Abstract
The candidate gene approach to pharmacogenetics is hypothesis driven, and anchored in biological plausibility. Whole genome scanning is hypothesis generating, and it may lead to new biology. While both approaches are important, the scientific community is rapidly reallocating resources toward the latter. We propose a step-wise approach to large-scale pharmacogenetic association studies that begins with candidate genes, then uses a pathway-based intermediate step, to inform subsequent analyses of data generated through whole genome scanning. Novel computational strategies are explored in the context of two clinically relevant examples, cholesterol synthesis and lipid signaling.
INTRODUCTION
Genotype-phenotype association studies are being performed on an unprecedented scale. They are being conducted in cohorts that are disease-based, treatment-based, practice-based, and/or population-based. They typically follow one of two strategies: (1) a candidate gene approach (which tends to be hypothesis-driven), or (2) a genome scanning approach (which tends to be hypothesis generating). It was initially argued that genome scanning would lead to the identification of numerous causative genetic polymorphisms - both genic and intergenic - with previously unappreciated biological function, yielding insight into a myriad of novel cellular signaling pathways. While this expectation has partly been realized (e.g., elucidation of the role of INSIG2 in the development of obesity) [Herbert, A. et al. 2006], it remains unclear whether the resulting data will justify the large amount of resources currently being redirected toward genome scanning in populations.
The strengths and limitations of genome-wide association studies have recently been reviewed by Dupuis and O’Donnell [2007]. There exists both optimism [Couzin, J. and Kaizer, J. 2007] and skepticism [Shriner, D. et al. 2007; Williams, S.M. et al. 2007] about whether a dense map of single-nucleotide polymorphisms (SNPs) from across the human genome will lead to a panacea of new genetic risk factors for common diseases. The datafiles generated by genome scanning are typically enormous, and strategies for dealing with multiple testing remain unresolved, particularly for large scan platforms. Even with less stringent strategies to correct for multiple testing, it remains difficult for investigators to identify gene variants that are consistently associated with phenotypic changes. This is in part attributable to the clinical heterogeneity of most common diseases. Furthermore, many of the genetic factors associated with common disease have very small effect size.
Misclassification bias and lack of standardized phenotypes may confound replication of associations identified through genome scanning. Variability in sample size, differing infrastructure, and unquantified environmental factors also lead to ambiguous results. As Williams, S.M. et al. [2007] point out, genes such as PPAR-γ may not have been convincingly identified as a genetic risk factor for type 2 diabetes in genome-wide association studies [Scott, L.J. et al. 2007;Ziggini et al. 2007] without an overwhelming amount of prior genetic and biological evidence suggesting the importance of these genes.
Unlike genome scanning, the candidate gene approach works with small numbers of polymorphisms in a biological framework that provides an interpretive context for association. Even in the absence of a single main effect, variant alleles contributing to phenotypic variability can often be identified by considering gene-gene interaction (epistasis) specifically within the context of a well characterized biological pathway [Moore, J.H. and Ritchie, M.D. 2004; Moore, J.H. 2005; Moore 2007a; 2007b; 2007c]; i.e., a congenitally altered enzyme may have no impact on phenotype unless it occurs in the context of another congenitally variant enzyme in the same pathway(s). Therefore, the importance of the suspect allele may only be recognized if it is characterized through the application of an analytical approach that considers epistasis. Nonetheless, the identification of epistasis is computationally intense, and the computational time is proportional to the number of genes being evaluated.
Thus, both candidate gene studies and genome wide association studies contain inherent challenges. To address these challenges, we explore a stepwise approach that combines information generated from candidate genes (i.e., a gene-centric foundation) with growing knowledge about biological pathways (i.e., a pathway-based framework), to inform the analysis of whole genome association studies (i.e., genome-wide data).
MODEL PATHWAYS
Epistasis is a ubiquitous component of the genetic architecture underlying complex phenotypic traits such as disease onset, disease progression, and treatment outcome [Moore, J.H. 2003] [Moore, J.H. 2005; Wilke, R.A. et al. 2005]. The detection of epistasis in large association studies is both statistically and computationally difficult due to the dimensionality associated with putting multiple genotypes together. Often, an enormous number of potential polymorphism combinations need to be evaluated. Further, the complexity increases when an individual polymorphism exhibits epistasis in the absence of independent main effects. Therefore, a number of new computational methods for detecting, characterizing and interpreting gene-gene interactions are needed [Thornton-Wells, T.A. et al. 2004].
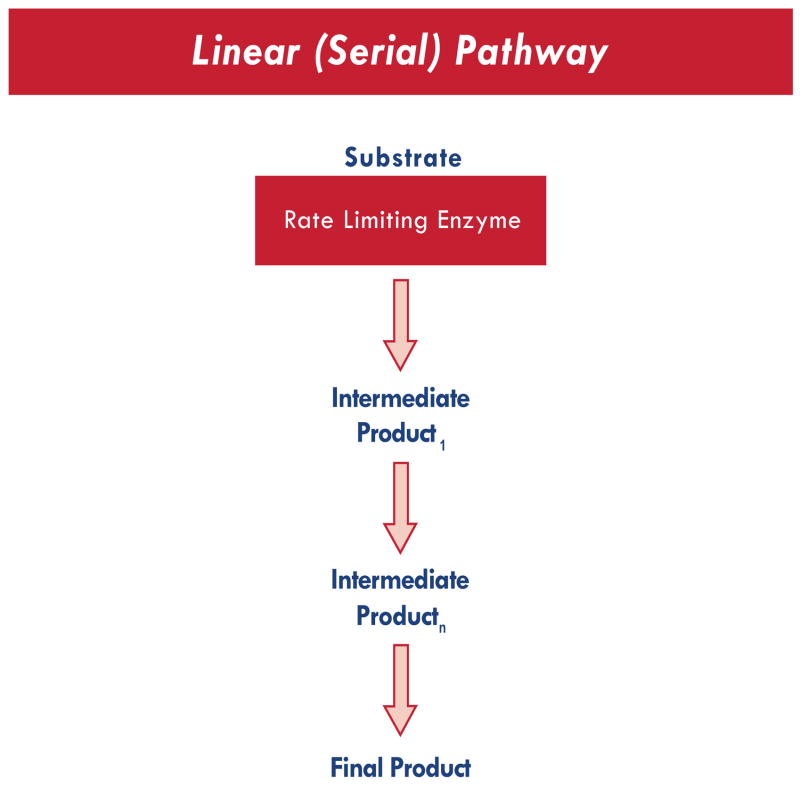
The robust nature of biosynthetic pathways suggests that the development of novel methods for detecting epistasis may be optimized if the analyses can be informed by existing biological knowledge. One way to implement this may be to view individual candidate gene products as contributing to biological pathways that are either linear or spatial. A simple linear biosynthetic pathway is illustrated in Fig. (1).
Fig. (1).
Linear Pathway, functionally analogous to a serial circuit.
In concept, this pathway represents a Serial Circuit. A single rate limiting enzyme (produced by a single gene) leads to the production of a linear array of metabolic intermediates (each with limited biological activity) and a final product with known biological relevance. A number of analytical strategies can be utilized for the characterization of epistasis within the context of such a linear pathway. Examples include combinatorial partitioning methods (CPM) and multifactor dimensionality reduction (MDR). MDR currently represents one of the most widely used computational methods available for characterizing epistasis [Ritchie, M.D. 2001; Moore, J.H. 2004; 2005; 2006; 2007a; 2007b].
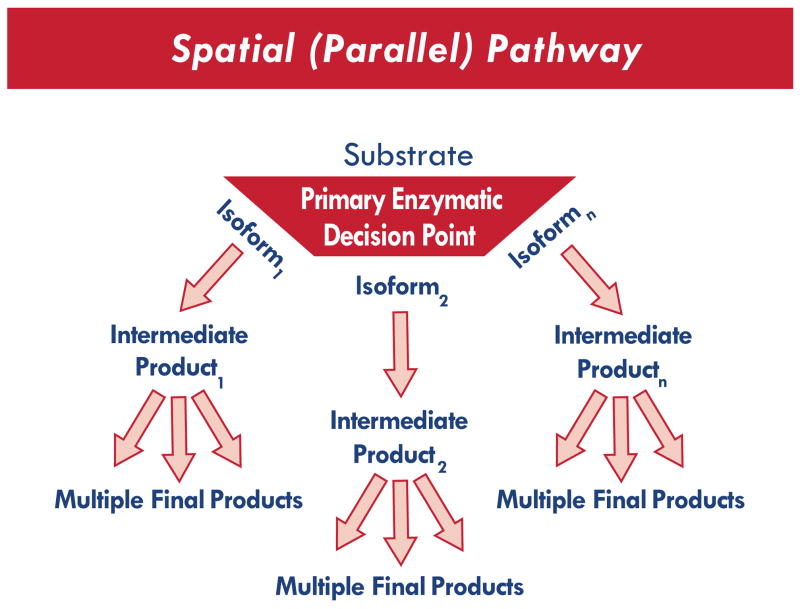
Simple linear biosynthetic pathways such as that shown in Fig. (1) typically lack signal amplification; i.e., only one product molecule will be generated by every molecule of substrate entering the sequence. In such a situation, the candidate gene most likely to determine phenotype would be the gene for the rate limiting enzyme. Nonetheless, this would not preclude polymorphisms in the genes encoding the more distal enzymes from altering phenotype through an interaction with polymorphisms in the proximal enzymes. To capture such an epistatic interaction, analytical methods may need to weight the candidate genes according to the relative proximity of each respective gene product to the rate-limiting enzyme in the pathway. For a simple linear pathway, one way to approach this would be to assign likelihood coefficients for each candidate gene based upon the position of that gene in the series. However, biological pathways are typically far more robust. Oftentimes, a substrate may undergo any of a number of potential enzymatic conversions, determined in part by the relative affinity of that substrate for each respective enzyme. As suggested in Fig. (2), it may be more advantageous to weight each candidate gene based upon previously determined in vitro enzyme affinity. Thus, prior biological knowledge is required regarding the pathway and the gene products.
Fig. (2).
Spatial Pathway, functionally analogous to a parallel circuit.
Fig. (2) represents a complex spatial metabolic network. In concept, this is a Parallel Circuit. Although a single rate limiting enzyme controls the flow of substrate into this model, the substrate can undergo an array of metabolic conversions leading to a complex network of intermediate products (each with variable biological activity). The primary pathway and the final product are determined by enzyme affinity at the decision point. In this example, the decision point represents a “choice” between three isoforms. However, it is important to note that although enzyme affinity is determined genetically, the effects of this genetic variability may not become evident until a group of subjects is challenged with a drug (i.e., gene-environment interaction). This is illustrated in Fig. (3) and Fig. (4).
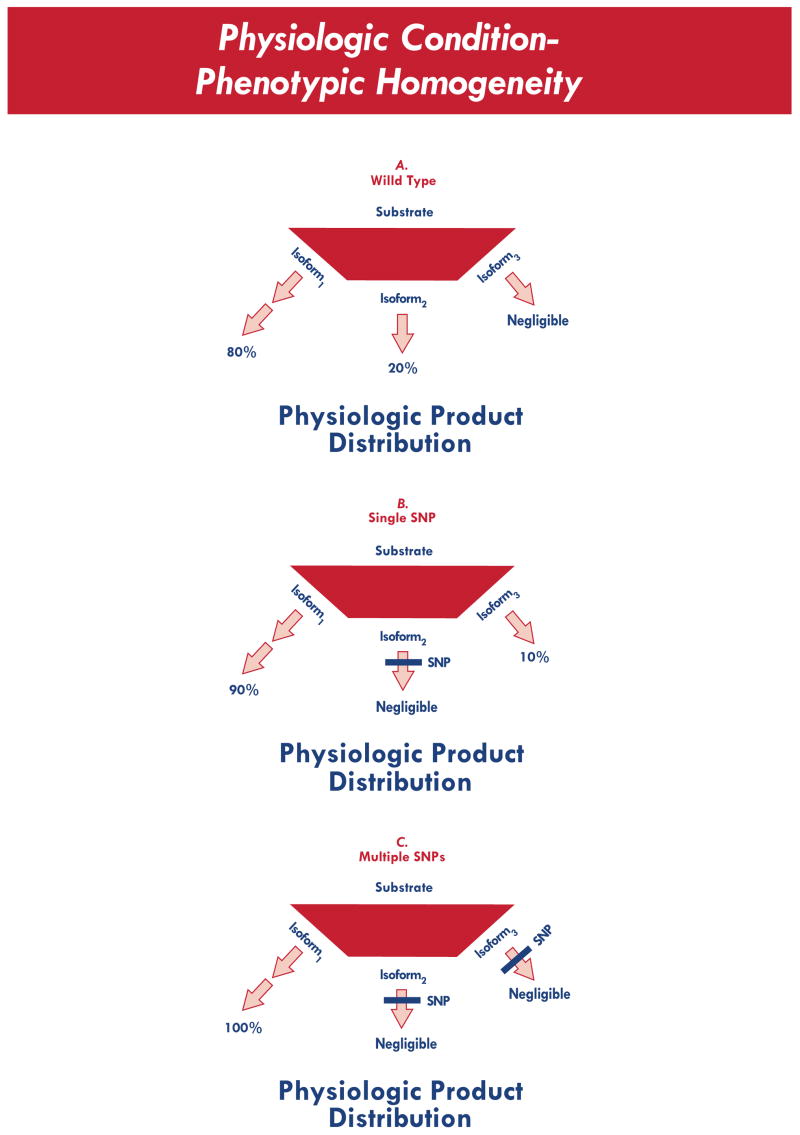
Fig. (3).
Physiologic product distribution, for an enzymatic decision pointwhere substrate affinity for isoform 1 > isoform 2 > isoform 3. Despite the presence of SNPs in key enzymes, the relative product distribution is largely unchanged under physiologic conditions (i.e., relative phenotypic homogeneity).
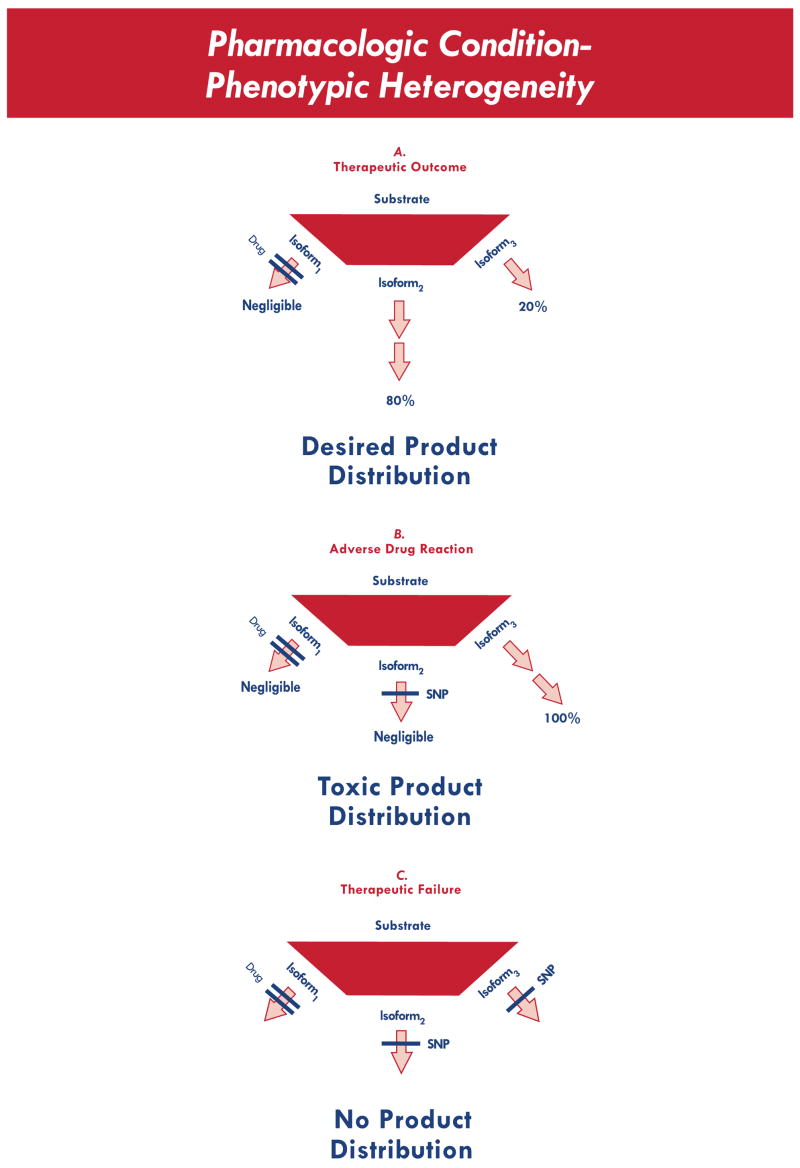
Fig. (4).
Alteration in product distribution, for a drug that selectively inhibits isoform 1. Three possible clinical outcomes are represented, corresponding to the presence of zero SNPs, one SNP, or two SNPs. In the presence of a drug, SNPs that are otherwise clinically silent now become relevant, leading to a drug-dependent change in phenotype (i.e., phenotypic heterogeneity).
The scenario in Fig. (3) treats the three “decision point” enzymes as genetically variant. The first panel (top) represents a physiologic product distribution in the context of no genetic polymorphism(s). The second (middle) and third (bottom) panels represent product distribution for polymorphisms altering the activity of one or more decision point enzymes. Even with relative differences in substrate affinity (isoform 1 > isoform 2 > isoform 3), these polymorphisms would likely have only a limited impact on clinical phenotype. In the context of drug exposure, however, the impact of these polymorphisms becomes much more evident. The net effect is a drug-induced change in phenotype that varies widely according to genotype. The difference is illustrated in Fig. (4).
In this scenario, a drug will lead to either the desired outcome (top) if given to a subject with no decision point polymorphism, or an adverse drug reaction (middle) if given to a subject with one decision point polymorphism, or a therapeutic failure (bottom) if given to a subject with multiple decision point polymorphisms. Hence, in the context of pharmacogenetic association studies, gene-environment interactions (with drug as the environmental perturbation) and gene-gene interactions (epistatic interaction between enzymes) must both be considered. To leverage such a study for success, the analytical strategy must be designed to identify such complex relationships. Within this context, the likelihood of success increases if the strategy is informed by pathway-specific data.
CLINICAL EXAMPLES
To illustrate the clinical utility of these models, we summarize two biosynthetic pathways known to impact the pharmacological management of coronary artery disease (CAD), the leading cause of death in the U.S. [Thom, T. et al. 2006]. The first application explores cholesterol synthesis in the context of an idealized linear pathway. In the clinical arena, this pathway is commonly manipulated pharmacologically by a class of drugs referred to as the statins (HMG CoA reductase inhibitors). The second application reviews a more complex array of lipid signaling molecules derived through the biotransformation of membrane-bound arachidonic acid. In the clinical setting, this pathway is manipulated pharmacologically by a variety of agents, including the non-steroidal anti-inflammatory drugs (NSAIDs).
EXAMPLE 1 - Cholesterol Synthesis as a Linear (Serial) Pathway
Under the conditions pictured in Fig. (1), it may be tempting to speculate that genetic polymorphisms affecting any of the enzymes within this linear sequence would be equally likely to alter phenotype. The available data, however, suggest that this may not be the case. As discussed above, one explanation might be that linear biosynthetic pathways tend to lack signal amplification; i.e., only one product molecule is generated for every single molecule of substrate entering the sequence. In such a situation, the candidate gene most likely to determine phenotype may actually be the gene for the rate limiting enzyme. Consider the situation for lipid-lowering therapy with the statins.
Multiple large clinical trials have demonstrated that statins reduce the incidence of both primary and secondary coronary artery disease in patients at risk. Each of the six currently available statin drugs is highly efficacious (See Sidebar 1) [Downs, J.R. et al. 1998; Shepherd, J. et al. 1995; Herd, J.A. et al. 1997; Pedersen, T.R. et al. 1998; Sever, P.S. et al. 2003; Shepherd, J. et al. 2003]. Recently, a number of retrospective association studies have been conducted with the specific purpose of elucidating genetic factors underlying the differential lipid lowering efficacy of statins at the population level. Cholesterol and Pharmacogenetics (CAP) was designed to address this issue [Simon, J.A. et al. 2006]. American patients of African and European ancestry were treated with simvastatin for 6 weeks. Patients of European ancestry had a larger reduction in LDL-cholesterol (−3 mg/dl) and a higher increase in HDL-cholesterol (+1 mg/dl; p<0.001). This effect was independent of other covariates. Older patients and smokers had larger treatment-induced changes in LDL-cholesterol; women had larger treatment-induced changes in HDL-cholesterol [Simon, J.A. et al. 2006]. This cohort has recently undergone whole genome scanning, and the resulting genotype-phenotype datasets will likely require extensive analyses, based both upon statistics and known biology.
The rate-limiting step in the de novo production of cholesterol is HMG CoA Reductase. This enzyme represents a biochemical portal of entry for simple 2-, 3-, and 4-carbon molecules (e.g., acetate, via acetyl CoA) into an essentially linear string of anabolic enzymes, leading to the production of cholesterol as their final product [Tolbert 2003; Thompson, P.D. et al. 2003]. This biosynthetic sequence may therefore be interpreted in the context of the idealized linear schematic shown in Fig. (1). In the case of cholesterol synthesis, the rate limiting enzyme generates a variety of multi-carbon metabolic intermediates commonly referred to as isoprenoids [Lewis, K.A. et al. 2005]. Under such conditions, it is tempting to hypothesize that genetic polymorphisms affecting any of the enzymes within this linear sequence would be equally likely to alter drug outcomes related to statin therapy. The available data, however, do not support this hypothesis [Mangravite, L.M. et al. 2006].
In a retrospective study of 148 SNPs in 10 candidate genes, Chasman and colleagues identified 2 intronic SNPs in the HMG CoA Reductase gene that were associated with the magnitude of LDL cholesterol lowering by pravastatin (40 mg/day) [Chasman, D.I. et al. 2004]. While their study also included candidate gene SNPs from enzymes occurring more distally within this biosynthetic pathway (e.g., squalene synthase), the impact of these SNPs was marginal. One possible reason for the marginal nature of this effect could be the position of the enzyme within the biosynthetic pathway. This is consistent with our earlier claim that the candidate gene most likely to determine phenotype would be the gene for the rate limiting enzyme. In the case of cholesterol synthesis, this enzyme is also the most proximal enzyme (HMG CoA reductase). In order to determine whether polymorphisms in the genes encoding the more distal enzymes (such as squalene synthase) are capable of altering phenotype, analytical strategies need the capability to interrogate the data for epistasis. Even in the absence of a single main effect, polymorphisms in the distal enzymes could have clinical meaning (i.e., altering lipid lowering efficacy of the statins) through a combinatorial interaction with polymorphisms in the more proximal enzymes. To our knowledge this possibility has not yet been evaluated within the biosynthetic pathway for cholesterol. Work published recently by Murthy suggests that this may be worth testing [Murthy, S. et al. 2005].
EXAMPLE 2 - Eicosanoid Signaling as a Spatial (Parallel) Pathway
The biotransformation of arachidonic acid (AA) represents a well characterized spatial network with profound impact on cardiovascular disease outcomes. AA is liberated from membrane phospholipids by acyl hydrolases. Free AA then serves as a substrate for a variety of tissue oxygenases. As shown in Figs. (2) –(4), this can be viewed as an enzymatic decision point. Cycoloxygenases convert AA to a series of lipids capable of modulating inflammation, vascular tone and platelet function. Conversely, lipoxygenases convert AA to a complex, combinatorial set of immune modulating lipids. Most of these lipid-derived products are referred to as eicosanoids, because they are derived from essential fatty acids containing a 20-carbon backbone. Aspirin (ASA) and other nonsteroidal anti-inflammatory drugs (NSAIDs) are widely used, clinically, to modify the relative production of eicosanoids. Each NSAID alters the production of lipid signaling molecules derived from AA through the inhibition of cyclooxygenases (COXs). Aspirin, in particular, is highly efficacious in the treatment of coronary artery disease (See Sidebar 2). However, recent clinical observations have revealed that COX inhibitors can either decrease or increase subject risk for the development of coronary artery disease (See Sidebar 2) [Spektor, G. and Fuster, V. 2005; McGettigan, P. and Henry, D. 2006; Capone, M.L. et al. 2007]. This observed difference in outcome is likely related to the differential binding affinity of each NSAID for either of two separate, well-characterized COX isoforms (COX1 and COX2).
In general, the clinical impact of genetic variability in AA metabolism can be modeled through the application of spatial metabolic schema (as sown in Figs. (2)–(4). The idea that the respective decision point enzymes vary genetically is not new. A promoter SNP in the COX-2 gene (G-765C) has previously been associated with the frequency of adverse cardiovascular events in a study comparing 864 subjects with first myocardial infarction or ischemic stroke to 864 hospitalized controls [Cipollone, F. et al. 2004]. In this study, heterozygosity (GC genotype) was 2.41 times more frequent among controls than cases (43.3% vs 17.9%; P<.001), and homozygosity for the minor allele (CC genotype) was 5.81 times more frequent in controls than cases (6.4% vs 1.1%; P =.04) [Cipollone, F. et al. 2004].
The mechanism by which this gene variant attenuates development of cardiovascular disease remains unclear. It may involve the COX2-dependent expression of key matrix-digesting proteases by macrophages [Cipollone, F. et al. 2004; Wang, X.M. et al. 2006]. It is conceivable, however, that the clinical importance of this SNP may be amplified in the context of drugs known to modulate “decision point” enzymes involved in eicosanoid synthesis. Fig. (4) shows one model whereby such a SNP could have an amplified (or a dampened) impact on clinical phenotype, during the introduction of different NSAIDs. To effectively capture such relationships in large population-based studies, an analytical strategy must contain an interpretation framework that simultaneously considers multiple layers of complexity. We therefore propose the following pathway-based approach.
STEP-WISE APPROACH
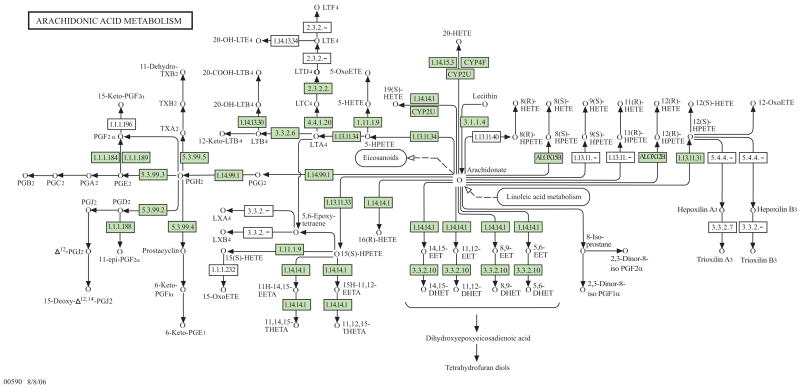
Large-scale pharmacogenetic studies need analytical strategies that embrace, rather than ignore, biological complexity. Computational methods are therefore being extended to address both divergence and redundancy in signaling pathways. For example, Lou et al. [2007] recently developed a parameterization of MDR that allows adjustment for discrete and continuous covariates. Thornton-Wells, T.A. [2006] developed and evaluated a Bayesian clustering algorithm that shows promise for clustering cases based on their genetic background prior to association analysis using methods such as MDR. These and other analytical advances will facilitate the development and application of a computational framework for pharmacogenetic association studies that considers the complexity within known biological pathways. The synthesis of eicosanoids, for example, represents a biosynthetic network which is large, robust, and in some cases redundant. Fig. (5) illustrates the current knowledge regarding this network, based upon data available in one of the larger publicly available pathway databases, the Kyoto Encyclopedia of Genes and Genomes (KEGG) [Kanehisa, M. 1997; Ogata, H. et al. 1999;http://www.genome.jp/keg].
Fig. (5). Potential complexity of a robust signaling network.
Current knowledge is summarized regarding the eicosanoid signaling network, based upon data available in one of the larger publicly available pathway databases (the Kyoto Encyclopedia of Genes and Genomes; KEGG). Several different classes of lipid-derived signaling molecules are represented within this network. They include prostaglandins, prostacyclin, thromboxane, hydroperoxyeicosatetraenoic acids, leukotrienes, hydroxyeicosatetraenoic acids, and epoxyeicosatrienoic acids. Prostaglandins are 20-carbon unsaturated carboxylic acids containing a cyclopentane ring. Prostaglandin (PG) synthesis can be summarized as follows. AA is converted to PGG by a cylcooxygenase, and then to PGH by a peroxidase. Both endoperoxides (PGG and PGH) are chemically unstable. In most tissues, they are subsequently converted to two hydroxyl ketones (PGD and PGE), interrelated by the activity of tissue isomerases. In some tissues, a specific hydroxyl ketone, PGE2, can be further converted to the PGFα prostaglandin series (1,3-diols) by a 9-keto reductase. PGA, PGB, PGC are lab artifacts. Prostacyclin (PGI2) and thromboxane (TXA2) are also synthesized from an endoperoxide, PGH2, via prostacyclin synthase and thromboxane synthase respectively. The relative balance of each is highly tissue specific. While both PGI2 and TXA2 are biologically active, both are rapidly inactivated to stable compounds (6-keto PGF 1α and TXB2). Leukotrienes are synthesized through a different branch within this network. At the “decisions point,” AA is oxidized to a series of hydroperoxyeicosatetraenoic acids or HPETEs (5-HPETE and 12-HPETE). HPETEs are unstable (analogous to PGG and PGH), and further metabolized by a variety of enzymes to form the leukotrienes [Hines and McCarver, 2006]. Through the combined activity of 5-lipoxygenase (ALOX-5) and 5-lipoxygenase activating protein (FLAP), AA is converted to an unstable intermediate called leukotriene A4 (LTA4). LTA4 is subsequently converted to a series of cysteinyl leukotrienes by a glutathione S-transferase (LTC4), a γ-glutamyl transpeptidase (LTD4), and a dipeptidase (LTE4). In general, these cysteinyl leukotrienes induce relaxation of vascular smooth muscle and constriction of bronchiolar smooth muscle. They have also been shown to alter endothelial function, immune function and vascular permeability.
Pharmacogenetic association studies designed to characterize the impact of drugs that interact with such pathways will require analytical strategies that incorporate network models, such as those represented in Figs. (3) –(4) [Thornton-Wells, T.A. et al. 2004], into larger genome-wide data-sets generated from subjects exposed to each respective drug. In this setting, a stepped analytical approach would offer the most informative strategy for utilizing the resulting data. We propose the three-step procedure illustrated in Fig. (6). This approach uses expert biological and statistical knowledge to increase the value of a genome-wide study, by placing analytical emphasis on candidate genes and pathways.
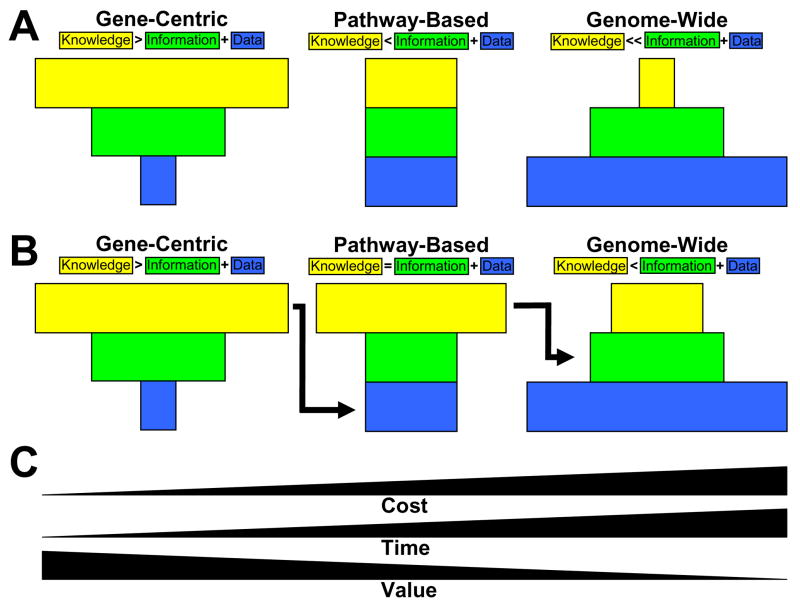
Fig. (6). Step-wise approach to pharmacogenetic association studies.
Illustrated are the relationships between the amount of genotype data collected (blue rectangles), the amount of information generated from statistical and computational analysis (green rectangles) and the amount of knowledge about genetic architecture that is generated from interpreting data analysis results (yellow rectangles) for 1) a gene-centric approach that focuses on one or several candidate genes selected on the basis of their biochemical properties, 2) a pathway-based approach that looks at candidate genes in a particular biochemical pathway and 3) a genome-wide approach that considers a dense map of single-nucleotide polymorphisms (SNPs) that capture most of the variability in the genome. (A) Here, genome-wide association studies carried out independently of gene-centric and pathway-based results are considered agnostic to prior biological and analytical knowledge. In this paradigm, the amount of knowledge gained from a genome-wide association study is very small in proportion to the amount of data and information that are generated. This is due to the high level of noise inherent to data where the number of variables greatly outnumbers the sample size. (B) In this paradigm, knowledge gained from gene-centric studies is used to help pick the pathways and the genes that will be considered in a pathway-based approach. Further, the knowledge gained from pathway-based studies is used to help interpret genome-wide data analysis results. Here, the amount of knowledge gained from the genome-wide association study is improved over that provided by the purely agnostic approach outlined in (A). (C) The genome-wide association study is more expensive and more time consuming than either of the other two approaches. This is especially true with respect to the greatly increased amount of time that it takes to carry out the quality control, data management, data analysis and results interpretation. Candidate gene studies therefore provide greater value, defined as knowledge gained by data generated. We propose that that the analysis and interpretation of a genome-wide association study will be most successful when carried out once the gene-centric and pathway-based approaches have been fully explored. This will ultimately increase the value of the genome-wide association study.
This figure illustrates the relationship between the amount of genetic data collected (blue), the amount of information generated from an analysis of the data (green), and the amount of knowledge such an analysis yields (yellow). These three parameters are shown in relative proportion for large pharmacogenetic association studies, according to each potential type of study design: gene-centric (left panel), pathway-based (middle panel) and genome-wide (right panel). Note that the ratio of knowledge gained to information generated is highest for the gene-centric (leftward) approach, where there is usually good experimental data available, and where the maximum number of analytical methods can be used to ask many different questions about genetic architecture. Note also that the ratio of knowledge gained to information generated is lowest for the genome-wide (rightward) approach, where it is extremely difficult to generate and interpret analytical results. The arrows demonstrate how these strategies could be combined as a step-wise process. In this way, each step (gene, path and genome) could be used to inform the next.
Based upon the relative study strengths illustrated in this schematic, we propose that the first step of any large pharmacogenetic association study should be the selection of one or several candidate genes that can be thoroughly characterized, analyzed and interpreted. We then propose that knowledge gained from this gene-centric approach should be used to select and limit relevant biological pathways. Using prior knowledge to guide pathway selection should yield better results and in turn yield more knowledge about the genetic architecture of the complex trait. The knowledge gained from pathway-based analysis can then be used along with other expert knowledge in public databases to facilitate the interpretation of the genome-wide results. As shown at the bottom of Fig. (6), this will likely improve the value of a time-consuming and costly approach to genetic analysis.
DISCUSSION
Historically, pharmacogenetic association studies have focused on one or more polymorphisms in a single gene selected based upon its biological role in the trait of interest. It has now become routine to measure most, if not all, of the relevant variations in a single candidate gene. This ‘gene-centric’ approach to association studies offers the opportunity to study a single gene in great depth, and it facilitates an understanding of the candidate gene’s role in the genetic architecture of a complex trait such as treatment outcome. Extending this idea to multiple genes within a given pathway yields additional ‘pathway-based’ knowledge, and this knowledge is likely to provide a clearer picture of the role of any one gene, and its interacting partners, in the trait of interest. Both of these approaches (gene-centric and pathway-based approaches) are of general interest because they are hypothesis driven, and because they often reflect the current state of the literature regarding the biochemistry and physiology of the trait being characterized.
The analysis of data from candidate gene studies typically requires the modeling of 10 to 100 polymorphisms, and the analysis of pathway-based data involves several hundred polymorphisms. From a computational point of view these are tractable problems since it is possible to enumerate all combinations of polymorphisms in an analysis of gene-gene interaction. The paradigm changes, however, when the number of polymorphisms begins to exceed several hundred. The largest available supercomputers are not fast enough to enumerate and analyze all combinations of three, four and five polymorphisms from a total list of one million. Further, if one could analyze all possible combinations, the interpretation of the results would be overwhelming. Given these two limitations, how should one confront the complexity we know exists in genome-wide association studies?
We propose that genome-wide association studies using analytical methods that address genetic interactions (gene-gene, gene-environment) and phenotypic heterogeneity are most likely to succeed if expert knowledge about the biology of a given trait is taken into consideration. First, knowledge about gene function can be used to help guide an algorithm as it picks polymorphisms to evaluate, using analytical methods such as MDR. Moore and White [2006 MDR. Moore and White [2007] and Greene et al. [2007] have shown that incorporating expert knowledge into a stochastic search algorithm significantly improves the ability of MDR to identify gene-gene interactions in the context of genome-wide association data. Second, expert knowledge will also be critical for the interpretation of the large volume of statistical results that will be generated from any analysis of gene-gene interaction.
Within such a context, our ability to separate the signal from the noise will depend critically upon our ability to identify and use pre-existing knowledge about gene regulation, biochemical pathways, protein-protein interactions, cellular biology and integrated physiological systems. Beyond pathway-based information, these analytical strategies will also benefit from knowledge of evolutionary selection, conservation across multiple species, and growing data from studies of eQTL (expression quantitative trait loci) [Goring, H.H. et al. 2007]. Fortunately, there are a number of highly useful bioinformatics tools that are now available for genetic and epidemiologic studies of complex clinical endpoints [Moore, J.H. 2007]. Consider for example the Kyoto Encyclopedia of Genes and Genomes (KEGG) database that stores and makes available knowledge on genes and their pathways [Kanehisa, M. 1997; Ogata, H. et al. 1999; http://www.genome.jp/keg]. The Pathway component of KEGG currently stores knowledge on more than 42,937 pathways generated from 307 reference pathways. Another valuable resource is the Gene Ontology (GO) project that has created a controlled vocabulary to describe genes and gene products in any organism in terms of their biological processes, cellular components and molecular functions [Ashburner, M. et al. 2000; Gene Ontology Consortium 2006; http://www.geneontology.org]. GO descriptions and KEGG pathways are both captured and summarized in the U.S. National Center for Biotechnology Information (NCBI) databases [http://www.ncbi.nlm.nih.gov/].
Pathway-based information from KEGG, gene-function information from GO, and other resources such as the Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB) [Hewett, M. et al. 2002; Altman, R.B. 2007; http://www.pharmgkb.org] can be used to inform the analysis of whole genome scans in several different ways. For example, a subset of polymorphisms in a particular pathway may not individually show significance after correction for multiple testing. However, it is possible that the number of genes significant at an uncorrected level is greater than would be expected by chance given the size of pathway. This kind of biologically meaningful pattern could only be revealed using prior knowledge about the pathway. In practice this type of analysis can be carried out using freely available bioinformatics tools such as the Exploratory Visual Analysis (EVA) database and software that organizes statistical results by pathway, GO and chromosomal location, for example, so that more subtle biological patterns can be quickly identified [Reif, D.M. et al. 2005; Reif, D.M. and Moore, J.H. 2006]. One challenge in applying tools such as EVA to genome-wide data is the assignment of intergenic or anonymous SNPs to specific genes so that they can then be tied to a pathway or gene function. Linkage disequilibrium information from the haplotype map and other related sources will be useful for this.
In the future, these tools will position investigators in the field of pharmacogenetics to better characterize the clinical impact of functionally relevant genetic variability within any given biosynthetic pathway. The resulting models should allow statistical geneticists to develop analytical strategies that weight individual components of each pathway appropriately, as the scientific community integrates candidate gene data with pathway-based knowledge to inform the computational approaches to genome-wide datasets.
Sidebar 1. Statins and Cardiovascular Risk
There are six clinically available statins [Tobert, J.A. 2003]. Lovastatin, the first drug approved for clinical use within this class, has been shown to lower the incident rate for initial coronary events (RR = 0.63 versus placebo [95% CI 0.50–0.79], p <0.001) [Downs, J.R. et al. 1998]. All statins inhibit the rate-limiting enzyme in the de novo production of cholesterol (HMG CoA Reductase), and the degree of cardiovascular risk reduction appears to be similar for each drug within the class. Like lovastatin, the relative risk for atherosclerotic coronary artery disease can be reduced to approximately 70% through the clinical administration of pravastatin [Shepherd, J. et al. 1995], fluvastatin [Herd, J.A. et al. 1997], simvastatin [Pedersen, T.R. et al. 1998], atorvastatin [Sever, P.S. et al. 2003], or rosuvastatin [Shepherd, J. et al. 2003].
Currently, the clinical community is moving toward more aggressive lipid lowering with higher doses of statins. In the Treating to New Targets (TNT) trial, more than 10,000 patients with coronary artery disease were randomized to low-dose atorvastatin (10 mg/day) or high-dose atorvastatin (80 mg/day). At follow-up (median of 4.9 years), subjects in the low-dose treatment group had mean LDL-cholesterol levels of 101mg/dl, whereas subjects in the low-dose treatment group had mean LDL-cholesterol levels of 77mg/dl. For the composite end point (death from coronary artery disease, nonfatal nonprocedure-related myocardial infarction, resuscitation after cardiac arrest, and fatal or nonfatal stroke), there was a further reduction in risk (hazard ratio, 0.78 [95% CI 0.69 to 0.89], p<0.001) in the high-dose group when compared to the low-dose group [LaRosa, J.C. et al. 2005]. Since intensive LDL cholesterol lowering may be associated with a reduction in atheroma volume [Nissen, S.E. et al. 2004; 2006], the use of these drugs is likely to increase.
Sidebar 2. NSAIDs and Cardiovascular Risk
Aspirin (ASA) is highly efficacious in the treatment of coronary artery disease. In a meta-analysis of >100 randomized trials, ASA has been shown to reduce the frequency of non-fatal cardiovascular events by approximately 30% in patients at risk [Antithrombotic Trialists Collaboration 2002;Patrono 2004]. The role of ASA in reducing cardiovascular mortality remains less clear [de Gaetano, G. 2001; Berger, J.S. et al. 2006]. The therapeutic benefit of ASA is derived through its irreversible inhibition of cyclooxegenase (COX) enzyme activity in a variety of tissues. While ASA interacts with COX1 and COX2, its ability to inhibit COX1 is 50- to 100-fold more potent than its ability to inhibit COX2 [Patrono 2004]. ASA therefore reduces the COX1-dependent production of TXA2 by platelets [Ouellet, M. et al. 2001]. Although ASA also reduces the COX2-dependent production of PGI2 by the vascular endothelium [Patrono 2004], the net effect is a relative shift toward a less thrombogenic microenvironment. The change is manifest as a reduction in coronary risk.
This may not be the case for other cyclooxegenase inhibitors. Examples include ibuprofen (an older non steroidal anti-inflammatory drug) and rofecoxib (a newer non steroidal anti-inflammatory drug). Both subclasses attenuate the production of TXA2 much less effectively than ASA [Ouellet, M. et al. 2001]. The cardiovascular impact of these non steroidal anti-inflammatory drugs appears to be influenced largely by the relative affinity of each for the COX2 enzyme [Capone, M.L. et al. 2007]. While ibuprofen appears to have similar affinity for COX1 and COX2, other older non steroidal anti-inflammatory drugs (such as meloxicam or diclofenac) are 10 to 30-fold more potent at the COX2 enzyme, and the newer non steroidal anti-inflammatory drugs (such as rofecoxib and valdecoxib) are 100 to 300-fold more potent at the COX2 enzyme [Capone, M.L. et al. 2007]. Correspondingly, these latter compounds have a greater ability to inhibit the COX2-dependent synthesis of PGI2 by the vascular endothelium [Capone, M.L. et al. 2007]. The COX2-specific properties of these drugs may therefore explain recent clinical observations that some newer non steroidal anti-inflammatory drugs appear to increase the frequency of cardiovascular events in subjects at risk (e.g., for rofecoxib, RR= 1.35, 95% CI, 1.15–1.59) [McGettigan, P. and Henry, D. 2006].
Acknowledgments
Grant support: U01HG004608, U01HL069757-06, R01A I59694, R01LM009102, R01HD047447 and P20RR018787.
ABBREVIATIONS
- AA
Arachidonic acid
- ALOX-5
5-lipoxygenase
- ASA
Aspirin
- CAD
Coronary artery disease
- CAP
Cholesterol and pharmacogenetics trial
- CI
Confidence interval
- COX
Cyclooxygenase
- CPM
Combinatorial partitioning methods
- eQTL
Expression quantitative trait loci
- EVA
Exploratory visual analysis
- GO
Gene Ontology
- HDL
High density lipoprotein
- HPETE
Hydroperoxyeicosatetraenoic acid
- INSIG2
Insulin-induced gene 2
- KEGG
Kyoto encyclopedia of genes and genomes
- LDL
Low density lipoprotein
- LTA4
Leukotriene A4
- MDR
Multifactor dimensionality reduction
- NCBI
National center for biotechnology information
- NSAIDs
Non-steroidal anti-inflammatory drugs
- PG
Prostaglandin
- PGI2
Prostacyclin
- PharmGKB
Pharmacogenetics and pharmacogenomics knowledge base
- PPAR-γ
Peroxisome proliferator-activated receptor gamma
- RR
Relative risk
- SNP
Single-nucleotide polymorphism
- TNT
Treating to new targets trial
- TXA2
Thromboxane
Footnotes
DUALITY/CONFLICTS OF INTERESTS
None declared.
References
- Altman RB. PharmGKB: a logical home for knowledge relating genotype to drug response phenotype. Nat Genet. 2007;39:426. doi: 10.1038/ng0407-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antithrombotic Trialists’ Collaboration. Collaborative meta-analysis of randomised trials of antiplatelet therapy for prevention of death, myocardial infarction, and stroke in high risk patients. BMJ. 2002;324(7329):71–86. doi: 10.1136/bmj.324.7329.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JS, Roncaglioni MC, Avanzini F, Pangrazzi I, Tognoni G, Brown DL. Aspirin for the primary prevention of cardiovascular events in women and men: a sex-specific meta-analysis of randomized controlled trials. JAMA. 2006;295(3):306–313. doi: 10.1001/jama.295.3.306. [DOI] [PubMed] [Google Scholar]
- Capone ML, Tacconelli S, Di Francesco L, Sacchetti A, Sciulli MG, Patrignani P. Pharmacodynamics of cyclooxygenase inhibitors in humans. Prostaglandins Other Lipid Mediat. 2007;82(1–4):85–94. doi: 10.1016/j.prostaglandins.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291(23):2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- Cipollone F, Toniato E, Martinotti S, Fazia M, Iezzi A, Cuccurullo C, Pini B, Ursi S, Vitullo G, Averna M, Arca M, Montali A, Campagna F, Ucchino S, Spigonardo F, Taddei S, Virdis A, Ciabattoni G, Notarbartolo A, Cuccurullo F, Mezzetti A. Identification of New Elements of Plaque Stability (INES) Study Group. A polymorphism in the cyclooxygenase 2 gene as an inherited protective factor against myocardial infarction and stroke. JAMA. 2004;291(18):2221–2228. doi: 10.1001/jama.291.18.2221. [DOI] [PubMed] [Google Scholar]
- Couzin J, Kaiser J. Genome-wide association. Closing the net on common disease genes. Science. 2007;316(5826):820–822. doi: 10.1126/science.316.5826.820. [DOI] [PubMed] [Google Scholar]
- de Gaetano G. Low-dose aspirin and vitamin E in people at cardiovascular risk: a randomised trial in general practice. Collaborative Group of the Primary Prevention Project. Lancet. 2001;357:89. doi: 10.1016/s0140-6736(00)03539-x. [DOI] [PubMed] [Google Scholar]
- Downs JR, Clearfield M, Weis S, Whitney E, Shapiro DR, Beere PA, Langendorfer A, Stein EA, Kruyer W, Gotto AM., Jr Primary prevention of acute coronary events with lovastatin in men and women with average cholesterol levels: results of AFCAPS/TexCAPS. Air Force/Texas Coronary Atherosclerosis Prevention Study. JAMA. 1998;279(20):1615–1622. doi: 10.1001/jama.279.20.1615. [DOI] [PubMed] [Google Scholar]
- Dupuis J, O’Donnell CJ. Interpreting results of large-scale genetic association studies. JAMA. 2007;297:529–531. doi: 10.1001/jama.297.5.529. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, White BC, Moore JH. An expert knowledge-guided mutation operator for genome-wide genetic analysis using genetic programming. Lect Notes Bioinform. 2007;4774:30–40. [Google Scholar]
- Göring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, Mahaney MC, Almasy L, MacCluer JW, Kissebah AH, Collier GR, Moses EK, Blangero J. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312(5771):279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- Herd JA, Ballantyne CM, Farmer JA, Ferguson JJ, 3rd, Jones PH, West MS, Gould KL, Gotto AM., Jr Effects of fluvastatin on coronary atherosclerosis in patients with mild to moderate cholesterol elevations (Lipoprotein and Coronary Atherosclerosis Study [LCAS]) Am J Cardiol. 1997;80(3):278–286. doi: 10.1016/s0002-9149(97)00346-9. [DOI] [PubMed] [Google Scholar]
- Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, Klein TE. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163–5. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–6. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- LaRosa JC, Grundy SM, Waters DD, Shear C, Barter P, Fruchart JC, Gotto AM, Greten H, Kastelein JJ, Shepherd J, Wenger NK Treating to New Targets (TNT) Investigators. Intensive lipid lowering with atorvastatin in patients with stable coronary disease. N Engl J Med. 2005;352(14):1425–1435. doi: 10.1056/NEJMoa050461. [DOI] [PubMed] [Google Scholar]
- Lewis KA, Holstein SA, Hohl RJ. Lovastatin alters the isoprenoid biosynthetic pathway in acute myelogenous leukemia cells in vivo. Leuk Res. 2005;29:527–33. doi: 10.1016/j.leukres.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80(6):1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangravite LM, Thorn CF, Krauss RM. Clinical implications of pharmacogenomics of statin treatment. Pharmacogenomics J. 2006;6(6):360–374. doi: 10.1038/sj.tpj.6500384. [DOI] [PubMed] [Google Scholar]
- McGettigan P, Henry D. Cardiovascular risk and inhibition of cyclooxygenase: a systematic review of the observational studies of selective and nonselective inhibitors of cyclooxygenase 2. JAMA. 2006;296(13):1633–1644. doi: 10.1001/jama.296.13.jrv60011. [DOI] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56(1–3):73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Moore JH, Ritchie MD. The challenges of whole-genome approaches to common diseases. JAMA. 2004;291(13):1642–1643. doi: 10.1001/jama.291.13.1642. [DOI] [PubMed] [Google Scholar]
- Moore JH. A global view of epistasis. Nat Genet. 2005;37(1):13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- Moore JH, White BC. Exploiting expert knowledge in genetic programming for genome-wide genetic analysis. In: Runarsson ThP, Beyer H-G, Burke E, Merelo-Guervós JJ, Whitley LD, Yao X., editors. Lect Notes Comput Sci. Vol. 4193. 2006. pp. 969–977. [Google Scholar]
- Moore JH. Genome-wide analysis of epistasis using multifator dimensionality reduction: feature selection and construction in the domain of human genetics. In: Zhu X, Davidson I, editors. Knowledge Discovery and Data Mining: Chanllenges and Realities. IGI Global; 2007b. pp. 17–30. [Google Scholar]
- Moore JH, White BC. Genome-wide genetic analysis using genetic programming: The critical need for expert knowledge. In: Riolo R, Soule T, Worzel B, editors. Genetic Programming Theory and Practice IV. Springer; 2007c. pp. 11–28. [Google Scholar]
- Moore JH. Bioinformatics. J Cell Physiol. 2007a;213(2):365–369. doi: 10.1002/jcp.21218. [DOI] [PubMed] [Google Scholar]
- Murthy S, Tong H, Hohl RJ. Regulation of fatty acid synthesis by farnesyl pyrophosphate. J Biol Chem. 2005;280(51):41793–804. doi: 10.1074/jbc.M504101200. [DOI] [PubMed] [Google Scholar]
- Nissen SE, Tuzcu EM, Schoenhagen P, Brown BG, Ganz P, Vogel RA, Crowe T, Howard G, Cooper CJ, Brodie B, Grines CL, DeMaria AN, REVERSAL Investigators. Effect of intensive compared with moderate lipid-lowering therapy on progression of coronary atherosclerosis: a randomized controlled trial. JAMA. 2004;291(9):1071–1080. doi: 10.1001/jama.291.9.1071. [DOI] [PubMed] [Google Scholar]
- Nissen SE, Nicholls SJ, Sipahi I, Libby P, Raichlen JS, Ballantyne CM, Davignon J, Erbel R, Fruchart JC, Tardif JC, Schoenhagen P, Crowe T, Cain V, Wolski K, Goormastic M, Tuzcu EM, ASTEROID Investigators. Effect of very high-intensity statin therapy on regression of coronary atherosclerosis: the ASTEROID trial. JAMA. 2006;295(13):1556–1565. doi: 10.1001/jama.295.13.jpc60002. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet M, Riendeau D, Percival MD. A high level of cyclooxygenase-2 inhibitor selectivity is associated with a reduced interference of platelet cyclooxygenase-1 inactivation by aspirin. Proc Natl Acad Sci USA. 2001;98(25):14583–14588. doi: 10.1073/pnas.251543298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen TR, Olsson AG, Faergeman O, Kjekshus J, Wedel H, Berg K, Wilhelmsen L, Haghfelt T, Thorgeirsson G, Pyörälä K, Miettinen T, Christophersen B, Tobert JA, Musliner TA, Cook TJ. Lipoprotein changes and reduction in the incidence of major coronary heart disease events in the Scandinavian Simvastatin Survival Study (4S) Circulation. 1998;97(15):1453–1460. doi: 10.1161/01.cir.97.15.1453. [DOI] [PubMed] [Google Scholar]
- Reif DM, Dudek SM, Shaffer CM, Wang J, Moore JH. Exploratory visual analysis of pharmacogenomic results. Pac Symp Biocomput. 2005:296–307. [PubMed] [Google Scholar]
- Reif DM, Moore JH. Visual analysis of statistical results from microarray studies of human breast cancer. Oncol Rep. 2006;15:1043–1047. doi: 10.3892/or.15.4.1043. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Mulfifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever PS, Dahlöf B, Poulter NR, Wedel H, Beevers G, Caulfield M, Collins R, Kjeldsen SE, Kristinsson A, McInnes GT, Mehlsen J, Nieminen M, O’Brien E, Ostergren J ASCOT investigators. Prevention of coronary and stroke events with atorvastatin in hypertensive patients who have average or lower-than average cholesterol concentrations, in the Anglo-Scandinavian Cardiac Outcomes Trial-Lipid Lowering Arm (ASCOT-LLA): a multicentre randomised controlled trial. Lancet. 2003;361:1149–1158. doi: 10.1016/S0140-6736(03)12948-0. [DOI] [PubMed] [Google Scholar]
- Shepherd J, Cobbe SM, Ford I, Isles CG, Lorimer AR, MacFarlane PW, McKillop JH, Packard CJ. Prevention of coronary heart disease with pravastatin in men with hypercholesterolemia. West of Scotland Coronary Prevention Study Group. N Engl J Med. 1995;333(20):1301–1307. doi: 10.1056/NEJM199511163332001. [DOI] [PubMed] [Google Scholar]
- Shepherd J, Hunninghake DB, Barter P, McKenney JM, Hutchinson HG. Guidelines for lowering lipids to reduce coronary artery disease risk: a comparison of rosuvastatin with atorvastatin, pravastatin, and simvastatin for achieving lipid-lowering goals. Am J Cardiol. 2003;91(5A):11C–17C. doi: 10.1016/s0002-9149(03)00004-3. [DOI] [PubMed] [Google Scholar]
- Shriner D, Vaughan LK, Padilla MA, Tiwari HK. Problems with genome-wide association studies. Science. 2007;316(5833):1840–1842. doi: 10.1126/science.316.5833.1840c. [DOI] [PubMed] [Google Scholar]
- Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M, Krauss RM. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97(6):843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- Spektor G, Fuster V. Drug insight: cyclooxygenase 2 inhibitors and cardiovascular risk--where are we now? Nat Clin Pract Cardiovasc Med. 2005;2(6):290–300. doi: 10.1038/ncpcardio0214. [DOI] [PubMed] [Google Scholar]
- Thom T, Haase N, Rosamond W, Howard VJ, Rumsfeld J, Manolio T, Zheng ZJ, Flegal K, O’Donnell C, Kittner S, Lloyd-Jones D, Goff DC, Jr, Hong Y, Adams R, Friday G, Furie K, Gorelick P, Kissela B, Marler J, Meigs J, Roger V, Sidney S, Sorlie P, Steinberger J, Wasserthiel-Smoller S, Wilson M, Wolf P for the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics: 2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2006;113:e85–e151. doi: 10.1161/CIRCULATIONAHA.105.171600. [DOI] [PubMed] [Google Scholar]
- Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20(12):640–7. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Thornton-Wells TA, Moore JH, Haines JL. Dissecting trait heterogeneity: a comparison of three clustering methods applied to genotypic data. BMC Bioinformatics. 2006;7:204. doi: 10.1186/1471-2105-7-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PD, Clarkson P, Karas RH. Statin-induced myopathy. JAMA. 2003;289:1681–1690. doi: 10.1001/jama.289.13.1681. [DOI] [PubMed] [Google Scholar]
- Tobert JA. Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors. Nat Rev Drug Discov. 2003;2(7):517–26. doi: 10.1038/nrd1112. [DOI] [PubMed] [Google Scholar]
- Wang XM, Wu TX, Lee YS, Dionne RA. Rofecoxib regulates the expression of genes related to the matrix metalloproteinase pathway in humans: implication for the adverse effects of cyclooxygenase-2 inhibitors. Clin Pharmacol Ther. 2006;79(4):303–315. doi: 10.1016/j.clpt.2005.12.306. [DOI] [PubMed] [Google Scholar]
- Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15(6):415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- Wilke RA, Reif DM, Moore JH. Combinatorial pharmacogenetics. Nat Rev Drug Discov. 2005;4(11):911–918. doi: 10.1038/nrd1874. [DOI] [PubMed] [Google Scholar]
- Williams SM, Canter JA, Crawford DC, Moore JH, Ritchie MD, Haines JL. Problems with genome-wide association studies. Science. 2007;316(5833):1840–1842. [PubMed] [Google Scholar]