts as a multiple turnover enzyme and targets predominantly viral RNAs without major negative impact on host gene expression or host cell viability, because of overwhelming abundance of viral siRNAs over cellular small RNAs in an infected cell. Both plants and animals encode multiple members in the AGO/PIWI family. In D. melanogaster cells infected with FHV, viral siRNAs are loaded in AGO2, and an essential role of AGO2 in antiviral silencing has been established in both cell culture and living fruit flies. Loading of viral siRNAs in A. thaliana AGO1, AGO2, and AGO5 has also been reported, although a specific role for any of these AGOs in antiviral silencing remains to be established. C. elegans encodes 27 members in the AGO/PIWI family, at least two of which have been implicated in antiviral silencing. Thus, although a natural virus for C. elegans is not known, it is possible that the largest expansion of the AGO/-PIWI family in C. elegans may represent in part an evolutionary adaptation to virus infection.
Recent studies have shown that bacteria and archaea also encode an RNA-based immune system against viruses (89–92). In this system, bacteria and archaea hosts acquire short viral sequences (26-72 bp) and integrate them as spacers within clusters of regularly interspaced short palindromic repeats (CRISPRs). CRISPRs are flanked by a characteristic set of CRISPR-associated (cas) genes and transcripts of CRISPRs are processed into small RNAs to guide specific antiviral defense. However, the eukaryotic RNAi machinery is not found in prokaryotes and many questions on the mechanism of the RNA-based immunity in prokaryotes remain to be addressed. How do prokaryotes acquire the virus-specific spacer sequences? What are the specific functional roles of these Cas proteins? Which nuclease is involved in the biogenesis of CRISPR small RNAs? Do CRISPR small RNAs direct specific antiviral defense by guiding cleavages of viral RNAs or modification of viral DNA? Do bacteriophages encode suppressors to interfere with either the biogenesis or activity of CRISPR small RNAs?
Acknowledgements
The research projects on antiviral silencing in the authors’ lab are supported by the National Institute of Allergy and Infectious Diseases (AI052447) and the National Research Initiative of the USDA cooperative State Research, Education, and Extension Service (2007-35319-18325).
References
- 1.Leulier F, Lemaitre B. Toll-like receptors — taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 2.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matranga C, Zamore PD. Small silencing RNAs. Curr Biol. 2007;17:R789–R793. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji X. The mechanism of RNase III action: how dicer dices. Curr Top Microbiol Immunolnol. 2008;320:99–116. doi: 10.1007/978-3-540-75157-1_5. [DOI] [PubMed] [Google Scholar]
- 7.Pertzev AV, Nicholson AW. Characterization of RNA sequence determinants and antideterminants of processing reactivity for a minimal substrate of Escherichia coli ribonuclease III. Nucleic Acids Res. 2006;34:3708–3721. doi: 10.1093/nar/gkl459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baulcombe D. RNA silencing. Trends Biochem Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 15.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 17.Siomi MC, Saito K, Siomi H. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 2008;582:2473–2478. doi: 10.1016/j.febslet.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 19.Li HW, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 20.Venter PA, Schneemann A. Recent insights into the biology and biomedical applications of Flock House virus. Cell Mol Life Sci. 2008;65:2675–2687. doi: 10.1007/s00018-008-8037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 22.Lingel A, Simon B, Izaurralde E, Sattler M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6:1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol. 2005;79:7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 26.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 27.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 28.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 29.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 32.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramachandran V, Chen X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;13:368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin H. Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 2008;582:2679–2684. doi: 10.1016/j.febslet.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 38.Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fusaro AF, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blevins T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz-Pendon JA, Ding SW. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu Rev Phytopathol. 2008;46:303–326. doi: 10.1146/annurev.phyto.46.081407.104746. [DOI] [PubMed] [Google Scholar]
- 42.Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 44.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci USA. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Catalanotto C, et al. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol. 2004;24:2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J Virol. 2008;82:2613–2619. doi: 10.1128/JVI.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 48.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 51.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barth S, et al. Epstein—Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 54.Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho T, Pallett D, Rusholme R, Dalmay T, Wang H. A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J Virol Methods. 2006;136:217–223. doi: 10.1016/j.jviromet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 56.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 57.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernandez-Pinzon I, Yelina NE, Schwach F, Studholme DJ, Baulcombe D, Dalmay T. SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J. 2007;50:140–148. doi: 10.1111/j.1365-313X.2007.03043.x. [DOI] [PubMed] [Google Scholar]
- 59.Yoo BC, et al. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 63.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 64.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 65.Hutvagner G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Lett. 2005;579:5850–5857. doi: 10.1016/j.febslet.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 66.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′-terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 68.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 69.Morel JB, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pantaleo V, Szittya G, Burgyan J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J Virol. 2007;81:3797–3806. doi: 10.1128/JVI.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Omarov RT, Ciomperlik JJ, Scholthof HB. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc Natl Acad Sci USA. 2007;104:1714–1719. doi: 10.1073/pnas.0608117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ge Q, et al. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schubert S, et al. Strand-specific silencing of a picornavirus by RNA interference: evidence for the superiority of plus-strand specific siRNAs. Antiviral Res. 2007;73:197–205. doi: 10.1016/j.antiviral.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 75.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Curaba J, Chen X. Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem. 2008;283:3059–3066. doi: 10.1074/jbc.M708983200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 80.Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schiebel W, et al. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell. 1998;10:2087–2101. doi: 10.1105/tpc.10.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 83.Muangsan N, Beclin C, Vaucheret H, Robertson D. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 2004;38:1004–1014. doi: 10.1111/j.1365-313X.2004.02103.x. [DOI] [PubMed] [Google Scholar]
- 84.Dalmay T, Hamilton A, Mueller E, Baulcombe DC. Potato virus X amplicons in arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell. 2000;12:369–379. doi: 10.1105/tpc.12.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol. 2005;79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc Natl Acad Sci USA. 2004;101:6297–6302. doi: 10.1073/pnas.0304346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donaire L, Barajas D, Martinez-Garcia B, Martinez-Priego L, Pagan I, Llave C. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J Virol. 2008;82:5167–5177. doi: 10.1128/JVI.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 91.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;82:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 92.Young RF., 3rd Molecular biology. Secret weapon. Science. 2008;321:922–723. doi: 10.1126/science.1162910. [DOI] [PMC free article] [PubMed] [Google Scholar]