Abstract
Parathyroid hormone-related protein (PTHrP) plays a primary role in the development of humoral hypercalcemia of malignancy (HHM) that occurs in the majority of patients with adult T-cell leukemia/lymphoma (ATLL) due to human T-cell lymphotropic virus type-1 (HTLV-1) infection. We previously showed that ATLL cells constitutively express high levels of PTHrP via activation of promoters P2 and P3, resulting in HHM. In this study, we characterized a nuclear factor-κB (NF-κB) binding site in the P2 promoter of human PTHrP. Using electrophoretic mobility shift assays, we detected a specific complex in Tax-expressing human T cells composed of p50/c-Rel, and two distinct complexes in ATLL cells consisting of p50/p50 homodimers and a second unidentified protein(s). Chromatin immunoprecipitation assays confirmed in vivo binding of p50 and c-Rel on the PTHrP P2 promoter. Using transient co-transfection with NF-κB expression plasmids and PTHrP P2 luciferase reporter-plasmid, we showed that NF-κB p50/p50 alone and p50/c-Rel or p50/Bcl-3 cooperatively upregulated the PTHrP P2 promoter. Furthermore, inhibition of NF-κB activity by Bay 11-7082 reduced PTHrP P2 promoter-initiated transcripts in HTLV-1-infected T cells. In summary, the data demonstrated that transcriptional regulation of PTHrP in ATLL cells can be controlled by NF-κB activation and also suggest a Tax-independent mechanism of activation of PTHrP in ATLL.
Keywords: ATLL, HTLV-1, PTHrP, NF-κB, transcriptional regulation
Introduction
Adult T-cell leukemia/lymphoma (ATLL) is a highly aggressive malignancy of peripheral helper T cells associated with human T-cell lymphotropic virus type-1 (HTLV-1) infection.1 About 80% of ATLL patients develop humoral hypercalcemia of malignancy (HHM), a life-threatening paraneoplastic syndrome, seen in a wide variety of cancers in addition to ATLL.2 In HHM, increased circulating parathyroid hormone-related protein (PTHrP) stimulates the parathyroid hormone-1 receptor to induce osteoclastic bone resorption and increase calcium reabsorption in kidneys, resulting in hypercalcemia.3 In addition to its role in the induction of HHM, PTHrP has been shown to be involved in the regulation of cell proliferation and apoptosis in a wide variety of normal and neoplastic tissues.4
Transcriptional regulation of the human PTHrP gene is achieved by three distinct promoters identified as P1, P2 and P3. Promoters P1 and P3 (previously called the P2 promoter) contain a typical TATA box,5–7 while P2 is a GC-rich promoter region located immediately upstream of exon 3 (also named exon 1c) with the transcription initiation site located 11 nucleotides upstream of the exon 3 splice acceptor site. Several studies have shown that P2/P3 promoter usage is prevalent in many cancers such as breast cancer and bone cancer in addition to HTLV-1-positive and ATLL cells.8–11 Furthermore, Brandt et al.12 reported a preferential usage of P2 over P1 and P3 promoters in different cancer cell lines examined. Although this stimulation and activity of the PTHrP P2 promoter was identified 10 years ago, little is known regarding the transcriptional mechanisms involving cis-acting regulatory sequences within this promoter.
HTLV-1-Tax is a potent transcriptional activator that not only drives the transcription of all HTLV-1 transcripts from the viral LTR but also activates numerous cellular genes through activation of nuclear factor-κB (NF-κB), cyclic AMP response element-binding protein (CREB/ATF) and serum response factor (SRF).13–15 NF-κB, a cytokine-inducible family of transcription factors, controls expression of genetic networks important in cell survival, proliferation, inflammation and T-cell transformation. Although tightly regulated in normal T cells, NF-κB is constitutively activated in ATLL and Tax-expressing T cells.16 While Tax-mediated NF-κB activation serves as a critical step in the induction of T-cell transformation by HTLV-1, the presence of constitutively active NF-κB in freshly isolated ATLL cells that lack detectable Tax expression, implicates a crucial role for NF-κB in the multistep process of leukemogenesis.17
The NF-κB family is composed of five structurally related protein subunits that can be divided into two groups: (1) p65/RelA, RelB and c-Rel contain a well-defined transactivation domain; and (2) p50 and p52, which are generated by proteolytic processing from their precursors, p105/NF-κB 1 and p100/NF-κB 2, respectively, and lack transactivation domains. Although the predominant complex in most cells is p50/p65, various dimeric transcription factor complexes can form between the family members.18 Heterodimers containing p52 or p50 combined with p65, c-Rel or RelB are capable of activating transcription. NF-κB dimers bind target gene regulatory regions through a wide variety of binding sites that generally match a 5′-GGGRNNYYCC-3′ consensus (R, purine; Y, pyrimidine; N, any base). Although their functions often overlap, NF-κB achieves target gene specificity in part through preferential binding of different subunit combinations to numerous similar DNA sequences.19
NF-κB is expressed in virtually all cell types, but in unstimulated cells the NF-κB homo- and heterodimers are sequestered in the cytoplasm in an inactive form complexed with one of the members of the family of regulatory IκBs, including IκB-α, IκB-β, IκB-γ and Bcl-3. IκB molecules are subject to phosphorylation, subsequent degradation and release of active NF-κB upon reception of signals that lead to NF-κB activation.20 In contrast to IκBα and IκBβ, which specifically interact with dimers containing p65 or c-Rel, Bcl-3 specifically interacts with p50 or p52 homodimers and contains N- and C-terminal regions that can act as transactivation domains.21,22 Although the cellular function of Bcl-3 is still unclear, various studies have suggested that Bcl-3 acts to increase transcription from NF-κB responsive promoters by either (1) acting as an antirepressor by removing homodimers from the κB sites so that transactivating NF-κB dimers can bind, (2) forming a complex with homodimers at κB sites and acting as a transactivator or (3) enhancing homodimer binding to κB sites. Interestingly, Bcl-3 can enhance p50 or p52 homodimer binding to DNA, without being a stable component of the complex.23,24
We have previously reported that the upregulation of PTHrP in ATLL and HTLV-1-infected T cells is mediated by the PTHrP P2 and P3 promoters and that the P3 promoter is regulated by the ETS signaling pathway. The relative PTHrP P2 promoter usage was determined to be 16, 4, 7 and 40 copies (per 104 copies of β2M) in HT-1RV, SLB-1, MT-2 and RV-ATL cells, respectively, compared to the 47, 86, 104 and 644 copies (per 104 copies of β2M) of total PTHrP.25 Since ATLL cells display constitutive expression of NF-κB and PTHrP and since sequence analysis of the PTHrP P2 promoter revealed that it contained potential binding sites for NF-κB, we tested in this report the hypothesis that PTHrP gene is a direct target of the transcription factor, NF-κB. We identified an NF-κB binding site within the human PTHrP P2 promoter region that is responsible for NF-κB-mediated stimulation of PTHrP promoter activity. We also demonstrated and characterized the formation of different protein complexes on the NF-κB-binding site located within the second promoter (P2) of the human PTHrP gene in HTLV-1-infected and ATLL cells. We present evidence that transactivation of the PTHrP P2 promoter can occur in a subunit-dependent manner by p50/c-Rel and p50/Bcl-3. Finally, we showed that inhibition of NF-κB by the Bay 11-7082 decreased P2 promoter-initiated transcription. Our data demonstrated that transactivation of the PTHrP P2 promoter in HTLV-1-infected and ATLL cells occurs by activation of NF-κB in a Tax-independent manner.
Materials and methods
Sequence analysis
Human, mouse, rat and dog upstream PTHrP P2 promoter sequences (GenBank accession: NM 002820, NM 008970, NM 012636 and NM 001003303, respectively) were analyzed for potential NF-κB binding sites using MatInspector (Genomatix Software GmbH, Munchen, Germany), with ‘weight matrices’ as the search parameter.
Animals and inoculation
Immunodeficient SCID-NOD (NOD CB17-PRKDC-SCID/J) mice (Jackson Lab, Bar Harbor, ME, USA) were maintained under specific pathogen-free conditions in the animal facility of the College of Veterinary Medicine at The Ohio State University. Male mice aged 5 weeks were used as recipients and injected intraperitoneally with 4 × 107 RV-ATL cells or 3 × 107 Met-1 cells suspended in RPMI 1640 medium. The source of the RV-ATL and Met-1 cells (a kind gift of Dr Feuer and Dr Waldman, respectively) was previously described.26,27 RV-ATL cells were harvested by peritoneal lavage 28 days post-inoculation. Met-1 cells were harvested from peritoneal tumors approximately 60 days post-inoculation.
Cell lines
MT-2 and SLB-1 are in vitro-transformed HTLV-1-positive cell lines that were obtained by co-cultivating lymphocytes from healthy donors with leukemic cells from ATLL patients.28 RV-ATL and Met-1 cells are leukemic cells derived from ATLL patients. HT-1RV cells were obtained by superinfecting RV-ATL cells with HTLV-1.29 IMM-1 cells are interleukin-2 (IL-2)-dependent immortalized cells obtained by in vitro co-cultivation of peripheral blood mononuclear cells from a healthy human donor with irradiated SLB-1 cells. MT-2 and SLB-1 cells were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS) and the HT-1RV and RV-ATL cells in RPMI 1640 with 20% heat-inactivated (56°C, 30 min) FBS, l-glutamine (2mM), penicillin (50 U/ml) and streptomycin (50 µg/ml) (Invitrogen, Carlsbad, CA, USA) at 37°C and 5% CO2. NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 10mM l-glutamine.
Long-term immortalization assay
Human peripheral blood mononuclear cells were isolated from the blood of healthy donors by centrifugation over Ficoll-Paque (Amersham, Piscataway, NJ, USA) and cultured in RPMI 1640 supplemented with 20% FBS, 10 U/ml IL-2 (Boehringer Mannheim, Mannheim, Germany), 2mm glutamine and antibiotics. Irradiated SLB-1 cells (106) were co-cultivated with 2 × 106 freshly isolated peripheral blood mononuclear cells with 10 U/ml IL-2 in 24-well culture plates. The presence of HTLV-1 expression was confirmed by detection of p19 Gag protein in the culture supernatant at weekly intervals using a commercially available enzyme-linked immunosorbent assay (ELISA) kit (Zeptometrix, Buffalo, NY). Cells from a donor that continued to proliferate after 30 weeks of co-culture in the presence of exogenous IL-2 were identified as HTLV-1-immortalized cells and were named IMM-1 cells.
Electrophoretic mobility shift assays and supershifts
Nuclear extracts were prepared from RV-ATL, MET-1, HT1-RV, MT-2, SLB-1, IMM-1 and Jurkat cells using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Pierce, Rockford, IL, USA). For NF-κB binding activity on the PTHrP P2 promoter, 1–5 µl of each nuclear extract (7.5 µg of protein) was incubated in 18 µl total reaction volume, containing 10mM Tris-HCl, pH 7.5, 50mm NaCl, 1mm EDTA, 1mm dithiothreitol, 10% glycerol, 1 µg of poly(dI-dC)-poly(dI-dC) (Amersham Biosciences, Piscataway, NJ, USA) and 0.4 µg/µl bovine serum albumin for 15 min at room temperature. The reaction mixture was then incubated with 50 F moles of a Cy5 5′-end-labeled double-stranded PTHrP P2 promoter wild-type oligonucleotide (sense strand: 5′-TCATTCCCGGCTCGGGGCTCCCCTCCACT CGCTCG-3′; κB site is underlined) alone or with 25-fold excess of unlabeled wild-type or mutant P2 oligonucleotides for 15 min at room temperature. Samples were analyzed by electrophoresis using 4% nondenaturing polyacrylamide gels with 1 × TGE buffer (25mm Tris, 189mm glycine and 1mm EDTA) containing 5% glycerol. The gels were scanned with a Typhoon 9410 Variable Mode Imager (Amersham Biosciences) to detect the Cy5 fluorescence. Consensus NF-κB binding activity was measured using 5 µg of the nuclear extracts with 2 × 104 c.p.m. of a 32P-labeled oligonucleotide probe containing a κB site from class I major histocompatibility complex (MHC) promoter (5′-CAGGGCTGGGGATTCCCCATCTCCA CAGTTTCACTTC-3′; κB site is underlined), as described previously.30 For supershift experiments, 4 µl each of p50 (H-119; sc-7178), p50 NLS (sc-114), p65 (sc-109), c-Rel (sc-71), Bcl-3 (C-14; sc-185) antibodies (Santa Cruz, Santa Cruz, CA, USA) was incubated with 7.5 µg nuclear extract for 10 min at room temperature before the addition of buffers and oligonucleotides.
Chromatin immunoprecipitation-PCR and quantitative PCR
Chromatin immunoprecipitation (ChIP) was performed with antibodies against Bcl-3 (C-14), c-Rel (C) and p50 (H-119) using the ChIP assay kit (Upstate, Charlottesville, VA, USA). The specific sequence from immunoprecipitated and input DNA was detected by PCR using primers for PTHrP P2 promoter region: (forward, 5-GCCACCTCTTTGCGACTAGCT-3) and (reverse, 5-GGTTGGAGGCGAGTTGAAAAC-3). The annealing temperature was 58°C and the amplicon size was 91 bp. Quantitative PCR monitored with SYBRGreen was performed using the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) as described previously.31 Experimental ChIP-PCR values were normalized against values obtained by a standard curve constructed by input DNA (5–0.008%, fivefold dilution, R2>0.99) with the same primer set. Quantitation of transcription factor binding was expressed as enrichment ratio of antibody over IgG control, and each error bar represents standard deviation calculated from triplicates.
Plasmids and site-directed mutagenesis
A human PTHrP P2-luciferase reporter gene construct was derived by cloning the human P2 promoter −1030 (SmaI) to −611 (Sau3A) fragment into pGL-2 basic vector (Promega, Madison, WI, USA). For site-directed mutagenesis, a SacI–ApaI restriction fragment from the human PTHrP P2 promoter was subcloned into the SacI–ApaI sites of pCR2.1TOPO plasmid (Invitrogen). The NF-κB site within the PTHrP P2 promoter was mutated by PCR-amplifying the entire PTHrP-P2/pCR2.1TOPO plasmid using the Expand Long Template PCR System (Roche, Indianapolis, IN, USA) and sense (5′-CTCACATCCACTCG CTCG-3′) and antisense (5′-CACAGAGCCGGGAATGAG-3') oligonucleotides that contained the desired mutations (mutated nucleotides are underlined), as described previously.32 A PCR product of appropriate size (~3.9 kb) was obtained. This PCR product was blunt-ended with T4 DNA polymerase (New England Biolabs, Ipswich, MA, USA), kinased with T4 polynucleotide kinase (New England Biolabs), circularized with T4 DNA ligase (New England Biolabs) and transformed into DH5α cells. Once clones containing the desired mutations were obtained and confirmed by DNA sequencing, the mutated P2 promoter was subcloned back into pGL2/PTHrP-P2/Luc plasmid.
Western blotting
Approximately 40 µg of the nuclear and cytoplasmic lysates were separated on a 12% Tris-glycine SDS-polyacrylamide gel electrolysis gel. Protein was transferred to a nylon membrane and then probed with primary antibodies specific for Tax (168A51-42; Tab 176; NIH AIDS Research & Reference Reagent Program), NF-κB p50 (Epitomics, Burlingame, CA, USA), p65, c-Rel, IκB-α, Bcl-3 (same as used in supershifts) and β-Actin (Sigma, St Louis, MO, USA) followed by incubation with goat anti-mouse or goat anti-rabbit (Promega) horseradish peroxidase-conjugated secondary antibodies. The signal was detected by chemiluminescence using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Boston, MA, USA).
Transfections
To investigate the effect of NF-κB on PTHrP transcriptional regulation, NIH3T3 cells at 60% confluence were co-transfected with 1 µg of either wild-type pGL2/PTHrP-P2/Luc, mutant pGL2/PTHrP-P2/Luc or pGL2 constructs in the presence or absence of expression vectors for NF-κB p50 (50–500 ng), p65 (500 ng), c-Rel (50–500 ng) or Bcl-3 (10–500 ng) using Lipofectamine Plus reagent (Invitrogen) and harvested after 48 h of transfection. pcDNA-3.1 was used as a ‘filler’ plasmid so that the total amount of DNA would be the same in all transfection groups. The plasmid pβgal-Control Vector (250 ng) was included in each transfection and served as an internal control to correct for transfection efficiency. Luciferase activity was measured with the Luciferase Assay System (Promega) using 40 µl of lysate. Simultaneously, β-galactosidase activity was measured with the Luminescent β-Galactosidase Detection Kit II (BD Biosciences, San Jose, CA, USA).
RT-PCR, Bay 11-7082 treatment and real time RT-PCR
Total RNA was extracted using TRIZOL reagent (Invitrogen). Approximately 2.5 µg of RNA was reverse transcribed with Superscript Reverse Transcriptase (Invitrogen). The cDNA was amplified with specific oligonucleotide primers for Bcl-3, and β2-microglobulin using Platinum Taq DNA polymerase (Invitrogen), as described previously.33,34 The PCR products were analyzed by electrophoresis on a 1% agarose gel. For Bay 11-7082 treatment, 2 × 106 cells were treated with Bay 11-7082 (Alexis Biochemical Corporation, CA, USA) or vehicle control (dimethyl sulfoxide). To measure the total PTHrP and P2 promoter-initiated transcripts, 1 µg RNA was reverse-transcribed and submitted for real-time RT-PCR analysis using TaqMan Gene Expression assays (4331182 and 364171-B4 respectively; Applied Biosystems). β2M (4333766; Applied Biosystems) was used as a reference gene.
Statistical analysis
Pooled-variance t-tests were used to analyze the data from transfection assays. Analysis of variance (ANOVA) with Dunnett’s tests was used to analyze the data from titration with p50, c-Rel, Bcl-3 and Bay 11-7082 treatment and vehicle control groups. Means and standard deviations are plotted in the figures. The mean differences and 95% confidence intervals are available upon request. A P-value of <0.05 was considered significant.
Results
Identification of NF-κB sites in the PTHrP P2 promoter
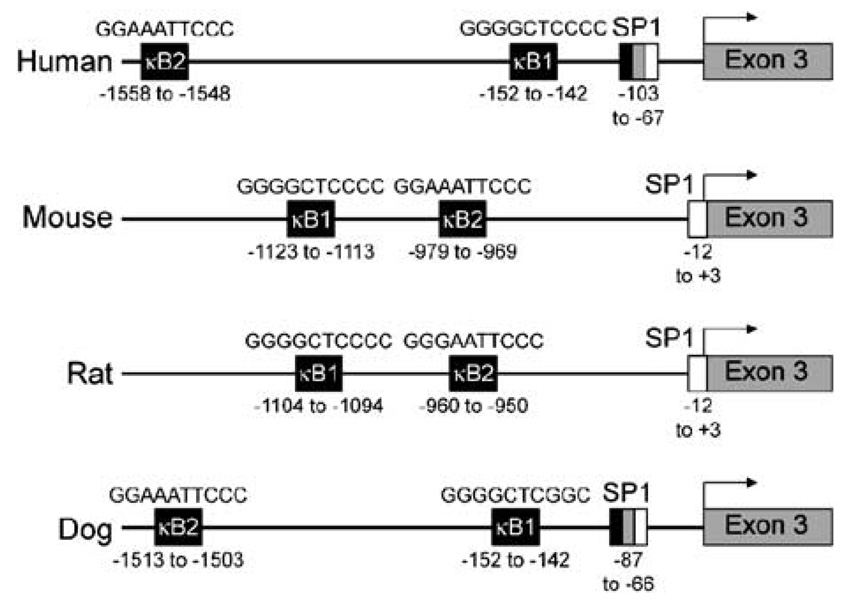
Following sequence analysis of the human PTHrP gene (described in the Materials and methods section), we identified two putative NF-κB binding sequences in the 5′-regulatory region beginning at positions −1558 to −1548 (GGAAATTCCC) and −152 to −142 (GGGGCTCCCC) nucleotides from the transcription start site for exon 3 (Figure 1). These sites represented excellent matches to the 5′-GGGRNNYYCC-3′ consensus NF-κB sequence.35 Putative κB sites almost identical to the human PTHrP κB sites were also present in the mouse and the rat PTHrP promoter regions (Figure 1). The κB binding sequence (−1513 to −1503) was present in dog but the sequence proximal to the transcription start site (−152 to −142) was a partial consensus. In this study, we focused on the κB1 sequence (−152 to −142) due to its proximity to the transcription start site for exon 3 in the human PTHrP gene.
Figure 1.
Schematic representation of human, mouse, rat and dog PTHrP P2 promoter regions. The diagram illustrates the highly conserved nature of the NF-κB sequences in the PTHrP P2 promoter of humans, mice, rats and dogs. The NF-κB binding sequence in the human promoter studied in this report was found at −152 to −142 nucleotides upstream from transcriptional start site (indicated by arrows) in exon 3.
Tax and IκB-α expression in HTLV-1-infected T cells and ATLL cells
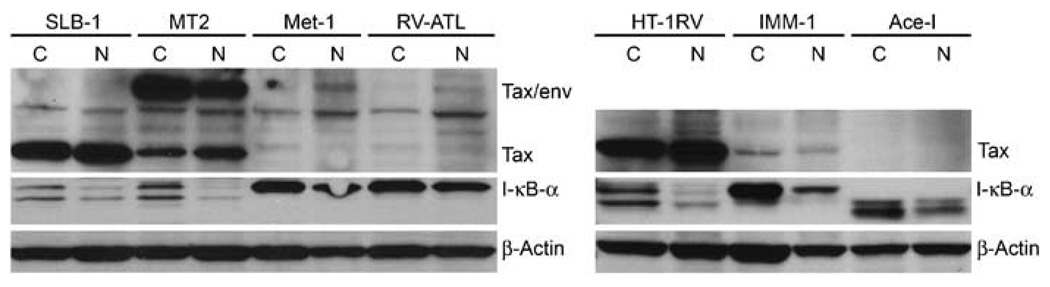
In resting cells, NF-κB is retained in the cytoplasm due to binding to specific NF-κB inhibitors of the IκB family. Following activation of cells by various stimuli, signal transduction cascades lead to the degradation of IκB-α and translocation of NF-κB into the nucleus. Since Tax is known to activate the NF-κB pathway, we measured the levels of Tax expression in various cell lines (Figure 2). Three of the HTLV-1-infected cell lines (MT-2, SLB-1 and HT-1RV) have very high levels of Tax expression. In addition to the 40-kDa Tax band, a 69-kDa band, which is a fusion between the envelope and the Tax-coding sequence, is seen in MT-2 cells as described previously.36 IMM-1 cells have a lower Tax protein expression compared to the other HTLV-1-infected cell lines. There was no detectable Tax protein expression in RV-ATL and MET-1 cells. A canine prostate carcinoma cell line, Ace-1, was used as a negative control for Tax. Also, in agreement with prior studies,16 the protein level of IκB-α was significantly lower in three of the four HTLV-1-infected cell lines (SLB-1, MT-2 and HT-1RV) compared to ATLL cells (MET-1 and RV-ATL) (Figure 2). Imm-1 cells have a higher level of IκB-α protein compared to the other HTLV-1-infected cell lines, indicating a lower IκB-α turnover in these cells and likely lower NF-κB activation. These results suggested a direct relationship between Tax, IκB-α expression and NF-κB activity in T cells.
Figure 2.
HTLV-1 Tax expression was inversely proportional to IκB-α. Western blot analysis was performed on cytoplasmic (C) and nuclear (N) lysates from HTLV-1-infected and ATLL cells with antibodies against Tax and IκB-α proteins. Ace-1, a canine prostate carcinoma cell line was used as a negative control. β-Actin was used as loading control.
Constitutive NF-κB binding activity in HTLV-1-infected T-cell lines and ATLL cells
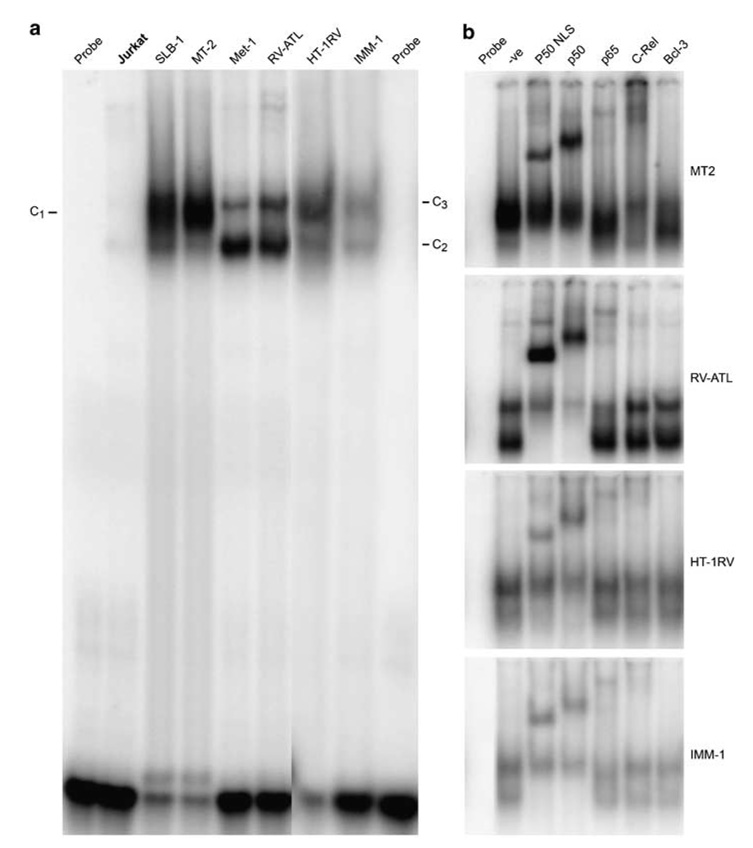
To measure the NF-κB binding activity in HTLV-1-infected and ATLL cells, nuclear extracts from these cells were subjected to electrophoretic mobility shift assays (EMSA) with a 32P-labeled, double-stranded oligonucleotide probe containing a κB site from the class I MHC promoter. No NF-κB-specific protein DNA complexes were detected in HTLV-1-negative Jurkat T cells. In contrast, enhanced NF-κB binding activity was detected in the nuclear extracts from HTLV-1-infected T-cell lines and ATLL cells (Figure 3a, lanes 3–8). The NF-κB binding activity in MT-2 and SLB-1 extracts consisted of a single complex (C1), while there were two complexes in MET-1, RV-ATL, HT-1RV and IMM-1 extracts (C2 and C3). The complexes in MT-2 cell extracts were composed predominantly of p50 and c-Rel as antibodies against p50 and c-Rel produced a shifted complex (Figure 3b, lanes 3, 4 and 6). The lower complex (C2) in MET-1, RV-ATL, HT-1RV and IMM-1 extracts consisted of p50/p50 (Figure 3b, lanes 3 and 4). The upper complex (C3) in RV-ATL and IMM-1 extracts contained p50 and p65 (Figure 3b, lanes 3–5) while the HT-1RV extracts contained p50, p65 and c-Rel (Figure 3b, lanes 3–6).
Figure 3.
NF-κB binding activity on a consensus κB binding site in various cell lines. (a) Approximately 5 µg of nuclear lysates from the cells were incubated with a [32P]-radiolabeled oligonucleotide corresponding to MHC class 1 gene κB sequence and analyzed by EMSA. The complexes are indicated as C1, C2 and C3. (b) In supershift assays, antibodies specific for each NF-κB subunit (indicated above the lane) were incubated with the nuclear extracts from MT-2, RV-ATL, HT-1RV and IMM-1 cells before addition of the probe.
NF-κB binds to the sequences in the 5′ regulatory region of PTHrP
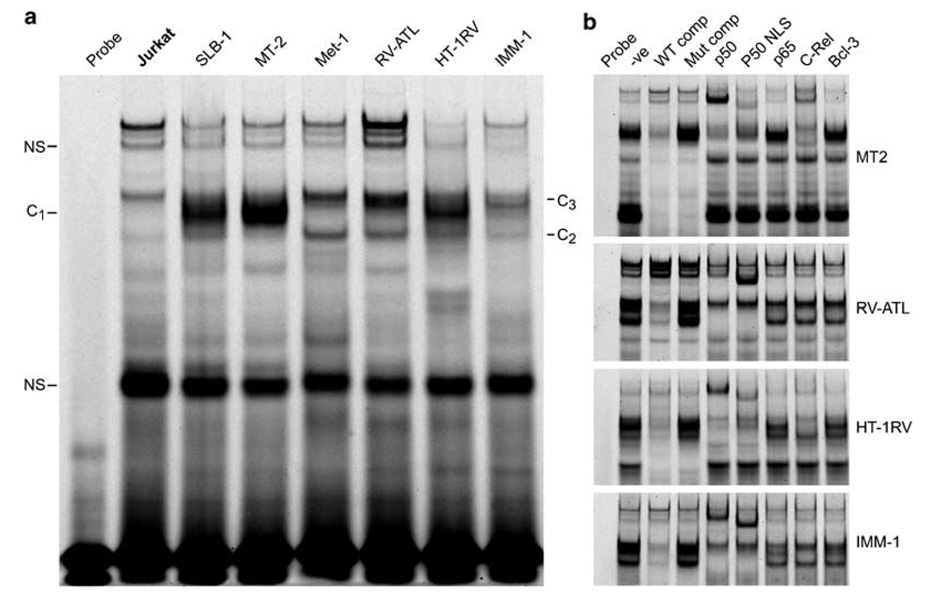
To determine if the sequences in the PTHrP P2 promoter region were authentic NF-κB binding sites, a Cy5-labeled, double-stranded oligonucleotide probe spanning the putative NF-κB binding site was tested by EMSA for binding of NF-κB proteins (Figure 4). A single complex (C1) formed in MT-2 and SLB-1 extracts, whereas two complexes (C2 and C3) formed in MET-1, RV-ATL, HT-1RV and IMM-1 extracts (Figure 4a). The complexes in MT-2 cells were composed of p50 and c-Rel since antibodies specific to these proteins produced a supershift (Figure 4b, lanes 5, 6 and 8). The lower complex (C2) in RV-ATL, HT-1RV, MET-1 (data not shown) and IMM-1 extracts contained only p50. The upper complex (C3) consisted predominantly of p50 and c-Rel in HT-1RV extracts, while the complexes in RV-ATL, MET-1 and IMM-1 extracts did not supershift with antibodies to p50, p52 (not shown), p65, c-Rel, RelB (not shown) or Bcl-3. All of the above complexes formed specifically on the NF-κB binding site, since competition with an excess of unlabeled wild-type probe eliminated the complex (Figure 4b, lanes 3), but an excess of an unlabeled probe containing a mutated NF-κB site did not eliminate the complex (Figure 4b, lanes 4). Additional complexes observed are nonspecific since they were out-competed by the mutated competitor.
Figure 4.
Nuclear NF-κB proteins specifically bound to an oligonucleotide containing the NF-κB binding sequences from the human PTHrP P2 promoter. (a) A Cy5 5′-end-labeled double-stranded PTHrP P2 promoter wild-type oligonucleotide was analyzed by EMSA using 7.5 µg of protein from nuclear extracts of various cell lines. The complex formation is indicated as C1, C2, C3 and the specificity of binding was evaluated using a mutant -κB binding site as described in Materials and methods. The additional bands in the gel were not characterized and considered nonspecific (NS). (b) Antibody-mediated shift interference or supershifts were performed using 7–15 µg of protein from the nuclear extracts with specific NF-κB antibodies as indicated.
ChIP demonstrated binding of NF-κB p50 and c-Rel to the PTHrP P2 promoter in vivo
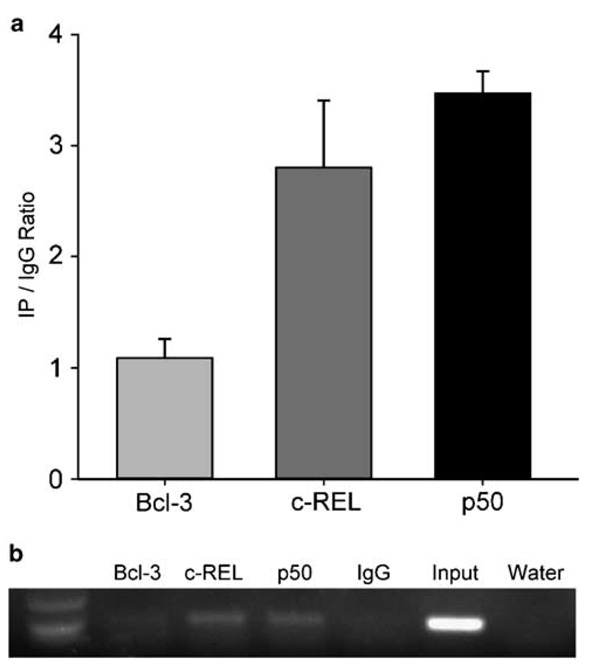
To determine if NF-κB transcription factors bind to the PTHrP P2 promoter in vivo, we performed a ChIP assay with antibodies against p50, c-Rel and Bcl-3. IgG was used as a negative control. As shown in Figure 5a, the PTHrP P2 promoter was occupied by NF-κB p50 and c-Rel in MT-2 cells as measured by ChIP with real-time PCR quantification. As an additional measure, gel electrophoresis of the PCR product after a limited number of cycles gave an independent visual confirmation of the binding activity on the PTHrP P2 promoter (Figure 5b).
Figure 5.
NF-κB p50 and c-Rel bound to the PTHrP P2 promoter in vivo: ChIP was performed in MT-2 cells with antibodies to p50, c-Rel, Bcl-3 and IgG. (a) Real-time PCR quantification of transcription factor binding was expressed as enrichment ratio of antibody over IgG control, and each error bar represents standard deviation calculated from triplicates. (b) PCR amplification of a 91-bp product from the PTHrP P2 promoter genomic DNA is shown on a 2% agarose gel.
Transactivation of the PTHrP κB site by the NF-κB family
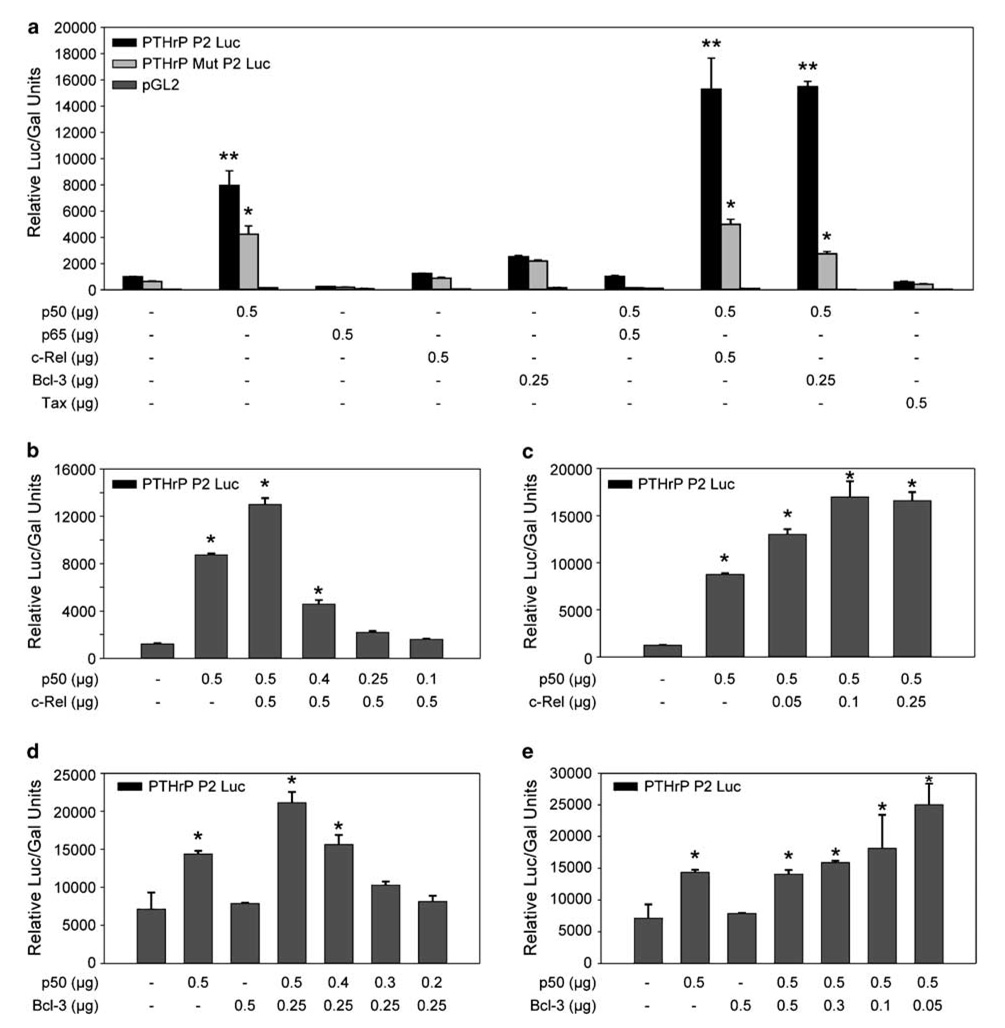
To determine which NF-κB subunits transactivated the PTHrP κB site, expression plasmids for four κB proteins, p50, p65, c-Rel and Bcl-3, were co-transfected in NIH3T3 cells with either wild-type pGL2/PTHrP-P2/Luc, mutant pGL2/PTHrP-P2/Luc or pGL2 reporter plasmids. As shown in Figure 6a, expression of p65, c-Rel, Bcl-3 or Tax by themselves did not increase the expression of luciferase activity. Interestingly, p50 alone or in combination with c-Rel or Bcl-3 strongly transactivated the PTHrP P2 promoter. Mutation of the κB binding sequence significantly reduced luciferase activity compared to the wild-type, showing that the transactivation was occurring through the κB site. The effect of p50 and c-Rel were directly proportional to their concentration (Figures 6b and c). Interestingly, the cooperative effect of Bcl-3 with p50 was inversely related to Bcl-3 concentration (Figures 6d and e). While low levels of Bcl-3 cooperated with p50 in transactivating PTHrP, higher levels of Bcl-3 nullified the effect.
Figure 6.
PTHrP P2 promoter can be transactivated by NF-κB in a subunit-dependent manner: Relative luciferase activity in NIH3T3 cells transfected with (a) 1µg of either wild-type pGL2/PTHrP-P2/Luc, mutant pGL2/PTHrP-P2/Luc or pGL2 constructs in the presence or absence of expression vectors for NF-κB p50, p65, c-Rel, Bcl-3, p50/p65, p50/c-Rel or p50/Bcl-3 or Tax; (b) 1µg wild-type pGL2/PTHrP-P2/Luc with or without 0.5 µg of NF-κB c-Rel expression vector and 0.1–0.5 µg of p50 expression vector; (c) 1µg wild-type pGL2/PTHrP-P2/Luc with or without 0.5 µg of NF-κB p50 expression vector and 0.05–0.25 µg of c-Rel expression vector; (d) 1µg wild-type pGL2/PTHrP-P2/Luc with or without 0.2–0.5 µg of NF-κB p50 expression vector and 0.25–0.5 µg of Bcl-3 expression vector; (e) 1µg wild-type pGL2/PTHrP-P2/Luc with or without 0.05–0.5 µg of NF-κB Bcl-3 expression vector and 0.5 µg of p50 expression vector. The quantity of the expression plasmids is indicated in µg. Bars represent the mean ± s.d. of three independent samples. (**) indicates significant differences between various groups compared to PTHrP P2 Luc alone (P<0.05). (*) indicates significant differences between mutant and wild-type (P<0.05).
HTLV-1-infected T cells and ATLL cells expressed Bcl-3 and other NF-κB family members
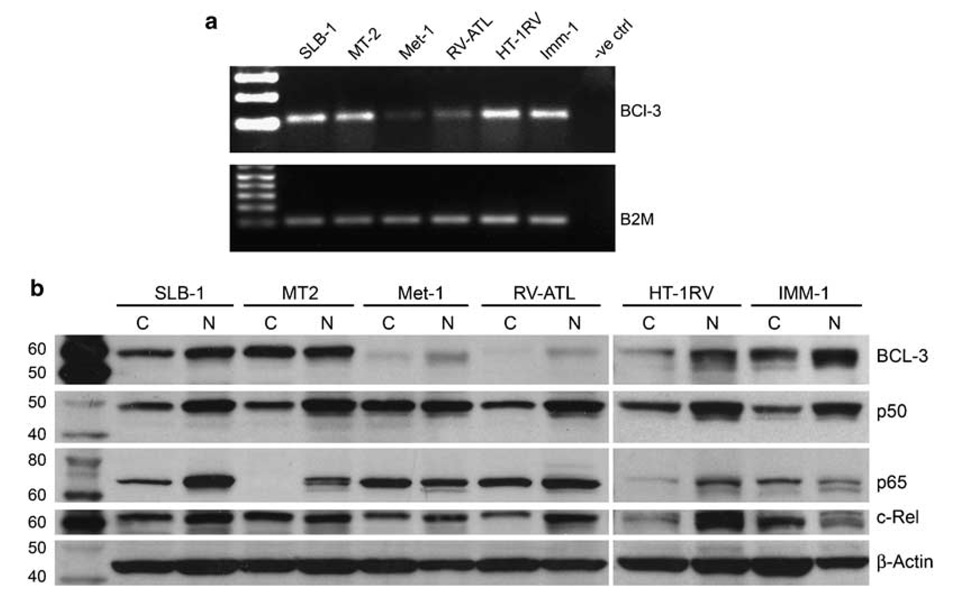
p50 is known to transactivate genes in association with Bcl-3. Following our observations that p50 alone and p50/Bcl-3 upregulated PTHrP, we measured the levels of Bcl-3 expression in HTLV-1-infected and ATLL cells. As shown in Figure 7a, Tax-expressing cells have higher levels of Bcl-3 mRNA expression compared to the ATLL cells. To determine the subcellular localization of Bcl-3 and other NF-κB members in these cells, western blotting was performed on cytoplasmic and nuclear extracts (Figure 7b). Cytoplasmic and nuclear fractions of MT-2, SLB-1, HT-1RV and IMM-1 cells had high levels of Bcl-3 protein. In contrast, lower levels of Bcl-3 protein were present in MET-1 and RV-ATL cells, but it was predominantly localized in the nucleus. This was in agreement with the RT-PCR data. The presence of other NF-κB family members was confirmed by western blot analysis of the cytoplasmic and nuclear fractions, with β-actin serving as a loading control. p50 and c-Rel were present in similar amounts in all the cells, while p65 was expressed in the highest levels in SLB-1, MT-2, MET-1 and RV-ATL cells.
Figure 7.
Bcl-3 expression was high in HTLV-1-infected T cells compared to ATLL cells: (a) RT–PCR was performed on total RNA isolated from various cells as indicated on the top row using primers specific for Bcl-3. β2-Microglobulin was used as a loading control. (b) Western blot analysis was performed on the cytoplasmic and nuclear lysates of various cells and probed with antibodies specific for Bcl-3, p50, p65 and c-Rel. β-Actin was used as protein loading control.
Inhibition of NF-κB by Bay 11-7082 decreased PTHrP mRNA expression in HTLV-1-infected T cells
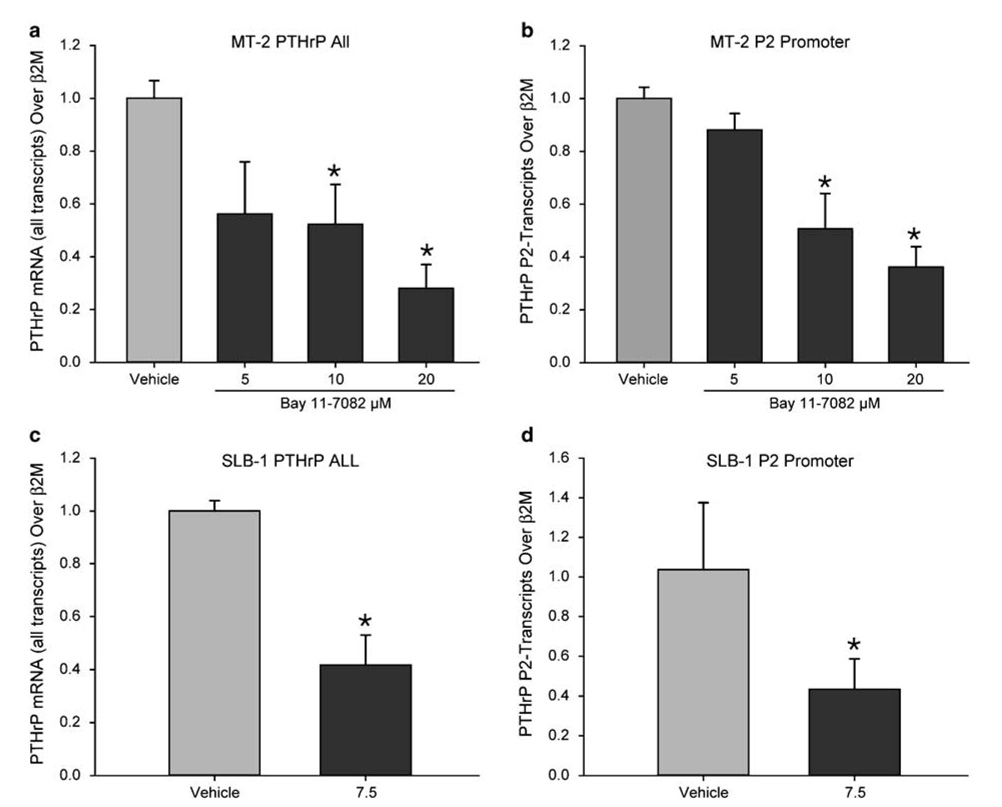
Since NF-κB is constitutively activated in HTLV-1-infected T cells, we wanted to determine the effect of inhibition of NF-κB on PTHrP expression. For this purpose, we used Bay 11-7082, a specific IκB phosphorylation inhibitor, which is known to reduce NF-κB activity. As shown in Figures 8a and c, inhibition of NF-κB activity by Bay 11-7082 significantly decreased PTHrP mRNA expression in MT-2 and SLB-1 cell lines at 3 and 6 h, respectively. Analysis of the promoter-initiated transcripts revealed that Bay 11-7082 reduced P2 promoter-initiated transcripts as expected (Figures 8b and d). Bay 11-7082 treatment reduced PTHrP starting at 1 h following treatment (data not shown). Reduction of PTHrP expression occurred at higher concentrations (20 µm) and at a later time point (24 h) in RV-ATL cells (data not presented). Inhibition of NF-κB activity with PS-341, a potent proteasome inhibitor, inhibited PTHrP P2-promoter initiated transcripts and the total PTHrP expression in RV-ATL cells (data not shown).
Figure 8.
Inhibition of the NF-κB activity in HTLV-1-infected T cells by Bay 11-7082 decreased PTHrP mRNA expression. Total RNA was extracted from MT-2 cells treated with 5, 10 and 20 µm for 3 h and SLB-1 cells treated with 7.5 µm Bay 11-7082 or vehicle alone for 6 h. Quantitative real time RT–PCR was performed using primers specific for PTHrP all transcripts and PTHrP P2 promoter-initiated transcripts using TaqMan Gene Expression Assays. In MT-2 cells, Bay 11-7082 decreased (a) total PTHrP (b) and the P2 promoter-initiated transcripts in a dose-dependent manner. In SLB-1 cells 11-7082 significantly decreased (c) total PTHrP and (d) the P2 promoter-initiated transcripts at 6 h of treatment. The data were normalized to β2-microglobulin (β2-M). Bars represent the mean±standard deviation of three independent samples (*P<0.05).
Discussion
PTHrP is expressed in a wide variety of tissues and is known to be dysregulated in several cancers. Increasing evidence suggests a role for PTHrP in proliferation and apoptosis besides its central role in the pathogenesis of HHM.37 However, transcriptional regulation of this complex gene is poorly understood. In this study, we tested the hypothesis that the PTHrP gene is a direct target for NF-κB and report three major observations: (1) sequence, EMSA and competition analyses of the PTHrP P2 promoter sequences identified NF-κB binding sequences in the human PTHrP P2 promoter in vitro that was further confirmed as an authentic NF-κB binding site in vivo by ChIP assay; (2) NF-κB activated the human PTHrP P2 promoter in a subunit-specific manner as determined by an NF-κB over-expressing model in 3T3 cells; (3) inhibition of NF-κB by Bay 11-7082 in HTLV-1-infected T cell decreased P2 promoter-initiated transcripts.
The GC-rich PTHrP P2 promoter has been shown to be active in a variety of cells, including HTLV-1-infected and ATLL cells.38 We identified two putative NF-κB-binding sequences in the human PTHrP P2 promoter that are located 1558 (κB2) and 152 (κB1) nucleotides upstream from the transcription initiation start site. Vasavada et al.39 have shown that the SmaI/Sau3A fragment of the P2 promoter had significant activity in renal carcinoma cells. We identified and characterized the κB binding site (κB 1) within this fragment of the promoter. In addition, both the κB1 and κB2 sites are also present in the P2 promoter regions of mice and rats, although their order is reversed compared to humans. While the κB1 sequence appears to have two point mutations in the dog, the κB2 sequence is present and the location of both these elements is the same as in humans. The presence of these transcription elements across different species is consistent with the concept that NF-κB is important in modulating the expression of PTHrP.
Although tightly regulated in normal physiologic cellular responses, NF-κB is constitutively activated in various malignancies such as solid tumors, lymphomas and leukemias, including ATLL.40 HTLV-1 Tax, in addition to mediating transcription from the viral promoter, activates numerous cellular genes by interacting with NF-κB, CREB/ATF, SRF and NF-AT pathways. Tax-mediated NF-κB activation is crucial for transformation of T cells by HTLV-1.41 However, the presence of constitutively activated NF-κB pathway in ATLL cells, despite the lack of detectable Tax expression, suggests that Tax may be needed to initiate but not to maintain NF-κB activation. Although several studies have shown that Tax transactivates PTHrP, particularly the P3 promoter,42–44 it is not clear whether Tax is required for PTHrP expression because ATLL cells that do not express detectable Tax have high levels of PTHrP. In this study, we have compared a variety of cell lines with different levels of Tax expression to understand the role of Tax and NF-κB in the regulation of PTHrP. In vitro-transformed MT-2 and SLB-1 cells express high tax and low PTHrP. ATLL cells (RV-ATL and MET-1) express PTHrP in the absence of Tax. HT-1RV cells, obtained by superinfecting RV-ATL cells with HTLV-1, express very high levels of Tax and low PTHrP.45 Our newly developed in vitro immortalized cells (IMM-1) express low levels of Tax and moderate levels of PTHrP (data not shown). Our data are in agreement with previous studies where Tax-expressing T cells have a very low level of IκBα compared to ATLL cells (RV-ATL and MET-1) indicating that Tax significantly accelerated loss of I-κBα.
EMSA and competition analyses of the PTHrP κB1 sequence showed specific complexes formed between NF-κB transcription factors and P2 oligonucleotides in extracts from ATLL and the HTLV-1 cell lines tested. It is a well-known feature of NF-κB signaling that different NF-κB dimers can preferentially bind functionally distinct DNA-binding sites. Tax has been shown to activate the c-Rel gene and induce predominantly c-Rel-containing complexes.46 Consistent with these observations, the complex formed in extracts from SLB-1 and MT-2 cells, which express high levels of Tax, contained p50 and c-Rel as the major DNA-binding components. This finding was confirmed by the presence of a similar complex with a known κB containing probe (from MHC class I gene promoter) in these cells. Interestingly, there were two distinct complexes in the ATL, HT-1RV and surprisingly even in IMM-1 cells. The lower complexes (C1) in extracts from ATL, HT-1RV and IMM-1 cells were composed of p50 alone. The upper complex (C2) in extracts from in HT-1RV cells was composed of p50/c-Rel, confirming our previous observation that Tax induces the formation of p50/c-Rel-containing complexes. On the other hand, the upper complex in extracts from ATL and IMM-1 cells is intriguing since it did not supershift with antibodies against p50, p65, c-Rel, Bcl-3, RelB (data not shown) or p52 (data not shown). Our attempts to identify these proteins by proteomics and mass spectrometry have been unsuccessful; therefore, the proteins in this complex remain unidentified. ChIP assays confirmed binding of NF-κB p50 and c-Rel on the PTHrP P2 promoter in vivo. Finally, the differences in the complexes formed in these cells potentially explain the differences in the level of PTHrP P2 promoter transactivation in these cells.
Our transfection studies showed that the human PTHrP P2 promoter can be activated in a subunit-dependent manner by p50/p50 alone, or by a combination of p50/c-Rel or p50/Bcl-3, but not by p65 or p50/p65. Most of the transcriptional effect was attributable to an intact NF-κB site as demonstrated by a significant decrease in luciferase activity in 3T3 cells transfected with constructs bearing a disrupted κB sequence. We were surprised by the activation of PTHrP by p50/50 alone since p50 does not have a transactivation domain. There have been conflicting reports regarding the role of p50/p50 in transcription. While some studies support the ability of p50 to transactivate, particularly in the presence of Bcl-3, others have shown that p50 homodimers act as repressors by binding κB sites that would normally be activated by p50/p65 heterodimers. Mori et al.16 have shown that leukemic cells from ATL patients have abundant p50/p50 homodimers; however, p50 was not able to activate transcription from a p50/p65-inducible construct containing five repeats of a κB motif from the IL-2Rα gene. Interestingly, p50 homodimers are capable of binding all κB binding motifs with similar efficiency but can probably transactivate only a subset of these such as those found in the Bcl-2 and H-2K genes.47–49 It is possible that κB binding motifs like those found in Bcl-2, H-2K and PTHrP promoters may provide a sequence with which p50 forms an activating rather than a repressing complex. Evidence in support of this hypothesis was provided recently by Schreiber et al.50 showing that p50 homodimers can interact with the RNA polymerase II at the promoters for genes such as CSF2, ICAM1 and TNF and likely maintain basal level of target gene activity in unstimulated cells. Alternatively, transcriptional activity of p50 may be cell type-dependent or dependent on undefined cofactors.
Although p50 and p52 have no defined transactivating domains, the association with Bcl-3 can lead to the conversion of these homodimers to positive regulators of gene expression. In addition, it has been shown that the formation of Bcl-3–p50 homodimer–DNA complexes depends on Bcl-3 concentration and phosphorylation.51 Furthermore, it has been shown that Bcl-3 can increase the quantity of p50 homodimers that translocate to the nucleus.52 In light of the observations that p50 activation can be induced by Bcl-3, we tested the possible association of Bcl-3 with p50 in transactivating PTHrP. First, we studied the expression of Bcl-3 in HTLV-1-infected and ATLL cells. Interestingly, Tax-expressing cells have higher levels of Bcl-3 compared to ATLL cells. Our data from transfection experiments have shown that a higher p50/Bcl-3 ratio resulted in activation of the promoter, while a lower p50/Bcl-3 ratio (higher concentrations of Bcl-3) resulted in a dramatic reduction of transactivation. This correlated well with our data where ATLL cells having lower Bcl-3 levels expressed high levels of PTHrP and Tax-expressing cells that had higher Bcl-3 levels expressed low levels of PTHrP. These data suggest that Bcl-3 may mediate its effect on PTHrP indirectly or at higher concentrations Bcl-3 dissociates from complexes with p50. Based on the results from Bcl-3 expression analysis and transfection assays, it is likely that the low level of Bcl-3 expression in ATLL cells contributed to the transactivation of the promoter while the high levels of Bcl-3 in Tax-expressing cells inhibited the promoter. This may help explain the differences in the transactivation of the PTHrP P2 promoter in ATL and Tax-expressing T cells.
Bay 11-7082 has been shown to be a specific NF-κB inhibitor by inhibiting the phosphorylation of the inhibitor IκB.53 Treatment of HTLV-1-infected T cells not only reduced the P2 promoter-initiated transcripts but also total PTHrP expression providing additional support to our hypothesis that PTHrP is a direct target of NF-κB signaling. PS-341, a potent and selective proteasome inhibitor reduced NF-κB activity and total PTHrP expression and the P2 promoter-initiated transcripts in ATLL cells (data not shown). These results support a therapy for attempting to reduce NF-κB levels in ATLL patients.
In conclusion, our results support a direct role for NF-κB in the regulation of PTHrP. Since NF-κB is known to often mediate its effects through other transcription factors including AP-1, SP-1 and the Ets family of proteins and because binding sites for these factors and others exist within the P2 promoter, it will be important to establish the functional interaction between these factors and NF-κB in the regulation of PTHrP.
Acknowledgements
This work was supported by the National Cancer Institute (CA100730 and CA77911). MVPN was supported by the Glenn C Barber Fellowship from the College of Veterinary Medicine, The Ohio State University; TR and SS were supported by the National Center for Research Resources (RR00168) and the NCRR T32 (RR07073), respectively.
References
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broadus AE, Mangin M, Ikeda K, Insogna KL, Weir EC, Burtis WJ, et al. Humoral hypercalcemia of cancer. Identification of a novel parathyroid hormone-like peptide. N Engl J Med. 1988;319:556–563. doi: 10.1056/NEJM198809013190906. [DOI] [PubMed] [Google Scholar]
- 3.Grill V, Martin TJ. Hypercalcemia of malignancy. Rev Endocr Metab Disord. 2000;1:253–263. doi: 10.1023/a:1026597816193. [DOI] [PubMed] [Google Scholar]
- 4.Wysolmerski JJ, Stewart AF. The physiology of parathyroid hormone-related protein: an emerging role as a developmental factor. Annu Rev Physiol. 1998;60:431–460. doi: 10.1146/annurev.physiol.60.1.431. [DOI] [PubMed] [Google Scholar]
- 5.Mangin M, Ikeda K, Dreyer BE, Broadus AE. Identification of an upstream promoter of the human parathyroid hormone-related peptide gene. Mol Endocrinol. 1990;4:851–858. doi: 10.1210/mend-4-6-851. [DOI] [PubMed] [Google Scholar]
- 6.Campos RV, Wang C, Drucker DJ. Regulation of parathyroid hormone-related peptide (PTHrP) gene transcription. Mol Endocrinol. 1992;6:1642–1652. doi: 10.1210/mend.6.10.1280327. [DOI] [PubMed] [Google Scholar]
- 7.Suva LJ, Mather KA, Gillespie MT, Webb GC, Ng KW, Winslow GA, et al. Structure of the 5' flanking region of the gene encoding human parathyroid-hormone-related protein (PTHrP) Gene. 1989;77:95–105. doi: 10.1016/0378-1119(89)90363-6. [DOI] [PubMed] [Google Scholar]
- 8.Vasavada RC, Wysolmerski JJ, Broadus AE, Philbrick WM. Identification and characterization of a GC-rich promoter of the human parathyroid hormone-related peptide gene. Mol Endocrinol. 1993;7:273–282. doi: 10.1210/mend.7.2.8469240. [DOI] [PubMed] [Google Scholar]
- 9.Bouizar Z, Spyratos F, De Vernejoul MC. The parathyroid hormone-related protein (PTHrP) gene: use of downstream TATA promotor and PTHrP 1–139 coding pathways in primary breast cancers vary with the occurrence of bone metastasis. J Bone Miner Res. 1999;14:406–414. doi: 10.1359/jbmr.1999.14.3.406. [DOI] [PubMed] [Google Scholar]
- 10.Southby J, Murphy LM, Martin TJ, Gillespie MT. Cell-specific and regulator-induced promoter usage and messenger ribonucleic acid splicing for parathyroid hormone-related protein. Endocrinology. 1996;137:1349–1357. doi: 10.1210/endo.137.4.8625910. [DOI] [PubMed] [Google Scholar]
- 11.Richard V, Luchin A, Brena RM, Plass C, Rosol TJ. Quantitative evaluation of alternative promoter usage and 3' splice variants for parathyroid hormone-related protein by real-time reverse transcription-PCR. Clin Chem. 2003;49:1398–1402. doi: 10.1373/49.8.1398. [DOI] [PubMed] [Google Scholar]
- 12.Brandt DW, Bruns ME, Bruns DE, Ferguson JE, Burton DW, Deftos LJ. The parathyroid hormone-related protein (PTHrP) gene preferentially utilizes a GC-rich promoter and the PTHrP 1-139 coding pathway in normal human amnion. Biochem Biophys Res Commun. 1992;189:938–943. doi: 10.1016/0006-291x(92)92294-8. [DOI] [PubMed] [Google Scholar]
- 13.Good L, Maggirwar SB, Sun SC. Activation of the IL-2 gene promoter by HTLV-I tax involves induction of NF-AT complexes bound to the CD28-responsive element. EMBO J. 1996;15:3744–3750. [PMC free article] [PubMed] [Google Scholar]
- 14.Ballard DW, Bohnlein E, Lowenthal JW, Wano Y, Franza BR, Greene WC. HTLV-I tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science. 1988;241:1652–1655. doi: 10.1126/science.241.4873.1652. [DOI] [PubMed] [Google Scholar]
- 15.Duyao MP, Kessler DJ, Spicer DB, Sonenshein GE. Transactivation of the c-myc gene by HTLV-1 tax is mediated by NFkB. Curr Top Microbiol Immunol. 1992;182:421–424. doi: 10.1007/978-3-642-77633-5_53. [DOI] [PubMed] [Google Scholar]
- 16.Mori N, Fujii M, Ikeda S, Yamada Y, Tomonaga M, Ballard DW, et al. Constitutive activation of NF-kappaB in primary adult T-cell leukemia cells. Blood. 1999;93:2360–2368. [PubMed] [Google Scholar]
- 17.Hironaka N, Mochida K, Mori N, Maeda M, Yamamoto N, Yamaoka S. Tax-independent constitutive IkappaB kinase activation in adult T-cell leukemia cells. Neoplasia. 2004;6:266–278. doi: 10.1593/neo.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.May MJ, Ghosh S. Signal transduction through NF-kappa B. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 19.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 21.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, et al. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 22.Franzoso G, Bours V, Park S, Tomita-Yamaguchi M, Kelly K, Siebenlist U. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-kappa B-mediated inhibition. Nature. 1992;359:339–342. doi: 10.1038/359339a0. [DOI] [PubMed] [Google Scholar]
- 23.Bundy DL, McKeithan TW. Diverse effects of BCL3 phosphorylation on its modulation of NF-kappaB p52 homodimer binding to DNA. J Biol Chem. 1997;272:33132–33139. doi: 10.1074/jbc.272.52.33132. [DOI] [PubMed] [Google Scholar]
- 24.Caamano JH, Perez P, Lira SA, Bravo R. Constitutive expression of Bc1-3 in thymocytes increases the DNA binding of NF-kappaB1 (p50) homodimers in vivo. Mol Cell Biol. 1996;16:1342–1348. doi: 10.1128/mcb.16.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard V, Nadella MV, Green PL, Lairmore MD, Feuer G, Foley JG, et al. Transcriptional regulation of parathyroid hormone-related protein promoter P3 by ETS-1 in adult T-cell leukemia/lymphoma. Leukemia. 2005;19:1175–1183. doi: 10.1038/sj.leu.2403787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feuer G, Zack JA, Harrington WJ, Jr, Valderama R, Rosenblatt JD, Wachsman W, et al. Establishment of human T-cell leukemia virus type I T-cell lymphomas in severe combined immunodeficient mice. Blood. 1993;82:722–731. [PubMed] [Google Scholar]
- 27.Phillips KE, Herring B, Wilson LA, Rickford MS, Zhang M, Goldman CK, et al. IL-2Ralpha-Directed monoclonal antibodies provide effective therapy in a murine model of adult T-cell leukemia by a mechanism other than blockade of IL-2/IL-2Ralpha interaction. Cancer Res. 2000;60:6977–6984. [PubMed] [Google Scholar]
- 28.Stewart SA, Feuer G, Jewett A, Lee FV, Bonavida B, Chen IS. HTLV-1 gene expression in adult T-cell leukemia cells elicits an NK cell response in vitro and correlates with cell rejection in SCID mice. Virology. 1996;226:167–175. doi: 10.1006/viro.1996.0643. [DOI] [PubMed] [Google Scholar]
- 29.Mori N, Gill PS, Mougdil T, Murakami S, Eto S, Prager D. Interleukin-10 gene expression in adult T-cell leukemia. Blood. 1996;88:1035–1045. [PubMed] [Google Scholar]
- 30.Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS., Jr NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Dirksen WP, Mohamed SA, Fisher SA. Splicing of a myosin phosphatase targeting subunit 1 alternative exon is regulated by intronic cis-elements and a novel bipartite exonic enhancer/silencer element. J Biol Chem. 2003;278:9722–9732. doi: 10.1074/jbc.M207969200. [DOI] [PubMed] [Google Scholar]
- 33.Nishikori M, Maesako Y, Ueda C, Kurata M, Uchiyama T, Ohno H. High-level expression of BCL3 differentiates t(2;5)(p23;q35)-positive anaplastic large cell lymphoma from Hodgkin disease. Blood. 2003;101:2789–2796. doi: 10.1182/blood-2002-08-2464. [DOI] [PubMed] [Google Scholar]
- 34.Holz LE, Jakobsen KP, Van Snick J, Cormont F, Sewell WA. Dexamethasone inhibits IL-9 production by human T cells. J Inflamm (Lond) 2005;2:3. doi: 10.1186/1476-9255-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glasgow JN, Wood T, Perez-Polo JR. Identification and characterization of nuclear factor kappaB binding sites in the murine bcl-x promoter. J Neurochem. 2000;75:1377–1389. doi: 10.1046/j.1471-4159.2000.0751377.x. [DOI] [PubMed] [Google Scholar]
- 36.Jeang KT, Derse D, Matocha M, Sharma O. Expression status of Tax protein in human T-cell leukemia virus type 1-transformed MT4 cells: recall of MT4 cells distributed by the NIH AIDS Research and Reference Reagent Program. J Virol. 1997;71:6277–6278. doi: 10.1128/jvi.71.9.6277-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard V, Lairmore MD, Green PL, Feuer G, Erbe RS, Albrecht B, et al. Humoral hypercalcemia of malignancy: severe combined immunodeficient/beige mouse model of adult T-cell lymphoma independent of human T-cell lymphotropic virus type-1 tax expression. Am J Pathol. 2001;158:2219–2228. doi: 10.1016/S0002-9440(10)64694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard V, Luchin A, Brena RM, Plass C, Rosol TJ. Quantitative evaluation of alternative promoter usage and 3' splice variants for parathyroid hormone-related protein by real-time reverse transcription-PCR. Clin Chem. 2003;49:1398–1402. doi: 10.1373/49.8.1398. [DOI] [PubMed] [Google Scholar]
- 39.Vasavada RC, Wysolmerski JJ, Broadus AE, Philbrick WM. Identification and characterization of a GC-rich promoter of the human parathyroid hormone-related peptide gene. Mol Endocrinol. 1993;7:273–282. doi: 10.1210/mend.7.2.8469240. [DOI] [PubMed] [Google Scholar]
- 40.Peloponese JM, Yeung ML, Jeang KT. Modulation of nuclear factor-kappaB by human T cell leukemia virus type 1 Tax protein: implications for oncogenesis and inflammation. Immunol Res. 2006;34:1–12. [PubMed] [Google Scholar]
- 41.Sun SC, Yamaoka S. Activation of NF-kappaB by HTLV-I and implications for cell transformation. Oncogene. 2005;24:5952–5964. doi: 10.1038/sj.onc.1208969. [DOI] [PubMed] [Google Scholar]
- 42.Dittmer J, Pise-Masison CA, Clemens KE, Choi KS, Brady JN. Interaction of human T-cell lymphotropic virus type I Tax, Ets1, and Sp1 in transactivation of the PTHrP P2 promoter. J Biol Chem. 1997;272:4953–4958. doi: 10.1074/jbc.272.8.4953. [DOI] [PubMed] [Google Scholar]
- 43.Mori N, Ejima E, Prager D. Transactivation of parathyroid hormone-related protein gene expression by human T-cell leukemia virus type I tax. Eur J Haematol. 1996;56:116–117. doi: 10.1111/j.1600-0609.1996.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 44.Ikeda K, Inoue D, Okazaki R, Kikuchi T, Ogata E, Matsumoto T. Parathyroid hormone-related peptide in hypercalcemia associated with adult T cell leukemia/lymphoma: molecular and cellular mechanism of parathyroid hormone-related peptide overexpression in HTLV-I-infected T cells. Miner Electrolyte Metab. 1995;21:166–170. [PubMed] [Google Scholar]
- 45.Richard V, Nadella MV, Green PL, Lairmore MD, Feuer G, Foley JG, et al. Transcriptional regulation of parathyroid hormone-related protein promoter P3 by ETS-1 in adult T-cell leukemia/lymphoma. Leukemia. 2005;19:1175–1183. doi: 10.1038/sj.leu.2403787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li CC, Ruscetti FW, Rice NR, Chen E, Yang NS, Mikovits J, et al. Differential expression of Rel family members in human T-cell leukemia virus type I-infected cells: transcriptional activation of c-rel by Tax protein. J Virol. 1993;67:4205–4213. doi: 10.1128/jvi.67.7.4205-4213.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurland JF, Kodym R, Story MD, Spurgers KB, McDonnell TJ, Meyn RE. NF-kappaB1 (p50) homodimers contribute to transcription of the bcl-2 oncogene. J Biol Chem. 2001;276:45380–45386. doi: 10.1074/jbc.M108294200. [DOI] [PubMed] [Google Scholar]
- 48.Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 1993;7:1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- 49.Lin R, Gewert D, Hiscott J. Differential transcriptional activation in vitro by NF-kappa B/Rel proteins. J Biol Chem. 1995;270:3123–3131. doi: 10.1074/jbc.270.7.3123. [DOI] [PubMed] [Google Scholar]
- 50.Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci USA. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bundy DL, McKeithan TW. Diverse effects of BCL3 phosphorylation on its modulation of NF-kappaB p52 homodimer binding to DNA. J Biol Chem. 1997;272:33132–33139. doi: 10.1074/jbc.272.52.33132. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe N, Iwamura T, Shinoda T, Fujita T. Regulation of NFKB1 proteins by the candidate oncoprotein BCL-3: generation of NF-kappaB homodimers from the cytoplasmic pool of p50-p105 and nuclear translocation. EMBO J. 1997;16:3609–3620. doi: 10.1093/emboj/16.12.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori N, Yamada Y, Ikeda S, Yamasaki Y, Tsukasaki K, Tanaka Y, et al. Bay 11-7082 inhibits transcription factor NF-kappaB and induces apoptosis of HTLV-I-infected T-cell lines and primary adult T-cell leukemia cells. Blood. 2002;100:1828–1834. doi: 10.1182/blood-2002-01-0151. [DOI] [PubMed] [Google Scholar]