Abstract
Exosomes are naturally occurring nanovesicles that can be tailored to display a broad range of drug targets, including G protein-coupled receptors. Such vesicles provide a new source of complex membrane proteins that are maintained in their native conformation. Given the difficulties to isolate receptors for drug target validation and discovery, receptor presentation on exosome emerges as a promising new tool for drug screening. The potential of this technology is illustrated here with recombinant exosomes presenting the somatostatin receptor 2 as an example. The receptor-containing vesicles were identified as exosomes since they also bear Lactadherin, a hallmark of exosome nanovesicles. The amount of somatostatin receptor 2 on exosomes was similar to the amount of the most abundant known exosome membrane proteins. The receptor was functional and similar in size to the form found on cell surface. Finally, recombinant exosomes were used in several assay formats that exemplify their capacity as a new receptor presentation platform for drug discovery. These include the induction and detection of antibody as well as screening of antibody repertoires without the need to purify membrane proteins.
Keywords: nanovesicles, exosome, GPCR, drug screening, antibody induction
Introduction
G protein-coupled receptors (GPCRs) constitute the largest family of receptors in the human genome (Pierce et al 2002). A large proportion of drugs currently on the market targets members of this family of receptors (Hopkins and Groom 2002; Klabunbe and Hessler 2002). Not surprisingly, this pharmacologically important group is the subject of continued interest with two thirds of drugs in development involving direct binding to GPCRs or interfering with GPCR-coupled pathways (Med Ad. News 2004). GPCRs share several common features, including the coupling of their signal transduction via G proteins and a structure with seven transmembrane domains (Gether 2000; Jacoby et al 2006; Lundstrom 2006). The latter renders this family of receptors difficult to produce and purify in large quantities (Helenius and Simons 1975; Sarramegna et al 2003; Lundstrom 2006). Previous drug development focused primarily on a limited number of GPCR family members on a gene by gene-based approach as new genes were identified and their product successfully isolated. The growing size and number of library of compounds available for drug screening and the recent identification of several hundreds of new GPCR sequences by genome sequencing has recently transformed the field of GPCR-targeted drug discovery (Pierce et al 2002). A large number of these new receptors are orphan receptors for which ligands are still unknown. Reagents such as monoclonal antibodies are needed to characterize and validate novel GPCRs as potential drug targets. There is also considerable interest in developing technologies and tools that facilitate the preparation and isolation of GPCR for high throughput screening of small molecule libraries (Lundstrom 2006). Ideally, such technologies should enable isolating material without a denaturing step such as solubilization with detergent. They should maintain GPCR in their native conformation and environment, ie, in a lipid bilayer structure and preferably derived from cells and tissues in which GPCRs are potentially pharmacologically relevant. Finally, these technologies should yield homogenous preparations and be applicable to many members of the GPCR family. In this regard, we have recently reported on a technology called Exosome Display that enables the presentation of soluble proteins, extracellular domains of receptors, as well as full-length membrane proteins on naturally occurring exosome nanovesicles (Delcayre, Estelles, et al 2005). This technology is amenable to drug screening studies while alleviating the drawbacks of existing approaches used to isolate membrane proteins.
Exosome nanovesicles of 50–100 nm are formed in intracellular vesicular bodies of most cells and released in the extracellular milieu following fusion of the vesicular body and plasma membranes (Johnstone 1992; Denzer et al 2000; Thery et al 2002). Exosome Display relates to two modes of protein transfer to exosomes (Delcayre, Estelles et al 2005). The first mode of transfer applies to soluble proteins and extracellular domains of receptors. It involves the C1C2 domain of Lactadherin which mediates the specific targeting of fusion proteins to the exosome compartment. The second mode applies to full-length membrane proteins, including GPCRs. Unexpectedly, Exosome Display of receptors does not require any sequence modification; however, the subcellular distribution of unmodified membrane proteins is not restricted to the exosomal vesicles since full-length receptors are also found on the plasma membrane. The mechanism of membrane protein trafficking to exosomes is unclear. We have found that overexpression of membrane proteins results in the distribution of a significant number of receptor molecules per exosomes, even if they do not occur there naturally (Delcayre, Estelles et al 2005). Exosomes, notably dendritic cell-derived exosomes (Dexo-somes), have drawn considerable interest because of their immunological properties (Zitvogel et al 1998; Thery et al 1999, 2002; Lamparski et al 2002; Vincent-Scheinder et al 2002; Andre et al 2004). Their studies culminated with the evaluation of patient-derived Dexosomes for the treatment of cancer (Delcayre, Shu et al 2005). Two Phase I clinical trials of autologous Dexosomes therapy for non-small cell lung (NSCL) and melanoma cancer patients, respectively, were completed that established the feasibility and safety of this approach (Morse et al 2005; Escudier et al 2005). As new exosome properties and technologies unveil, a growing number of possible applications is emerging in the fields of vaccine (Delcayre and Le Pecq 2006), autoimmune diseases (Abusamra et al 2005; Kim et al 2005; Taylor et al 2006), and transplantation (Morelli 2006; Peche et al 2006). Here, the potential of Exosome Display as a promising receptor presentation platform for GPCR-targeted drug discovery was evaluated. The technology was applied to the generation and screening of antibodies in various assay formats using the somatostatin receptor 2 (SSTR-2) as an example.
Material and methods
Material
All cell lines were purchased from ATCC except D2SC-1 and Phoenix (E and A) cells that were kindly provided by Dr. Riccardo and Dr. Nolan (Stanford University, Palo Alto, CA), respectively. Human Embryonic Kidney 293-F cells were maintained in 293-SFM II medium (Invitrogen, San Diego, CA) supplemented with fetal bovine serum (FBS; 2%) except for exosome production where cells were transferred to a chemically defined CD293 medium (Invitrogen). Mouse A20 and YAC cells were maintained in ADCF medium (HyClone, Logan, UT) supplemented with FBS (2%). For exosome production, cells were transferred to ADCF medium without serum. Mouse D2SC-1 and EL4 cells were maintained in AIMV (Invitrogen) supplemented with FBS (2%). Again, for exosome production, cells were transferred to protein free-AIMV medium. Phoenix cells were grown in DMEM-high glucose supplemented with 10% FBS. Commercially available antibodies were purchased from Pharmingen, San Diego (PE-conjugated anti-CD19 antibody; PE-conjugated anti-rat Ig antibody; Alexa-conjugated Streptavidin), and Roche, Palo Alto (Rat anti-HA tag antibody; Horse Radish Peroxidase-conjugated anti-HA tag antibody). The polyclonal anti-Lactadherin antibody was a gift from Dr. Sebastian Amigorena (Institut Curie, Paris, France). Biotinyl Somatostatin-28 was purchased from Bachem Bioscience (King of Prussia, PA).
Cloning of SSTR-2 and preparation of recombinant cell lines
HA-tagged SSTR-2 was prepared using a previously reported method (Koller et al 1997). Briefly, SSTR-2 cDNA was prepared by RT-PCR of human brain RNA (Clonetech, Palo Alto, CA) using primers designed for cloning into HApC3.1, as previously described (Delcayre, Estelles, et al 2005). HA-SSTR-2 insert was then subcloned into the retroviral shuttle vector pBABE MN IRES GFP to produce recombinant retrovirus as previously reported (Swift et al 1999). For this, Phoenix cells (2 × 106) were transfected with 3 μg pBABE MN-HASSTR2-IRES GFP using Fugene 6 (Roche, Palo Alto, CA) according to the manufacturer’s recommendations. Forty eight hours post-transfection, culture supernatants were collected, filtered through 0.45 μm-filters and used to infect 5 × 105 target cells. Forty eight hours post-infection, cells were immunostained using a purified rat anti-HA antibody followed by anti-rat antibody coupled to PhycoErythrin (PE). Cells were isolated by fluorescent-activated sorting (FACS) and expanded. Viral particles produced by Phoenix E cells were used to transduce mouse cell lines, while 293-F cells were transduced with viral particles obtained from Phoenix A cells.
Exosome preparation
Cells were transferred into protein-free medium as described above and exosomes were produced during 5-day cultures. Exosomes were prepared as previously described (Lamparski et al 2002) with some modifications (Delcayre, Estelles, et al 2005). Briefly, the culture supernatant was cleared from cell debris and large vesicles by centrifugation at 2,000 rpm for 10 minutes followed by 0.8 and 0.2 μm filtrations. Supernatant concentration and buffer-exchange to PBS was performed by ultrafiltration/diafiltration using a fiber cartridge with a 500 kD cut-off. The concentrated fraction was subjected to ultracentrifugation on a 30%-sucrose cushion in PBS/D2O and exosomes were recovered by collecting the sucrose-containing layer. The exosome fraction was further concentrated and diafiltered to yield a 500X-concentrated solution of purified exosomes. Exosomes were aliquoted and stored at −80 °C until use. Protein content of purified exosomes was determined using a Bradford assay (Pierce, Rockford, IL). Exosomes prepared following this method appear as a homogeneous population of vesicles 50–100 nm in diameter when evaluated by electron microscopy (Lamparski et al 2002).
Cross-capture and adsorption ELISA
ELISA was performed as previously described (Lamparski et al 2002) with some modifications (Delcayre, Estelles, et al 2005). Briefly, for adsorption ELISA, exosomes were directly coated to the wells of microtitration plates placed overnight at 37 °C. The plate was washed and blocked in PBS/0.05% Tween 20 and incubated for one hour at room temperature with anti-HA antibody coupled to HRP. Bound antibodies were detected using an ECL Western blot detection reagent (Amersham Bioscience, Piscataway, NJ, USA) and measuring light emission on a Wallac Microbeta 1450 Trilux plate reader (PerkinElmer Life Sciences Inc., Boston, MA, USA). For capture ELISA, the wells of microtitration plates were coated with monoclonal anti-CD81 or anti-HA antibodies (2 μg/mL). Following PBS washing and PBS/0.05% Tween 20 blocking steps, serial dilutions of exosomes were added to each well and incubated overnight at room temperature. After washing, bound exosomes were detected with either biotinylated anti-CD81 antibody followed with streptavidin-HRP conjugate, anti-HA antibody conjugated to HRP, or polyclonal anti-Lactadherin antibody followed with anti-rabbit antibodies coupled to HRP (Jackson ImmunoResearch Laboratories, West Grove, PA). ELISA measurement was performed as above using ECL Western blot detection reagent.
Western blot analysis of exosomes and membrane fractions
Exosomes were prepared as described above. Crude plasma membranes were prepared by homogenizing cells resuspended in 10 mM Tris/HCl pH7.6, 2mM MgCl2, and a cocktail of protease inhibitors (Roche, Palo Alto, CA), using a glass homogenizer. Homogenized cells were spun at 500 g for 5 minutes at 4 °C and the supernatant was retained. The procedure was repeated with the pellet and both supernatants were pooled and spun at 10,000 g for 30 minutes. The pellet was resuspended in 50 mM Tris/HCl, pH7.6, 7mM MgCl2, protease inhibitors, and 2mg/ml DβM-n-Dodecyl-β-maltoside. The resulting solution was retained as the plasma membrane fraction. For Western blot analysis, fifty μg of membrane fraction and 5 μg of purified exosomes were diluted 1:1 in 2X SDS-PAGE sample buffer and heat-denatured for 5 min. Following electrophoresis, samples were transferred to PVDF membrane that were then blocked by incubation into PBS/0.05% Tween 20/6% non-fat milk for 1 hour at RT. The presence of an HA tag on the membrane was evaluated by probing with an anti-HA antibody conjugated to HRP. Bound antibodies were then detected using CN/DAB chromogenic substrate (Pierce, Rockford, IL).
SSTR-2 ligand binding assay
SSTR-2/YAC cells were washed with PBS and resuspended in FACS staining buffer (PBS-4% FBS) containing different amounts of Biotinyl-Somatostatin-28 (Bachem Bioscience, King of Prussia, PA). Following a 1-hour incubation at 4 °C, excess ligand was removed by washing cells in staining buffer. Bound ligand was detected by flow cytometry after incubation of the cells with Streptavidin coupled to PE. For competition assay, incremental amounts of exosomes were added to cells during incubation with 7.5 nM SSTR-2 ligand. The ligand concentration was determined empirically based on the concentration range that yielded binding within the linear trendline of the assay.
Induction and analysis of anti-SSTR-2 antibody response
Mice were immunized with recombinant SSTR-2/exosomes by subcutaneous injections in both footpads. Mice received 5 μg of SSTR-2/YAC exosomes in PBS in each footpad and immunizations were performed at 1-week intervals for the first two injections and at two week intervals thereafter. Induction of anti-SSTR-2 antibodies was evaluated by adsorption ELISA as described above using serum from blood samples collected 1 week following the second injection. The detection of anti-SSTR-2 antibody-producing cells was performed by flow cytometry using popliteal lymph nodes and spleen cells of mice one, two, or five days following the last immunization. For this analysis, single cell suspensions (5 × 105 cells) were preincubated for 15 min. on ice with 25 μg of parental 293 exosomes. Biotinylated recombinant SSTR-2/293 exosomes (1 μg) prepared as previously described (10) were then added to the samples and incubation continued for 45 min. Cells bearing exosomes and CD19+-cells were detected by staining with Streptavidin-Alexa and anti-CD19-PE antibody conjugates, respectively, followed by FACS analysis.
Results
Recombinant cells release exosomes containing the somatostatin receptor, SSTR-2
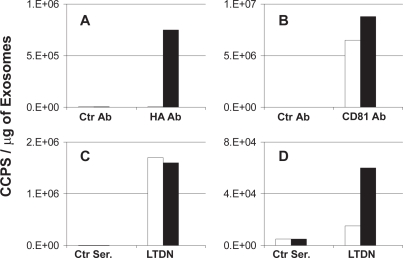
The expression of GPCRs on vesicles released in the supernatant of recombinant cells was previously examined by cross-capture ELISA with anti CD81 and anti-HA antibodies (Delcayre, Estelles, et al 2005). To fully identify these vesicles as exosomes, assays were repeated using antibodies against Lactadherin, a hallmark protein of exosomes (Delcayre and Le Pecq 2006), and SSTR-2 as a model for GPCR. Recombinant SSTR-2/293 cells were cultured in protein-free medium and exosomes released in the extracellular medium were purified as described in Material and Methods. The display of recombinant SSTR-2 on 293 exosomes was demonstrated in cross-capture ELISA measuring the physical association between CD81, the HA-tag of SSTR-2 and Lactadherin. As shown in Figure 1A, specific binding of anti-HA antibody to vesicles captured with an anti-CD81 antibody was detected when using exosomes produced by recombinant SSTR-2 cells. No signal was detected when using the same antibody combination with exosomes from parental cells. In contrast, identical signals were obtained when detecting CD81 on parental and recombinant exosomes supporting that exosomes from both recombinant and parental cells contained similar amounts of CD81 molecules (Figure 1B). The identification of vesicles as true exosomes was then established in cross-capture assays using an anti-lactadherin antibody for detection. Indeed, unlike CD81 that can be found in different subcellular compartments, Lactadherin is found exclusively on exosomes but not on the surface of the cells used in this assay (Delcayre, Estelles, et al 2005). As shown in Figure 1C, similar amounts of Lactadherin were detected on both parental and recombinant exosomes captured via their CD81. In contrast, specific binding of the anti-Lactadherin antibody to vesicles captured with the anti-HA antibody was detected when using exosomes produced by recombinant SSTR-2 cells only (Figure 1D). Background signal was detected when using the same antibody combination with exosomes from parental cells. Overall, this data indicate that recombinant SSTR-2 is released in a particulate form in association with other membrane proteins like CD81. The CD81 and GPCR-containing vesicles are exosomes since they also contain Lactadherin, which is a specific marker of exosomes produced by these cells.
Figure 1.
Cross-capture ELISA detecting SSTR-2 on exosomes. The wells of microtitration plates were coated with either monoclonal anti-CD81 antibody (panel A, B and C) or monoclonal anti-HA antibody (panel D). Exosomes from 293 cells (empty bars) and from recombinant SSTR-2/293 cells (black bars) were added to the wells and antigens on captured exosomes were detected with (A) Monoclonal anti-HA antibody (HA Ab) and control isotype-matching antibody (Ctr Ab), (B) monoclonal anti-CD81 antibody (CD81 Ab) and control isotype-matching antibody (Ctr Ab) and (C) and (D) polyclonal anti-Lactadherin antibody (LTDN) and control pre-immune serum (Ctr Ser.) Bound antibodies were detected with species-specific antibodies conjugated to HRP. Cross-capture of serial dilution of exosomes in duplicate was performed for each antibody pairs. Data represent the slope of the linear trendline obtained for each exosome serial dilutions used.
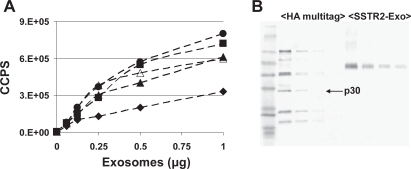
To verify that release of GPCR-containing exosomes by recombinant 293 cells is a general phenomenon, recombinant cell lines of various types were generated. For each cell line tested, infection was performed with the same retrovirus and all cell lines were placed in appropriate protein-free media during the exosome production phase. As shown in Figure 2A, all recombinant cell lines tested released exosomes containing HA-tagged SSTR-2. Exosomes from the recombinant mouse cell line A20, EL4 and YAC displayed similar amounts of SSTR-2 per microgram of vesicles as recombinant 293 cell-derived exosomes. Lower amounts were detected in D2SC-1-derived exosomes. No HA-tagged component was revealed in exosomes from parental cell lines. The number of GPCR per exosome was estimated by Western blot analysis using a HA multitag quantization marker (Figure 2B). According to the manufacturer’s description and the exosome amounts used for the western blot, anti-HA tag antibody detected approximately 1 to 3 ng of HA tag per μg of exosomes. Based on the molecular weight of SSTR-2 and previous quantization of Dexosome markers, it can be estimated that there are 10 to 30 molecules of SSTR-2 per exosome. This estimate of multiple copies of SSTR2 per exosomes was corroborated by a different quantization method using exosome-coated beads analyzed by FACS. This method measures the amount of markers on exosome-coated beads using a calibration bead kit for marker-specific antibodies (Quantum Simply Cellular kit; Bangs Laboratories, Fishers, IN). The number of exosomes per bead is estimated as previously described using an occupancy prediction of 50% (Feder 1980). Such occupancy rate was corroborated by electronic microscopy analysis of exosome-coated beads (Clayton et al 2001). This approach revealed values of 5 to 10 copies of SSTR-2 per exosome. As a comparison, the most abundant marker detected on Dexosomes is MHC-Class II which represents approximately 3% to 6% of the exosome surface proteins. Quantization by flow cytometry of exosome-coated beads revealed values of 30–60 molecules of MHC class II per Dexosome (Roulon et al in preparation). Hence, the number of SSTR-2 molecules on exosomes is significant and near that of the most abundant known Dexosome markers.
Figure 2.
Detection of SSTR-2 on recombinant exosomes from diverse origins. (A) The wells of microtitration plates were coated with incrementing amounts of exosomes derived from recombinant A20 (filled triangles), D2SC-1 (filled diamonds), YAC (filled circles), EL4 (open triangles) and 293 (filled squares) cells expressing SSTR-2. A monoclonal anti-HA antibody conjugated to HRP was used to detect SSTR-2. Each data point represents the mean of duplicates. Background counts below 1.E4 were detected when wells were coated with exosomes from parental cell lines (data not shown). (B) Western blot analysis of HA multitag markers (4, 2, 1 and 0.5 ng of p30) and recombinant SSTR-2/YAC exosomes (5, 2.5, 1.25, 0.625 μg exosomes). HA-containing proteins were detected with a monoclonal anti-HA antibody conjugated to HRP and a chromogenic substrate.
Recombinant exosomes contain the mature and functional form of SSTR-2
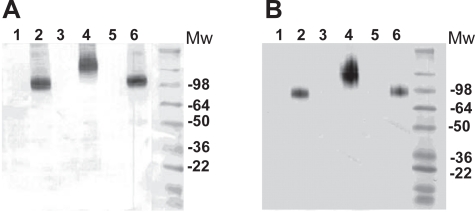
To further characterize SSTR-2 on exosomes, the biochemical properties of SSTR-2 found on exosomes and plasma membranes from different cell lines were analyzed by electrophoresis in reducing and denaturing conditions. As shown in Figure 3A, the electrophoretic migration of SSTR-2 from recombinant A20 and YAC cells was similar, whereas a larger size protein was detected in exosomes from recombinant EL4 cells. These apparent molecular weight variations reflect more likely differences in post-translational modification or glycosylation of SSTR-2 in the recombinant cell lines tested. Remarkably, the same electrophoretic profile was detected when analyzing plasma membrane fractions. Figure 3B shows that like for exosomes, SSTR-2 in plasma membrane from recombinant EL4 was larger in size than SSTR-2 on plasma membrane of both recombinant A20 and YAC cells. However, for each cell line, the same protein size was detected in both subcellular compartments. Similar data was observed in 293 cells (data not shown). These findings support that regardless of the glycosylation state provided by the cells used to express recombinant SSTR-2, exosomes more likely contain the mature form of the GPCR that is also found at the cell surface. It should be noted that the size of proteins detected by Western blot and shown in Figure 3 is >2-fold higher than the expected size of SSTR-2. It is therefore likely that dimeric forms of SSTR-2 were detected in this experiment.
Figure 3.
SDS-PAGE analysis of recombinant SSTR-2 expressed by various cell lines. SSTR-2 in exosomes (panel A) and plasma membranes (panel B) were analyzed by SDS-PAGE followed by Western blot analysis using a monoclonal anti-HA antibody conjugated to HRP and chromogenic substrate. Material loaded on the gel was derived from parental and recombinant YAC cells (lane 1 and 2), EL4 cells (lane 3 and 4) and A20 cells (lane 5 and 6).
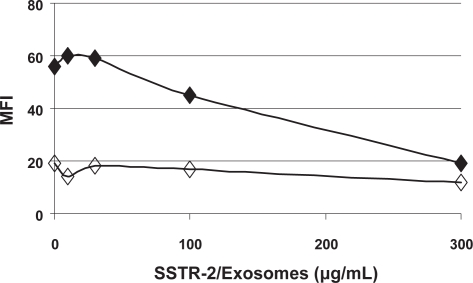
We then evaluated whether SSTR-2 on exosomes can bind its native ligand, somatostatin. An assay was performed to measure the binding of biotynilated somatostatin-28 (Biot-SST) to recombinant YAC cells expressing SSTR-2. Bound Biot-SST was detected by FACS analysis of YAC cells stained with Streptavidin-Phycoerithrein conjugate. The concentration of Biot-SST for which linear dose-dependent binding was reached was determined empirically and the assay was repeated in the presence of recombinant exosomes expressing SSTR-2. As shown in Figure 4, Biot-SST at 7.5 nM resulted in a specific staining of recombinant YAC cells with an MFI of ~50. Decreased staining was observed when 100 micrograms of recombinant exosomes expressing SSTR-2 was added to the sample. Binding of Biot-SST to YAC cells was at background level in the presence of 300 micrograms of exosomes. No significant decrease of Biot-SST binding was detected when using parental exosomes (data not shown). These data demonstrate that SSTR-2 on exosomes can compete with SSTR-2 at the cell surface for its binding to Somatostatin. Hence, mature SSTR-2 on exosomes is also in a native conformation enabling ligand binding.
Figure 4.
Binding of Biotinylated SST-28 to recombinant SSTR-2/YAC cells. SSTR-2/YAC cells were incubated with (filled diamonds) or without (empty diamonds) biotinylated SST-28 in the presence of incremental amounts of SSTR-2/YAC exosomes. Cells stained with Streptavidin-PE were analyzed by FACS. The assay was repeated successfully twice and data of one representative experiment are shown.
Recombinant exosomes are suitable immunogens for the induction of anti-GPCR antibodies
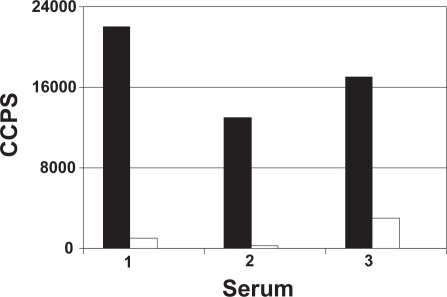
The potency of recombinant exosomes as a novel immunogen platform for the induction of anti-GPCR antibodies was evaluated in mice. In order to minimize anti-exosome responses, the experiment was performed in a syngeneic environment where exosome-producing cell line and animals have the same genetic haplotype. To fully appreciate the potency of the approach, mice were immunized with low amounts of SSTR-2. No adjuvant other than the exosome vesicle itself was added and antibody induction was evaluated after two immunizations only. Mice were treated twice at one week intervals with 10 μg of SSTR-2/YAC exosomes. Based on the estimated number of SSTR-2 molecules per exosome, approximately 100 to 200 ng of SSTR-2 was injected at each treatment in the mice receiving recombinant exosomes. Antibody induction was detected using an ELISA in which exosomes were directly adsorbed to the wells of a microtitration plate. To avoid the detection of non-specific anti-YAC exosome antibodies in the serum of immunized mice, the ELISA was performed using recombinant and parental exosomes from a different cell type. As shown in Figure 5, a specific signal was detected when the serum of mice immunized with recombinant YAC exosomes were applied to the wells coated with SSTR-2/293 exosomes but not parental 293 exosomes. Although the detection of anti-SSTR-2 antibody induction required low serum dilutions, a specific response could be detected in all three mice tested. The modest but specific antibody titer detected is more likely due to the immunization regimen used where antibody induction was evaluated upon exposure to low amounts of SSTR-2. Remarkably, detectable antibody induction was achieved using only 1%–2% of a standard exosome preparation from the supernatant of a 1-Liter culture.
Figure 5.
Detection of anti-SSTR-2 antibody in serum of mice immunized with SSTR-2/YAC exosomes. Serum of three immunized mice was added to the wells of microtitration plate coated with exosomes from parental (empty bars) or recombinant SSTR-2 293 cells. Bound antibodies were detected with an anti-mouse IgG antibody conjugated to HRP and chemiluminescent substrate. Data represent the mean of duplicate wells. Background signal obtained with serum of non-immunized mice was subtracted.
Recombinant exosomes are suitable for the screening of GPCR-binding compounds
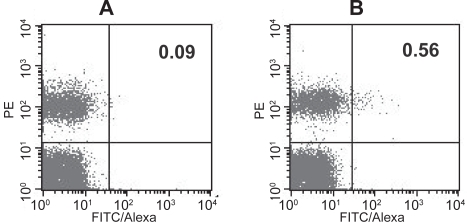
The possibility of using recombinant exosomes for the screening of GPCR-binding compounds was first illustrated above using an ELISA-based screening assay. The versatility of exosomes as a source of material for screening repertoires of GPCR-binding entities was further demonstrated in a different assay format identifying cells that express antibodies reacting with GPCRs. For this approach, the immunization regimen described above was extended with several additional boosts. Local lymph nodes were isolated on the day following the last injection. Single cell suspensions were prepared and incubated with biotinylated 293 exosomes containing SSTR-2 in the presence of an excess of parental 293 exosomes to saturate putative non-specific interactions. A PE-conjugated anti-CD19 antibody was also added to the cell suspension to identify B-cells and cells reacting with biotinylated exosomes were detected with Streptavidin-Alexa conjugates. As shown in Figure 6, 0.56% of CD19-positive cells from lymph nodes of immunized mice reacted with SSTR2-containing exosomes whereas CD19-negative cells from the same organ did not. In contrast, background level (0.09%) of double positive cells was detected when the assay was performed using lymph nodes of naïve mice. There was also no significant difference between cell populations from naïve and immunized mice when biotinylated parental 293 exosomes were used in the assay (data not shown). Hence, the specific staining of CD19-positive cells reflects more likely the interaction of SSTR-2 with antibodies at the surface of cells induced upon immunization. These data demonstrate that recombinant exosomes could be used to identify and isolate antigen-specific antibody-producing cells.
Figure 6.
Detection of anti-SSTR-2 antibody producing B-cells in Lymph nodes of mice immunized with SSTR-2/YAC exosomes. Lymph node cells from naïve mice (A) and mice immunized with SSTR-2/YAC exosomes (B) were incubated with biotinylated SSTR-2/293 exosomes and a 25X excess of parental 293 exosomes. Cells were then stained with Streptavidin-Alexa and anti-CD19-PE antibody conjugates and analyzed by FACS. Numbers in the upper right quadrants indicate the percent of Alexa/PE double positive cells. The assay was repeated successfully multiple times and data of one representative experiment are shown.
Discussion
Exosomes are naturally occurring nanovesicles with a broad spectrum of biological activities in line with their protein compositions. Although the antigen presentation and immune modulation properties of these vesicles have attracted a lot of interest for their use in cancer therapy (Thery et al 2002; Delcayre, Shu et al 2005), a growing field of biotechnology applications is emerging as new discoveries unfold (Delcayre and Le Pecq 2006). Notably, Exosome Display is a novel technology enabling to manipulate the protein content of exosomes that opens many possibilities. These include applications in the field of drug target validation and screening of libraries of compounds binding to drug targets. As an example, we used SSTR-2 to characterize the biochemical properties of the receptor released with exosomes and show that the latter can be utilized in various assay formats for drug discovery.
The presence of particle-bound SSTR-2 in the culture supernatant of recombinant cells was established as previously described for CXCR4 and CCR7 (Delcayre, Estelles, et al 2005), by cross capture ELISA linking SSTR-2 to CD81-containing vesicles. The vesicles were further iden-tified here as exosomes in cross-capture ELISA linking SSTR-2 to Lactadherin, a hallmark of exosomes. Indeed, Lactadherin produced by cells other than mammary cells has been shown to be released in association with exosomes (Oshima et al 2002; Miyasaka et al 2004; Veron et al 2005; Delcayre and Le Pecq 2006). Moreover, Lactadherin is not found on the surface of healthy cells while many membrane proteins found on the cell surface are not present on Lactadherin-containing vesicles (Delcayre and Le Pecq 2006). This further distinguishes Lactadherin-containing vesicles as exosomes which originate in an intracellular compartment from other vesicles budding from the cell surface which could also contain GPCRs.
Exosome Display of SSTR-2 was achieved on exosomes derived from various cell types and resulted in the presentation of 5–30 molecules per vesicles. This figure is remarkably high and is similar to the number previously established for the most abundant proteins present in dendritic cell-derived exosomes. Hence, the detection of GPCRs in exosomes is specific and unlikely due to contamination by other vesicles. The latter generally accounts for a fraction of the preparation and component quantity much below 1 molecule per vesicle. So far, the specific detection of significant amounts of recombinant GPCRs on exosomes has been successfully attempted with five GPCR models, including SSTR-2, CCR7, CXCR4, CD97 and CCR5. Surprisingly, trafficking of these receptors to the exosome compartment did not require any sequence modification. This is in contrast with Exosome Display of soluble antigen and extracellular domains of membrane proteins which is achieved via specific targeting of chimeric proteins fused to an exosome localization domain (Delcayre, Estelles et al 2005). The mechanism leading to the GPCR distribution in exosomes is unknown. It is non-exclusive in that the receptors were also found in the plasma membrane. In addition, the level of GPCR expression in the vesicles varied with the model used and was proportional to the overall level of GPCR expression in recombinant cells (data not shown).
An important feature of SSTR-2 on exosomes is its similarity with the receptor form found on the cell surface. As expected, we detected different forms of SSTR-2 produced by various cell lines that most likely result from cell-specific post-translational modification. In the cell lines tested here, the form of SSTR-2 found on exosomes mirrored that found on the surface of the matching exosome-producing cell line. This indicates that the fully post-translationally modified form of the receptor traffics to the exosome compartment for release. Finally, we showed that SSTR-2 on exosomes is functional in its ability to bind to its native ligands. This was demonstrated in competition experiment where SSTR-2-containing exosomes could block binding of biotinylated SST-28 to its receptor on SSTR-2-expressing cells. Further experiments will be required to determine whether the affinity of SSTR-2 ligand for its receptor is similar when the latter is presented on exosomes or on cell surfaces. However, this data indicates that SSTR-2 on exosomes is most likely properly folded and oriented in the lipid bilayer to allow for binding to ligands.
Recombinant exosomes represent an ideal source of material where receptors are in their native conformation. A robust and reliable procedure has been established to reproducibly isolate and characterize exosomes (Le Pecq 2005; Patel et al 2005). The same procedure can be performed regardless of the nature of the recombinant receptor expressed on exosomes and of the cell type used to produce exosomes. The purification process used is devoid of any denaturing steps and the receptors are maintained in their native conformation throughout. The resulting material is homogenous as compared to membrane preparation. Indeed, the latter may contain large amounts of endogenous GPCRs which could temper with assays. As for membrane preparation, an added advantage of preparing recombinant exosomes as a source of GPCR material is that it provides the flexibility to isolate GPCRs produced by relevant cells, ie, where the receptor occurs naturally. In contrast, the preparation of recombinant GPCRs in large amount requires using specific cell expression systems with unique post-translational modification pathways. The use of such heterologous systems may therefore result in the production of a different form of the receptor than the one desired for drug discovery studies (Lundstrom 2005). Also, GPCRs on exosomes are physically linked to a particle which can be labeled via diverse standard methods targeting specific proteins or lipids. Hence, easy detection of particles can be achieved without modifying the GPCR itself, which alleviates the risk of encroaching on receptor properties.
The potential of recombinant exosomes as a novel GPCR platform for drug discoveries was illustrated with the induction of anti-SSTR-2 antibodies and screening of anti-SSTR-2 antibody repertoire using SSTR-2 exosomes as sole source of GPCRs. The induction of anti-SSTR-2 antibodies in animals was detected following two injections only. The immunogen used comprised recombinant exosomes derived from syngeneic mouse cells and resuspended in PBS with no added adjuvant. Based on the estimated number of SSTR-2 molecules per vesicle, the immunogen contained approximately 100–200 ng of SSTR-2 at each treatment. Although additional boosts and higher amounts of immunogen would most likely have resulted in increased antibody titers, the conditions used were sufficient to induce a response detectable by ELISA using recombinant exosomes as antigen. This data confirm that exosomes exhibit potent adjuvant activity that renders antigens highly immunogenic. They also support that anti-GPCR antibody induction and detection can be performed without the need to purify receptors. Standard ELISA for antibody detection were carried out using ~ 250 ng of recombinant exosomes per well of a 96-well microtitration plate. A 1-Liter culture supernatant which generally yields ~1 to 2 mg of exosomes could be used to coat up to 8000 wells. Hence, this approach is realistically scalable to also provide the material required to screen hybridoma supernatants for the isolation of monoclonal antibodies. The flexibility of the exosome-based GPCR tool to adapt to various formats was further established in soluble phase assay using biochemically labeled exosomes and a library of antibody-expressing cells. In this assay, the library of antibody repertoire consisted of a suspension of cells from lymph nodes of animals immunized with SSTR-2 exosomes. We showed that CD-19+ cells could be stained in an antigen-specific manner. This staining is more likely due to an antibody-antigen interaction and therefore enabled the identification of anti-SSTR-2 antibody-producing cells. Combined to cell sorting, this approach may enable the isolation of antigen-specific antibody producing cells from a repertoire library. The detection of anti-SSTR-2 antibody producing cells was dependent of the time at which organs were collected after the last immunization. Indeed, antigen-specific staining was maximal at day 1 and lost at day 5 post immunization. The presence of anti-SSTR-2 antibody producing cells was also detected in spleens of immunized animals; however, optimal detection occurred at day-5 post-immunization (data not shown). Further studies will be required to determine the precise subtype of B-cells identified with this approach. Regardless, our findings demonstrate that exosome-based GPCR tools are readily suitable for the generation of antibodies and screening of antibody repertoires without the need to purify receptors. This is notably of vital importance for the validation of orphan receptors. Moreover, screening assays can easily be adapted to repertoires of different origins (natural, recombinant, or synthetic) and compositions (antibodies or small molecules).
Overall, exosomes containing GPCRs as source and form of receptors compares favorably to purified proteins or membrane preparations. In addition to the assay format reported here, the different solid support platforms recently described for GPCR drug screening such as protein microarrays (Fang et al 2002) and parmagnetic proteoliposomes (Mirzabekov et al 2000) could also be adapted to utilize recombinant exosomes as source of GPCRs instead of membrane preparations or purified receptors. Given the size of exosomes, it can be estimated that 16 × 10−3 fmol of SSTR-2 can be deposited in a protein array well of 100 μm when using exosomes bearing ten SSTR-2 molecules per vesicles (100 nm diameter vesicle, 10 SSTR-2 per exosomes and 50% occupancy). This quantity is similar to previous estimates when using membrane preparations (Hong et al 2006). Hence the exosome-based GPCR platform provides an enriched source of GPCR maintained in their native conformation that should facilitate GPCR drug discovery and warrant further evaluation in this field.
References
- Abusamra AJ, Zhong Z, Zheng X, et al. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35:169–73. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Andre F, Chaput N, Schartz NE, et al. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. 2004;172:2126–36. doi: 10.4049/jimmunol.172.4.2126. [DOI] [PubMed] [Google Scholar]
- Clayton A, Court J, Navabi H, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247:163–74. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- Delcayre A, Estelles A, Sperinde J, et al. Exosome Display technology: applications to the development of new diagnostics and therapeutics. Blood Cells Mol Dis. 2005;35:158–68. doi: 10.1016/j.bcmd.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Delcayre A, Shu H, Le Pecq JB. Dendritic cell-derived exosomes in cancer immunotherapy: exploiting nature’s antigen delivery pathway. Expert Rev Anticancer Ther. 2005;5:537–47. doi: 10.1586/14737140.5.3.537. [DOI] [PubMed] [Google Scholar]
- Delcayre A, Le Pecq JB. Exosomes as novel therapeutic nanodevices. Curr Opin Mol Ther. 2006;8:31–8. [PubMed] [Google Scholar]
- Denzer K, Kleijmeer MJ, Heijnen HF, et al. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci, 113 Pt. 2000;19:3365–74. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- Escudier B, Dorval T, Chaput N, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase I clinical trial. J Transl Med. 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Frutos AG, Lahiri J. G-protein-coupled receptor microarrays. Chembiochem. 2002;3:987–91. doi: 10.1002/1439-7633(20021004)3:10<987::AID-CBIC987>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Feder J. Random sequential adsorption. J Theor Biol. 1980;87:237–54. [Google Scholar]
- Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- Helenius A, Simons K. Solubilization of membranes by detergents. Biochim Biophys Acta. 1975;415:29–79. doi: 10.1016/0304-4157(75)90016-7. [DOI] [PubMed] [Google Scholar]
- Hong Y, Webb BL, Pai S, et al. G-protein-coupled receptor microarrays for multiplexed compound screening. J Biomol Screen. 2006;11:435–8. doi: 10.1177/1087057106287139. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Jacoby E, Bouhelal R, Gerspacher M, et al. The 7 TM G-protein-coupled receptor target family. ChemMedChem. 2006;1:761–82. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- Johnstone RM. The Jeanne Manery-Fisher Memorial Lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochem Cell Biol. 1992;70:179–90. doi: 10.1139/o92-028. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lechman ER, Bianco N, et al. Exosomes derived from IL-10-treated dendritic cells can suppress inflammation and collagen-induced arthritis. J Immunol. 2005;174:6440–8. doi: 10.4049/jimmunol.174.10.6440. [DOI] [PubMed] [Google Scholar]
- Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–44. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Koller KJ, Whitehorn EA, Tate E, et al. A generic method for the production of cell lines expressing high levels of 7-transmembrane receptors. Anal Biochem. 1997;250:51–60. doi: 10.1006/abio.1997.2190. [DOI] [PubMed] [Google Scholar]
- Lamparski HG, Metha-Damani A, Yao JY, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270:211–26. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- Le Pecq JB. Dexosomes as a therapeutic cancer vaccine: from bench to bedside. Blood Cells Mol Dis. 2005;35(2):129–35. doi: 10.1016/j.bcmd.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Lundstrom K. Structural genomics of GPCRs. Trends Biotechnol. 2005;23:103–8. doi: 10.1016/j.tibtech.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Lundstrom K. Latest development in drug discovery on G protein-coupled receptors. Curr Protein Pept Sci. 2006;7:465–70. doi: 10.2174/138920306778559403. [DOI] [PubMed] [Google Scholar]
- Med, Ad, News Staff World’s best-selling medicines. Med Ad News. 2004;23:60–4. [Google Scholar]
- Mirzabekov T, Kontos H, Farzan M, et al. Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat Biotechnol. 2000;18:649–54. doi: 10.1038/76501. [DOI] [PubMed] [Google Scholar]
- Miyasaka K, Hanayama R, Tanaka M, et al. Expression of milk fat globule epidermal growth factor 8 in immature dendritic cells for engulfment of apoptotic cells. Eur J Immunol. 2004;34:1414–22. doi: 10.1002/eji.200424930. [DOI] [PubMed] [Google Scholar]
- Morelli AE. The immune regulatory effect of apoptotic cells and exosomes on dendritic cells: its impact on transplantation. Am J Transplant. 2006;6:254–61. doi: 10.1111/j.1600-6143.2005.01197.x. [DOI] [PubMed] [Google Scholar]
- Morse MA, Garst J, Osada T, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3:9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Aoki N, Kato T, et al. Secretion of a peripheral membrane protein, MFG-E8, as a complex with membrane vesicles. Eur J Biochem. 2002;269:1209–18. doi: 10.1046/j.1432-1033.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- Patel S, Mehta-Damani A, Shu H, et al. An analysis of variability in the manufacturing of dexosomes: implications for development of an autologous therapy. Biotechnol Bioeng. 2005;92:238–49. doi: 10.1002/bit.20596. [DOI] [PubMed] [Google Scholar]
- Peche H, Renaudin K, Beriou G, et al. Induction of tolerance by exosomes and short-term immunosuppression in a fully MHC-mismatched rat cardiac allograft model. Am J Transplant. 2006;6:1541–50. doi: 10.1111/j.1600-6143.2006.01344.x. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol. Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Sarramegna V, Talmont F, Demange P, et al. Heterologous expression of G-protein-coupled receptors: comparison of expression systems from the standpoint of large-scale production and purification. Cell Mol Life Sci. 2003;60:1529–46. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, Lorens J, Achacoso P, et al. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293 T-cell-based systems. Current Protocols in Immunology, Unit 10. 1999;28(Suppl 31) doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T-cell signaling. J Immunol. 2006;176:1534–42. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- Thery C, Regnault A, Garin J, et al. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Veron P, Segura E, Sugano G, et al. Accumulation of MFG-E8/lactadherin on exosomes from immature dendritic cells. Blood Cells Mol Dis. 2005;35:81–8. doi: 10.1016/j.bcmd.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Vincent-Schneider H, Stumptner-Cuvelette P, Lankar D, et al. Exosomes bearing HLA-DR1 molecules need dendritic cells to efficiently stimulate specific T-cells. Int Immunol. 2002;14:713–22. doi: 10.1093/intimm/dxf048. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]