Abstract
Based on comparative genome analyses, the increases in protein-coding gene number could not account for the increases of morphological and behavioral complexity of higher eukaryotes. Transcriptional regulations, alternative splicing and the involvement of non-coding RNA in gene expression regulations have been credited for the drastic increase of transcriptome complexity. However, an emerging theme of another mechanism that contributes to the formation of alternative mRNA 3′-ends is alternative polyadenylation (APA). First, recent studies indicated that APA is a wide spread phenomenon across the transcriptomes of higher eukaryotes and being regulated by developmental and environmental cues. Secondly, our characterization of the Arabidopsis polyadenylation factors suggested that plant polyadenylation has also evolved to regulate the expression of specific genes by means of APA and therefore the specific biological functions. Finally, Phylogenetic analyses of eukaryotic polyadenylation factors from several organisms revealed that the number of polyadenylation factors tends to increase in higher eukaryotes, which provides the potential for their functional differentiation in regulating gene expression through APA. Based on above evidence, we, thus, hypothesize that APA, serving as an additional mechanism, contributes to the complexity of higher eukaryotes.
Key words: alternative polyadenylation (APA), alternative processing, mRNA 3′-end formation, Arabidopsis
Prior to the genome era, prevalent assumption was that the more complicated an organism was, the more protein-coding genes the organism would possess. However, the sequencing of multiple genomes revealed that the volume of protein-coding genes was not correlated with the complexity of the organisms.1 Not only so, the proportion of protein-coding sequences even tends to decrease in the organism's genome when its complexity increases.2,3 How, then, do the higher eukaryotes gain their complexity? Two major explanations have been proposed. One states that although the number of protein-coding genes is not significantly increased, the transcriptions of those protein-coding genes in higher eukaryotes are much more intensively regulated in term of both transcription level and the forms of their transcripts.1,4 The other explanation is that the transcription of protein-coding genes counted only a minor portion of the whole transcriptomes while the most part of the transcriptomes consists of non-coding RNAs. In this case, non-coding RNAs are assumed to play a variety of functional and/or structural roles and therefore contribute to the complexity of the higher eukaryotes.3,5 An array of evidence support both mechanisms,1,3–5 therefore, either might partially hold the truth.
For the first explanation, enriching the transcriptomes by regulating the protein-coding genes, there are two major mechanisms being well appreciated. One is that protein-coding genes are much intensively regulated at transcription level in higher eukaryotes. This hypothesis is rooted in the observation that higher eukaryotes have the increased number of transcription factors.1 Although the increase is not dramatic (e.g., yeast contains one transcription factor per 20 genes while human contains one for every ten genes),1 the potential combinatorial effects of those transcription factors in regulating gene expression could be huge. In consistent with the combinatorial transcription regulation, the genes in higher eukaryotes tend to have much longer promoters,1 which could serve as the action site of multiple transcription factors. Another well appreciated mechanism to regulate gene expressions of higher eukaryotes is the alternative splicing by which multiple transcripts, which may or may not encode different peptide isoforms, could be generated from a single gene.6 Similar to alternative splicing, APA could also produce multiple transcripts from a single gene. Yet, the role of APA in enriching the transcriptomes of higher eukaryotes has not been well appreciated. In this paper, we will discuss and present evidence of that APA, serving as an additional mechanism, does add another layer of complexity to the transcriptome of higher eukaryotes.
The role of APA in regulating gene expression and its physiological relevance has long been recorded. Two classical examples are of the transcription of calcitonin/CGRP and immunoglobulin genes.7 However, it has not been appreciated until recently that APA affects the transcriptome of higher eukaryotes as a general mechanism. A globe survey of human transcriptome suggested that more than 60% of the human genes encode multiple transcripts derived from APA.8,9 APA has been estimated to occur in at least 25% of genes in Arabidopsis10, and about 50% of genes in rice.11 Further, the transcript isoforms derived from APA vary significantly across different human tissues.12 In a recent study, Brain-specific APAs were identified and being regulated by a neuron-specific splicing factor, Nova.13 Those investigations provide evidence pointing to not only on how general the APA is in the transcriptome of higher eukaryotes but that the APA could be physiologically relevant in development.
While the evidence clearly indicates that APA is a general phenomenon, it is not clear how higher eukaryotes evolve to explore the house-keeping process, polyadenylation, to regulate the expression of specific genes by means of APA. To gain insight into the evolution mechanism, we performed phylogenetic analyses of the polyadenylation factors from several eukaryotes including lower eukaryote (yeast and green algae), higher eukaryotes (human, Arabidopsis and rice) and an eukaryote in between (Lycophyte). Since only have the polyadenylation factors of yeast and human been experimentally defined by in vitro biochemical assays,7 we started from those factors to extract the candidate homologues of the eukaryotes. We used two strategies to collect the candidate homologues: (1) directly extract candidates from YOGY database;14 and (2) extract similar peptide sequences by blasting their genomes individually. Candidates gathered from both strategies were combined and redundant entries were manually removed. The non-redundant whole collection of candidate homologs for each polyadenylation factor were subject to phylogenetic analysis using parsimony and bootstrap programs within PAUP 4.0 package. The true orthologs of each yeast and human polyadenylation factor were judged by whether they form a clade with the polyadenylation factor. With these analyses, we found that the number of polyadenylation factors tends to increase in higher eukaryotes (Fig. 1). The number of polyadenylation factors ranges from 24 to 27 in higher eukaryotes (human, 24; rice, 26; Arabidopsis, 27), twice of that of lower eukaryotes (13 for both yeast and algae). The number in Lycophyte (18) falls in between (Fig. 1), consistent with its evolutionary position among these eukaryotes. This increased number of orthologs in the range of eukaryotes provides a potential for their functional differentiation and thus a potential for some of them being specifically adapted to playing a role in the APA mechanism.
Figure 1.
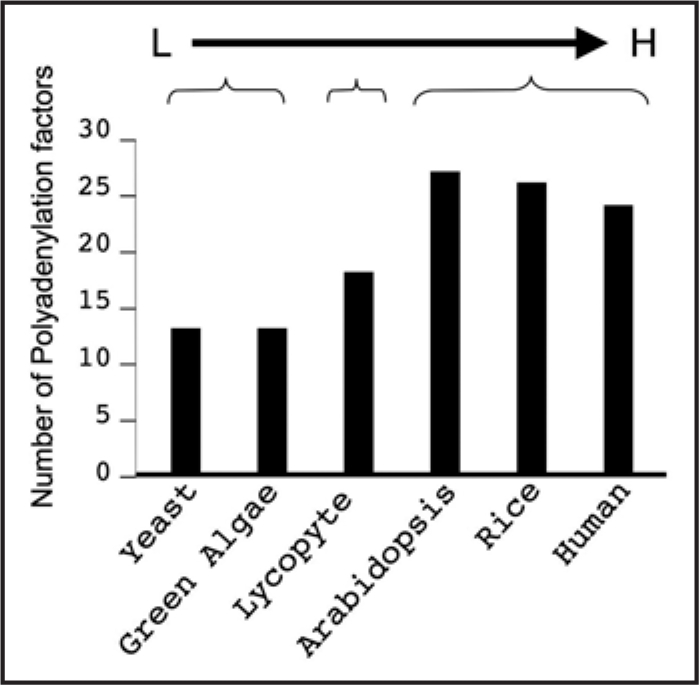
The increased number of core polyadenylation factors in a range of eukaryotes. The polyadenylation factors were identified based on phylogenetic analyses from lower (L) to higher (H) eukaryotes. Yeast (Saccharomyces cerevisiae), Human (Homo sapiens), Arabidopsis (Arabidopsis thaliana), Rice (Oryza sativa Japonica), Lycophyte (Selaginella moellendorffii) and Green algae (Chlamydomonas reinhardtii).
Indeed, characterization of Arabidopsis mutants of these polyadenylation factors supports above hypothesis since most of the paralogs showed functional differentiation.15–19 For example, there are two genes encoding Arabidopsis orthologs of yeast Clp1p or human hClp1, AtCLPS3 and AtCLPS5. Lost-of-function analyses revealed that CLPS3 functions in the female gametophyte transmission and is required for early embryo development.20 In addition, overexpression of CLPS3 specifically cause altered meristem differentiation and early flowering time.20 However, T-DNA knockout mutants of CLPS5 showed no visible phenotype (Xing et al. unpublished observation). In the case of yeast Pcf11p and human hPcf11, there are four Arabidopsis paralogs, PCFS1, PCFS2, PCFS4 and PCFS5.21 PCFS1 was most closely related to PCFS5, yet the T-DNA knockout mutant of PCFS1 showed no visible phenotype while PCFS5 was essential for plant viability (Xing et al. unpublished observation). Different from both PCFS1 and PCFS5, the knockout mutants of PCFS4 showed delayed flowering time and slightly altered leaf shape.21 How do PCFS4 and CLPS3, two polyadenylation factors, gain their specific biological functions? Molecular studies on the role of PCFS4 and CLPS3 in controlling flowering time revealed that PCFS4 and CLPS3, working together with another polyadenylation factor FY, regulated the APA of pre-mRNA of FCA, a promoter of flowering time. Therefore, the function of PCFS4 and CLPS3 in regulating flowering time is mediated by APA. It will be interesting to see whether and how the overall APAs in Arabidopsis transcriptome are affected by PCFS4 and CLPS3. In addition to polyadenylation factors, other components were also found to participate in APA mediated gene regulation such as FCA. It is reasonable to expect that there would be additional factors involved in this process.
Acknowledgements
This research was supported by US NSF Arabidopsis 2010 Program (MCB 0313472 to Q.Q.L.) and, in part, by a NIH grant (1R15GM077192-01A1 to Q.Q.L.), and a grant from Ohio Plant Biotech Consortium (to Q.Q.L. and D.X.).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8345
References
- 1.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 2.Szymanski M, Barciszewski J. Regulatory RNAs in mammals. Handb Exp Pharmacol. 2006:45–72. doi: 10.1007/3-540-27262-3_3. [DOI] [PubMed] [Google Scholar]
- 3.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 4.Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 6.Lareau LF, Green RE, Bhatnagar RS, Brenner SE. The evolving roles of alternative splicing. Curr Opin Struct Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucl Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, et al. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol. 2004;22:1006–1011. doi: 10.1038/nbt992. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, Ji G, Haas BJ, Wu X, Zheng J, Reese GJ, Li QQ. Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucl Acids Res. 2008;36:3150–3161. doi: 10.1093/nar/gkn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penkett CJ, Morris JA, Wood V, Bahler J. YOGY: a web-based, integrated database to retrieve protein orthologs and associated Gene Ontology terms. Nucl Acids Res. 2006;34:330–334. doi: 10.1093/nar/gkl311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing D, Zhao H, Li QQ. Arabidopsis CLP1-SIMILAR PROTEIN3, an ortholog of human polyadenylation factor CLP1, functions in gametophyte, embryo and postembryonic development. Plant Physiol. 2008;148:2059–2069. doi: 10.1104/pp.108.129817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing D, Zhao H, Xu R, Li QQ. Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03455.x. In press. [DOI] [PubMed] [Google Scholar]
- 17.Xu R, Ye X, Quinn Li Q. AtCPSF73-II gene encoding an Arabidopsis homolog of CPSF 73 kDa subunit is critical for early embryo development. Gene. 2004;324:35–45. doi: 10.1016/j.gene.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Xu R, Zhao H, Dinkins RD, Cheng X, Carberry G, Li QQ. The 73 kD subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol Biol. 2006;61:799–815. doi: 10.1007/s11103-006-0051-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, et al. A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE. 2008;3:2410. doi: 10.1371/journal.pone.0002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing D, Zhao H, Li QQ. Arabidopsis CLPS3, an ortholog of human polyadenylation factor CLP1, functions in gametophyte, embryo and post-embryonic development. Plant Physiol. 2008;29:29. doi: 10.1104/pp.108.129817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing D, Zhao H, Xu R, Li QQ. Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. 2008;54:899–910. doi: 10.1111/j.1365-313X.2008.03455.x. [DOI] [PubMed] [Google Scholar]


