Abstract
Imatinib mesylate is a potent, molecularly targeted therapy against the oncogenic tyrosine kinase BCR-ABL. Although imatinib mesylate has considerable efficacy against chronic myeloid leukemia (CML), advanced-stage CML patients frequently become refractory to this agent. The bone marrow is the predominant microenvironment of CML and is a rich source of both soluble factors and extracellular matrices, which may influence drug response. To address the influence of the bone marrow microenvironment on imatinib mesylate sensitivity, we used an in vitro coculture bone marrow stroma model. Our data show culturing K562 cells, in bone marrow stroma-derived conditioned medium (CM), is sufficient to cause resistance to BCR-ABL inhibitors. Drug resistance correlated with increased pTyrStat3, whereas no increases in pTyrStat5 were noted. Moreover, resistance was associated with increased levels of the Stat3 target genes Bcl-xl, Mcl-1, and survivin. Finally, reducing Stat3 levels with small interfering RNA sensitized K562 cells cultured in CM to imatinib mesylate-induced cell death. Importantly, Stat3 dependency was specific for cells grown in CM, as reducing Stat3 levels in regular growth conditions had no effect on imatinib mesylate sensitivity. Together, these data support a novel mechanism of BCR-ABL-independent imatinib mesylate resistance and provide preclinical rationale for using Stat3 inhibitors to increase the efficacy of imatinib mesylate within the context of the bone marrow microenvironment.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized cytogenetically by the presence of the Philadelphia chromosome, which results from the reciprocal translocation of chromosomes 9 and 22 [t(9:22); refs. 1–3]. The identification of BCR-ABL as the transforming event in CML provided an ideal target for drug discovery. Imatinib mesylate, emerged as a lead candidate from a drug discovery program for inhibiting BCR-ABL tyrosine kinase inhibitors and has proven to be a very effective agent for the treatment of BCR-ABL leukemias (4–6). However, despite the success of imatinib mesylate, overtime some CML patients become refractory to further treatment (particularly those with advanced-stage disease) and almost all patients have detectable levels of BCR-ABL-positive cells, indicating that imatinib mesylate does not eliminate minimal residual disease (5).
Due to the development of drug resistance, an active area of research is focused on the development of second-generation compounds that can circumvent resistant mechanism associated with imatinib mesylate. Specifically, addressing BCR-ABL mutation-mediated imatinib mesylate resistance led to the development and clinical use of more potent second-generation BCR-ABL inhibitors, such as the selective inhibitor nilotinib (AMN107) and the dual BCR-ABL/SRC kinase inhibitor dasatinib (BMS354825; refs. 7, 8). However, recent studies have shown that these second-generation inhibitors also failed to achieve sustained responses in imatinib mesylate-resistant CML blast crisis patients (9–11). These results support the emergence of BCR-ABL-independent resistant mechanisms during the progression of the disease.
The bone marrow microenvironment, which is critical for long-term hematopoiesis and the maintenance and regulation of stem cells and their progeny, is a rich source of paracrine- and autocrine-derived growth factors and cytokines. We reported previously that adhesion to fibronectin was sufficient to protect K562 cells from imatinib mesylate-induced cell death (12, 13). In this report, we sought to address the potential role of bone marrow stroma cells in mediating resistance to BCR-ABL inhibitors. The bone marrow microenvironment is a rich source of extracellular matrices and provides an environment with high local concentrations of cytokines and growth factors. Thus, to further address the contribution of soluble factors derived from the bone marrow microenvironment in mediating resistance to BCR-ABL inhibitors in CML, we used the human stromal cell line, HS-5, to produce conditioned medium (CM). Previous studies showed that HS-5 cells are able to produce cytokines involved in the support of the ex vivo expansion of both immature and mature progenitors cells (14, 15). Additionally, some of those cytokines, including interleukin-6, granulocyte-macrophage colony-stimulating factor, and vascular endothelial growth factor, reported to be expressed in HS-5 cells, are capable of activating Stat3. Stat3 is a member of a family of seven proteins (1–4, 5a, 5b, and 6) that are involved in cell proliferation, angiogenesis, and cell survival. Increased activation of Stat3 has been associated with malignant cell transformation of numerous human cancers and drug-resistant tumors (16–19). Moreover, Stat3 governs signal transduction in growth factor-mediated control of hematopoiesis and myeloid cell differentiation (18).
In this study, we showed that stable soluble factors secreted by HS-5 cells were sufficient to cause resistance to imatinib mesylate, nolotinib, and dasatinib. We also determined that CM increased the clonogenic survival of K562 cells following imatinib mesylate treatment. Moreover, exposure of K562 and KU812 cells to CM caused increased pTyr705 phosphorylation of Stat3. Furthermore, in K562 cells, increased pStat3 levels correlated with increased expression of Stat3-regulated genes Bcl-xl, Mcl-1, and survivin following imatinib mesylate treatment. Finally, reducing Stat3 levels with small interfering RNA (siRNA) resulted in increased imatinib mesylate-induced apoptosis when K562 cells were cultured in CM. Taken together, our data indicate that soluble factor(s), which activates Stat3 within the bone marrow microenvironment, is sufficient to cause resistance to BCR-ABL inhibitors and may contribute to the failure of BCR-ABL inhibitors to eradicate minimal residual disease. These data delineate a novel mechanism of imatinib mesylate resistance that was only operative in the context of the bone marrow model and provide preclinical rationale for using Stat3 inhibitors to increase the efficacy of imatinib mesylate within the context of the bone marrow microenvironment.
Materials and Methods
Cell Cultures
Human CML K562 and KU812 cells (obtained from the American Type Culture Collection) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 1% L-glutamine, and penicillin/streptomycin [regular medium (RM); Life Technologies] at 37°C in 5% CO2 in a humidified incubator. The human stromal cell line HS-5 (also obtained from the American Type Culture Collection) was maintained under the same conditions.
Generation of CM
CM was generated by culturing 5 × 105 HS-5 cells/mL in RM overnight in a humidified atmosphere at 37°C with 5% CO2 to achieve 75% to 80% confluency. Subsequently, the medium was removed and the cells were incubated in fresh medium for 3 h. The supernatant was then collected and cleared of contaminating cells by centrifuging at 2,000 rpm for 5 min. Supernatant was aliquoted and stored at −80°C for future use.
Drugs and Reagents
Imatinib mesylate and nilotinib (AMN107), both supplied by Novartis Pharma, were dissolved in DMSO as a 10 mmol/L stock solution and stored in aliquots at −20°C. Dasatinib, supplied by Bristol-Myers Squibb, was treated similarly.
Preparation of Lysates andWestern Blotting
K562 or KU812 cells were cultured at a density of 4 × 105/mL in either RM or CM for various time points in the presence or absence of either a matched vehicle control (DMSO) or drug (imatinib mesylate or dasatinib). Subsequently, cells were pelleted and lysed in radio-immunoprecipitation assay lysis buffer supplemented with phosphatase and protease inhibitors (1 mmol/L Na3VO4, 1 mmol/L NaF, 1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/mL leupeptin, and 10 μg/mL aprotinin). Insoluble materials were removed by centrifugation at 4°C for 20 min at 15,000 × g. Antibodies used for Western blotting were anti-Stat3, anti-Stat3 (Tyr705), anti-Bcl-xl, anti-Mcl-1, anti-survivin, and pSrc (all from Cell Signaling Technologies).
Apoptosis Assay
Apoptosis was measured using the Annexin V-FITC apoptosis detection kit (Alexis Biochemicals) according to the manufacturer’s recommendations. Data were acquired using CellQuest Pro software version 4.0.2 (BD Biosciences) and analyzed using FlowJo software version 7.2.2 (Tree Star).
siRNATransfection
K562 cells cultured in RM were transiently transfected with either Stat3 siRNA (siGENOME ON-TARGETplus SMARTpool reagent; Dharmacon) or a control siRNA (ON-TARGETplus siCONTROL Nontargeting reagent; Dharmacon) using Amaxa Nucleofector methodology (Amaxa). Briefly, 1 × 106 K562 cells were transfected per manufacturer’s instructions. Thirty-six hours post-transfection, the process was repeated to ensure continued suppression of Stat3 levels. Transfected K562 cells (2.0 × 105/mL) were then cultured in RM or CM for 3 h treated with 250 nmol/L imatinib mesylate or a vehicle control (DMSO). Cell death was measured using Annexin V apoptosis assay as described previously.
Statistical Analysis
A preliminary examination of the data was done using descriptive summary analysis and the Anderson-Darling statistic. Significance testing of the dose-response apoptosis assays was done using analysis of covariance. The Student’s t test was used for the siRNA experiments to compare the log-transformed fold change values, with the null hypothesis being a log (fold change) = 0 (or conversely the fold-change = 1). A significance level of 0.05 was considered statistically significant for all tests. The geometric means of the four ratios for siRNA experiments and the 95% confidence intervals were also calculated (see Supplementary Data).4
Results
CM Protects CML Cells from Cell Death Induced by Multiple BCR-ABL Inhibitors
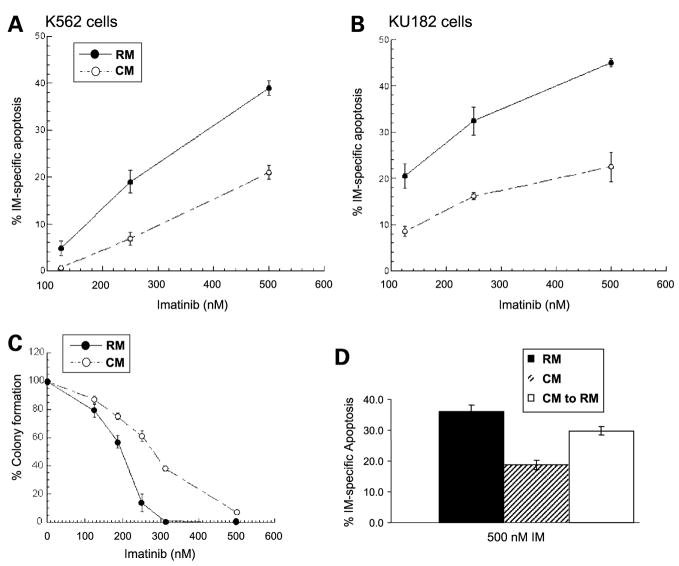
As shown in Fig. 1A and B, exposing either K562 or KU182 cells to CM for 3 h before drug treatment was sufficient to inhibit apoptosis induced by imatinib mesylate. In addition to inhibiting apoptosis, we did a clonogenic assay to determine whether CM increased the ability of K562 cells to divide and form colonies. As shown in Fig. 1C, CM increases the clonogenic survival of K562 cells following imatinib mesylate treatment compared with cells treated in RM. These data indicate that not only does CM protect from apoptosis or cell death but also results in a higher percentage of cells capable of dividing and repopulating. Finally, as shown in Fig. 1D, removal of K562 cells from CM into regular RPMI partially reverses resistance to imatinib mesylate-induced cell death.
Figure 1.
HS-5-derived CM protects K562 and KU182 CML cells from death induced by imatinib mesylate. K562 (A) or KU182 (B) cells were cultured in either RM or CM for 3 h and before the addition of various concentrations of imatinib mesylate or 0.1% DMSO for 36 h. Cell death was measured using Annexin V apoptosis assay followed by fluorescence-activated cell sorting analysis. % Drug-specific apoptosis was calculated by subtracting the background cell death in control DMSO-treated cells from drug treated cell death. A, CM significantly protects K562 cells from imatinib mesylate-mediated cell death (P < 0.05, analysis of covariance). B, CM significantly protects KU182 cells from imatinib mesylate-mediated cell death (P < 0.05, analysis of covariance). C, CM increases the clonogenic survival of K562 CML cells treated with imatinib mesylate. K562 cells were cultured in RM or CM and treated with increasing concentrations of imatinib mesylate as depicted. After 6 h incubation at 37°C, 5% CO2, cells were cultured in 0.3% agar made of RM or CM and containing their respective imatinib mesylate concentrations. Cells were allowed to incubate at 37°C, 5% CO2 for 10 d. Cell colonies (>50 cells) were counted. Representative figure in triplicate (n = 3 independent experiments). D, removal of CM partially reverses resistance to imatinib mesylate. K562 cells were grown in either CM or RM for 3 h. Following 3 h, appropriate samples were removed from CM and placed in RM for an additional 3 h. Cells were treated with 500 nmol/L imatinib mesylate for 36 h and apoptosis was measured by Annexin V positivity. Representative figure in triplicates.
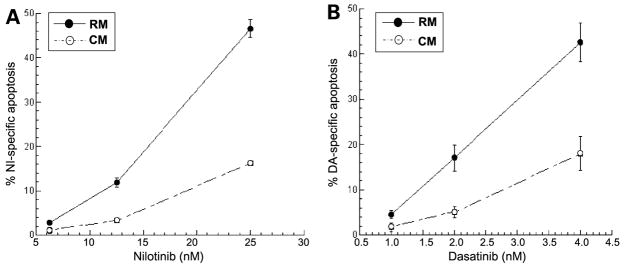
Dasatinib and nilotinitib represent second-generation BCR-ABL inhibitors, both of which are more potent compared with imatinib mesylate and show activity against most of the well-characterized BCR-ABL mutants. Both imatinib mesylate and nilotinib bind the inactive conformation of BCR-ABL, although nilotinib binds with a higher affinity (8). In contrast, dasatinib binds the active conformation of BCR-ABL kinase and is a less selective kinase inhibitor targeting Src kinase family members as well (7). As shown in Fig. 2A and B, CM protects from both nilotinib- and dasatinib-induced cell death. These data indicate that developing more potent BCR-ABL inhibitors will not circumvent resistance associated with exposure of CML to cytokines produced by the bone marrow micro-environment.
Figure 2.
Cells were allowed to incubate for either 48 h (nilotinitib) or 24 h (dasatinib). CM significantly protects K562 cells from (A) nilotinib-mediated cell death (P < 0.05, analysis of covariance) and (B) dasatinib-mediated cell death (P < 0.05, analysis of covariance). Representative graph in triplicates (n = 2 independent experiments).
CM Activates Stat3 and Correlates with Increased Expression of Stat3 Target Genes
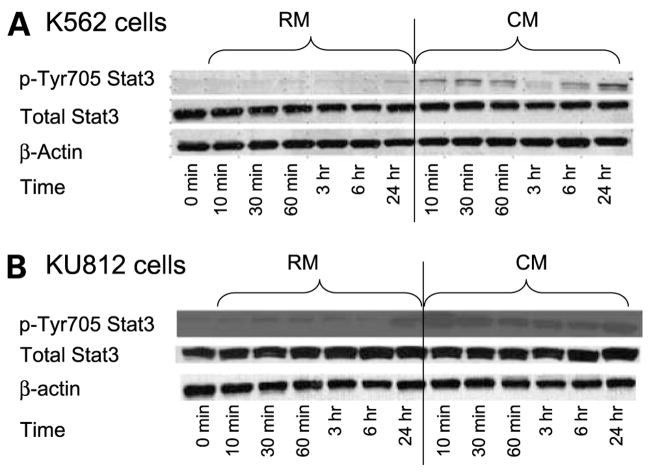
Many of the growth factors and cytokines reported to be expressed in the HS-5 cell line are known activators of Stat3, including granulocyte-macrophage colony-stimulating factor, G-CSF, interleukin-6, and vascular endothelial growth factor (20–23). Thus, we initially asked whether culturing K562 cells in CM caused an increase in phosphorylation of Stat3. As shown in Fig. 3A and B, a rapid and sustained increase in pTyr705Stat3 levels is observed in both K562 and KU182 cells grown in CM.
Figure 3.
Stat3 pTyr705 is increased in CM. A, K652 cells were cultured in RM or CM for the time points indicated. Cells were collected, lysed, and analyzed for either pTyr705 or total Stat3 via Western blotting. β-Actin was used as a loading control. B, KU812 cells were cultured in RM or CM for the time points indicated. Cells were collected, lysed, and analyzed for either pTyr705 or total Stat3 via Western blotting. β-Actin was used as a loading control. Representative figure (n = 3 independent experiments).
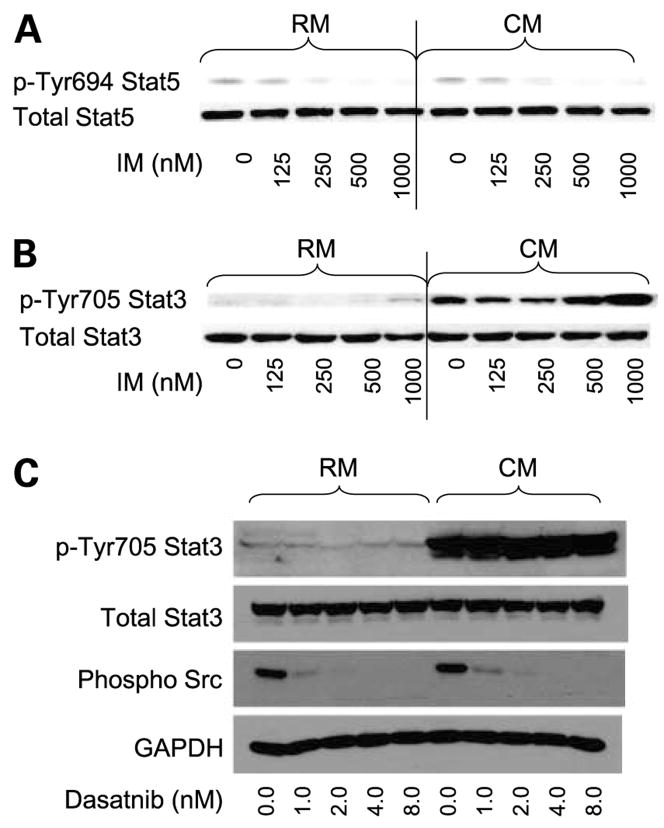
In contrast, culturing K562 cells in CM did not increase the basal levels of pStat5. Furthermore, pStat5 levels were equally inhibited regardless of the culture condition (see Fig. 4). Moreover, treatment with either imatinib mesylate or dasatinib did not inhibit pStat3 levels. Together, these data indicate that Stat3 activation in CM is independent of Src and BCR-ABL activity.
Figure 4.
A, K562 cells grown in CM show equal levels of pTyrStat5 and equal inhibition of pTyrStat5 levels following 48 h of imatinib mesylate treatment compared with cells treated in RM. B, treating K562 cells with imatinib mesylate for 48 h did not reduce the levels of pStat3 when K562 cells were cultured in CM. C, treating K562 cells with dasatinib inhibited levels of pSrc family members but did not attenuate the levels of pStat3 when K562 cells were cultured in CM. Representative figure (n = 3 independent experiments).
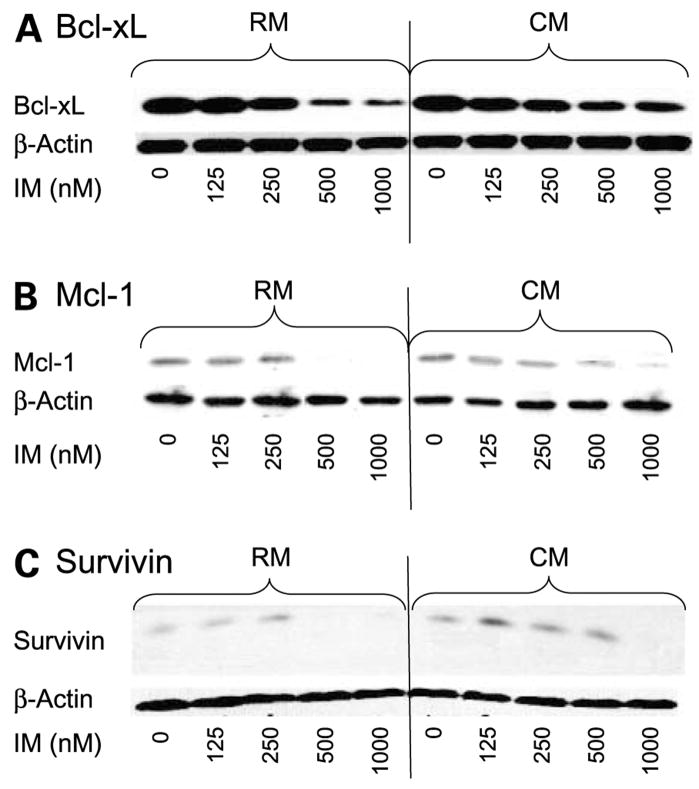
Our next question was to determine whether increased activation of Stat3 corresponded with increased expression of targets that, depending on the cellular context, can be regulated by either Stat5 or Stat3. Previous reports have indicated that inhibition of BCR-ABL results in decreased expression of Stat5-regulated genes including Bcl-xl (24). As shown in Fig. 5, when cells are cultured in CM, Bcl-xl, Mcl-1, and survivin expression are sustained compared with cells treated in RM. Specifically, we observed that, in the presence of 500 nmol/L imatinib mesylate, Bcl-xl, Mcl-1, and survivin were increased by 4.19 ± 1.82, 3.48 ± 1.84, and 7.0 ± 3.41, respectively (n = 3 independent experiments) when cells were cultured in CM compared with RM. These data suggest that activation of Stat3 may be responsible for persistent expression of Stat-regulated genes despite inhibition of Stat5 following imatinib mesylate treatment.
Figure 5.
Effects of CM on Stat3 downstream targets in CM. K562 CML cells were treated with imatinib mesylate in a dose-dependent manner for 48 h. Western blot analysis was done with specific antibodies to Bcl-xl, Mcl-1, and survivin as described in Materials and Methods. A, Bcl-xl; B, Mcl-1; C, survivin. β-Actin was used as a loading control. Representative figure (n = 3 independent experiments).
Reducing Stat3 Levels with siRNA Increases Sensitivity to Imatinib Mesylate in CM
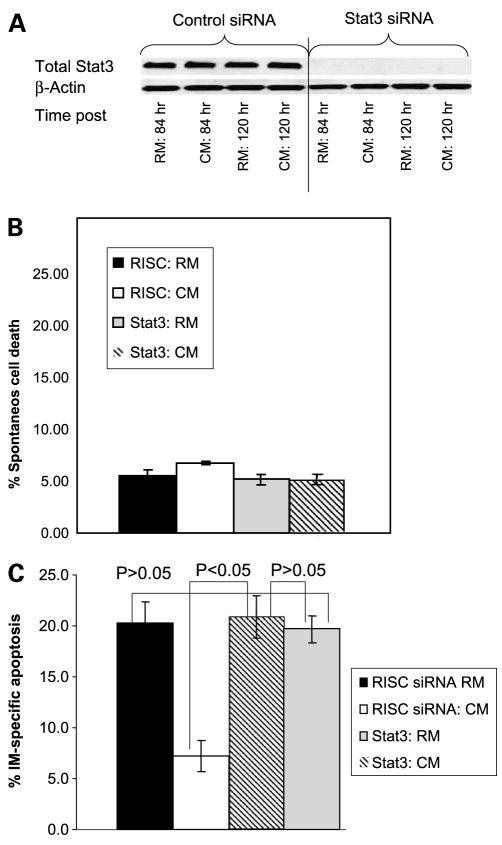
siRNA technology was used to determine the causative role for activation of Stat3 with respect to mediating resistance to imatinib mesylate when K562 cells were cultured in CM. As shown in Fig. 6A, Stat3 was reduced at 84 h and remained reduced for 120 h. Based on these time-course experiments, apoptotic assays were designed as follows: 84 h following imatinib mesylate treatment, control siRNA and Stat3 siRNA-transfected cells were treated with either 250 nmol/L imatinib mesylate or vehicle control for 36 h (120 h after transfection with siRNA) and apoptosis was detected by Annexin V binding. Because some tumor types require Stat3 for survival, we first analyzed whether reducing Stat3 levels with siRNA was sufficient to induce apoptosis when cells were cultured in either RM or CM. As shown in Fig. 6B, reducing Stat3 levels (120 h time point) with siRNA did not cause apoptosis in either growth condition. However, reducing Stat3 levels sensitized K562 to imatinib mesylate-induced cell death when K562 cells were grown in CM but not in RM (see Fig. 6C for a representative figure and Supplementary Fig. S14 for combined representation of 5 independent experiments). Taken together, these data indicate that Stat3 contributes to imatinib mesylate resistance only when cells are cultured within the context of bone marrow stroma-derived CM. Furthermore, these data suggest that Stat3 can compensate for BCR-ABL survival signals and thus represents a potential BCR-ABL-independent mechanism of drug resistance.
Figure 6.
Reducing Stat3 levels with siRNA reverses resistance to imatinib mesylate when K562 cells are grown in CM. K562 cells were cultured in either RM or CM and treated with either siRNA to Stat3 or RISC control siRNA. A, Stat3 knockdown was confirmed using Western blotting and Stat3 was noted to be maximally decreased at 84 h and remained reduced for at least 120 h (n = 5 independent experiments). B, reducing Stat3 levels was not sufficient to cause cell death in cells cultured in RM or CM. Mean ± SD of 5 independent experiments. C, reducing Stat3 levels enhances sensitivity to imatinib mesylate when K562 cells are cultured in CM. Apoptosis of K562 cells cultured in RM or CM, treated with either Stat3 siRNA or RISC control siRNA, in the presence or absence of 250 nmol/L imatinib mesylate was done using the Annexin V detection of apoptotic cells and fluorescence-activated cell sorting analysis. % Specific apoptosis was calculated by subtracting background cell death from imatinib mesylate-mediated cell death. Representative figure in triplicates (n = 5 independent experiments).
Discussion
The emergence of drug resistance continues to limit the success in finding a cure for hematologic malignancies. CML represents a disease that is initially driven by the well-established oncogenic event resulting in the expression of the BCR-ABL fusion oncogene. This uniform transforming event common to CML results in an ideal disease model to delineate the emergence of drug resistance. Clinical data arising from studies targeting BCR-ABL inhibitors indicate that, although BCR-ABL inhibitors are very effective, targeted therapy using a single agent does not eliminate minimal residual disease as detected by quantitative reverse transcription-PCR. These findings are consistent with clinical data indicating that patients that discontinue imatinib mesylate rapidly relapse to a tumor burden, which is as great or greater than before treatment, suggesting that imatinib mesylate does not induce cell death in the stem cell or the population capable of self-renewal (25, 26).
A predominate mechanism of BCR-ABL resistance is the emergence of clones possessing point mutations within the kinase domain of BCR-ABL. However, recent evidence indicates that mutations of BCR-ABL occur in approximately 40% to 60% of imatinib mesylate-resistant patients (27). These data indicate that BCR-ABL-independent mechanisms can contribute to resistance and indicate the need to identify and target BCR-ABL-independent pathways. BCR-ABL activates a multitude of pathways including mitogen-activated protein kinase, AKT, and Stat5 that can contribute to cell survival. However, these pathways can certainly be activated and essentially reconstituted by other external signals including growth factors, cytokines, and extracellular matrices. For example, a recent report indicates that granulocyte-macrophage colony-stimulating factor can cause a BCR-ABL-independent Stat5 signaling in CML cells (28). Additionally, we recently showed that adhesion to fibronectin inhibits cell death induced by BCR-ABL inhibitors, a finding that correlates with reduced Bim levels (12, 13).
In this study, we sought to extend the observation using a single extracellular matrix into a model that consists of exposure to multiple soluble factors produced by bone marrow stroma cells. The HS-5 cell line is described as fibroblastic and has been shown to support the ex vivo expansion of progenitor cells (15). In this report, we chose to further examine the role of stable soluble factors in conferring resistance to BCR-ABL inhibitors. Using medium (RPMI + 10% fetal bovine serum) conditioned by HS-5 cells for 3 h was sufficient to cause resistance to BCR-ABL inhibitors. Furthermore, CM could be stored at −70°C and resistance was not decreased, indicating that the soluble factor(s) conferring resistance was stable (data not shown). HS-5 cells are known to produce several cytokines including interleukin-6, vascular endothelial growth factor, and granulocyte-macrophage colony-stimulating factor, all of which are known activators of Stat3. In this report, we show that exposure of either K562 or KU812 cells to CM derived from HS-5 cells results in a rapid and sustained increase in Tyr705Stat3 levels. The addition of imatinib mesylate does not reduce pStat3 levels, indicating that activation of Stat3 is independent of BCR-ABL activity. Furthermore, phosphorylation of Stat3 occurs independently of Src family kinases, as dasatinib does not attenuate pStat3 levels when K562 cells are cultured in CM. Further studies are warranted to delineate the factor(s) responsible for Stat3 activation when CML cells are cultured in bone marrow stroma-derived CM.
BCR-ABL has been shown previously to activate Stat5 and Stat1 independently of activation of Janus-activated kinases (29). Furthermore, a dominant-negative Stat5 was shown to inhibit colony formation of K562 cells and blocking BCR-ABL was shown to inhibit Stat5-dependent DNA binding and Stat5 target genes Bcl-xl and Mcl-1 (24, 30–34). Bcl-xl, Mcl-1, and cyclin D1 represent genes that can be regulated by either Stat5 or Stat3 activation. Thus, we postulate that, within the bone marrow microenvironment, Stat3 can compensate for BCR-ABL-dependent activation of Stat5-dependent genes, which becomes critical for survival when Stat5 is blocked by BCR-ABL inhibitors. In support of this hypothesis, our data indicated, when cells were treated with imatinib mesylate, the levels of Bcl-xl, survivin, and Mcl-1 were increased when cells were cultured in CM compared with cells treated in RM. Together, these data suggest that, depending on culture conditions, K562 cells have plasticity with respect to oncogene or, more specifically, Stat dependency. More studies are warranted to determine whether other survival pathways known to be regulated by extracellular signals such as AKT and mitogen-activated protein kinase may contribute to imatinib mesylate resistance when CML cells are cultured in a bone marrow stroma model.
Finally, in this report, we were able to show that reducing Stat3 levels with siRNA enhanced the sensitivity of K562 cells to imatinib mesylate-induced cell death. Importantly, the increase in sensitivity was only noted when K562 cells were cultured in CM. We believe these data provide the necessary rationale for further studies in primary patient specimens, and delineation of whether small molecules targeted at Stat3 can enhance the efficacy of BCR-ABL inhibitors using in vivo models. In conclusion, these results show the importance of coculture models that consider the tumor microenvironment for the identification of novel targets to enhance the efficacy of existing targeted drug therapies.
Supplementary Material
Acknowledgments
Grant support: National Cancer Institute grant CA5R01CA122065-02 (L.A. Hazlehurst).
We thank the Flow Cytometry Core at the H. Lee Moffitt Cancer Center and Research Institute.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
References
- 1.Bartram CR, de Klein A, Hagemeijer A, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–80. doi: 10.1038/306277a0. [DOI] [PubMed] [Google Scholar]
- 2.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–9. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 3.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–82. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 4.Buchdunger E, Zimmermann J, Mett H, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–4. [PubMed] [Google Scholar]
- 5.Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 6.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–6. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 7.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 8.Weisberg E, Manley PW, Breitenstein W, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–41. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Druker BJ. Circumventing resistance to kinase-inhibitor therapy. N Engl J Med. 2006;354:2594–6. doi: 10.1056/NEJMe068073. [DOI] [PubMed] [Google Scholar]
- 10.Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–51. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 11.Talpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–41. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 12.Damiano JS, Hazlehurst LA, Dalton WS. Cell adhesion-mediated drug resistance (CAM-DR) protects the K562 chronic myelogenous leukemia cell line from apoptosis induced by BCR/ABL inhibition, cytotoxic drugs, and γ-irradiation. Leukemia. 2001;15:1232–9. doi: 10.1038/sj.leu.2402179. [DOI] [PubMed] [Google Scholar]
- 13.Hazlehurst LA, Argilagos RF, Dalton WS. β1 integrin mediated adhesion increases Bim protein degradation and contributes to drug resistance in leukaemia cells. Br J Haematol. 2007;136:269–75. doi: 10.1111/j.1365-2141.2006.06435.x. [DOI] [PubMed] [Google Scholar]
- 14.Graf L, Iwata M, Torok-Storb B. Gene expression profiling of the functionally distinct human bone marrow stromal cell lines HS-5 and HS-27a. Blood. 2002;100:1509–11. doi: 10.1182/blood-2002-03-0844. [DOI] [PubMed] [Google Scholar]
- 15.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85:997–1005. [PubMed] [Google Scholar]
- 16.Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22:4150–65. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 17.Garcia R, Bowman TL, Niu G, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 18.Coffer PJ, Koenderman L, de Groot RP. The role of STATs in myeloid differentiation and leukemia. Oncogene. 2000;19:2511–22. doi: 10.1038/sj.onc.1203479. [DOI] [PubMed] [Google Scholar]
- 19.Catlett-Falcone R, Landowski TH, Oshiro MM, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–15. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 20.Bartoli M, Gu X, Tsai NT, et al. Vascular endothelial growth factor activates STAT proteins in aortic endothelial cells. J Biol Chem. 2000;275:33189–92. doi: 10.1074/jbc.C000318200. [DOI] [PubMed] [Google Scholar]
- 21.Brizzi MF, Aronica MG, Rosso A, Bagnara GP, Yarden Y, Pegoraro L. Granulocyte-macrophage colony-stimulating factor stimulates JAK2 signaling pathway and rapidly activates p93fes, STAT1 p91, and STAT3 p92 in polymorphonuclear leukocytes. J Biol Chem. 1996;271:3562–7. doi: 10.1074/jbc.271.7.3562. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty A, White SM, Schaefer TS, Ball ED, Dyer KF, Tweardy DJ. Granulocyte colony-stimulating factor activation of Stat3α and Stat3β in immature normal and leukemic human myeloid cells. Blood. 1996;88:2442–9. [PubMed] [Google Scholar]
- 23.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 264:95–8. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 24.Horita M, Andreu EJ, Benito A, et al. Blockade of the Bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of Bcl-xL. J Exp Med. 2000;191:977–84. doi: 10.1084/jem.191.6.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortes J, O’Brien S, Kantarjian H. Discontinuation of imatinib therapy after achieving a molecular response. Blood. 2004;104:2204–5. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- 26.Michor F, Hughes TP, Iwasa Y, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–70. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 27.Guilhot F, Apperley J, Kim DW, et al. Dasatinib induces significant hematologic and cytogenetic responses in patients with imatinib-resistant or -intolerant chronic myeloid leukemia in accelerated phase. Blood. 2007;109:4143–50. doi: 10.1182/blood-2006-09-046839. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Cai D, Brendel C, et al. Adaptive secretion of granulocyte-macrophage colony-stimulating factor (GM-CSF) mediates imatinib and nilotinib resistance in BCR/ABL+ progenitors via JAK-2/STAT-5 pathway activation. Blood. 2007;109:2147–55. doi: 10.1182/blood-2006-08-040022. [DOI] [PubMed] [Google Scholar]
- 29.Carlesso N, Frank DA, Griffin JD. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med. 1996;183:811–20. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Groot RP, Raaijmakers JA, Lammers JW, Jove R, Koenderman L. STAT5 activation by BCR-Abl contributes to transformation of K562 leukemia cells. Blood. 1999;94:1108–12. [PubMed] [Google Scholar]
- 31.Huang M, Dorsey JF, Epling-Burnette PK, et al. Inhibition of Bcr-Abl kinase activity by PD180970 blocks constitutive activation of Stat5 and growth of CML cells. Oncogene. 2002;21:8804–16. doi: 10.1038/sj.onc.1206028. [DOI] [PubMed] [Google Scholar]
- 32.Aichberger KJ, Mayerhofer M, Krauth MT, et al. Identification of mcl-1 as a BCR/ABL-dependent target in chronic myeloid leukemia (CML): evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonucleotides. Blood. 2005;105:3303–11. doi: 10.1182/blood-2004-02-0749. [DOI] [PubMed] [Google Scholar]
- 33.de Groot RP, Raaijmakers JA, Lammers JW, Koenderman L. STAT5-dependent cyclinD1 and Bcl-xL expression in Bcr-Abl-transformed cells. Mol Cell Biol Res Commun. 2000;3:299–305. doi: 10.1006/mcbr.2000.0231. [DOI] [PubMed] [Google Scholar]
- 34.Dumon S, Santos SC, Debierre-Grockiego F, et al. IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene. 1999;18:4191–9. doi: 10.1038/sj.onc.1202796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.