Abstract
Transfer RNA-like structures (TLSs) that are sophisticated functional mimics of tRNAs are found at the 3′-termini of the genomes of a number of plant positive strand RNA viruses. Three natural aminoacylation identities are represented: valine, histidine, and tyrosine. Paralleling this variety in structure, the roles of TLSs vary widely between different viruses. For Turnip yellow mosaic virus, the TLS must be capable of valylation in order to support infectivity, major roles being the provision of translational enhancement and down-regulation of minus strand initiation. In contrast, valylation of the Peanut clump virus TLS is not essential. An intermediate situation seems to exist for Brome mosaic virus, whose RNAs 1 and 2, but not RNA3, need to be capable of tyrosylation to support infectivity. Other known roles for certain TLSs include (i) the recruitment of host CCA nucleotidyltransferase as a telomerase to maintain intact 3′ CCA termini, (ii) involvement in the encapsidation of viral RNAs, and (iii) presentation of minus strand promoter elements for replicase recognition. In the latter role, the promoter elements reside within the TLS but are not functionally dependent on tRNA mimicry. The phylogenetic distribution of TLSs indicates that their evolutionary history includes frequent horizontal exchange, as has been observed for protein-coding regions of plant positive strand RNA viruses.
Keywords: aminoacylation, eEF1A, translational enhancement, tRNA mimicry, CCA nucleotidyltransferase, positive strand RNA viruses
1. Introduction
The genomes of positive strand RNA viruses simultaneously serve as mRNAs for expressing viral genes and as templates for RNA replication. Some such genomes of eukaryote-infecting viruses have the 5′ and 3′ terminal features — m7GpppN cap and poly(A) tail, respectively — characteristic of their hosts’ mRNAs. These features are known to play important roles in translation and RNA stability (reviewed in Dreher, 1999; Gale et al., 2000; Gallie, 1998). However, many positive strand RNA viral genomes lack one or both of these features, necessitating novel ways of ensuring the translatability and stability of the viral RNA. At the same time, 3′-terminal sequences of positive strand RNA viral genomes are intimately involved in initiating RNA replication, necessitating coordination between the RNA’s translation and replication roles (Dreher, 1999). Among the viral 3′ termini faced with these functions, one of the most distinctive types of is the tRNA-like structure (TLS) found on a number of plant viral genomes.
The first tRNA-like structure to be identified in viral genomes was the TLS at the 3′ end of the Turnip yellow mosaic virus (TYMV) genome (Pinck et al., 1970; Yot et al., 1970). This remains the best studied viral TLS and also the one that most closely resembles canonical tRNAs in terms of its structure and biochemical properties. Other viral TLSs are structurally more complex yet possess key biochemical properties of tRNAs, while still others are structurally related to viral TLSs but have only vestigial tRNA-like functional properties. Three basic types of 3′-terminal TLS have been described, exemplified by the TLSs in the genomes of TYMV, Tobacco mosaic virus (TMV), and Brome mosaic virus (BMV). These TLSs are each capable of specific aminoacylation, with valine, histidine or tyrosine, respectively, and related TLSs are represented in a number of virus genera (Table 1). The group of viral 3′-terminal TLSs is thus quite diverse. Although the biological roles of viral TLSs are far from being fully understood, the available evidence indicates that these roles are also diverse.
Table 1.
Plant viruses whose genomes terminate at the 3′-end in aminoacylatable TLSs
| Genus | Virus | Amino acid bound |
|---|---|---|
| Tymovirus | Turnip yellow mosaic virus (TYMV) | valine |
| Andean potato latent virus (APLV) | valine | |
| Belladonna mottle virus (BeMV) | valine | |
| Cacao yellow mosaic virus (CYMV) | valine | |
| Clitoria yellow vein virus (CYVV) | valine | |
| Eggplant mosaic virus (EMV) | valine | |
| Kennedya yellow mosaic virus (KYMV) | valine | |
| Okra mosaic virus (OkMV) | valine | |
| Ononis yellow mosaic virus (OYMV) | valine | |
| Wild cucumber mosaic virus (WCMV) | valine | |
| Nemesia ring necrosis virus (NeRNV)1 | histidine | |
| Furovirus | Soil-borne wheat mosaic virus (SBWMV)2 | valine |
| Pomovirus | Beet soil-borne virus (BSBV)2 | valine |
| Potato mop-top virus (PMTV)2 | valine | |
| Pecluvirus | Indian peanut clump virus (IPCV)2 | valine |
| Peanut clump virus (PCV)2 | valine | |
| Tobamovirus | Tobacco mosaic virus (TMV) | histidine |
| Cucumber green mottle mosaic virus (CGMMV) | histidine | |
| Green tomato atypical mosaic virus (GTAMV) | histidine | |
| Satellite tobacco mosaic virus (STMV)3 | histidine | |
| Sunnhemp mosaic virus (SHMV)4 | valine | |
| Bromovirus | Brome mosaic virus (BMV) | tyrosine |
| Broad bean mottle virus (BBMV) | tyrosine | |
| Cowpea chlorotic mottle virus (CCMV) | tyrosine | |
| Cucumovirus | Cucumber mosaic virus (CMV) | tyrosine |
| Hordeivirus | Barley stripe mosaic virus (BSMV) | tyrosine |
| Poa semilatent virus (PSLV)5 | tyrosine |
Unless otherwise indicated, the references for aminoacylation studies can be found in Mans et al. (1991). Other references are:
This review will focus on studies relating to the biological roles of the plant viral 3′-terminal TLSs during viral infection. The three key tRNA-like biochemical properties of the plant viral TLSs, which can readily be assessed with in vitro experiments, are their abilities to be aminoacylated, to form a ternary complex with EF1 translation elongation factors, and for their 3′-CC variants to serve as substrates for 3′-adenylation by CCA-nucleotidyltransferase (CCA-NTase, also known as CTP, ATP:tRNA nucleotidyltransterase). Other RNA elements that structurally mimic tRNAs but have much more limited functional (and sometimes structural) mimicry exist in viral and cellular mRNAs but will not be discussed here. Examples of such elements in viral RNAs include the dicistrovirus-type internal ribosome entry site (IRES) elements, which load directly into the P site of ribosomes and thereby mimic the function of the initiator tRNA ternary complex (Costantino et al., 2008) and the T-stem-like motif in the intergenic region of BMV RNA3 that is capable of tRNA-like base modification and that is involved in RNA recruitment to RNA replication (Baumstark and Ahlquist, 2001). Previous reviews that present more detailed information on the structural and biological properties of the plant viral TLSs that will be discussed here include those by (Fechter et al., 2001b; Florentz and Giegé, 1995; Giegé et al., 1993; Haenni et al., 1982; Hall, 1979; Mans et al., 1991). Reviews that have addressed the biological roles of TLSs include those by (Dreher, 1999; Fechter et al., 2001b; Haenni and Chapeville, 1997).
2. tRNA mimicry of valine-, histidine- and tyrosine-specific TLSs
The key experiments that can test the role of aminoacylation and tRNA mimicry require a knowledge of the features that control interaction with tRNA-specific enzymes or factors. In the case of the aminoacyl-tRNA synthetases, those features constitute the set of so-called identity elements. The main identity elements responsible for the aminoacylation of each of the three types of plant viral TLSs are now known.
2.1. Valine-specific TLSs
2.1.1. The TYMV TLS: valine identity concentrated in the anticodon
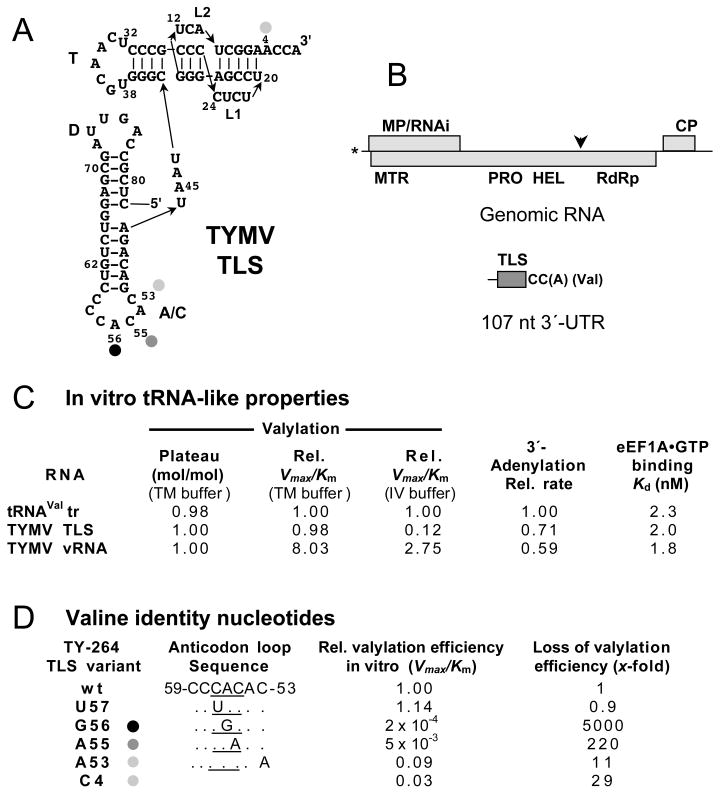
The TYMV TLS is an extraordinarily efficient tRNA mimic. This is the plant viral TLS that most closely resembles the structure of tRNA, with readily identifiable acceptor-T and anticodon-D arms that join to form an L-conformation like that of tRNAs. Solution structure-probing experiments were pivotal in recognizing the importance of a pseudoknot in forming the acceptor-T arm, and in the ability of the TLS to form the L-conformation (reviewed in Giegé et al., 1993). The TLS is comprised of the 3′ 82 nucleotides of the genome (Fig. 1) (Joshi et al., 1982a), which can be valylated in vitro by wheat germ valyl-tRNA synthetase (ValRS) with about the same kinetic efficiency as wheat germ tRNAVal made by in vitro transcription (Dreher and Goodwin, 1998). Nevertheless, full-length virion RNA and a 264 nt RNA containing the TLS are valylated with superior kinetics (Fig. 1D) (Dreher and Goodwin, 1998), emphasizing the free accessibility of the TLS in the context of the overall folding of the c. 6-kb genomic RNA.
Fig. 1. The TYMV TLS is a highly efficient mimic of tRNAVal.
A Sequence and structure of the 82-nt-long TLS of TYMV RNA (Rietveld et al., 1982). The pseudoknot-containing amino acid acceptor stem and the T-, D- and anticodon (A/C) domains are clear structural analogs of the four arms of canonical tRNAs. A CAC valine anticodon in present. The nucleotides that serve as valine identity elements are indicated with adjacent dots (black strongest, light grey weakest). Note that it is conventional to number the nucleotides in TLSs from the 3′-end. B. Diagram of the TYMV genome showing the three coding regions for expressing the movement protein/RNAi suppressor (MP/RNAi); replication protein, with methyltranserase (MTR), proteinase (PRO), helicase (HEL) and RNA-dependent RNA polymerase (RdRp) domains; and coat protein (CP). The asterisk signifies a 5′-cap structure; the arrowhead indicates the site of proteolytic maturation cleavage. The cartoon of the 3′-UTR features the TLS and the 3′-terminus, which lacks the A residue in most virion RNAs; the TLS serves as translational enhancer and the 3′-CC(A) serves as the initiation box controlling minus strand synthesis (see text). C. Summary of in vitro tRNA-like properties of the TYMV TLS, assayed with wheat germ enzymes and eEF1A (Dreher and Goodwin, 1998), compared to a lupine tRNAVal transcript. The TLS was studied in 83-nt long RNA (TLS), 264-nt long RNA (TY-264) or in the 6.3 kb virion RNA (vRNA). TM buffer has low ionic strength, while IV buffer has higher ionic strength and is similar to the conditions used for in vitro translation. Plateau refers to the extent of aminoacylation (moles valine per mole RNA). D. Contributions to valine identity by anticodon loop nucleotides, studied with wheat germ ValRS (Dreher et al., 1992), and by A4, studied with yeast ValRS (Florentz et al., 1991).
Studies with wheat germ ValRS revealed a set of identity elements in the TYMV TLS that closely parallel those in tRNAVal (Dreher et al., 1992; Florentz et al., 1991). The most important identity nucleotide is A56 in the middle of the anticodon, followed by C55 also in the anticodon (Fig. 1A, C). As befitting the fact that valine is encoded by 4 codons that differ in the third position, the wobble (C57) position of the anticodon is not an identity element (Fig. 1D). Additional, weaker contributions to identity come from the 3′-most nucleotide (C53) of the anticodon loop and the ‘discriminator base’ (A4) adjacent to the 3′-CCA. The importance of A56, C55 and C53 in the anticodon loop as identity elements was confirmed with in vitro selection experiments beginning with RNAs with partially randomized sequences (Wientges et al., 2000).
TYMV RNA is functionally specific for aminoacylation with valine; aminoacylation of RNAs from some virion preparations with other amino acids is considered to be the result of contaminating cellular tRNAs (van Belkum et al., 1987a). That said, it has been demonstrated that TYMV RNA, and probably all viral TLSs that contain a pseudoknotted acceptor stem, possess some histidine identity that can be detected under appropriate in vitro conditions (Rudinger et al., 1992; 1997). This is unlikely to be relevant in vivo, because substantial histidylation in buffers that approximate in vivo conditions requires substrate levels of purified yeast histidyl-tRNA synthetase (HisRS), and because no histidylation above background levels could be observed with a preparation of mixed wheat germ aminoacyl-tRNA synthetases capable of quantitative histidylation of tRNAHis (Dreher and Goodwin, 1998).
2.1.2. 3′-Adenylation and translation elongation factor interaction with TYMV RNA
Most molecules of TYMV RNAs cannot initially be valylated, because some 85% of virion RNAs terminate in 3′-CC (Giegé et al., 1978). These RNAs can be 3′-adenylated — and thereby become competent for valylation — by host CCA-NTase, an enzyme that is involved both in the initial maturation of newly synthesized tRNAs and in the maintenance of an intact 3′-CCA, which undergoes steady turnover in the cell (Deutscher, 1982). The 3′-CC form of TYMV RNA can be as efficiently 3′-adenylated by wheat germ CCA-NTase as the 3′-CC form of a plant tRNAVal transcript (Dreher and Goodwin, 1998).
Aminoacylated tRNAs form aminoacyl-tRNA•EF1A•GTP ternary complexes to allow delivery to the ribosome for participation in protein synthesis. Valylated TYMV RNAs ranging in length from the minimal TLS to genomic RNA were bound by wheat germ eEF1A•GTP with Kd’s of about 2 nM (Fig. 1D)(Dreher and Goodwin, 1998). These complexes were about as stable as those formed with mature and unmodified transcript plant valyl-tRNAVal (Kd’s of 1–2 nM), in contrast to the weak complex formed with non-aminoacylated tRNA (Kd = 15 μM) (Dreher and Goodwin, 1998; Dreher et al., 1999).
2.1.3. Other tRNA-like activities associated with TYMV RNA
At least three additional properties associated with tRNAs have been identified for TYMV RNA. Some of these have also been observed with other viral TLSs (reviewed in Mans et al., 1991). Ribonuclease P, which cleaves precursor tRNA molecules to generate the 5′-termini of tRNAs, cleaves at the 3′-end of the TYMV TLS acceptor-T arm helix (Guerrier-Takada et al., 1988). Valyl-TYMV RNA is recognized by E. coli peptidyl-tRNA hydrolase (formerly called N-acetylaminocyl-tRNA hydrolase), which catalyses removal of the aminoacyl group (Yot et al., 1970). Finally, certain base modifications can be introduced into the TYMV TLS by tRNA modifying enzymes (Becker et al., 1998; Brulé et al., 1998). To date, there is no evidence that these properties are relevant during an infection.
2.1.4. Other valine-specific TLSs of tymoviruses and furo-like viruses
RNAs from several other tymoviruses have been shown to be capable of valylation (Dreher and Goodwin, 1998; van Belkum et al., 1987a). In each of these, a TLS similar to that of TYMV is present, although two structural variants of the acceptor-T arm exist. Alongside the TYMV type, in which the 12 bp arm is built of 4 bp, 3 bp and 5 bp segments (Fig. 1A), is the type present in the Eggplant mosaic virus (EMV) TLS built of 3 bp, 3 bp and 6 bp segments. Both types of TLS are highly efficiently 3′-adenylated and valylated, but the 3/3/6 acceptor-T stem seems to be correlated with weaker eEF1A•GTP binding (Kd >50 nM) (Dreher and Goodwin, 1998; Goodwin and Dreher, 1998).
The sequencing of genomes from the Furovirus, Pomovirus and Pecluvirus genera of fungus-borne rod-shaped (furo-like) viruses unexpectedly revealed the presence of tymoviral-like TLSs (Koenig et al., 1996; Shirako and Wilson, 1993). Several of these have been shown to possess tRNA mimicry properties similar to those of tymoviral TLSs (Goodwin and Dreher, 1998). Both the 4/3/5 and 3/3/6 forms of the acceptor-T arm are represented. A different, remarkable structural variation is found in the pecluviral RNAs (Peanut clump, PCV, and Indian peanut clump, IPCV, viruses), in the form of a c. 40 nt insertion placed between the two halves (acceptor-T arm and anticodon-D arm) of the TLS. Despite this insertion, these RNAs can be very efficiently valylated, though they interact poorly with eEF1A•GTP (Goodwin and Dreher, 1998).
2.2. The Histidine-specific TLSs of TMV and other tobamoviruses
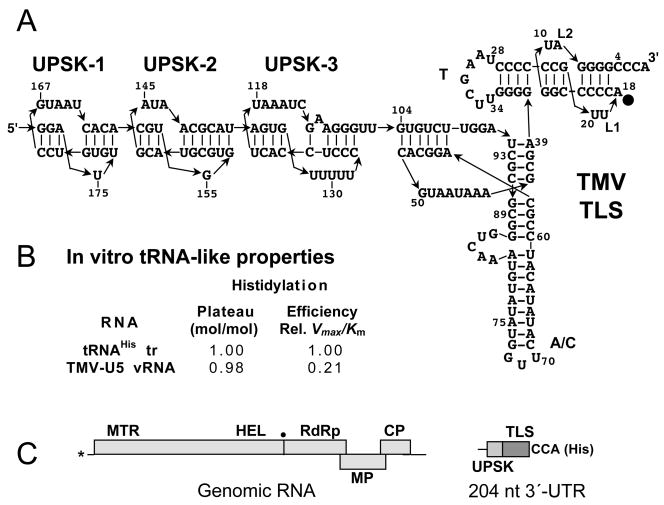
The TMV TLS has an acceptor-T arm that is built in a similar way to that of TYMV, except that the arm is only 11 bp long (Fig. 2A). The rest of the TLS involves a considerably more complex structure than the TYMV TLS and has rather weak similarity to tRNA, except for a GUU histidine anticodon (GUG in some strains). Structure-probing experiments led to the recognition that the 104-nt-long TLS core (Felden et al., 1996; Rietveld et al., 1984) sits adjacent to a series of distinctive upstream pseudoknots that are strongly conserved among tobamoviruses (van Belkum et al., 1985), as well as in many hordeiviruses, furoviruses, pomoviruses and even in two tymoviruses (summarized in Koenig et al., 2005; Leathers et al., 1993).
Fig. 2. The histidine-specific TMV TLS.
A Sequence and structure of the 104-nt-long TLS of TMV RNA (Rietveld et al., 1984) and the adjacent upstream pseudoknots (UPSK 1–3) (van Belkum et al., 1985). The TMV TLS has obvious acceptor and T stem analogs and an anticodon (A/C) domain with a GUU histidine anticodon; 98-UGGA-95 is thought to serve as a D-loop analog (Felden et al., 1996). Nucleotide A18 in the acceptor stem is thought to be the major histidine identity element (Rudinger et al., 1997). B. Comparison of histidylation efficiency between TMV-U5 RNA and a yeast tRNAHis transcript, determined with yeast HisRS (Felden et al., 1994). C. Diagram of the TMV genome with domains labeled as in Fig. 1. The dot indicates the location of a suppressible stop codon that allows RdRp expression by read-through. The major domains in the 3′-UTR are the TLS and the upstream pseudoknot domain (UPSK). The minus strand promoter overlaps the TLS; the UPSK provides translational enhancement (see text).
Early studies suggested that RNAs of different TMV strains have different capacities for histidine charging (Garcia-Arenal, 1988; Hall, 1979). The most detailed kinetic studies with TMV RNA have been conducted with TMV-U5 RNA and partially purified yeast histidyl-tRNA synthetase (HisRS) (Felden et al., 1994). TMV-U5 RNA is a strong tRNA mimic that could be fully histidylated with similar Km and approaching the kinetic efficiency (Vmax/Km) of a yeast tRNAHis transcript (Fig. 2B). The major histidine identity element in eukaryotic (yeast) tRNAHis is an additional residue at the 5′-end that stacks onto the acceptor stem, positioned opposite the discriminator base (Nameki et al., 1995; Rudinger et al., 1994). Modest identity resides in the anticodon, and the GUG to GUU anticodon mutation results in a 4-fold drop in Vmax/Km (Nameki et al., 1995). Based on model studies with TYMV-derived minihelices (acceptor-T arm RNAs), it has been proposed that A18 in the L1 linking strand of the TMV TLS acceptor stem serves the role of the extra 5′ residue of tRNAHis (Fig. 2A)(Rudinger et al., 1994; 1997). Evidence that the anticodon-like arm is recognized as such by HisRS has been provided by the observation that the enzyme protects access by structural probes to residues in this arm (Garcia-Arenal, 1988), but no mutagenesis studies have been reported.
TMV RNA from which the 3′-terminal A has been removed can be adenylated with CCA-NTase from E. coli, yeast or sheep liver (Joshi et al., 1985). As deduced by a “damage-selection” approach involving limited chemical modification before 3′-adenylation, nucleotides in the T-loop and near the 3′-end are important for the recognition of TMV RNA by CCA-NTase (Hegg et al., 1990). Histidyl-TMV RNA is known to form a ternary complex with wheat germ eEF1A (Litvak et al., 1973), though the stability of the complex has not been determined.
Similar TLS structures have been modeled from the 3′-UTRs of several tobamoviral RNAs (Felden et al., 1996; Rietveld et al., 1984) and in Satellite tobacco mosaic virus RNA (STMV) (Felden et al., 1994; Gultyaev et al., 1994). Because only limited analyses have been reported, it is not known to what extent the tRNA-like properties of TMV RNA are conserved in other tobamoviral TLSs.
2.3. Tyrosine-specific TLSs
2.3.1. The BMV TLS: a TLS with a long-distance pseudoknot
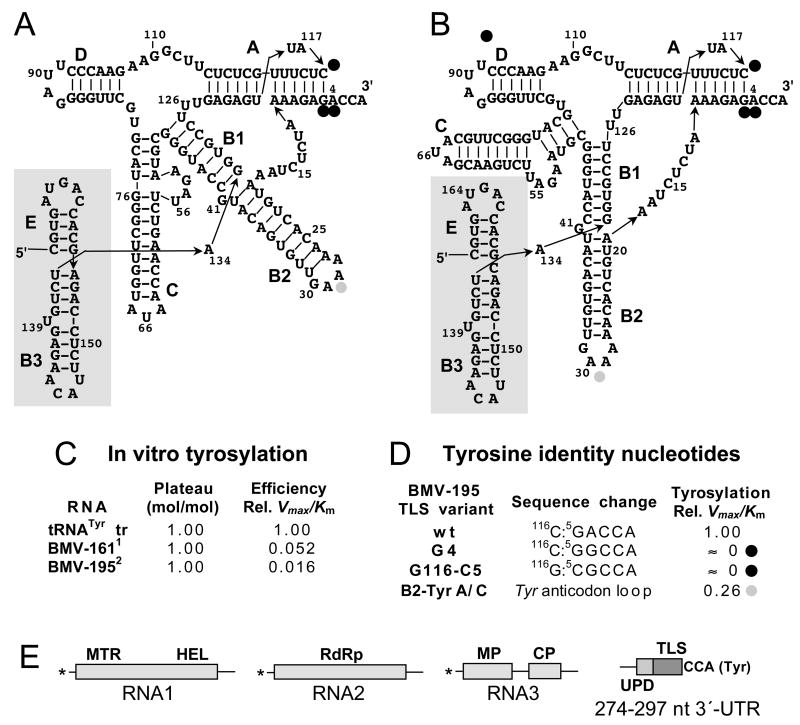
The BMV TLS is the structurally most complex of the viral TLSs. Several studies have sought to understand the structure of the BMV TLS, which has been diagrammed in different ways (Fig. 3A, B) as investigators have attempted to deduce the closest correspondence to the structure of tRNATyr. The difficulties have been (i) that the upstream extent of the TLS has been uncertain, (ii) this longest of plant viral TLSs includes multiple stem-loop domains, and (iii) there has been no clear identification of analogs of the anticodon and D stems. The structural studies have focused on chemical and enzymatic structure probing and subsequent modeling, checked against the results of structure/function mutational studies; no physical methods of structural analysis, such as NMR or X-ray crystallography, have been reported to date.
Fig. 3. The tyrosine-specific BMV TLS.
A, B Sequence of the 133-nt-long TLS of BMV RNA3 and adjacent upstream stem-loops. The structure in (A) was originally proposed by (Rietveld et al., 1983), who suggested that arm C with its AUA tyrosyl anticodon serves as the analog of the anticodon domain of tRNA. The current structural model proposed by (Fechter et al., 2001a) in (B) shows arm B2 as the analog of the anticodon. The major tyrosine identity elements are present at the 3′-end of the acceptor stem helix, and only minor tyrosine identity is contributed by the B2 loop (Fechter et al., 2001a). Stems C and D can be considered as insertions into the basic tRNA-like structure; the terminal AUA loop of stem C serves as the core element of the minus strand promoter, and stem D is absent from some bromoviral TLSs. Stems B3 and E (shaded) are outside the core TLS, but mutations in B3 impair tyrosylation (see text). C. Comparison of in vitro tyrosylation by yeast TyrRS of two forms of BMV RNA with an in vitro-generated yeast tRNATyr; 1 BMV-161 is a 161 nt-long 3′-fragment (Ω161) produced from RNA4 by limited ribonuclease cleavage (Perret et al., 1989); 2 BMV-195 is a 195 nt RNA3 transcript (Fechter et al., 2001a). D. Contributions to tyrosine identity by nucleotides at the end of the acceptor stem helix, and by the B2 loop, studied with yeast TyrRS (Fechter et al., 2001a). E. Diagram of the tripartite BMV genome with domains labeled as in Fig. 1. The major domains in the 3′-UTR are the TLS and the upstream domain (UPD), both of which are strongly conserved among the three genomic RNAs. The core minus strand promoter resides in stem C within the TLS; the 3′-UTR serves as a translational enhancer, but the responsible domain has not been identified (see text).
The features that are clearly counterparts to features of canonical tRNAs are restricted to the acceptor-T arm (Fig. 3), making the assignment of an anticodon-like domain difficult. Although an AUA tyrosine anticodon sequence is present in the terminal loop of stem C, this was found by mutagenesis and in vitro functional studies to play no role in tyrosylation (Dreher et al., 1984). Rather, the AUA loop serves as the core element of the promoter controlling minus strand RNA synthesis (Chapman and Kao, 1999; Dreher and Hall, 1988a; Kim et al., 2000).
According to our current understanding, which draws on structure-probing, structural modeling and structure-function experiments, the BMV TLS comprises the 3′ 134 nts of BMV RNA (Fechter et al., 2001a; Joshi et al., 1983)(Fig. 3C). In yeast tRNATyr the strongest tyrosine identity nucleotides are an A discriminator base and G-C first base-pair (GA immediately upstream of the 3′-CCA and a C base-paired to the G) (Fechter et al., 2000). Mutation of these three bases in BMV RNA3 abolishes tyrosylation, validating their critical function in directing charging (Fig. 3)(Fechter et al., 2001a). The sequence of the anticodon loop analog, loop B2 (-GAAA-), has only a minor influence in tyrosylation (Fig. 3B, D)(Fechter et al., 2001a). Thus, although the anticodon does confer substantial tyrosine identity in yeast tRNATyr (Fechter et al., 2000), the tyrosylation of BMV RNA is accomplished without this feature being present.
BMV RNA can be aminoacylated in vitro to high levels with tyrosine by TyrRS from plants or yeast (Fechter et al., 2001a; Kohl and Hall, 1974; Perret et al., 1989), though the reaction is sensitive to buffer composition. The aminoacylation kinetics (Fig. 3C) do not appear to be as close to those of the cognate tRNA as is the case for the TYMV or TMV TLSs, although the kinetics for a homologous system with plant TyrRS compared to plant tRNATyr have not yet been reported. Nevertheless, tyrosylation efficiency and tRNA mimicry are clearly high enough to be biologically functional, since tyrosylated genomic RNAs have been observed during infections in barley protoplasts (Loesch-Fries and Hall, 1982).
BMV RNA from which the 3′-A has been removed is a substrate for bacterial or plant CCA-NTase (Dreher and Hall, 1988b; Joshi et al., 1983), and the tyrosylated RNA interacts with wheat germ eEF1A•GTP (Bastin and Hall, 1976), but no quantitative measures of these properties are available.
2.3.2. BMV-like TLSs of other viruses: Bromoviridae and hordeiviruses
BMV-like TLS structures can be drawn for the members of the Bromovirus and Cucumovirus genera of the Bromoviridae, although the TLS of Broad bean mottle bromovirus (BBMV) lacks arm D (Ahlquist et al., 1981; Joshi et al., 1983). Tyrosylation has been observed for Cowpea chlorotic mottle (CCMV) and BBMV bromovirus RNAs and for Cucumber mosaic cucumovirus (CMV) RNA (Kohl and Hall, 1974). Tomato aspermy virus (TAV) is another cucumovirus whose RNAs have BMV-like TLSs. However, TAV RNA is not tyrosylatable (Joshi, 1986), perhaps because of critical sequence differences in the identity positions at the 3′-end of the acceptor arm (3′-UCCCA end with A base-paired to the U). While the TAV TLS is unable to bind tyrosine, the existence of overall tRNA mimicry is demonstrated by the strong 3′-adenylation activity of a 3′-CC form of the RNA by E. coli CCA-NTase (Joshi, 1986). Minor sequence differences exist between the TLSs of the three genomic RNAs for each of these viruses.
Although the Hordeivirus genus does not belong to the Bromoviridae, the RNAs of two hordeiviruses have been shown to be tyrosylatable: Barley stripe mosaic virus (BSMV) (Agranovsky et al., 1981) and Poa semilatent virus (PSLV) (Agranovsky et al., 1992). These RNAs have TLSs similar to that of BBMV, missing arm D; the core TLS appears to be only about 95 nt long. As for BMV, it has been shown that tyrosylated full-length BSMV RNAs are present during infections in barley (Loesch-Fries and Hall, 1982). Notably, the most important tyrosine identity elements identified in Fig. 3 at the end of the acceptor-T arm (Fechter et al., 2001a) are present in all of the tyrosylatable TLSs.
An intriguing situation has been proposed for Alfalfa mosaic virus (AMV) and members of the related Ilarvirus genus of the Bromoviridae. These viruses share the property of requiring coat protein molecules to interact with a series of stem-loops present near the 3′-end in order to activate the genome for infectivity, apparently by enhancing genome translation (Bol, 2005). Evidence suggests that the 3′-end is capable of an alternative tRNA-like folding that has some resemblance to the BMV TLS (Olsthoorn et al., 1999); this is the form of the RNA recognized by the AMV replicase for minus strand RNA synthesis. The RNAs of the alfamo- and ilarviruses do not end in 3′-CC(A), but the detection of weak 3′-adenylation of AMV RNA by yeast CCA-NTase supports the existence of a structure with some tRNA characteristics (Olsthoorn et al., 1999).
3. Biological importance of aminoacylation
The role of aminoacylation has been tested with aminoacylation-defective mutants in three systems, the valine-specific TYMV and PCV and the tyrosine-specific BMV. These studies have indicated that aminoacylation holds different significance for different viruses.
3.1. Valylation of TYMV RNA is required for virus amplification
If the TYMV TLS, and in particular the full-length virion genomic RNA, is such an excellent substrate for 3′-adenylation and valylation (Fig. 1), one would expect the viral RNA to be valylated in vivo. Suggestive, though not compelling, evidence for this has been obtained by detecting valylated RNA oligomers released by ribonuclease T1 that corresponded to the 3′-end of TYMV RNA and not plant tRNAVal (Joshi et al., 1982b). Strong evidence that valylation is required (and therefore does occur) has been provided by genetic experiments with anticodon mutants that are essentially incapable of valylation (Tsai and Dreher, 1991).
A direct correlation was observed between virus amplification (assessed as genomic RNA and coat protein accumulation) and valylatability (assessed in vitro with wheat germ ValRS) for a set of 18 TYMV RNAs with TLS mutations that were inoculated in Chinese cabbage protoplasts and plants (Tsai and Dreher, 1991). Mutations at A56 resulted in virus that was barely capable of gene expression and replication in protoplasts and which was unable to establish a systemic infection in plants. Additional support for the importance of valylation was provided by the discovery that the loss of infectivity as a result of the C55→A mutation (CAA anticodon) could be suppressed by the C57→U mutation (UAA anticodon) (Tsai and Dreher, 1992). TLS RNA with the UAA anticodon could be fully valylated in vitro, with a kinetic efficiency (Vmax/Km) 16-fold higher than that of the initial mutant, though still 13-fold lower than the wild-type TLS.
To test whether aminoacylation specifically with valine or in a more general sense is required, attempts were made to switch the amino acid acceptor identity of TYMV RNA. Alteration of the C55 and C53 valine identity nucleotides to U55 and A53 as found in plant tRNAMet (Dreher et al., 1996) created an RNA with the set of major methionine identity nucleotides (Schulman and Pelka, 1988). Although these mutations were sufficient to switch aminoacylation from valine to methionine under standard in vitro aminoacylation conditions, only weak methionylation occurred under higher ionic strength conditions as found in vivo (Dreher et al., 1996). Methionylation under those conditions required additional mutation to destabilize the acceptor stem pseudoknot by shortening the linking strand L1. The combined anticodon loop and acceptor stem mutations produced TYMV genomes with methionine rather than valine identity, and these genomes were infectious to Chinese cabbage (although somewhat attenuated) (Dreher et al., 1996).
These experiments demonstrated that there is no specific requirement for aminoacylation of TYMV RNA with valine, and that aminoacylation with methionine — and maybe other amino acids — is compatible with the infection cycle. This means that there is no special role for valyl-tRNA synthetase in viral biology (e.g., as a replication cofactor) and that the significance of aminoacylation should be sought either in the protection of the free 3′-OH of the terminal A residue of the viral RNA or in a downstream step, such as interaction with eEF1A•GTP. Ternary complexes of similar stability were formed with wheat germ eEF1A•GTP and RNAs containing either valylated or methionylated forms of the TYMV TLS (Matsuda and Dreher, 2004).
3.2. Valylation of TYMV RNA is required for maximal enhancement of translational gene expression
Current understanding of mRNA translation in eukaryotes is that the most active form of an mRNA is the “closed-loop” cyclized form that results from contacts between eIF4 initiation factors bound to the 5′-cap and PABP bound to the 3′-poly(A) tail (Gallie, 1998; Kahvejian et al., 2005; Sachs et al., 1997). Translational enhancers equivalent to a poly(A) tail would be expected, and have been found, in viral mRNAs lacking a poly(A) tail (Dreher and Miller, 2006; Gallie, 1998). Experiments studying translational expression in plant protoplasts from luciferase reporter mRNAs have shown that the TYMV 3′-UTR serves as an expression enhancer (Matsuda et al., 2004a; Matsuda and Dreher, 2004). Translational enhancement mapped principally to the TLS, although the upstream-adjacent pseudoknot was important in optimum enhancement, probably by serving in a non-specific way as a spacer to ensure presentation of the TLS in an accessible format (Matsuda and Dreher, 2004). The TLS enhanced translational expression synergistically with the 5′-cap (Matsuda and Dreher, 2004), suggesting physical communication between the two extremities of the RNA. As with a poly(A) tail, the enhancement of translational expression by the TYMV TLS appears to be mostly due to enhanced translational efficiency, though increased mRNA stability is also a factor (Matsuda and Dreher, 2004).
No translational enhancement was observed with TYMV RNA constructs that lack an accessible 3′-CCA end to allow aminoacylation (Gallie and Kobayashi, 1994). A requirement for aminoacylation was directly demonstrated by observing strongly enhanced translation from RNAs with TYMV TLSs capable of aminoacylation with valine or methionine, but only weak enhancement by RNAs with discrete mutations that prevent aminoacylation (Matsuda and Dreher, 2004). The major role for aminoacylation and, by inference, the subsequent eEF1A binding that is necessary for infectivity may thus be to ensure early gene expression. A definitive role for eEF1A in translational enhancement has not yet been established because of a lack of mutations that specifically and strongly exclude eEF1A interaction. Neither is there any information on how eEF1A interaction might allow communication with the 5′-end to enhance translation. The TYMV TLS is able to enhance translation in essentially the same manner from luciferase mRNAs with a non-viral 5′-UTR (Matsuda and Dreher, 2004) or the TYMV 5′-UTR with its dual initiation sites (Matsuda et al., 2004a), but this has not yet been directly tested with genomic TYMV RNA. Note that, as is common with the poly(A) and other 3′ translational enhancers (Gallie, 1998), the TYMV TLS does not enhance translation in cell-free extracts (Matsuda and Dreher, 2007).
3.3. Valylation of TYMV RNA provides a way to regulate ribosome and replicase traffic on the RNA
Positive strand RNA viruses face the potential problem of the same genomic RNA supporting ribosome traffic 5′→3′ and replicase traffic in the opposite direction. Regulation aimed at managing this traffic has been uncovered for Qβ bacteriophage (Kolakofsky and Weissmann, 1971) and poliovirus (Gamarnik and Andino, 1998). Evidence suggests that eEF1A interaction with the TYMV TLS also allows such regulation (Matsuda et al., 2004b). Minus strand initiation by TYMV replicase occurs opposite the 3′-C of the genome (Singh and Dreher, 1997), adjacent to the valylated CCA end that is strongly bound by eEF1A•GTP. In vitro experiments with TYMV replicase demonstrated that decreased levels of minus strand synthesis occurred in the presence of eEF1A•GTP using aminoacylated TLS RNA templates, but not using non-aminoacylated templates, which fail to interact with eEF1A (Matsuda et al., 2004b). Valylated RNAs thus simultaneously experience enhanced translation function and blocked replication, ensuring unfettered translation early in the infection when new translation begins to increase the levels of replicase. Perhaps as replicase levels increase, replication complexes are at some point able to displace eEF1A from the TLS. This shift would then decrease the translational efficiency of the RNA and help to clear ribosomes from the RNA and allow it to move to the membranous sites of replication in the peripheral vesicles of the chloroplast (Matthews, 1991).
3.4. Valylation of Peanut clump virus RNA is not required but provides a slim advantage
The similarity between the PCV and TYMV TLSs and the conservation of valine identity elements between these RNAs provided an opportunity to test the role of valylation with PCV. This was of particular interest because RNA1 of strain P02A has an intact valine anticodon, but RNA2 has a deletion of the middle anticodon nucleotide (Manohar et al., 1993), the strongest valine identity element. This suggested the possibility that, in multipartite viruses, aminoacylation may be important for one RNA component (e.g., the component encoding essential early genes for assembling replicase complexes) but not for another.
Variants of PCV-P02A RNAs 1 and 2 with anticodon sequences that either supported efficient valylation or were incapable of valylation were similarly infectious to Nicotiana benthamiana plants (Matsuda et al., 2000). RNAs accumulated to similar levels in systemically infected leaves, and the input TLS sequences were unchanged. In direct competition experiments involving coinoculation of valylatable and nonvalylatable variants of RNA1 or RNA2, the valylatable RNAs outcompeted but did not entirely replace the nonvalylatable RNAs during systemic spread in individual plants (Matsuda et al., 2000). Thus, in the PCV system, there is a weak advantage conferred by valylatability for both RNAs. The existence of the anticodon mutation in strain P02A indicates that viability does not mandate valylation for RNA2. Nevertheless, sequencing of further PCV isolates has revealed intact anticodons in all cases, indicating that PCV-P02A is an unusual case with a mutation that may not be stable in the long term.
An explanation for the different importance of valylation in the TYMV and PCV systems may lie in the fact that valyl-PCV RNA interacts poorly with eEF1A•GTP. This would prevent a translational enhancer function for the TLS by mechanism described for TYMV. Translational enhancement does not need to be provided by features placed at the very 3′-end, as demonstrated by studies with TMV RNA (see below). It may be that the PCV TLS does not provide translational enhancement, and that this role has moved upstream to another feature in the rather long PCV 3′-UTR.
3.5. Tyrosylation of BMV RNA is needed for RNAs 1 and 2 but not RNA3
The role of tyrosylation has been investigated in the tripartite BMV system using two mutations (Δ5′ and 5′AGA) that were found to strongly decrease tyrosylation, with no affect on 3′ adenylation and no or minor effect on in vitro minus strand RNA synthesis (Dreher and Hall, 1988a; 1988b). Later studies have shown that these mutations in stem B3 fall outside the actual TLS (Fig. 3B) and have also described the strong tyrosine identity nucleotides in the acceptor stem (Fechter et al., 2001a) (Fig. 3). Mutations in these identity elements have not yet been utilized for experiments investigating the role of tyrosylation.
RNA3 with the Δ5′ or 5′AGA mutations replicated to normal levels in plants in the presence of wild type RNAs 1 and 2, and the input mutations were retained in progeny RNAs (Dreher et al., 1989). Thus, aminocylation of RNA3 is not needed and the replication of BMV RNAs does not mechanistically depend on aminoacylation, consistent with the central role of stem C in minus strand promotion (Chapman and Kao, 1999); stem C is an insertion element in the TLS with no analogy to the features of a tRNA. The same mutations introduced into RNAs 1 or 2 had different effects, especially in the case of mutation Δ5′, which lacks arm B3. Accumulation of all RNAs was strongly decreased in barley protoplasts when the Δ5′ mutation was carried in RNA1 (Rao, 2006b) or RNA2 (Rao and Hall, 1991). By contrast, inhibition of only RNA2 accumulation was observed in similar experiments in which RNA2 carried mutations that debilitated minus strand promoter activity (Rao and Hall, 1990). The trans-inhibition effects of certain TLS mutations carried on RNA1 or RNA2 (Rao, 2006b; Rao and Hall, 1991) and its relationship to the role of the tRNA-like properties of the TLS are not understood.
The dispensability of tyrosylation for RNA3 but sensitivity to tyrosylation mutations in RNAs 1 or 2 indicates that aminoacylation is not required for the process of BMV RNA synthesis. An involvement in enhancing translation, as observed for TYMV, would explain the greater importance for RNAs 1 and 2, which encode essential proteins needed to enable viral RNA synthesis. Strong expression enhancement in protoplasts from reporter mRNAs is indeed provided by the 3′ 200 nucleotides of BMV RNA3, as a result of both increased translational efficiency and mRNA half-life (Gallie and Kobayashi, 1994). Preliminary results suggest that tyrosylation is important for this effect (Matsuda and Dreher, unpub.). If tyrosylation does enhance translation in the BMV system, infectivity with tyrosylation-defective RNA3 may occur because sufficient translation from RNA3 and subgenomic RNA4 (made from RNA3) is supported despite the absence of 3′-enhancement; these RNAs may use another translation enhancer or be less dependent on 3′-enhancement than RNAs 1 and 2.
4. What are the selective advantages of tRNA mimicry?
The preceding discussion describing our current knowledge of the role of aminoacylation for TYMV suggests that translational enhancement and ribosome/replicase traffic control are the main functions provided by tRNA mimicry in the TYMV system. These may not be the only roles, however, and the TLSs of other viruses may serve other primary functions. The following sections describe additional or alternative TLS functions that have been studied; some hypothesized roles have not been supported by experimental results.
4.1. tRNA-like function on the ribosome?
Initial speculation over the role of TLSs during an infection explored a role closely following the function of tRNAs, which become aminoacylated, loaded into ternary complex with EF1A•GTP, and delivered to the ribosomal A site for the bound amino acid to participate in protein synthesis. This was considered as a potential way for viral RNAs to be competitively or preferentially translated (Hall, 1979; Litvak et al., 1973). Although some experiments in the 1970’s detected amino acid donation for ribosome-dependent peptide synthesis, other studies failed to do so, and it was concluded that transfer of the aminoacyl group via peptidyltransferase to nascent protein did not occur (reviewed in Haenni et al., 1982). The appearance of labeled valine from valyl-TYMV RNA into protein appeared to be the result of transfer to valyl-tRNA before incorporation.
More recently, experiments with TYMV RNA revived the possibility addressed in the earlier experiments (Barends et al., 2003). In this case, the TLS interaction with the ribosome was reported to support initiation in a manner that was independent of the 5′-cap, with the valine group from valyl-TYMV appearing at the N-terminus of viral protein. This scenario was proposed to pertain to the translation of only one of the two open reading frames expressed from TYMV RNA. Experiments were conducted in vitro with wheat germ extracts and N-terminal donation was deduced on the basis of [3H]valine release by dansyl chloride reactivity. The lack of positive identification of labeled viral protein by immunological or other means (Barends et al., 2003) left open the possibility that some other incorporation into TCA-precipitable material was being observed. One such possibility is suggested by the transfer of tyrosine originating from tyrosyl-BMV RNA to lysine amino groups (presumably through isopeptide bonds) on pre-existing proteins (Barends et al., 2004), a reaction similar to one previously identified with aspartyl-tRNA synthetase (Mejdoub et al., 1987).
In experiments similar to those with TYMV RNA, no evidence for amino acid donation to viral protein synthesis was observed with tyrosyl-BMV RNA (Barends et al., 2004) nor for histidyl-TMV or histidyl-NeRNV (tymovirus) RNAs (Rudinger-Thirion et al., 2006). Key aspects of the TYMV proposal (Barends et al., 2003) could not be repeated by (Matsuda and Dreher, 2007) using the same commercial wheat germ extracts, defined transcripts rather than ribonuclease H treatment to generate RNA variants lacking a TLS, and immunoprecipitation for positive product identification. In vivo studies also emphasized that TYMV gene expression occurs by conventional cap-dependent translation (Matsuda et al., 2004a), with bicistronic expression involving initiation coupling, a newly discovered variant of leaky scanning (Matsuda and Dreher, 2006). Suggestions that short reporter variants of TYMV RNA are translated via a different mode than full-length genomic RNAs (Rudinger-Thirion et al., 2006) are refuted by the observation of similar ribosomal toe-prints on TYMV-derived luciferase reporter mRNAs and genomic RNAs (Matsuda and Dreher, 2006; 2007).
There thus appears to be no credible current evidence for plant viral TLS function on the ribosome. Nevertheless, it would be premature to exclude all such possibilities, since no direct binding studies involving viral TLS-containing eEF1A•GTP ternary complexes and ribosomes have been reported. Some form of interaction at the ribosomal A site may occur in order to influence translation to favor the virus, an avenue of studies that deserves attention.
4.2. tRNA-like function in RNA replication as minus strand promoter?
The other main line of early speculation concerning the role of viral TLSs concerned minus strand initiation. tRNA-like properties were expected to contribute to the initiation of replication (Hall, 1979; Litvak et al., 1973), both because it occurs within the 3′-terminal TLS and by analogy to the makeup of Qβ replicase, which includes the translation factors EF-Tu and EF-Ts as subunits. In fact, it emerged that EF-Tu (EF1A) does not participate in Qβ replicase function by using its tRNA-associated properties (Stringfellow and Blumenthal, 1983). As discussed below, the accumulated evidence likewise indicates that minus strand synthesis of the plant viral TLS-bearing genomes does not occur via a mechanism that directly depends on their tRNA-like properties; although minus strand promoters are localized to the TLS, their function is not dependent on tRNA-associated properties.
These issues have been addressed with TYMV through extensive infectivity and cell-free replication studies. A number of chimeric TYMV genomes in which the TLS was replaced with heterologous sequences derived from the 3′-UTRs of other tymoviruses or TMV have proven to be infectious in Chinese cabbage plants (Filichkin et al., 2000; Goodwin et al., 1997; Skuzeski et al., 1996). Most telling was a chimera bearing the 3′-UTR of Erysiumum latent tymovirus (ErLV) (Filichkin et al., 2000; Goodwin et al., 1997), which possesses only vestigial tRNA character (Dreher and Goodwin, 1998) and essentially no sequence similarity to the TYMV TLS beyond the 3′-terminal CC(A). The TYMV replicase is clearly able to initiate minus strand synthesis and amplify genomes despite substantial variation in 3′-UTR sequences and the absence of tRNA mimicry. Enzymology studies using extracts with TYMV replicase activity made from infected plants have provided an explanation for this observation. In vitro minus strand initiation, which occurs opposite the 3′-C in the CCA terminus (Singh and Dreher, 1997), and subsequent template copying does not depend on the TLS (Gargouri-Bouzid et al., 1991), requiring only an accessible CCR (R = purine) initiation box (Deiman et al., 1998; 2000; Singh and Dreher, 1997; 1998). The lack of a mechanistic connection between promotion of RNA replication by TYMV replicase and tRNA mimicry is consistent with the inability to find eEF1A or valyl-tRNA synthetrase in TYMV replicase preparations (Joshi et al., 1986; Pulikowska et al., 1988), searches that were motivated by a proposed parallel with Qβ replicase. While template copying by TYMV replicase is not achieved by taking direct advantage of tRNA-like properties, the TLS may be considered an excellent format for making the 3′-CC(A) initiation box sterically accessible.
There have also been extensive studies with BMV on the involvement of the TLS in RNA replication. As with TYMV, minus strand initiation by BMV replicase occurs opposite the 3′-C in the -CCA terminus (Miller et al., 1986). In vitro transcription of RNAs containing the TLS was decreased by most of the mutations that also decreased tyrosylation or 3′-adenylation (Dreher and Hall, 1988a), suggesting a requirement for an intact TLS structure. The most inhibitory mutations, however, were those in the 3′-CCA initiation site and in the AUA sequence in the terminal loop of stem C (Fig. 3) (Bujarski et al., 1985; Dreher et al., 1984; Dreher and Hall, 1988b). Stem C, with its terminal and bulged loops, were subsequently shown to serve as the binding site for BMV replicase and to be capable of independently directing initiation to an accessible CCA 3′-end outside the context of the TLS (Chapman and Kao, 1999). Thus, as for TYMV, BMV minus strand synthesis is controlled by promoter elements embedded within the TLS but whose function do not directly rely on tRNA mimicry.
Minus strand synthesis has been less intensively studied with TMV. In vitro minus strand initiation by TMV-L replicase requires the 3′-CA and is sensitive to mutations disrupting both arms of the TLS, the central connecting region and an upstream pseudoknot (Osman et al., 2000). On the basis of competition experiments, the anticodon arm appears to be the major domain for replicase recognition, but features throughout the TLS and upstream pseudoknot domain also seem to be involved in replicase binding (Osman et al., 2000). As with the TYMV and BMV systems, there is no indication that aminoacylation is required for in vitro minus strand synthesis (Osman and Buck, 1996), but it remains uncertain whether minus strand synthesis and tRNA mimicry are independent in the TMV case as for TYMV and BMV.
4.3. CCA-NTase as 3′ telomerase
Because most of the TYMV RNA inoculum lacks the 3′-A necessary for aminoacylation, and since valylation is necessary for infectivity, 3′-adenylation by host CCA-NTase can be deduced as being an initial step in an infection. Note that, while no experiments have been conducted to definitively invoke a role for CCA-NTase instead of another host terminal transferase, the efficiency of TYMV RNA as a CCA-NTase substrate makes this enzyme the likely participant.
Evidence that more directly implicates CCA-NTase in maintaining intact 3′-CCA ends of BMV RNA has been obtained. Mutations in the RNA3 TLS that impair in vitro 3′-adenylation by CCA-NTase (coinoculated with wild type RNA1 and 2) led to decreased RNA amplification in protoplasts and plants(Dreher et al., 1989). Progeny RNAs 3 and 4 (subgenomic RNA produced from RNA3) had heterogeneous 3′-termini with the terminal A missing from a majority of RNAs. In a different experiment, RNA3 variants with mutations in the 3′-CA terminus, which simultaneously prevent tyrosylation and minus strand initiation (Dreher et al., 1984), replicated normally and without noticeable delay in protoplasts or plants (Rao et al., 1989). Progeny had acquired the correct 3′-CCA termini, suggesting that rapid turnover of the 3′-terminal nucleotides and repair by CCA-NTase had occurred, although it cannot be excluded that a replicase-dependent 3′-end repair mechanism using short aborted replication products from RNA1 or 2 as primers (Kao et al., 2001) could have contributed to terminus correction. Together, these studies suggest that CCA-NTase serves an important role as a telomerase in maintaining intact 3′-CCA termini. This is probably an important general role for TLSs, and even more important for tymoviruses, whose encapsidated RNAs lack a 3′-terminal A.
4.4. TLS as a 3′-translational enhancer and regulator of minus strand synthesis
Section 3.2 has described evidence that the TYMV TLS serves as a translational enhancer and in negatively regulating minus strand synthesis. It is not yet known how widespread these roles are among the various viruses with TLS-containing genomes. Experiments with chimeric TYMV genomes with switched TLSs indicate that the TLSs of two other tymoviruses adequately provide the functions needed for infectivity. TYMV genomes in which the TLS was replaced by the TLSs of Kennedya yellow mosaic (KYMV) or Eggplant mosaic (EMV) tymoviruses were both infectious, though attenuated, particularly in the case of the EMV chimera (Skuzeski et al., 1996). This is interesting, because both TLSs support efficient valylation, but the valyl-KYMV TLS binds eEF1A•GTP as tightly as the TYMV TLS while the valyl-EMV TLS binds considerably weaker (Dreher and Goodwin, 1998). The EMV chimera may have been more attenuated because of this difference. Spontaneous mutations that overcame much of the attenuation phenotype were located in the coding region of the TYMV movement protein/RNAi suppressor (Filichkin et al., 2000) rather than in the TLS, suggesting that other viral functions can adjust to some degree for differences in TLS function.
If the translational enhancement and regulation of minus strand synthesis roles of the TYMV TLS depend on the binding of eEF1A•GTP, the same roles should be provided by all TLSs that are capable of aminoacylation and ternary complex formation. If the prediction were correct, however, there is still the possibility that some viruses have acquired RNA elements that provide those functions in a different way, perhaps making the TLS contributions redundant. One can view this to be the case with TMV. Strong translational enhancement is provided by the 3′-untranslated region, but most of the enhancement originates from the pseudoknot domain immediately upstream of the TLS (Gallie et al., 1991; Gallie and Walbot, 1990; Leathers et al., 1993). The TLS itself provided only a minor supportive role, although this may have been underestimated if its function (as with TYMV) were aminoacylation-dependent, since the RNA constructs tested were not specifically designed with correct 3′-CCA termini. The feature with the strongest influence on translational enhancement is the pseudoknot immediately upstream of the TLS (UPSK-3; Fig. 2A)(Leathers et al., 1993), which is also essential for viral amplification in protoplasts and plants (Chandrika et al., 2000; Takamatsu et al., 1990). Intriguingly, this pseudoknot has been observed to bind eEF1A in a GTP-dependent way but independent of the TLS and aminoacylation (Zeenko et al., 2002). It is not known whether the eEF1A interaction is involved in translational enhancement, but such a role would parallel the postulated role of eEF1A binding to the TYMV TLS.
4.5. The TLS and RNA encapsidation
Experiments with BMV, for which capsid assembly and encapsidation can occur in vitro from purified components, have shown that the TLS is needed for RNA encapsidation (Choi et al., 2002). An overall tRNA-like structure is needed, based on studies with a mutation that disrupts the acceptor stem pseudoknot. The TLS need not be tyrosylated, however, and short CMV and TMV TLS-containing RNAs, as well as cellular tRNAs, are also effective in supporting encapsidation. The encapsidation of each of the BMV RNAs requires a TLS, which can be present either in cis or in trans (Choi et al., 2002). When present in trans, the TLS RNAs do not become encapsidated, suggesting they provide a transient role as chaperones during capsid assembly.
In plants, RNA3 will only become encapsidated when a TLS is present in cis, but RNAs 1 and 2 do not have that requirement (Annamalai and Rao, 2007). In view of the ability of cellular tRNAs to replace the TLS in in vitro encapsidation assays, host tRNAs may support RNA1 and 2 packaging in vivo (Rao, 2006a).
A role in encapsidation is not a universal role for viral TLSs, since the TLS is not needed for in vitro packaging of the closely related Cowpea chlorotic mottle bromovirus RNAs (Annamalai and Rao, 2005) nor for TYMV genomic RNA (Cho and Dreher, 2006). For some RNAs, it has not yet been possible to separate an involvement of the TLS in encapsidation from a requirement for replication and packaging to be coupled during the infection; this has been observed for the BMV (Annamalai and Rao, 2006) and TYMV (Cho and Dreher, 2006) subgenomic RNAs.
5. TLSs are evolutionarily exchangeable modules
5.1. Distinct TLSs do not exist in monophyletic lineages
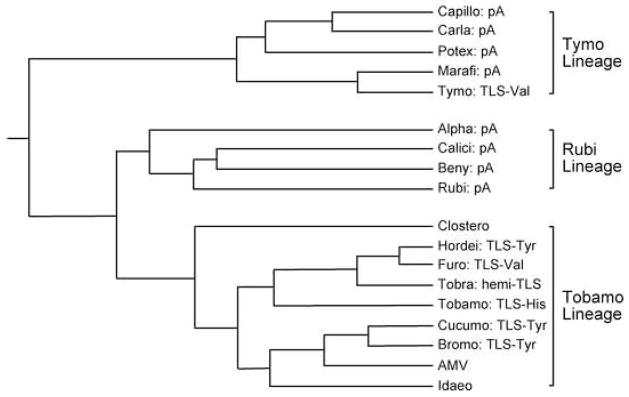
The viruses whose genomes have TLSs all belong to the Supergroup 3 lineage as classified according to viral RNA-dependent RNA polymerases (RdRp) (Koonin and Dolja, 1993). TLSs are represented in two of the three lineages in Supergroup 3, the Tymo and Tobamo lineages (Fig. 4). The Tobamo lineage includes no viruses whose genomes have poly(A) tails, but the full variety of other termini is represented: (i) TLSs of each of the three aminoacylation specificities, valine, histidine and tyrosine; (ii) TLS-related termini that have an acceptor-T arm but no anticodon-like domain, whose tRNA mimicry is restricted to 3′-adenylation by CCA-NTase (tobraviruses)(van Belkum et al., 1987b); (iii) termini with TLS-related structures that lack 3′-CCA ends and exhibit essentially no tRNA mimicry (AMV and ilarviruses) (Olsthoorn et al., 1999); (iv) termini with no known tRNA association (closteroviruses) (Fig. 4). In the Tymo lineage, the tymoviruses are the only viruses with TLS 3′-termini, all others possessing poly(A) tails. Surprisingly, not all tymoviruses have aminoacylatable TLSs, ErLV being an example of a tymovirus with only vestigial tRNA mimicry that is detectable as 3′-adenylation by CCA-NTase (Dreher and Goodwin, 1998). Evolution thus seems to rather readily allow the development or acquisition of different 3′-termini.
Fig. 4. Phylogenetic distribution of TLSs.
The occurrence of distinct classes of 3′-termini is shown for selected genera of the three lineages of viruses that have been classified to Supergroup 3 on the basis of their RdRp sequences (based on Koonin and Dolja, 1993). The 3′-termini are indicated as: pA, poly(A) tail; TLS with aminoacyl specificity indicated; hemi-TLS, to indicate an acceptor-T arm not capable of aminoacylation; no indication for viruses whose genomes have none of the above types of termini. Branch lengths are approximate.
There is clear evidence that TLSs have been exchanged between viruses as independent genome modules, in parallel with the prevalent horizontal gene exchange of open reading frames that has occurred during the evolution of positive strand RNA viruses (Dolja and Carrington, 1992). Among the Tobamo lineage, the hordeiviruses (tyrosyl-TLS) are more closely related to the furo-like viruses (valyl-TLSs) than the other viruses with similar TLSs (bromo- and cucumoviruses). The furo-like viruses and tymoviruses have essentially the same TLS termini but are in different lineages. Most strikingly, there are two instances of reciprocal exchanges between tymoviruses and tobamoviruses: Sunnhemp mosaic tobamovirus (formerly known as the cowpea strain of TMV, CcTMV) has a tymo-like valylatable TLS (Beachy et al., 1976), while Neresia ring necrosis tymovirus has a tobamo-like histidylatable TLS (Koenig et al., 2005). Both of these chimeric viruses are unique outliers in their genus with respect to TLS properties, and must represent recent genetic recombination events.
5.2. 3′-UTR function can be provided with and without tRNA mimicry
The importance of the TLS and its highly evolved tRNA mimicry in TYMV was discussed in section 3.1. The properties of engineered chimeric TYMV genomes with various elements replacing the TLS have indicated, surprisingly, that the TLS is rather readily replaceable with 3′-terminal modules that are incapable of aminoacylation (Filichkin et al., 2000; Goodwin et al., 1997). There certainly are strong restrictions on 3′-end compatibility (Skuzeski et al., 1996), but certain viral termini with appropriate properties are compatible. According to our understanding of TYMV TLS function, these properties would be (i) provision of an accessible 3′-CC(A) initiation site for minus strand synthesis, (ii) provision of translational enhancement, (iii) ability to down-regulate minus strand initiation, and (iv) maintenance of an intact 3′-terminus. Those properties can be provided in various ways, not only through an aminoacylatable TLS. For instance, several different 3′-translational enhancer elements have been described (Dreher and Miller, 2006; Gale et al., 2000; Gallie, 1998), including a poly(A) tail (Sachs et al., 1997), the pseudoknot domain upstream of the TMV TLS (Gallie and Walbot, 1990), the 3′-stem-loop of histone mRNAs (Ling et al., 2002), and the coat protein-bound 3′-terminus of AMV RNA (Krab et al., 2005). Since translational enhancers are thought to work through the recruitment of a bound protein, the opportunity for repressing minus strand synthesis should present itself with any potential translational enhancer. The presence of a bound protein should also stabilize the 3′-end against nuclease attack.
How the chimeric TYMV genomes TYMC-H, -XX and -YY (Goodwin et al., 1997) manage to function despite the absence of aminoacylation is not fully known. However, the 3′-termini of each of these genomes terminates in -CC(A). Unpublished experiments have shown that the ErLV-derived 3′-end of TYMC-H does provide translational enhancement (Matsuda and Dreher), while that role is expected to be provided in TYMC-XX and -YY by the TMV-derived upstream pseudoknots. Whether or to what extent (and how) negative regulation of minus strand synthesis might be provided by these chimeras is not known, but this issue might be flexible, as suggested by the viability of the EMV chimera described in section 4.4.
The understanding from studying these chimeric genomes is that the TYMV TLS is a modular genome element that has a stand-alone function that can be rather readily provided by a different cis-element. That insight is emphasized by the existence of a range of 3′ termini with varying tRNA mimicry among the Tymovirus genus (Dreher and Goodwin, 1998). The presence of a minus strand promoter element within the BMV TLS (Chapman and Kao, 1999) constitutes a genetic linkage between the RdRp gene and the TLS that should limit the evolutionary fluidity of the TLS and the rate of genetic exchange of the 3′-end. Indeed, the BMV-like TLS is widely conserved among the Bromoviridae (Ahlquist et al., 1981) and TWD, unpub. observations). Although the TMV minus strand promoter has also been reported to reside within, or overlap, the TLS (Osman et al., 2000), the chimeric nature of the Sunnhemp mosaic virus genome with its tymo-like TLS (Beachy et al., 1976) indicates some tolerance of 3′ module switching among the tobamoviruses.
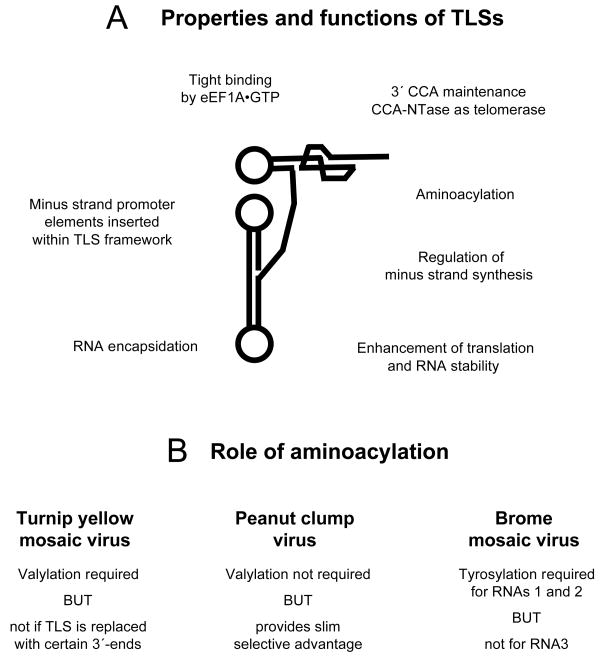
6. Conclusions: TLS variability in form and function
The information described in this review presents a picture of structurally varied TLSs (Figs. 1–3), and of varied functions provided by TLSs (Fig. 5). These can differ widely even between similar TLSs. For example, valylation of the TLS is necessary for the infectivity of TYMV, but not of PCV RNA. The roles of the TLSs present on the RNAs of multipartite genomes can vary from segment to segment. Thus, the TLS is required in cis for in vivo encapsidation of BMV RNA3, but not of RNAs 1 and 2.
Fig. 5. Varied functions of TLSs and significance of tRNA mimicry during infection.
A Plant viral TLSs have a number of properties and provide varied and different functions in different viral RNAs. B. The role of TLS aminoacylation differs between the three viruses in which its importance for infectivity has been assessed.
We have probably not yet determined all of the roles served by TLSs in the course of viral infections. Future discoveries that expand the repertoire of functions will add to the already remarkable properties of these highly sophisticated tRNA mimics that have coopted the properties of tRNAs from translation elongation to support viral replication.
Acknowledgments
Research on viral TLSs in the author’s laboratory over the years has benefited by grants from USDA, NSF, NIH and Oregon State University. The author is grateful for collaborations that have been conducted with colleagues, and especially for the contributions of members of the laboratory at Oregon State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agranovsky AA, Dolja VV, Gorbulev VG, Kozlov YV, Atabekov JG. Aminoacylation of barley stripe mosaic virus RNA: polyadenylate-containing RNA has a 3′-terminal tyrosine-accepting structure. Virology. 1981;113:174–187. doi: 10.1016/0042-6822(81)90146-x. [DOI] [PubMed] [Google Scholar]
- Agranovsky AA, Karasev AV, Novikov VK, Lunina NA, Loginov S, Tyulkina LG. Poa semilatent virus, a hordeivirus having no internal polydisperse poly(A) in the 3′ non-coding region of the RNA genome. J Gen Virol. 1992;73:2085–92. doi: 10.1099/0022-1317-73-8-2085. [DOI] [PubMed] [Google Scholar]
- Ahlquist P, Dasgupta R, Kaesberg P. Near identity of 3′-RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell. 1981;23:183–9. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- Annamalai P, Rao AL. Dispensability of 3′ tRNA-like sequence for packaging cowpea chlorotic mottle virus genomic RNAs. Virology. 2005;332:650–8. doi: 10.1016/j.virol.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Annamalai P, Rao AL. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J Virol. 2006;80:10096–108. doi: 10.1128/JVI.01186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai P, Rao AL. In vivo packaging of brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis-acting 3′ tRNA-like structure. J Virol. 2007;81:173–81. doi: 10.1128/JVI.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S, Bink HH, van den Worm SH, Pleij CW, Kraal B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–9. doi: 10.1016/s0092-8674(02)01256-4. [DOI] [PubMed] [Google Scholar]
- Barends S, Rudinger-Thirion J, Florentz C, Giegé R, Pleij CW, Kraal B. tRNA-like structure regulates translation of Brome mosaic virus RNA. J Virol. 2004;78:4003–10. doi: 10.1128/JVI.78.8.4003-4010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin M, Hall TC. Interaction of elongation factor 1 with aminoacylated brome mosaic virus and tRNA’s. J Virol. 1976;20:117–22. doi: 10.1128/jvi.20.1.117-122.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark T, Ahlquist P. The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TψC-stem loop and is modified in vivo. RNA. 2001;7:1652–70. [PMC free article] [PubMed] [Google Scholar]
- Beachy RN, Zaitlin M, Bruening G, Israel HW. A genetic map for the cowpea strain on TMV. Virology. 1976;73:498–507. doi: 10.1016/0042-6822(76)90411-6. [DOI] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Florentz C, Giegé R, Grosjean H. Pseudouridine and ribothymidine formation in the tRNA-like domain of turnip yellow mosaic virus RNA. Nucleic Acids Res. 1998;26:3991–7. doi: 10.1093/nar/26.17.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol JF. Replication of alfamo- and ilarviruses: role of the coat protein. Annual Reviews of Phytopathology. 2005;43:39–62. doi: 10.1146/annurev.phyto.43.101804.120505. [DOI] [PubMed] [Google Scholar]
- Brulé H, Grosjean H, Giegé R, Florentz C. A pseudoknotted tRNA variant is a substrate for tRNA (cytosine-5)-methyltransferase from Xenopus laevis. Biochimie. 1998;80:977–85. doi: 10.1016/s0300-9084(99)80003-0. [DOI] [PubMed] [Google Scholar]
- Bujarski JJ, Dreher TW, Hall TC. Deletions in the 3′-terminal tRNA-like structure of brome mosaic virus RNA differentially affect aminoacylation and replication in vitro. Proc Natl Acad Sci U S A. 1985;82:5636–40. doi: 10.1073/pnas.82.17.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika R, Rabindran S, Lewandowski DJ, Manjunath KL, Dawson WO. Full-length tobacco mosaic virus RNAs and defective RNAs have different 3′ replication signals. Virology. 2000;273:198–209. doi: 10.1006/viro.2000.0414. [DOI] [PubMed] [Google Scholar]
- Chapman MR, Kao CC. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J Mol Biol. 1999;286:709–20. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- Cho TJ, Dreher TW. Encapsidation of genomic but not subgenomic Turnip yellow mosaic virus RNA by coat protein provided in trans. Virology. 2006;356:126–35. doi: 10.1016/j.virol.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Choi YG, Dreher TW, Rao AL. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc Natl Acad Sci U S A. 2002;99:655–60. doi: 10.1073/pnas.022618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiman BALM, Koenen AK, Verlaan PWG, Pleij CWA. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998;72:3965–72. doi: 10.1128/jvi.72.5.3965-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiman BALM, Verlaan PWG, Pleij CWA. In vitro transcription by the turnip yellow mosaic virus RNA polymerase: a comparison with the alfalfa mosaic virus and brome mosaic virus replicases. J Virol. 2000;74:264–71. doi: 10.1128/jvi.74.1.264-271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. The Enzymes. XV. Academic Press, Inc; 1982. tRNA nucleotidyltransferase; pp. 183–215. [Google Scholar]
- Dolja V, Carrington J. Evolution of positive-strand RNA viruses. Seminars in Virology. 1992;3:315–326. [Google Scholar]
- Dreher TW. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu Rev Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- Dreher TW, Bujarski JJ, Hall TC. Mutant viral RNAs synthesized in vitro show altered aminoacylation and replicase template activities. Nature. 1984;311:171–5. doi: 10.1038/311171a0. [DOI] [PubMed] [Google Scholar]
- Dreher TW, Goodwin JB. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998;26:4356–64. doi: 10.1093/nar/26.19.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Hall TC. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988a;201:31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- Dreher TW, Hall TC. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. Sequence and structural requirements for aminoacylation and 3′-adenylation. J Mol Biol. 1988b;201:41–55. doi: 10.1016/0022-2836(88)90437-8. [DOI] [PubMed] [Google Scholar]
- Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–97. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Rao ALN, Hall TC. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J Mol Biol. 1989;206:425–38. doi: 10.1016/0022-2836(89)90491-9. [DOI] [PubMed] [Google Scholar]
- Dreher TW, Tsai CH, Florentz C, Giege R. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry. 1992;31:9183–9. doi: 10.1021/bi00153a010. [DOI] [PubMed] [Google Scholar]
- Dreher TW, Tsai CH, Skuzeski JM. Aminoacylation identity switch of turnip yellow mosaic virus RNA from valine to methionine results in an infectious virus. Proc Natl Acad Sci U S A. 1996;93:12212–6. doi: 10.1073/pnas.93.22.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Uhlenbeck OC, Browning K. Quantitative assessment of EF-1a-GTP binding to aminoacyl-tRNA, aminoacyl-viral RNA and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J Biol Chem. 1999;274:666–672. doi: 10.1074/jbc.274.2.666. [DOI] [PubMed] [Google Scholar]
- Fechter P, Giege R, Rudinger-Thirion J. Specific tyrosylation of the bulky tRNA-like structure of brome mosaic virus RNA relies solely on identity nucleotides present in its amino acid-accepting domain. J Mol Biol. 2001a;309:387–99. doi: 10.1006/jmbi.2001.4654. [DOI] [PubMed] [Google Scholar]
- Fechter P, Rudinger-Thirion J, Florentz C, Giege R. Novel features in the tRNA-like world of plant viral RNAs. Cell Mol Life Sci. 2001b;58:1547–61. doi: 10.1007/PL00000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechter P, Rudinger-Thirion J, Theobald-Dietrich A, Giege R. Identity of tRNA for yeast tyrosyl-tRNA synthetase: tyrosylation is more sensitive to identity nucleotides than to structural features. Biochemistry. 2000;39:1725–33. doi: 10.1021/bi992276t. [DOI] [PubMed] [Google Scholar]
- Felden B, Florentz C, Giege R, Westhof E. A central pseudoknotted three-way junction imposes tRNA-like mimicry and the orientation of three 5′ upstream pseudoknots in the 3′ terminus of tobacco mosaic virus RNA. RNA. 1996;2:201–12. [PMC free article] [PubMed] [Google Scholar]
- Felden B, Florentz C, McPherson A, Giege R. A histidine accepting tRNA-like fold at the 3′-end of satellite tobacco mosaic virus RNA. Nucleic Acids Res. 1994;22:2882–6. doi: 10.1093/nar/22.15.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Bransom KL, Goodwin JB, Dreher TW. The infectivities of turnip yellow mosaic virus genomes with altered tRNA mimicry are not dependent on compensating mutations in the viral replication protein. J Virol. 2000;74:8368–75. doi: 10.1128/jvi.74.18.8368-8375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentz C, Dreher TW, Rudinger J, Giegé R. Specific valylation identity of turnip yellow mosaic virus RNA by yeast valyl-tRNA synthetase is directed by the anticodon in a kinetic rather than affinity-based discrimination. Eur J Biochem. 1991;195:229–34. doi: 10.1111/j.1432-1033.1991.tb15698.x. [DOI] [PubMed] [Google Scholar]
- Florentz C, Giegé R. tRNA-like structures in plant viral RNAs. In: Söll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis, and Function. ASM Press; Washington, DC: 1995. pp. 141–163. [Google Scholar]
- Gale M, Jr, Tan SL, Katze MG. Translational control of viral gene expression in eukaryotes. Microbiol Mol Biol Rev. 2000;64:239–80. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Feder JN, Schimke RT, Walbot V. Functional analysis of the tobacco mosaic virus tRNA-like structure in cytoplasmic gene regulation. Nucleic Acids Res. 1991;19:5031–6. doi: 10.1093/nar/19.18.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Kobayashi M. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene. 1994;142:159–65. doi: 10.1016/0378-1119(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Walbot V. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 1990;4:1149–57. doi: 10.1101/gad.4.7.1149. [DOI] [PubMed] [Google Scholar]
- Gamarnik A, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes and Development. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Arenal F. Sequence and structure at the genome 3′ end of the U2-strain of tobacco mosaic virus, a histidine-accepting tobamovirus. Virology. 1988;167:201–6. doi: 10.1016/0042-6822(88)90070-0. [DOI] [PubMed] [Google Scholar]
- Gargouri-Bouzid R, David C, Haenni AL. The 3′ promoter region involved in RNA synthesis directed by the turnip yellow mosaic virus genome in vitro. FEBS Lett. 1991;294:56–8. doi: 10.1016/0014-5793(91)81342-6. [DOI] [PubMed] [Google Scholar]
- Giegé R, Briand JP, Mengual R, Ebel JP, Hirth L. Valylation of the two RNA components of turnip-yellow mosaic virus and specificity of the tRNA aminoacylation reaction. Eur J Biochem. 1978;84:251–6. doi: 10.1111/j.1432-1033.1978.tb12163.x. [DOI] [PubMed] [Google Scholar]
- Giegé R, Florentz C, Dreher TW. The TYMV tRNA-like structure. Biochimie. 1993;75:569–82. doi: 10.1016/0300-9084(93)90063-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JB, Dreher TW. Transfer RNA mimicry in a new group of positive-strand RNA plant viruses, the furoviruses: differential aminoacylation between the RNA components of one genome. Virology. 1998;246:170–8. doi: 10.1006/viro.1998.9193. [DOI] [PubMed] [Google Scholar]
- Goodwin JB, Skuzeski JM, Dreher TW. Characterization of chimeric turnip yellow mosaic virus genomes that are infectious in the absence of aminoacylation. Virology. 1997;230:113–24. doi: 10.1006/viro.1997.8475. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, van Belkum A, Pleij CW, Altman S. Novel reactions of RNase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988;53:267–72. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Gultyaev AP, van Batenburg E, Pleij CW. Similarities between the secondary structure of satellite tobacco mosaic virus and tobamovirus RNAs. J Gen Virol. 1994;75:2851–6. doi: 10.1099/0022-1317-75-10-2851. [DOI] [PubMed] [Google Scholar]
- Haenni AL, Chapeville F. An enigma: the role of viral RNA aminoacylation. Acta Biochim Pol. 1997;44:827–37. [PubMed] [Google Scholar]
- Haenni AL, Joshi S, Chapeville F. tRNA-like structures in the genomes of RNA viruses. Prog Nucleic Acid Res Mol Biol. 1982;27:85–104. doi: 10.1016/s0079-6603(08)60598-x. [DOI] [PubMed] [Google Scholar]
- Hall TC. Transfer RNA-like structures in viral genomes. Int Rev Cytol. 1979;60:1–26. doi: 10.1016/s0074-7696(08)61257-7. [DOI] [PubMed] [Google Scholar]
- Hegg LA, Kou M, Thurlow DL. Recognition of the tRNA-like structure in tobacco mosaic viral RNA by ATP/CTP:tRNA nucleotidyltransferases from Escherichia coli and Saccharomyces cerevisiae. J Biol Chem. 1990;265:17441–5. [PubMed] [Google Scholar]
- Joshi R, Haenni AL. Search for tRNA-like properties in tomato aspermy virus RNA. FEBS Letters. 1986;194:157–194. [Google Scholar]
- Joshi RL, Chapeville F, Haenni AL. Conformational requirements of tobacco mosaic virus RNA for aminoacylation and adenylation. Nucleic Acids Res. 1985;13:347–54. doi: 10.1093/nar/13.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi RL, Joshi S, Chapeville F, Haenni AL. tRNA-like structures of plant viral RNAs: conformational requirements for adenylation and aminoacylation. EMBO J. 1983;2:1123–7. doi: 10.1002/j.1460-2075.1983.tb01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi RL, Ravel JM, Haenni AL. Interaction of turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1 [alpha]. Search for a function. EMBO J. 1986;5:1143–8. doi: 10.1002/j.1460-2075.1986.tb04339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Chapeville F, Haenni AL. Length requirements for tRNA-specific enzymes and cleavage specificity at the 3′ end of turnip yellow mosaic virus RNA. Nucleic Acids Res. 1982a;10:1947–62. doi: 10.1093/nar/10.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Chapeville F, Haenni AL. Turnip yellow mosaic virus RNA is aminoacylated in vivo in Chinese cabbage leaves. EMBO J. 1982b;1:935–938. doi: 10.1002/j.1460-2075.1982.tb01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–13. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CC, Singh P, Ecker DJ. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–60. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- Kim CH, Kao CC, Tinoco I., Jr RNA motifs that determine specificity between a viral replicase and its promoter. Nat Struct Biol. 2000;7:415–23. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- Koenig R, Barends S, Gultyaev AP, Lesemann DE, Vetten HJ, Loss S, Pleij CW. Nemesia ring necrosis virus: a new tymovirus with a genomic RNA having a histidylatable tobamovirus-like 3′ end. J Gen Virol. 2005;86:1827–33. doi: 10.1099/vir.0.80916-0. [DOI] [PubMed] [Google Scholar]
- Koenig R, Beier C, Commandeur U, Luth U, Kaufmann A, Luddecke P. Beet soil-borne virus RNA 3--a further example of the heterogeneity of the gene content of furovirus genomes and of triple gene block-carrying RNAs. Virology. 1996;216:202–7. doi: 10.1006/viro.1996.0047. [DOI] [PubMed] [Google Scholar]
- Kohl RJ, Hall TC. Aminoacylation of RNA from several viruses: amino acid specificity and differential activity of plant, yeast and bacterial synthetases. J Gen Virol. 1974;25:257–61. doi: 10.1099/0022-1317-25-2-257. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, Weissmann C. Possible mechanism for transition of viral RNA from polysome to replication complex. Nat New Biol. 1971;231:42–6. doi: 10.1038/newbio231042a0. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Krab IM, Caldwell C, Gallie DR, Bol JF. Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J Gen Virol. 2005;86:1841–9. doi: 10.1099/vir.0.80796-0. [DOI] [PubMed] [Google Scholar]
- Leathers V, Tanguay R, Kobayashi M, Gallie DR. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol Cell Biol. 1993;13:5331–47. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR. The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol Cell Biol. 2002;22:7853–67. doi: 10.1128/MCB.22.22.7853-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak S, Tarrago A, Tarrago-Litvak L, Allende JE. Elongation factor-viral genome interaction dependent on the aminoacylation of TYMV and TMV RNAs. Nat New Biol. 1973;241:88–90. doi: 10.1038/newbio241088a0. [DOI] [PubMed] [Google Scholar]
- Loesch-Fries LS, Hall TC. In vivo aminoacylation of brome mosaic and barley stripe mosaic virus RNAs. Nature. 1982;298:771–773. [Google Scholar]
- Manohar SK, Guilley H, Dollet M, Richards K, Jonard G. Nucleotide sequence and genetic organization of peanut clump virus RNA 2 and partial characterization of deleted forms. Virology. 1993;195:33–41. doi: 10.1006/viro.1993.1343. [DOI] [PubMed] [Google Scholar]
- Mans RM, Pleij CW, Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur J Biochem. 1991;201:303–24. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- Matsuda D, Bauer L, Tinnesand K, Dreher TW. Expression of the two nested overlapping reading frames of turnip yellow mosaic virus RNA is enhanced by a 5′ cap and by 5′ and 3′ viral sequences. J Virol. 2004a;78:9325–35. doi: 10.1128/JVI.78.17.9325-9335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA in a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA. 2006;12:1338–49. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. Cap- and initiator tRNA-dependent initiation of TYMV polyprotein synthesis by ribosomes: evaluation of the Trojan horse model for TYMV RNA translation. RNA. 2007;13:129–37. doi: 10.1261/rna.244407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Dunoyer P, Hemmer O, Fritsch C, Dreher TW. The valine anticodon and valylatability of Peanut clump virus RNAs are not essential but provide a modest competitive advantage in plants. J Virol. 2000;74:8720–5. doi: 10.1128/jvi.74.18.8720-8725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Yoshinari S, Dreher TW. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004b;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- Matthews R. Plant Virology. 3. Academic Press, Inc; San Diego: 1991. [Google Scholar]
- Mejdoub H, Kern D, Giege R, Ebel JP, Boulanger Y, Reinbolt J. Covalent aspartylation of aspartyl-tRNA synthetase from bakers’ yeast by its cognate aspartyl adenylate: identification of the labeled residues. Biochemistry. 1987;26:2054–9. doi: 10.1021/bi00381a039. [DOI] [PubMed] [Google Scholar]
- Miller WA, Bujarski JJ, Dreher TW, Hall TC. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986;187:537–46. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- Nameki N, Asahara H, Shimizu M, Okada N, Himeno H. Identity elements of Saccharomyces cerevisiae tRNA(His) Nucleic Acids Res. 1995;23:389–94. doi: 10.1093/nar/23.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn RC, Mertens S, Brederode FT, Bol JF. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman TA, Buck KW. Complete replication in vitro of tobacco mosaic virus RNA by a template- dependent, membrane-bound RNA polymerase. J Virol. 1996;70:6227–34. doi: 10.1128/jvi.70.9.6227-6234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman TA, Hemenway CL, Buck KW. Role of the 3′ tRNA-like structure in tobacco mosaic virus minus-strand RNA synthesis by the viral RNA-dependent RNA polymerase In vitro. J Virol. 2000;74:11671–80. doi: 10.1128/jvi.74.24.11671-11680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret V, Florentz C, Dreher T, Giege R. Structural analogies between the 3′ tRNA-like structure of brome mosaic virus RNA and yeast tRNATyr revealed by protection studies with yeast tyrosyl-tRNA synthetase. Eur J Biochem. 1989;185:331–9. doi: 10.1111/j.1432-1033.1989.tb15120.x. [DOI] [PubMed] [Google Scholar]
- Pinck M, Yot P, Chapeville F, Duranton HM. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature. 1970;226:954–6. doi: 10.1038/226954a0. [DOI] [PubMed] [Google Scholar]
- Pulikowska J, Wojtaszek P, Korcz A, Michalski Z, Candresse T, Twardowski T. Immunochemical properties of elongation factors 1 of plant origin. Eur J Biochem. 1988;171:131–6. doi: 10.1111/j.1432-1033.1988.tb13768.x. [DOI] [PubMed] [Google Scholar]
- Rao AL. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006a;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- Rao AL. Sensitivity of brome mosaic virus RNA1 replication to mutations in the 3′ tRNA-like structure implies a requirement for sustained synthesis of replicase protein 1a. Arch Virol. 2006b;151:721–33. doi: 10.1007/s00705-005-0658-y. [DOI] [PubMed] [Google Scholar]
- Rao ALN, Dreher TW, Marsh LE, Hall TC. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc Natl Acad Sci U S A. 1989;86:5335–9. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao ALN, Hall TC. Requirement for a viral trans-acting factor encoded by brome mosaic virus RNA-2 provides strong selection in vivo for functional recombinants. J Virol. 1990;64:2437–2441. doi: 10.1128/jvi.64.5.2437-2441.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao ALN, Hall TC. Interference in trans with brome mosaic virus replication by RNA-2 bearing aminoacylation-deficient mutants. Virology. 1991;180:16–22. doi: 10.1016/0042-6822(91)90004-u. [DOI] [PubMed] [Google Scholar]
- Rietveld K, Linschooten K, Pleij CW, Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984;3:2613–2619. doi: 10.1002/j.1460-2075.1984.tb02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K, Pleij CW, Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 1983;2:1079–85. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K, Van Poelgeest R, Pleij CW, Van Boom JH, Bosch L. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982;10:1929–46. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J, Felden B, Florentz C, Giege R. Strategy for RNA recognition by yeast histidyl-tRNA synthetase. Bioorg Med Chem. 1997;5:1001–9. doi: 10.1016/s0968-0896(97)00061-8. [DOI] [PubMed] [Google Scholar]
- Rudinger J, Florentz C, Dreher T, Giege R. Efficient mischarging of a viral tRNA-like structure and aminoacylation of a minihelix containing a pseudoknot: histidinylation of turnip yellow mosaic virus RNA. Nucleic Acids Res. 1992;20:1865–70. doi: 10.1093/nar/20.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J, Florentz C, Giege R. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues -1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994;22:5031–7. doi: 10.1093/nar/22.23.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger-Thirion J, Olsthoorn RC, Giege R, Barends S. Idiosyncratic behaviour of tRNA-like structures in translation of plant viral RNA genomes. J Mol Biol. 2006;355:873–8. doi: 10.1016/j.jmb.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–8. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Schulman LH, Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988;242:765–8. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Shirako Y, Wilson TM. Complete nucleotide sequence and organization of the bipartite RNA genome of soil-borne wheat mosaic virus. Virology. 1993;195:16–32. doi: 10.1006/viro.1993.1342. [DOI] [PubMed] [Google Scholar]
- Singh RN, Dreher TW. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997;233:430–9. doi: 10.1006/viro.1997.8621. [DOI] [PubMed] [Google Scholar]
- Singh RN, Dreher TW. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and -CCR- boxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998;4:1083–95. doi: 10.1017/s1355838298980694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski JM, Bozarth CS, Dreher TW. The turnip yellow mosaic virus tRNA-like structure cannot be replaced by generic tRNA-like elements or by heterologous 3′ untranslated regions known to enhance mRNA expression and stability. J Virol. 1996;70:2107–15. doi: 10.1128/jvi.70.4.2107-2115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringfellow L, Blumenthal T. Q beta replicase containing a Bacillus stearothermophilus elongation factor. J Bacteriol. 1983;153:1083–7. doi: 10.1128/jb.153.2.1083-1087.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu N, Watanabe Y, Meshi T, Okada Y. Mutational analysis of the pseudoknot region in the 3′ noncoding region of tobacco mosaic virus RNA. J Virol. 1990;64:3686–93. doi: 10.1128/jvi.64.8.3686-3693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CH, Dreher TW. Turnip yellow mosaic virus RNAs with anticodon loop substitutions that result in decreased valylation fail to replicate efficiently. J Virol. 1991;65:3060–7. doi: 10.1128/jvi.65.6.3060-3067.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CH, Dreher TW. Second-site suppressor mutations assist in studying the function of the 3′ noncoding region of turnip yellow mosaic virus RNA. J Virol. 1992;66:5190–9. doi: 10.1128/jvi.66.9.5190-5199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Abrahams JP, Pleij CW, Bosch L. Five pseudoknots are present at the 204 nucleotides long 3′ noncoding region of tobacco mosaic virus RNA. Nucleic Acids Res. 1985;13:7673–86. doi: 10.1093/nar/13.21.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Bingkun J, Rietveld K, Pleij CWA, Bosch L. Structural similarities among valine-accepting tRNA-like structures in tymoviral RNAs and elongator tRNAs. Biochemistry. 1987a;26:1144–1151. [Google Scholar]
- van Belkum A, Cornelissen B, Linthorst H, Bol J, Pley C, Bosch L. tRNA-like properties of tobacco rattle virus RNA. Nucleic Acids Res. 1987b;15:2837–50. doi: 10.1093/nar/15.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientges J, Putz J, Giege R, Florentz C, Schwienhorst A. Selection of viral RNA-derived tRNA-like structures with improved valylation activities. Biochemistry. 2000;39:6207–18. doi: 10.1021/bi992852l. [DOI] [PubMed] [Google Scholar]
- Yot P, Pinck M, Haenni AL, Duranton HM, Chapeville F. Valine-specific tRNA-like structure in turnip yellow mosaic virus RNA. Proc Natl Acad Sci U S A. 1970;67:1345–52. doi: 10.1073/pnas.67.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeenko VV, Ryabova LA, Spirin AS, Rothnie HM, Hess D, Browning KS, Hohn T. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J Virol. 2002;76:5678–91. doi: 10.1128/JVI.76.11.5678-5691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]