Abstract
DNA damage is an important mechanism of toxicity for a variety of pollutants, and therefore, is often used as an indicator of pollutant effects in ecotoxicological studies. Here, we adapted a PCR-based assay for nuclear and mitochondrial DNA damage for use in an important environmental model, the Atlantic killifish (Fundulus heteroclitus). We refer to this assay as the large amplicon quantitative PCR (LA-QPCR) assay. To validate this method in killifish, DNA damage was measured in liver, brain, and muscle of fish dosed with 10 mg/kg benzo[a]pyrene. This exposure caused 0.4-0.8 lesions/10 kb. We also measured DNA damage in liver and muscle tissues from killifish inhabiting a Superfund site, confirming the utility of this method for biomonitoring. In both cases, damage levels were comparable in nuclear DNA (nDNA) and mitochondrial DNA (mtDNA). Since extensive nDNA sequence data are not readily available for many environmentally relevant species, but mitochondrial genomes are frequently fully sequenced, this assay can be adapted to examine mtDNA damage in virtually any species with little development. Therefore, we argue that this assay will be a valuable tool in assessing DNA damage in ecotoxicological studies.
Keywords: DNA damage, LA-QPCR assay, Fundulus heteroclitus, PAH, Benzo[a]pyrene, Elizabeth River
1. Introduction
DNA damage results from exposure to many contaminants, and is widely used as an indicator or biomarker of biological effects (van der Oost et al., 2003). In addition, DNA damage and repair is an important field of study within ecotoxicology (Theodorakis, 2001). The long amplicon quantitative PCR (LA-QPCR) assay, previously referred to as the QPCR assay, provides a sensitive way of assessing DNA damage and alterations to DNA that often lead to damage (Ayala-Torres et al., 2000). The assay measures the fraction of undamaged template DNA by comparing the amplification of very long PCR target (amplicon), under the assumption that lesions and/or structural alterations in the target genomic DNA template will block or slow the progression of the DNA polymerase used in the reaction (Kalinowski et al., 1992). We can then mathematically transform the difference in amplification to represent lesion frequencies. With parallel amplification of shorter PCR target within the amplified region of the longer target, we can normalize for DNA copy number and compare DNA damage in different DNA sources. The LA-QPCR assay can detect DNA strand breaks, adducts, and many other types of structural modifications such as those caused by oxidative damage. Therefore, it is potentially of particular utility in biomonitoring contexts where the specific types of DNA damage may not be well-defined or predictable ahead of time. It has been used successfully in human cell lines (Yakes and Van Houten, 1997; Van Houten et al., 2000) and laboratory model organisms (Chan et al., 2006; Meyer et al., 2007). In these models, the assay has been shown to have a high sensitivity, with a limit of detection of approximately 1 lesion per 105 nucleotide (Santos et al., 2006). This assay is a relatively simple method of detecting damage in nuclear DNA (nDNA) and mitochondrial DNA (mtDNA), and if adapted, would be valuable for detecting DNA damage in wildlife populations. However, the use of this assay in an environmental model has not been published.
The Atlantic killifish (Fundulus heteroclitus) is an estuarine fish species distributed throughout the coastal marshes along the North American Atlantic Coast. This species has a limited home range (Lotrich, 1975), and is considered to be very adaptable to diverse local environments and various stressors that are present in their habitats (Burnett et al., 2007). Different populations show resistance to pollutants such as metals (Weis and Weis, 1989), dioxin-like compounds and PCBs (Prince and Cooper, 1995a, 1995b; Nacci et al., 1999), and PAHs (Meyer et al., 2002; Ownby et al., 2002; Meyer and Di Giulio, 2003). Furthermore, they are easy to collect and maintain in the laboratory (Burnett et al., 2007). Thus, killifish have been widely utilized in laboratory toxicology (Eisler, 1986; Wassenberg et al., 2002), genetic adaptation research (Nacci et al., 2002), ecological studies (Weis and Weis, 1989; Nacci et al., 1999), and as a model organism for estuarine system monitoring (Eisler, 1986).
Several assays have been developed for assessing contaminant exposures in killifish (Binder et al., 1985; Van Veld et al., 1992; Greytak et al., 2005). Most of these assays are specific to individual or groups of compounds, and do not address the downstream and potentially higher level biological effects of contaminants on this species. Considering the role of killifish as an important environmental model, an assay that can easily detect general DNA damage would be a valuable tool for assessing contaminant effects in this organism.
In this study, we explored the potential of the LA-QPCR assay to measure PAH-induced genetic damage in Atlantic killifish. First, we tested the utility of this method for use in laboratory exposures by quantifying DNA damage in killifish that had been injected with the well-established genotoxin benzo[a]pyrene (BaP). Second, in an environmental study, we used LA-QPCR to look for evidence of genotoxicity in Atlantic killifish inhabiting a highly contaminated Superfund site.
2. Materials and Method
2.1. Killifish care
Adult killifish (Fundulus heteroclitus, Fundulidae, Cyprinodontiformes) were captured using baited minnow traps from King’s Creek, a tributary of York River in Gloucester County, VA. This is a relatively unpolluted site that we have used for several years as a reference site (Meyer et al., 2002; 2005). After capture, the fish were transported to the Duke University Ecotoxicology Laboratory. Fish were maintained in a recirculating system containing 23-25°C, 25 ppt artificial sea water (Instant Ocean®, Aquarium Systems, Forster & Smith, Rhinelander, WI, USA) with a 14:10 Light:Dark photoperiod. The fish were fed a mixed diet of Tetramin® Tropical Fish Food (Tetra Systems, Blacksburg, VA, USA) and brine shrimp (Artemia sp, Brine Shrimp Direct, Ogden, UT, USA).
2.2. BaP treatment and DNA isolation
Male killifish were moved to individual aerated tanks with 3 L of artificial seawater 24h before treatment. Ten fish were injected intraperitoneally with BaP in corn oil. Fish were injected with 5 μL/g wet mass of 10 mg/kg BaP. Additional fish were injected with 5 μL/g wet mass of corn oil as a carrier control. The fish were fed everyday and sacrificed 72h post-treatment. Brain, liver, and muscle tissues were dissected out and flash frozen in 20% glycerol, and stored at -80°C. Tissues were later ground in liquid nitrogen, and total DNA was extracted with the Genomic-tip 20/G kit (Qiagen Inc., Valencia, CA, USA) according to the manufacture’s protocol. In addition, fish from King’s Creek and from the Atlantic Wood Superfund site at the Elizabeth River in Portsmouth, VA were collected and sacrificed within 24h of capture. Previous studies have shown that populations from these two sites are genetically suited to be used in comparison studies (Mulvey et al., 2002; 2003). Liver and muscle from the fish were dissected out. The tissues were stored at -80°C until total DNA was isolated as described above. Since this assay relies upon the amplification of long stretches of DNA, it is critical that the DNA template be extracted as carefully as possible. The extracted DNA should not be exposed to phenol, and should be of high molecular weight. Additional protocol details are available in Santos et al. (2006).
2.3. Ultraviolet radiation C (UVC) exposure
Total DNA was isolated from the liver and brain of adult male Fundulus heteroclitus using the isolation methods described above. Equal amounts of DNA (50 μL of 3 ng/μL) were exposed to 0, 5, 10, and 20 J/m2 of ultraviolet radiation (254 nm; hereafter referred to as UVC) using either an ultraviolet lamp (UVLMS-38 EL Series 3UV Lamp, UVP, Upland, CA, USA) in conjunction with a UVX radiometer and UVX-25 sensor (UVP), or a CL-1000 Ultraviolet Crosslinker (UVP) with an emission peak at 254 nm. DNA was immediately frozen until further analysis.
2.4. Primer selection
Primers for large and small nuclear targets were designed for the cystic fibrosis transmembrane conductance regulator gene (CFTR, GenBank assession no. AY028263), and large and small mitochondrial targets were designed from a cDNA sequence for cytochrome c oxidase polypeptide VIa, mitochondria (CN984995). The CFTR gene was selected as it was the only published gene over 10kB long. Primers were designed using PRIMER3 (Rozen and Skaletsky, 2000). The primer sequences for the large mitochondrial target were obtained from Kim et al (2004). Primers and amplicaon sizes are described in Table 1. All primers were tested to confirm the amplification of a single band of the expected length.
Table 1.
Primers used for Fundulus heteroclitus LA-QPCR assay.
| Target | Primer sequences |
|---|---|
| Large nuclear target 11459 bp |
F: 5’- CAGCCGCCCGCAAATTCTCA -3’ R: 5’- CAGAATGCGGGCCTTGCTGA -3’ |
| Small nuclear target 234 bp |
F: 5’- GCCGCTGCCTTCATTGCTGT -3’ R: 5’- ATGAGCTGGGTGTGCGCTGA -3’ |
| Long mitochondrial target a 9416 bp |
F: 5’- TTGCACCAAGAGTTTTTGGTTCCTAAGACC -3’ R: 5’- GATGTTGGATCAGGACATCCCAATGGTGCA -3’ |
| Small mitochondrial target 264 bp |
F: 5’- ATCTGCATGGCCAACGCCTA -3’ R: 5’- GGCGGTGCCAGTTTCCTTTT –3’ |
Adapted from (Kim et al., 2004)
2.6. LA-QPCR
LA-QPCR was performed according to a protocol modified from Santos et al. (2006). This assay has previously been referred to as the QPCR for DNA damage assay; we have chosen to refer to it as the LA-QPCR assay to avoid confusion with quantitative PCR (qPCR), the abbreviation frequently used to refer to real-time PCR-based measurement of mRNA levels.
Briefly, 10 ng DNA (5 μL of 2 ng/μL DNA) from each sample was amplified with rTth polymerase (Applied Biosystems) using the primers described above. Small nDNA and mtDNA targets were amplified for normalization/verification of DNA concentration and to account for mitochondrial copy number, respectively. We optimized the elongation temperature, Mg(OAc)2 concentration, and cycle number for each PCR target. The PCR conditions for each set of primers are as follows. For all targets, final concentrations of 1x buffer (provided in the rTth polymerase kit), 100μg/mL of BSA, 200 μM of each dNTP, and 0.4 mM of each primer were added in the PCR mix. Water volume was adjusted to make the volume 50 μL for each reaction. For both short targets, 1.2 mM of Mg(OAc)2 was used in the PCR mix. The cycling conditions were 75°C for 2 min; 94°C for 1 min; 94°C for 15 s, 62°C for 45 s, and 72°C for 30s (repeated 24 cycles); and 72°C for 5 min. For the long nuclear target, 1.1 mM of Mg(OAc)2 was used in the PCR mix. The cycling conditions were 75°C for 2 min; 94°C for 1 min; 94°C for 15 s and 68°C for 12 min (repeated 24 cycles); and 72°C for 10 min. For the long mitochondrial target, 1.2 mM of Mg(OAc)2 was used in the PCR mix. The cycling conditions were 75°C for 2 min; 94°C for 1 min; 94°C for 15 s and 65°C for 12 min (repeated 16 cycles); and 72°C for 10 min. We added 5 μL of the rTth enzyme (diluted to 1 unit/μL) after 90 s of the 75°C incubation at the beginning of the reaction to initiate the amplification with “hot start.” PicoGreen dye (Invitrogen Corporation, Carlsbad, CA, USA) was used to quantify the template and PCR product. DNA concentrations were then converted to lesion frequencies per 10kB DNA by application of the Poisson distribution, as described by Ayala-Torres et al. (2000). This approach defines the control samples as undamaged, and generates a lesion frequency in experimental samples relative to the control samples, based on alterations in amplification efficiency and an assumption of random distribution of damage. With each PCR reaction, we included 5 ng of one of the control DNAs to monitor amplification quality. Only PCR products in which the amplification of 5 ng DNA was 40-60% of the control DNA (10 ng), indicating that the PCR reaction was quantitative, were used in the analysis.
2.7. Statistics
Statistical analyses were performed using SPSS, version 15.0 for Windows (SPSS Inc., Chigago, IL, USA). The assumption of normality was tested for all data sets using the Shapiro-Wilk’s test. Analysis of Variance (ANOVA) and Fisher’s Protected Least-Significant Differences (LSD) were used to test for differences among groups (α = 0.05).
3. Results
3.1. Adaptation of the LA-QPCR assay
To confirm the success of primer selection and condition optimization for this assay, we exposed purified total DNA in buffer from adult male killifish liver and brain to various doses of UVC and assessed damage (Figure 1). A dose-dependent increase in damage to nDNA and mtDNA exposed to various doses of UVC radiation was detected (p < 0.001), but no differences in damage were observed between mtDNA and nDNA at a given dose (p = 0.775). The increase in DNA damage fit the linear regression with r2 values of 0.84 for mtDNA and 0.81 for nDNA respectively, and the lesion frequencies detected were comparable to those obtained previously using DNA purified from human cells in culture or Caenorhabditis elegans at the same UV doses (Eischeid et al., 2008).
Figure 1. DNA damage in UVC-treated killifish DNA.
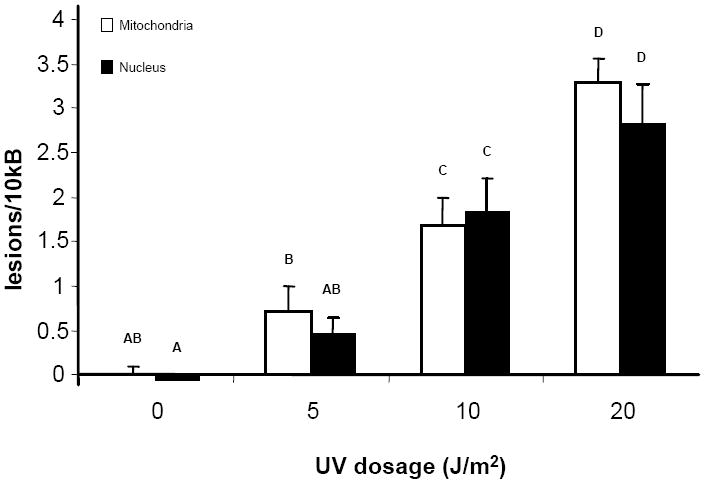
DNA isolated from adult killifish liver and brain was exposed to different doses of UVC. There are significant dose-dependent increases in both mitochondrial and nuclear DNA (p < 0.001), but no differences between mtDNA and nDNA at a given dose (p = 0.775). Letters indicate significant groupings (p < 0.05) according to Fisher’s LSD. n = 6 per treatment group. Error bars indicate standard error of means.
3.2. DNA damage in response to BaP
As expected, adult male killifish dosed with intraperitoneal (i.p.) injection of 10 mg/kg BaP showed increased levels of DNA damage relative to killifish dosed with corn oil for both mtDNA and nDNA (Figure 2) in all three tissues examined. Three-way ANOVA showed that there was a significant effect of treatment (p < 0.001). However, neither DNA source (mitochondria versus nucleus) nor tissue type significantly affected the result (p = 0.177 and p = 0.493 respectively). In addition, there was no interaction among any of the independent variables.
Figure 2. Levels of DNA damage in BaP treated adults.

DNA lesion frequencies were measured from adult male fish dosed with 10 mg/kg BaP. BaP treatment was the only significant factor according to three-way ANOVA (p < 0.001). Neither DNA source nor tissue significantly affected the result (p = 0.177 and p = 0.493 respectively). n = 10 per treatment group. Error bars indicate standard error of means.
3.3. Comparison of Elizabeth River and reference site killifish populations
Killifish from the Atlantic Wood Superfund site and King’s Creek (reference site) were sacrificed within 24h of capture, and lesion frequencies in mtDNA and nDNA from muscle and liver were examined (Figure 3). In this case, brain was not examined due to difficulties in acquiring sufficient amount of tissue. Three-way ANOVA showed that there was a significant effect of site (p < 0.001) and tissue type (p = 0.047). However, DNA source (nuclear or mitochondrial genome) was not significant (p = 0.839). There was also a significant interaction between tissue type and population (p = 0.033), reflecting the fact that the Elizabeth River killifish seemed to show more sensitivity to nDNA damage in muscle, but to mtDNA damage in liver. However, while statistically significant, this difference does not seem large enough to be of clear biological relevance.
Figure 3. Levels of DNA damage in Superfund site and reference Fundulus populations.

DNA lesion frequency was measured from liver and muscle of adult fish captured from a Superfund site and a reference site. Population and tissue type significantly affected the results (p < 0.001 and p = 0.047 respectively) according to three-way ANOVA. However, there was no difference between mtDNA and nDNA (p = 0.839). * denotes significant difference (p < 0.05) according to Fisher’s LSD. n = 5 per treatment. Error bars indicate standard error of means.
4. Discussion
We have shown for the first time that the LA-QPCR assay can be adapted to a widely studied environmental model. We detected significant increases in the frequency of DNA lesions after exposure to an environmentally relevant dose (10 mg/kg) of BaP as well as contaminants present at a Superfund site.
In our experiments with BaP-treated fish as well as with the field-caught fish, we detected no significant differences in damage in response to BaP treatment between mtDNA and nDNA. This was a surprise since previous data in mammalian cell culture studies indicated a much greater susceptibility of the mitochondrial genome to polycyclic aromatic hydrocarbon exposure (Allen and Coombs 1980, Backer and Weinstein 1980). We do not know the reason for this difference. It seems unlikely to be related to the requirement for metabolic activation: Backer and Weinsten used the reactive metabolite benzo[a]pyrene diol epoxide, rather than the parent benzo[a]pyrene, but Allen and Coombs used parent compounds that require activation, as we did. More likely candidates include in vitro vs in vivo differences (the studies cited above used cell culture systems), DNA damage detection methodology, or species differences. With aflatoxin B1, another lipophilic chemical that is activated to a DNA-reactive form by CYP proteins, Niranjan et al (1982) saw ~3-fold higher binding to mtDNA than nDNA in vivo. While this difference was still significant, it is much less than the in vitro differences reported by Allen and Coombs and Backer and Weinstein (40- to 500-fold). Furthermore, the vulnerability of mtDNA to this chemical showed species variation (Niranjan et al 1986), which may be in part related to mitochondrial enzyme differences leading to differential activation (Niranjan et al (1985). It will be interesting to further explore the relative vulnerabilities of the mitochondrial and nuclear genomes in fish; mtDNA has been shown to be more sensitive to various genotoxins than nDNA in mammalian studies (Backer and Weinstein, 1980; Balansky et al., 1996; Yakes and Van Houten, 1997; Sawyer and Van Houten, 1999).
However, another important implication of the equal or greater sensitivity of the mitochondrial genome to at least many pollutants is that unless a specific nuclear-coded gene needs to be targeted to assess DNA damage to specific genes, using the LA-QPCR assay with just mtDNA would be sufficient in many field studies. This is advantageous for many environmental models, such as the Atlantic killifish, as they generally do not have significant nuclear genome sequence data available, particularly the 10 kb or more of contiguous sequence needed to design primers for the LA-QPCR assay. In such models, it is much easier to design primers for mtDNA, since there is a tremendous amount of mtDNA sequence data available and “universal” primers have already been designed in conserved regions that will amplify the mtDNA of most vertebrates (Kocher et al., 1989). In fact, the primers that we used for Fundulus were initially designed for the javeline goby (Acanthogobius hasta) (Kim et al., 2004).
Interestingly, liver DNA showed more DNA damage in Superfund site killifish than did muscle DNA, although this difference was not observed in the acute exposure. This may be explained by the fact that the killifish population at the Superfund site has been chronically exposed to a complex mixture of chemicals including primarily several different PAHs, as well as PCP and metals (EPA, 2007). Therefore, this population is exposed continuously to a variety of genotoxic agents through their diet. In this case, liver would be one of the primary targets of the toxic effect. The differences in contaminant mixture, and/or time course and route of exposure, may be why we see differences in the DNA damage profile of Elizabeth River Fundulus relative to fish acutely exposed to BaP via i.p. injection. Thus, our data suggest that data from muscle tissue alone might not be as informative as other tissues such as liver. Therefore, an examination of several tissues may be necessary to correctly assess the genotoxic effects of pollutants.
Currently, there are several assays that measure DNA damage. DNA-adduct analysis by 32P-postlabelling method can be used to measure chemical-specific adducts. The method is considered the most sensitive in detecting PAH-adducts, but is expensive and time consuming (van der Oost et al., 2003). Flow cytometry (Barbee et al.,; Theodorakis, 2001; Goanvec et al., 2004), single-strand break assays (McFarland et al., 1999; Bolognesi et al., 2006) and the micronucleus test (Al-Sabti and Metcalfe, 1995; Hayashi et al., 1998; Arkhipchuk and Garanko, 2005; Cavas and Ergene-Gozukara, 2005; Bolognesi et al., 2006) are also used in laboratory and field studies as genotoxic indicators, but these assays detect gross chromosomal damage or abnormalities from clastogenic and aneugenic effects. The comet assay is one of the most widely used biomarkers of DNA damage in laboratory (Pandrangi et al., 1995; Belpaeme et al., 1996; Nacci et al., 1996; Devaux et al., 1997; Devaux et al., 1998) and field studies (Pandrangi et al., 1995; Devaux et al., 1998; Steinert et al., 2002; Lemos et al., 2005; Yang et al., 2006). However, there is still high study-to-study variation, and standardization of measurement is necessary to overcome this issue (Cotelle and Férard, 1999; Siu et al., 2004; Lemos et al., 2005).
Considering the issues concerning the assays described above, the adaptation of LA-QPCR assay will be an important and effective means to measure general DNA damage in the environment. With this assay, one can detect general lesions caused by a variety of pollution sources or complex mixtures. At the same time, one can target a specific gene or the entire mitochondrial genome for damage assessment. This ability to easily distinguish mtDNA damage from nDNA damage or total DNA damage is an important advantage of this assay, since there is increasing concern for the vulnerability of mitochondria to various pollutants (Backer and Weinstein, 1980; Sawyer and Van Houten, 1999), and for the lower DNA repair capability of the mitochondria for certain kinds of damage (Yakes and Van Houten, 1997; Larsen et al., 2005).
In conclusion, we have successfully adapted the LA-QPCR assay in an environmental model. This assay can be utilized as a sensitive method of detecting general nuclear and mitochondrial DNA damage, and has significant potential as a tool for biomonitoring. Therefore, we propose the use of the LA-QPCR assay for DNA damage for use in environmental assessments.
Acknowledgments
We thank Dr. Cole Matson, Lauren Battle, Bryan Clark, and Lindsey Van Tiem for laboratory assistances and advices. We also thank Dr. Andrew Whitehead and Dr. Sibel I. Karchner for their advices on primer selection. This research was supported by the Superfund Basic Science Research Center (P42 ES10356) and NIEHS Integrated Toxicology and Environmental Health Program (T32ES07031).
Footnotes
This paper is derived from a presentation given at the 4th Aquatic Animal Models of Human Disease Conference: hosted by Duke University’s Nicholas School of the Environment and Earth Sciences, and Duke’s Comprehensive Cancer Center, Durham, NC, USA, January 31 - February 3, 2008.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Sabti K, Metcalfe CD. Fish micronuclei for assessing genotoxicity in water. Mutat Res. 1995;343:121–135. doi: 10.1016/0165-1218(95)90078-0. [DOI] [PubMed] [Google Scholar]
- Arkhipchuk VV, Garanko NN. Using the nucleolar biomarker and the micronucleus test on in vivo fish fin cells. Ecotoxicol Environ Saf. 2005;62:42–52. doi: 10.1016/j.ecoenv.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Allen JA, Coombs MM. Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature. 1980;287:244–245. doi: 10.1038/287244a0. [DOI] [PubMed] [Google Scholar]
- Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- Backer JM, Weinstein IB. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- Balansky R, Izzotti A, Scatolini L, D’Agostini F, De Flora S. Induction by carcinogens and chemoprevention by N-acetylcysteine of adducts to mitochondrial DNA in rat organs. Cancer Res. 1996;56:1642–1647. [PubMed] [Google Scholar]
- Barbee GC, Barich J, Duncan B, Bickham JW, Matson CW, Hintze CJ, Autenrieth RL, Zhou G-D, McDonald TJ, Cizmas L, Norton D, Donnelly KC. In situ biomonitoring of PAH-contaminated sediments using juvenile Coho salmon (Oncorhynchus kisutch) Ecotoxicol Environ Saf. doi: 10.1016/j.ecoenv.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Belpaeme K, Delbeke K, Zhu L, KirschVolders M. Cytogenetic studies of PCB77 on brown trout (Salmo trutta fario) using the micronucleus test and the alkaline comet assay. Mutagenesis. 1996;11:485–492. doi: 10.1093/mutage/11.5.485. [DOI] [PubMed] [Google Scholar]
- Binder RL, Stegeman JJ, Lech JJ. Induction of cytochrome P-450-dependent monooxygenase systems in embryos and eleutheroembryos of the killfish Fundulus heteroclitus. Chem Biol Interact. 1985;55:185–202. doi: 10.1016/s0009-2797(85)80127-7. [DOI] [PubMed] [Google Scholar]
- Bolognesi C, Perrone E, Roggieri P, Sciutto A. Bioindicators in monitoring long term genotoxic impact of oil spill: Haven case study. Mar Environ Res. 2006;62:S287–S291. doi: 10.1016/j.marenvres.2006.04.047. [DOI] [PubMed] [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gomez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, Maclatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Rees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL. Fundulus as the premier teleost model in environmental biology: Opportunities for new insights using genomics. Comp Biochem Physiol Part D Genomics Proteomics. 2007;2:257–286. doi: 10.1016/j.cbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavas T, Ergene-Gozukara S. Micronucleus test in fish cells: A bioassay for in situ monitoring of genotoxic pollution in the marine environment. Environ Mol Mutagen. 2005;46:64–70. doi: 10.1002/em.20130. [DOI] [PubMed] [Google Scholar]
- Chan SS, Santos JH, Meyer JN, Mandavilli BS, Cook DL, Jr, McCash CL, Kissling GE, Nyska A, Foley JF, van Houten B, Copeland WC, Walker VE, Witt KL, Bishop JB. Mitochondrial toxicity in hearts of CD-1 mice following perinatal exposure to AZT, 3TC, or AZT/3TC in combination. Environ Mol Mutagen. 2007;48:190–200. doi: 10.1002/em.20191. [DOI] [PubMed] [Google Scholar]
- Cotelle S, Férard JF. Comet assay in genetic ecotoxicology: A review. Environ Mol Mutagen. 1999;34:246–255. [PubMed] [Google Scholar]
- Devaux A, Flammarion P, Bernardon V, Garric J, Monod G. Monitoring of the chemical pollution of the River Rhone through measurement of DNA damage and cytochrome P4501A induction in chub (Leuciscus cephalus) Mar Environ Res. 1998;46:257–262. [Google Scholar]
- Devaux A, Pesonen M, Monod G. Alkaline comet assay in rainbow trout hepatocytes. Toxicol In Vitro. 1997;11:71–73. doi: 10.1016/s0887-2333(97)00004-0. [DOI] [PubMed] [Google Scholar]
- Eischeid AC, Meyer JN, Linden KG. UV disinfection of adenoviruses: Molecular indications of DNA Damage Efficiency. In review. 2008 doi: 10.1128/AEM.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisler R. Use of Fundulus Heteroclitus in Pollution Studies. Am Zool. 1986;26:283–288. [Google Scholar]
- EPA. Proposed Plan: Atlantic Wood Industries Superfund Site Portsmouth, Virginia. US EPA Region 3. 2007 Available: http://www.epa.gov/reg3hwmd/super/sites/VAD990710410/planproposed/AWI_July_2007_Proposed_Plan_Text.pdf.
- Goanvec C, Theron M, Poirier E, Le Floch S, Laroche J, Nonnotte L, Nonnotte G. Evaluation of chromosomal damage by flow cytometry in turbot (Scophthalmus maximus L.) exposed to fuel oil. Biomarkers. 2004;9:435–446. doi: 10.1080/13547500400027001. [DOI] [PubMed] [Google Scholar]
- Greytak SR, Champlin D, Callard GV. Isolation and characterization of two cytochrome P450 aromatase forms in killifish (Fundulus heteroclitus): Differential expression in fish from polluted and unpolluted environments. Aquat Toxicol. 2005;71:371–389. doi: 10.1016/j.aquatox.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Ueda T, Uyeno K, Wada K, Kinae N, Saotome K, Tanaka N, Takai A, Sasaki YF, Asano N, Sofuni T, Ojima Y. Development of genotoxicity assay systems that use aquatic organisms. Mutat Res. 1998;399:125–133. doi: 10.1016/s0027-5107(97)00251-0. [DOI] [PubMed] [Google Scholar]
- Kalinowski DP, Illenye S, Van Houten B. Analysis of DNA damage and repair in murine leukemia L1210 cells using a quantitative polymerase chain-reaction assay. Nucleic Acids Res. 1992;20:3485–3494. doi: 10.1093/nar/20.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IC, Kweon HS, Kim YJ, Kim CB, Gye MC, Lee WO, Lee YS, Lee JS. The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene. 2004;336:147–153. doi: 10.1016/j.gene.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: Similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lemos NG, Dias AL, Silva-Souza AT, Mantovani MS. Evaluation of environmental waters using the comet assay in Tilapia rendalli. Environ Toxicol Pharmacol. 2005;19:197–201. doi: 10.1016/j.etap.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Lotrich VA. Summer home range and movements of Fundulus heteroclitus (Pisces, Cyprinodontidae) in a tidal creek. Ecology. 1975;56:191–198. [Google Scholar]
- McFarland VA, Inouye LS, Lutz CH, Jarvis AS, Clarke JU, McCant DD. Biomarkers of oxidative stress and genotoxicity in livers of field-collected brown bullhead, Ameiurus nebulosus. Arch Environ Contam Toxicol. 1999;37:236–241. doi: 10.1007/s002449900510. [DOI] [PubMed] [Google Scholar]
- Meyer J, Di Giulio R. Heritable adaptation and fitness costs in killifish (Fundulus heteroclitus) inhabiting a polluted estuary. Ecol Appl. 2003;13:490–503. [Google Scholar]
- Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Nacci DE, Di Giulio RT. Cytochrome P4501a (Cyp1a) in Killifish (Fundulus heteroclitus): Heritability of altered expression and relationship to survival in contaminated sediments. Toxicol Sci. 2002;68:69–81. doi: 10.1093/toxsci/68.1.69. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Volz DC, Freedman JH, Giulio RTD. Differential display of hepatic mRNA from killifish (Fundulus heteroclitus) inhabiting a superfund estuary. Aquat Toxicol. 2005;73:327–341. doi: 10.1016/j.aquatox.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Mulvey M, Newman MC, Vogelbein W, Unger MA. Genetic structure of Fundulus heteroclitus from PAH-contaminated and neighboring sites in the Elizabeth and York Rivers. Aquat Toxicol. 2002;61:195–209. doi: 10.1016/s0166-445x(02)00055-3. [DOI] [PubMed] [Google Scholar]
- Mulvey M, Newman MC, Vogelbein WK, Unger MA, Ownby DR. Genetic structure and mtDNA diversity of Fundulus heteroclitus populations from polycyclic aromatic hydrocarbon-contaminated sites. Environ Toxicol Chem. 2003;22:671–677. [PubMed] [Google Scholar]
- Nacci D, Coiro L, Champlin D, Jayaraman S, McKinney R, Gleason TR, Munns WR, Specker JL, Cooper KR. Adaptations of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Mar Biol. 1999;134:9–17. [Google Scholar]
- Nacci DE, Cayula S, Jackim E. Detection of DNA damage in individual cells from marine organisms using the single cell gel assay. Aquat Toxicol. 1996;35:197–210. [Google Scholar]
- Nacci DE, Champlin D, Coiro L, McKinney R, Jayaraman S. Predicting the occurrence of genetic adaptation to dioxinlike compounds in populations of the estuarine fish Fundulus heteroclitus. Environ Toxicol Chem. 2002;21:1525–1532. [PubMed] [Google Scholar]
- Niranjan BG, Bhat NK, Avadhani NG. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215:73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- Niranjan BG, Avadhani NG, DiGiovanni J. Formation of benzo[α]pyrene metabolites and DNA adducts catalyzed by a rat liver mitochondrial monooxygenase system. Biochem Biophys Res Commun. 1985;131:935–942. doi: 10.1016/0006-291x(85)91329-4. [DOI] [PubMed] [Google Scholar]
- Niranjan BG, Schaefer H, Ritter C, Avadhani NG. Protection of mitochondrial genetic system agains aflatoxin B1 binding in animals resistant to aflatoxicosis. Cancer Res. 1986;46:3637–3641. [PubMed] [Google Scholar]
- Ownby D, Newman M, Mulvey M, Vogelbein W, Unger M, Arzayus L. Fish (Fundulus heteroclitus) populations with different exposure histories differ in tolerance of creosote-contaminated sediments. Environ Toxicol Chem. 2002;21:1897–1902. [PubMed] [Google Scholar]
- Pandrangi R, Petras M, Ralph S, Vrzoc M. Alkaline single cell gel (comet) assay and genotoxicity monitoring using bullheads and carp. Environ Mol Mutagen. 1995;26:345–356. doi: 10.1002/em.2850260411. [DOI] [PubMed] [Google Scholar]
- Prince R, Cooper KR. Comparisons of the effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on chemically impacted and nonimpacted subpopulations of Fundulus hteroclitus. 1. TCDD toxicity. Environ Toxicol Chem. 1995a;14:579–587. [Google Scholar]
- Prince R, Cooper KR. Comparisons of the Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on chemically impacted and nonimpacted subpopulations of Fundulus heteroclitus. 2. Metabolic considerations. Environ Toxicol Chem. 1995b;14:589–595. [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the www for general users and for biologist programmers. Humana Press; Totowa, NJ: 2000. [DOI] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- Sawyer DE, Van Houten B. Repair of DNA Damage in Mitochondria. Mutat Res. 1999;434:161–176. doi: 10.1016/s0921-8777(99)00027-0. [DOI] [PubMed] [Google Scholar]
- Siu WHL, Cao J, Jack RW, Wu RSS, Richardson BJ, Xu L, Lam PKS. Application of the comet and micronucleus assays to the detection of B[a]P genotoxicity in haemocytes of the green-lipped mussel (Perna viridis) Aquat Toxicol. 2004;66:381–392. doi: 10.1016/j.aquatox.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Steinert SA, Streib-Montee R, Armstrong JL, Robertson G, Schlenk D, Roy L. DNA damage in multiple tissues from flatfish collected for coastal ocean monitoring off southern California. Mar Environ Res. 2002;54:532. [Google Scholar]
- Theodorakis CW. Integration of genotoxic and population genetic endpoints in biomonitoring and risk assessment. Ecotoxicology. 2001;10:245–256. doi: 10.1023/a:1016677629442. [DOI] [PubMed] [Google Scholar]
- van der Oost R, Beyer J, Vermeulen NPE. Fish bioaccumulation and biomarkers in environmental risk assessment: A review. Environ Toxicol Pharmacol. 2003;13:57–149. doi: 10.1016/s1382-6689(02)00126-6. [DOI] [PubMed] [Google Scholar]
- Van Houten B, Cheng S, Chen Y. Measuring gene-specific nucleotide excision repair in human cells using quantitative amplification of long targets from nanogram quantities of DNA. Mutat Res. 2000;460:81–94. doi: 10.1016/s0921-8777(00)00018-5. [DOI] [PubMed] [Google Scholar]
- Van Veld PA, Vogelbein WK, Smolowitz R, Woodin BR, Stegeman JJ. Cytochrome P4501A1 in hepatic lesions of a teleost fish (Fundulus heteroclitus) collected from a polycyclic aromatic hydrocarbon-contaminated site. Carcinogenesis. 1992;13:505–507. doi: 10.1093/carcin/13.3.505. [DOI] [PubMed] [Google Scholar]
- Wassenberg DM, Swails EE, Di Giulio RT. Effects of single and combined exposures to benzo(a)pyrene and 3,3’4,4’5-pentachlorobiphenyl on EROD activity and development in Fundulus heteroclitus. Mar Environ Res. 2002;54:279–283. doi: 10.1016/s0141-1136(02)00182-4. [DOI] [PubMed] [Google Scholar]
- Weis JS, Weis P. Tolerance and stress in a polluted environment- the case of mummichog. Bioscience. 1989;39:89–95. [Google Scholar]
- Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XA, Meier J, Chang L, Rowan M, Baumann PC. DNA damage and external lesions in brown bullheads (Ameiurus nebulosus) from contaminated habitats. Environ Toxicol Chem. 2006;25:3035–3038. doi: 10.1897/05-706r.1. [DOI] [PubMed] [Google Scholar]


