Abstract
Serotonin (5-HT) decreases NHE2 and NHE3 activities under acute conditions in human intestinal epithelial cells. Here, we have investigated the effects of 5-HT on expression of the human NHE3 gene and the mechanisms underlying its transcriptional regulation in differentiated C2BBe1 cells. Treatment of the human intestinal epithelial cell line, C2BBe1, with 5-HT (20 μM) resulted in a significant decrease in NHE3 mRNA and protein expression. In transient transfection studies, 5-HT repressed the NHE3 promoter activity by ~55%. The repression of the NHE3 promoter activity in response to 5-HT was accompanied by reduced DNA-binding activity of transcription factors Sp1 and Sp3 to the NHE3 promoter without alteration in their nuclear levels. Pharmacological inhibitors of protein kinase C reversed the inhibitory effect of 5-HT on the promoter activity. Our data indicate that 5-HT suppresses the transcriptional activity of the NHE3 promoter and this effect may be mediated by PKCα and modulation of DNA binding affinities of Sp1 and Sp3.
Keywords: Na+/H+ exchanger NHE3, serotonin, Sp1, Sp3, PKC, transcriptional regulation
Introduction
Serotonin (5-hydroxytryptamine, or 5-HT) is an important neurotransmitter predominantly present in the gut and brain. It is synthesized and secreted into the blood stream and intestinal lumen by enterochromaffiin cells in the gastrointestinal tract representing the largest store (~95%) of 5-HT in the body [1, 2]. Serotonin plays a critical role in regulating the gastrointestinal motility, secretion and absorption. Increased serotonin levels have been implicated in the pathophysiology of diarrhea associated with carcinoid syndrome [3], ulcerative colitis and irritable bowel syndrome [4]. We have recently shown that serotonin decreases NHE2 and NHE3 activities via 5-HT4 receptors in human intestinal epithelial cells under acute conditions [5]. However, the effects of serotonin on NHE3 gene expression are not known.
To date nine members of the NHE gene family have been identified and characterized in eukaryotic cells. The NHE gene family is involved in the regulation of a diverse range of biological functions including intracellular pH and cell volume regulation, and sodium absorption in the human intestine. NHE1, NHE2, NHE3 and NHE8 are expressed in the human gastrointestinal tract. In polarized cells, NHE1 is localized to the basolateral membrane while NHE2, NHE3 and NHE8 are localized to the apical membrane [6, 7]. The importance of the NHE3 isoform in electrolyte and fluid homeostasis has been demonstrated by reduction of Na+ absorption in NHE3 knockout studies [8] and by inhibition of its transport activity in diarrheal diseases [7].
We have previously shown that the NHE3 core promoter region, bp −95 to +5, which harbors a number of cis-elements including Sp1 binding sites is sufficient for the basal NHE3 promoter activity in C2BBe1 cells [9]. Our subsequent studies revealed that this promoter region plays a critical role for the NHE3 promoter activity in response to various stimuli. For example, the signaling pathways responsible for the effects of phorbol ester PMA, butyrate or IFN-γ/TNF-α, converge at an overlapping Sp1/Egr-1 motif in the core promoter region and each pathway triggers a distinct response [10–12]. Our current data show that serotonin represses the NHE3 transcription and the Sp1/Egr-1 motif is also responsible for the repression elicited by serotonin. Furthermore, we show that serotonin-response may be mediated by a PKCα signaling pathway and modulation of the DNA-binding activity of Sp1 and Sp3 to the Sp1/Egr-1 motif.
Materials and methods
Reagents
Most of the chemical reagents including serotonin hydrochloride were purchased from Sigma-Aldrich Inc., (St. Louis, MO), unless otherwise noted. Briefly, Dulbecco’s modified Eagle’s Medium, fetal bovine serum, and LipoFectamine™2000 were obtained from Invitrogen life Technologies Inc., (Carlsbad, CA). Protein kinase inhibitors obtained from Calbiochem (La Jolla, CA). Gel shift assay core system was from Promega (Madison, WI). The NHE3, Sp1, Sp3 and mouse monoclonal actin and tubulin antibodies, were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Promoter-Reporter Plasmids
Plasmids used for functional analysis of the NHE3 promoter activity were generated using pGL2-Basic (Promega) and have been previously described [9].
Cell Culture, Transfections and Reporter Assays
C2BBe1 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained as previously described [13]. For transient transfection studies, cells (1.5×105 cells/well) were seeded onto collagen coated 12-well plates and grown for 12 days. Differentiated cells were co-transfected with one of the NHE3-reporter constructs along with pSV-βgal as an internal control using LipoFectamine™2000 (Invitrogen). A total of 4.0 μg DNA at a ratio of 4:1 for experimental to pSV-βgal was used for each transfection. Cells were incubated with the DNA/transfection mixture for 6 h. Subsequently media was replaced with serum-reduced media (0.5% FBS) for 24 h and incubation continued in the presence or absence of serotonin. All serotonin treatments were for 16 h with, unless otherwise noted. Serotonin containing media were replenished after 8 hours for prolonged incubation periods. For signal transduction experiments, transfected cells were pre-incubated with the pathway specific inhibitors for 1 h prior to the addition of serotonin (20 μM) and incubation continued for additional 16 h. Luciferase activity was determined as described [12].
RNA Extraction and RT-PCR Analysis
Total RNA extraction, reverse transcription and polymerase chain reactions (PCR) were performed as described [12].
Western Blot Analysis
Total cell extracts or nuclear proteins were prepared from differentiated C2BBe1 cells as described [12]. For NHE3, 100 μg total cell lysate and for Sp1 and Sp3, 20 μg nuclear proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto PVDF membrane (Immobilon-P Millipore). The NHE3, Sp1 and Sp3 proteins were detected using NHE3, Sp1 and Sp3 antibodies, respectively. The blots were re-probed with either actin or tubulin mouse monoclonal antibodies as loading control. For immunoprecipitation studies, 200 μg total cell proteins from C2BBe1 cells were immunoprecipitated using the NHE3 (3H3) monoclonal antibody (kindly provided by Dr. O.W. Moe) and subjected to Western blot analysis using the NHE3 antibody from Santa Cruz Biotechnology as described above.
Gel Mobility Shift Assay (GMSA)
GMSAs were performed as described previously [10].
Statistical Analyses
Differences between the two groups were evaluated using Student’s t-test. P < 0.05 was used to indicate statistical significance.
Results
The dose- and time-dependent effect of serotonin on the expression of NHE3 mRNA and protein in C2BBe1 cells
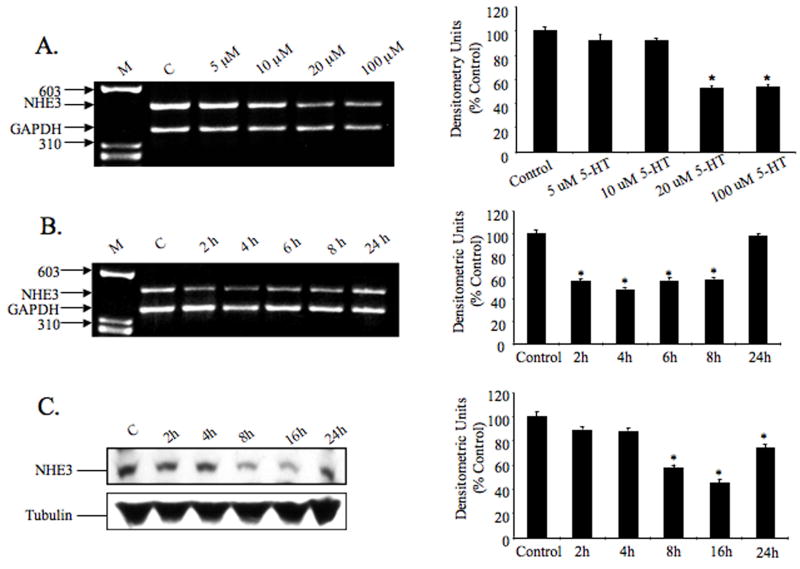
RT-PCR experiments were performed to investigate the effect of serotonin on NHE3 mRNA expression. Total RNA was obtained from differentiated C2BBe1 cells treated with or without serotonin and subjected to reverse transcription and subsequent PCR amplification using NHE3 and GAPDH gene-specific primers. As shown in Figure 1A, the NHE3 mRNA expression level decreased significantly in the presence of 20 and 100 μM Serotonin. By densitometric quantifications, the reduction in mRNA expression was estimated to be approximately 50% at 20 and 100 μM Serotonin (Fig. 1A, right panel).
Fig. 1.
Dose- and time-dependent effects of serotonin on the expression of the NHE3 mRNA in C2BBe1 cells. Differentiated serum-starved C2BBe1 cells treated with different doses of 5-HT for 4 h (A), or with 20 μM concentration for 0, 2, 4, 6, 8 and 24 h; total RNA was extracted and subjected to RT-PCR (B). GAPDH was used as an internal control. Effect of serotonin on the NHE3 protein levels examined by Western blot analysis (C), differentiated serum-starved cells were treated with serotonin for the indicated time period and total cell proteins were prepared and separated (100 μg/lane) by 10% SDS-polyacrylamide gel electrophoresis. NHE3 protein was detected using anti-NHE3 goat polyclonal antibody. The blot was re-probed with tubulin antibody. Densitometric analysis of each experiment is shown on the right panels. Data are presented as intensity of the experimental sample relative to the GAPDH intensity in the corresponding sample. The activity in the control was set at 100. Results were obtained from three separate experiments (mean ± S.D.). * P<0.05 compared to control.
To establish a time-course for the NHE3 mRNA reduction in response to serotonin, cells were exposed to 5-HT (20 μM) for various time intervals and NHE3 mRNA expression assessed by RT-PCR. Serotonin exposure led to decreased NHE3 mRNA expression in a time-dependent and transient manner with the maximum repression after 4 h and subsequent recovery by 24 h (Fig. 1B). Replenishing 5-HT during longer incubation periods showed no effect on the recovery of NHE3 mRNA at 24 h. Intestinal serotonin is inactivated by metabolic degradation after reuptake mediated by the serotonin transporter SERT. Inactivation of serotonin is important to limit its action both temporally and spatially. Moreover, enteric 5-HT receptors are subject to desensitization [14]. Therefore, it is possible that with continuous availability and prolonged exposure to 5-HT desensitization of 5-HT receptors may be responsible for blunting the signaling pathways mediating the effects of 5-HT on the NHE3 transcription in these cells.
To demonstrate a relationship between NHE3 mRNA and protein levels, we analyzed the total cell extracts from 5-HT treated cells by immunoblotings. As shown in Figure 1C, 5-HT treatment was associated with a time-dependent reduction in the level of NHE3 protein in the treated cells and correlated with the NHE3 mRNA expression in response to 5-HT. The authenticity of the signals detected in the immunoblot (Fig. 1C) was confirmed by using NHE3 (3H3) monoclonal antibody. In these studies, NHE3 antibody (Santa Cruz Bioteh) was used to detect NHE3 protein in Western blots of the immunoprecipitated proteins using the NHE3 (3H3) monoclonal antibody. Both antibodies detected a signal at 85 kDa (data not shown).
The effects of serotonin on the NHE3 promoter and identification of the serotonin-responsive region
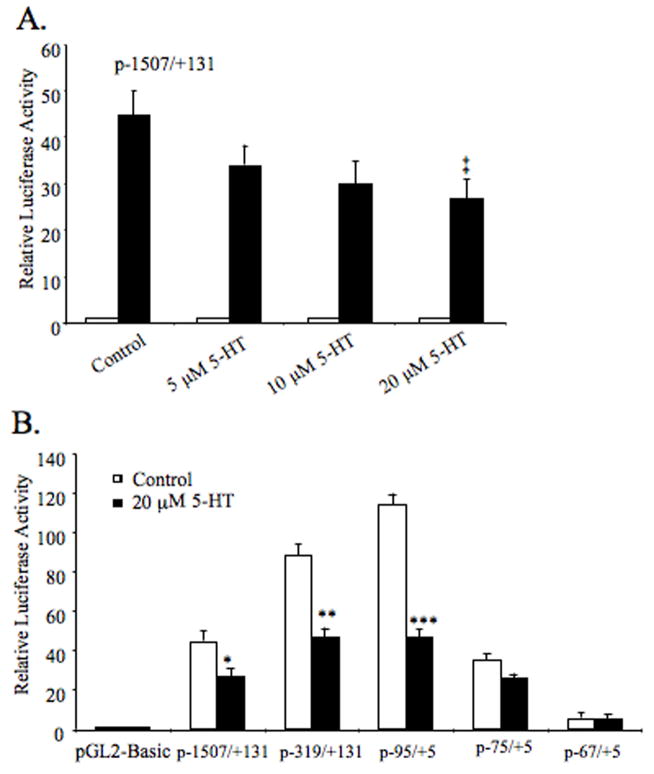
We next investigated whether the serotonin-induced repression of the NHE3 mRNA is impacted through its effects on the NHE3 promoter. The NHE3 promoter construct, p-1507/+131, was transiently transfected into C2BBe1 cells and reporter gene activity was analyzed in the presence of increasing concentrations of serotonin for 16 h. A gradual decrease in NHE3 promoter activity was observed with increasing 5-HT levels (Fig. 2A). The highest concentration (20 μM) led to a significant reduction in the reporter gene activity compared to the untreated control suggesting that the repressive effect of serotonin on the NHE3 mRNA expression is mediated by transcriptional regulation.
Fig. 2.
Functional analysis of the NHE3 promoter by luciferase assays and identification of serotonin-responsive region. Promoter construct p-1507/+131 was transiently transfected into differentiated C2BBe1 cells (A). The effect of indicated doses of serotonin on NHE3 promoter activity was determined by luciferase assays. The luciferase activities are presented relative to the normalized activity of the promoter-less pGL2-Basic. 5′deletion NHE3 promoter constructs were transiently transfected into C2BBe1 cells and treated with or without serotonin (20 μM) for 16 h (B). Values shown are mean ± S.E. obtained from three different experiments performed in triplicate assays on different days. Statistical differences from the control values were determined by Student’s t test (‡ P = 0.007; * P = 0.006; ** P = 0.0002; *** P = 0.0001).
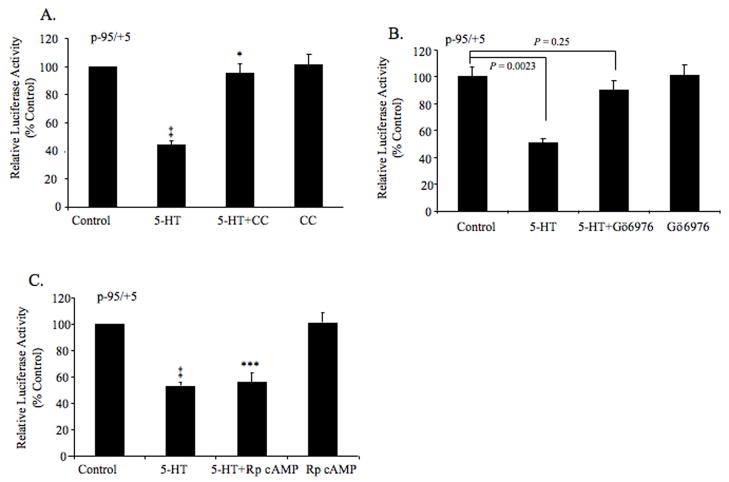
Next, we identified the serotonin-responsive region by functional analysis of various 5′-deletion constructs of p-1507/+131. The construct carrying bp −95/+5 was capable of conferring the repressive effects of 5-HT on the NHE3 promoter activity (~55% reduction) whereas, deletion of an additional 20-nucleotide led to a diminished basal promoter activity and disappearance of 5-HT response (Fig. 4B). Thus, these data suggest that the cis-element(s) required for 5-HT responsiveness must be localized in the region between bp −95 to −76.
Fig. 4.
Effects of protein kinase inhibitors on the serotonin-mediated repression of the NHE3 core promoter activity. Differentiated C2BBe1 cells were transiently transfected with the p-95/+5. After serum-starvation, cell were treated without (control) or with the chelerythrine chloride (CC) (A), GÖ6976 (B), or Rp-8-Br-cAMP (C), for 1 h prior to further incubation in the presence or absence of 5-HT for 16 h. The luciferase activities are presented as percentage of the control (n=3, mean ± S.E.). ‡, *** P < 0.0005; * P = 0.49.
Identification of proteins interacting with the serotonin-responsive region
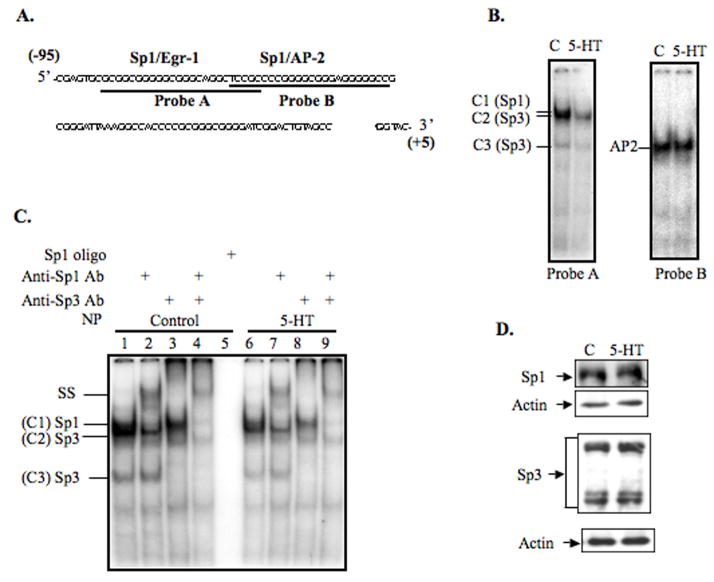
Previously, we have shown that sequence motifs composed of overlapping sites for Sp1/Egr-1 (bp −89 to −68), and Sp1/AP-2 (bp −73 to −49) could interact with transcription factors Sp1/Sp3 and AP-2, respectively, and control the constitutive NHE3 promoter activity [9, 10]. Our results (Fig. 2), suggested that serotonin might regulate the NHE3 promoter activity through Sp1/Egr-1 motif. To address this possibility, we performed GMSAs with probes A and B using nuclear proteins from the control and serotonin treated cells. As expected, in the control reactions nucleoprotein complexes were formed between probe A and transcription factors Sp1 and Sp3; and AP-2 and probe B (Fig. 3B). Interestingly, DNA-binding affinities of both the Sp1 and Sp3 to probe A were diminished by serotonin treatment, while no obvious alteration in AP-2 interaction with probe B was detected. Figure 3C shows a representative GMSA using probe A and nuclear proteins from untreated (lanes 1–5) and serotonin-treated (lanes 6–9) cells. Consistent with the presence of Sp1 and Sp3 proteins in these complexes, an unlabeled Sp1 specific oligonucleotide competed out all nucleoproteins (Fig. 3C, lane 5). In the presence of Sp1 antibody, a slow migrating band was formed (SS) and complex 1 (C1) was eliminated (Fig. 3C, lanes 2 and 7); and Sp3 antibody completely removed C2 and C3 complexes (Fig. 3C, lanes 3 and 8). A combination of Sp1 and Sp3 antibodies eliminated all these bands indicating that protein component of these complexes were Sp1 and Sp3 (Fig. 3C lane 4 and 9).
Fig. 3.
Effect of 5-HT on the DNA-binding activity of nuclear proteins to the NHE3 core promoter region. Nucleotide sequence of the NHE3 core promoter region (bp −95 to −5) is shown and probes used for GMSAs are indicated (A). DNA-binding of nuclear proteins from untreated and serotonin treated cells to the probes A and B (B). Detailed analysis of DNA-binding characteristics of probe A and nuclear proteins from untreated and 5-HT treated cells (C). Lanes 1–5 and 6–9 show complex formation between probe A and nuclear proteins from untreated or serotonin-treated cells, respectively. The binding specificity of these complexes was examined by competition assays where excess unlabeled Sp1-specific probe was used (lane 5). The identities of the proteins present in these complexes were established by supershift assays and are indicated by slow migrating supershift bands (SS). Effects of serotonin on the nuclear expression of Sp1 and Sp3 in C2BBe1 cells (D). Nuclear proteins (20 μg/lane) were separated by 10% SDS-polyacrylamide gel electrophoresis and Western blots were performed. Sp1 and Sp3 were detected using Sp1 and Sp3 antibodies, respectively. The blots were re-probed with actin antibody. C, control; Ab, antibody; NP, nuclear proteins.
To investigate whether reduced DNA-protein interactions observed between Sp1/Sp3 and NHE3 promoter were the result of decreased protein levels in the serotonin-treated nuclear extracts or their reduced DNA-binding affinities, Western blot analyses were performed. As demonstrated (Fig. 4D), Sp1 and Sp3 levels were not altered by 5-HT treatment. Thus reduced DNA-binding affinity of Sp1 and Sp3 to the NHE3 core promoter may be mediated via mechanisms other than the absolute quantity of the proteins.
Serotonin-induced repression of NHE3 promoter activity is mediated via activation of PKC pathway
Recently we reported that the acute treatment of intestinal epithelial cells with serotonin decreases the transport activity of both NHE2 and NHE3 via a process that involves the activation of PKCα [5]. To examine the possible regulatory effect of PKC on the NHE3 promoter activity in response to serotonin, C2BBe1 cells were transfected with the p−95/+5 and pretreated with PKC inhibitors, chelerythrine chloride (2 μM), or GÖ6976 (5 nM), followed by serotonin treatment. Inhibition of PKC pathway by chelerythrine chloride led to complete reversal of the suppressor effect of 5-HT on the NHE3 promoter activity (Fig. 4A). Consistent with the inhibition by chelerythrine chloride, GÖ6976, a PKCα-specific inhibitor, also blocked the 5-HT response (Fig. 4B). Neither one of these inhibitors by themselves had any effect on the NHE3 promoter activity. To assess the involvement of PKA, p-95/+5 transfected cells were treated similarly using PKA inhibitor Rp-8-Br-cAMP (0.5 mM). Inhibition of PKA pathway did not alter the serotonin-mediated repression of the NHE3 promoter activity (Fig. 4C). These data suggest that the suppressor effect of serotonin on the human NHE3 expression is mediated by PKCα-dependent pathway, but is PKA independent.
Discussion
In previous studies, we showed that the interactions of various transcription factors with the cis-elements present in the human NHE3 core promoter region mediate the basal and regulated NHE3 gene expression [9–12]. Here, we investigated the effects of serotonin on the NHE3 gene expression in the human intestinal epithelial cell line, C2BBe1. Serotonin is released from enterochromaffin cells in the intestine and plays an important role in intestinal function. Studies from our group showed that acute serotonin treatment repressed the NHE3 transport activity [5].
In this study, by 5′-deletion analyses of the NHE3 promoter-reporter constructs we localized the 5-HT responsive region to the sequences between bp −95 to −76 in the core promoter region. Previously, we have shown that transcription factors Sp1 and Sp3 interact with an overlapping Sp1/Egr-1 motif in this region and regulate the constitutive NHE3 expression under the basal growth conditions [10]. Here, we demonstrate that 5-HT treatment leads to reduced DNA-binding activity of Sp1 and Sp3 although their nuclear contents are not affected. Consistent with these results the NHE3 promoter activity and transcript levels are down-regulated upon 5-HT treatment and expression of NHE3 protein is diminished. Thus our data indicate that the Sp1/Egr-1 motif in the NHE3 core promoter region plays a dual regulatory role in expression of NHE3 gene; i.e., not only it can server as a transcriptional enhancer [10], but can also play a role as suppressor of transcription. Regulation of gene expression by Sp1 is complex and involves multiple mechanisms including direct protein-DNA binding and protein-protein interactions [15]. In addition, transactivation potential of Sp1 itself is dictated by various posttranslational modifications such as phosphorylation, acetylation, and glycosylation [16]. Previous studies have established that phosphorylation of Sp1 may decrease its DNA-binding affinity [15–17]. Chu, et. al., [18] have shown that exposure of the lung epithelial cells to protein phosphatase 1, reduces the DNA-binding activity of Sp1 and Sp3 to the α-ENaC2 promoter and leads to diminished promoter activity. This mode of gene regulation has also precedence in the NHE3 gene expression [12]. We have shown that chronic exposure of C2BBe1 cells to a combination of TNF-α and IFN-γ results in repression of the NHE3 promoter activity, transcript and protein levels. Furthermore, repression of NHE3 promoter activity was mediated via a PKA signaling pathway leading to phosphorylation of Sp1/Sp3 and their reduced interactions with the core promoter region [12].
We speculate that reduced Sp1/Sp3 DNA-binding affinity to the NHE3 promoter in response to 5-HT may be controlled by posttranslational modification of these transcription factors by phosphorylation. Earlier reports have shown the involvement of PKC isozymes in Sp1-mediated transcriptional regulation of the target genes [19, 20]. Using pathway specific pharmacological inhibitors we examined the contribution of the PKC signaling pathway to the serotonin-mediated inhibition of NHE3 promoter activity. Exposure to chelerythrine chloride, a general PKC inhibitor, led to a complete reversal of the suppressor effect of 5-HT on the NHE3 promoter activity. Similarly, GÖ6976, a PKCα-specific inhibitor, also prevented the 5-HT response. Therefore, we conclude that PKCα is involved in mediating the effect of serotonin on the NHE3 transcription. Alterations in Sp1 interactions with other proteins have also been documented to influence its DNA-binding affinity and transactivation potential [17, 21]. Therefore, we could not exclude the possibility that regulatory mechanisms other than interference with direct protein-DNA interaction might affect Sp1/Sp3 availability for interactions with the NHE3 promoter.
In conclusion, transcriptional down-regulation of the NHE3 gene may be an important mechanism responsible for the decreased expression of NHE3 in response to long-term exposure to 5-HT. This mode of regulation may play a critical role in repression of NHE3 in response to stimuli such as serotonin, IFN-γ, and TNF-α and may account for, in part, the underlying reduction in Na+/H+ exchange activity implicated in the onset of diarrhea associated with intestinal inflammatory disorders.
Acknowledgments
This work was supported in part by NIDDK awards R01-DK 33349 (JM), DK 54016 (PKD, KR), DK 81858 (PKD) P01-DK 06887 (PKD, JM and KR), and Dept. of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kohle C, Badary OA, Nill K, Bock-Hennig BS, Bock KW. Serotonin glucuronidation by Ah receptor- and oxidative stress-inducible human UDP-glucuronosyltransferase (UGT) 1A6 in Caco-2 cells. Biochem Pharmacol. 2005;69:1397–1402. doi: 10.1016/j.bcp.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Kawano H, Tsuji H, Nishimura H, Kimura S, Yano S, Ukimura N, Kunieda Y, Yoshizumi M, Sugano T, Nakagawa K, Masuda H, Sawada S, Nakagawa M. Serotonin induces the expression of tissue factor and plasminogen activator inhibitor-1 in cultured rat aortic endothelial cells. Blood. 2001;97:1697–1702. doi: 10.1182/blood.v97.6.1697. [DOI] [PubMed] [Google Scholar]
- 3.Zuetenhorst JM, Korse CM, Bonfrer JM, Peter E, Lamers CB, Taal BG. Daily cyclic changes in the urinary excretion of 5-hydroxyindoleacetic acid in patients with carcinoid tumors. Clin Chem. 2004;50:1634–1639. doi: 10.1373/clinchem.2004.032151. [DOI] [PubMed] [Google Scholar]
- 4.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, Moses PL. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657–1664. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Gill RK, Saksena S, Tyagi S, Alrefai WA, Malakooti J, Sarwar Z, Turner JR, Ramaswamy K, Dudeja PK. Serotonin inhibits Na+/H+ exchange activity via 5-HT4 receptors and activation of PKC alpha in human intestinal epithelial cells. Gastroenterology. 2005;128:962–974. doi: 10.1053/j.gastro.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Xu H, Chen R, Ghishan FK. Subcloning, localization, and expression of the rat intestinal sodium-hydrogen exchanger isoform 8. Am J Physiol Gastrointest Liver Physiol. 2005;289:G36–41. doi: 10.1152/ajpgi.00552.2004. [DOI] [PubMed] [Google Scholar]
- 7.Zachos NC, Tse M, Donowitz M. Molecular physiology of intestinal Na+/H+ exchange. Annu Rev Physiol. 2005;67:411–443. doi: 10.1146/annurev.physiol.67.031103.153004. [DOI] [PubMed] [Google Scholar]
- 8.Woo AL, Gildea LA, Tack LM, Miller ML, Spicer Z, Millhorn DE, Finkelman FD, Hassett DJ, Shull GE. In vivo evidence for interferon-gamma-mediated homeostatic mechanisms in small intestine of the NHE3 Na+/H+ exchanger knockout model of congenital diarrhea. J Biol Chem. 2002;277:49036–49046. doi: 10.1074/jbc.M205288200. [DOI] [PubMed] [Google Scholar]
- 9.Malakooti J, Memark VC, Dudeja PK, Ramaswamy K. Molecular cloning and functional analysis of the human Na(+)/H(+) exchanger NHE3 promoter. Am J Physiol Gastrointest Liver Physiol. 2002;282:G491–500. doi: 10.1152/ajpgi.00273.2001. [DOI] [PubMed] [Google Scholar]
- 10.Malakooti J, Sandoval R, Amin MR, Clark J, Dudeja PK, Ramaswamy K. Transcriptional stimulation of the human NHE3 promoter activity by PMA: PKC independence and involvement of the transcription factor EGR-1. Biochem J. 2006;396:327–336. doi: 10.1042/BJ20051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amin MR, Dudeja PK, Ramaswamy K, Malakooti J. Involvement of Sp1 and Sp3 in differential regulation of human NHE3 promoter activity by sodium butyrate and IFN-gamma/TNF-alpha. Am J Physiol Gastrointest Liver Physiol. 2007;293:G374–382. doi: 10.1152/ajpgi.00128.2007. [DOI] [PubMed] [Google Scholar]
- 12.Amin MR, Malakooti J, Sandoval R, Dudeja PK, Ramaswamy K. IFN-gamma and TNF-alpha regulate human NHE3 gene expression by modulating the Sp family transcription factors in human intestinal epithelial cell line C2BBe1. Am J Physiol Cell Physiol. 2006;291:C887–896. doi: 10.1152/ajpcell.00630.2005. [DOI] [PubMed] [Google Scholar]
- 13.Malakooti J, Dahdal RY, Dudeja PK, Layden TJ, Ramaswamy K. The human Na(+)/H(+) exchanger NHE2 gene: genomic organization and promoter characterization. Am J Physiol Gastrointest Liver Physiol. 2001;280:G763–773. doi: 10.1152/ajpgi.2001.280.4.G763. [DOI] [PubMed] [Google Scholar]
- 14.Martel F. Recent advances on the importance of the serotonin transporter SERT in the rat intestine. Pharmacol Res. 2006;54:73–76. doi: 10.1016/j.phrs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50:111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- 16.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 17.Rossi A, Mukerjee R, Ferrante P, Khalili K, Amini S, Sawaya BE. Human immunodeficiency virus type 1 Tat prevents dephosphorylation of Sp1 by TCF-4 in astrocytes. J Gen Virol. 2006;87:1613–1623. doi: 10.1099/vir.0.81691-0. [DOI] [PubMed] [Google Scholar]
- 18.Chu S, Cockrell CA, Ferro TJ. Expression of alpha-ENaC2 is dependent on an upstream Sp1 binding motif and is modulated by protein phosphatase 1 in lung epithelial cells. Biochem Biophys Res Commun. 2003;303:1159–1168. doi: 10.1016/s0006-291x(03)00497-2. [DOI] [PubMed] [Google Scholar]
- 19.Rafty LA, Khachigian LM. Sp1 phosphorylation regulates inducible expression of platelet-derived growth factor B-chain gene via atypical protein kinase C-zeta. Nucleic Acids Res. 2001;29:1027–1033. doi: 10.1093/nar/29.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao M, Zhang Y, Dufau ML. Protein kinase Calpha-induced derepression of the human luteinizing hormone receptor gene transcription through ERK-mediated release of HDAC1/Sin3A repressor complex from Sp1 sites. Mol Endocrinol. 2008;22:1449–1463. doi: 10.1210/me.2008-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu S, Ferro TJ. Sp1: regulation of gene expression by phosphorylation. Gene. 2005;348:1–11. doi: 10.1016/j.gene.2005.01.013. [DOI] [PubMed] [Google Scholar]