Abstract
Background
In 2001, the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program established Residual Tissue Repositories (RTR) in the Hawaii, Iowa, and Los Angeles Tumor Registries to collect discarded tissue blocks from pathologic laboratories within their catchment areas. To validate the utility of the RTR for supplementing SEER’s central database, we assessed human epidermal growth factor receptor-2 (HER2) and estrogen receptor expression (ER) in a demonstration project.
Materials
Using a prepared set of tissue microarrays (TMAs) residing in the Hawaii Tumor Registry (HTR), we performed standard immunohistochemistry. Breast cancers in the TMA were diagnosed in 1995, followed through 2006, and linked to SEER’s main database.
Results
The TMA included 354 cases, representing 51% of 687 breast cancers in the HTR (1995). The HTR and TMA cases were similar with respect to patient demographics and tumor characteristics. Seventy-six percent (76%, 268 of 354) of TMA cases were HER2+ and/or ER+, i.e., 28 HER2+ER−, 12 HER2+ER+, and 228 HER2−ER+. There were 67 HER2−ER− cases and 19 were unclassified. Age distributions at diagnosis were bimodal with dominant early-onset modes for HER2+ER−tumors and dominant late-onset modes for HER2−ER+ breast cancers. Epidemiologic patterns for concordant HER2+ER+ (double-positive) and HER2−ER−(double-negative) were intermediate to discordant HER2+ER− and HER2−ER+.
Conclusion
Results showed contrasting incidence patterns for HER2+ (HER2+ER−) and ER+ (HER2−ER+) breast cancers, diagnosed in 1995. Though sample sizes were small, this demonstration project validates the potential utility of the RTR for supplementing the SEER program.
Keywords: Immunohistochemical stains, Tissue microarrays, Human epidermal growth factor receptor-2 (HER2), Estrogen receptor (ER), Breast cancer incidence and survival, SEER
Introduction
In 1973, the National Cancer Institute established the Surveillance, Epidemiology, and End Results (SEER) program to collect incidence and population data by age at diagnosis, year of diagnosis, and geographic location (SEER Tumor Registry) for selected patient demographics and tumor characteristics. In 2001, SEER supplemented tumor registries in Hawaii, Iowa, and Los Angeles County to collect discarded formalin-fixed, paraffin-embedded tissue blocks from pathologic laboratories within their catchment areas [1]. SEER’s Residual Tissue Repository (RTR) creates the opportunity to complement its central database, provided that the discarded tissue blocks are representative of the entire registry, the material can be suitably prepared for analysis, and the statistical approaches relate data in the RTR to the entire registry.
For example, SEER has not collected all case-related data for all time periods. Age-at-diagnosis, Black–White race, histopathological subtype, and tumor grade were recorded from 1973 forward (1973+). Extent of disease codes (EOD-88 3rd edition) for tumor size and axillary lymph nodes (LN) were recorded from 1988 forward (1988+). Estrogen receptor (ER) was recorded from 1990 forward (1990+). Human epidermal growth factor receptor-2 (HER2) was never collected, but it could be measured in the RTR.
In this report, we validate the utility of SEER’s Residual Tissue Repository for assessing data missing in SEER, such as HER2 expression. We applied standard IHC stains to a prepared set of tissue microarrays (TMAs), residing in the Hawaii Tumor Registry (HTR). This demonstration project also provides initial SEER estimates for the breast cancer molecular subtypes defined by the distribution of HER2 (±) and/or ER (±).
Materals and methods
The NCI’s SEER program is a consortium of 17 regional cancer registries (http://seer.cancer.gov/), covering approximately 26% of the United States. Since 1973, SEER’s incidence and mortality rates are considered nationally represented. Ninety-four percent of SEER cancer cases are pathologically confirmed.
Pathology departments, generally retain their formalin-fixed, paraffin-embedded tissue blocks according to standard practice guidelines and regulations, but then may discard them for logistic and/or storage constraints. For this study, we used a set of breast cancer tissue microarrays (TMA) prepared from discarded tissue blocks within the catchment area of the Hawaii Tumor Registry (HTR) and linked to SEER’s main database [2]. The TMAs were constructed by the University of Hawaii Cancer Research Center in collaboration with the Department of Pathology, Tissue Microarray Facility of the University of Virginia Health System. TMAs were prepared using 4 tissue cores per tumor (0.6 mm in diameter, and 0.4 mm spacing between cores). Each tissue core was located on a TMA map with a TMA accession number.
The HTR’s TMA included 354 of 687 invasive female breast cancer cases, newly diagnosed in Hawaii in 1995 and followed through December 2006. Patient records were anonymized by SEER. This project was conducted with IRB approvals from the University of Hawaii (#11444) and the National Institutes of Health (OHSR #3122).
Immunohistochemistry (IHC)
Standard immunohistochemistry with antigen retrieval prior to antibody incubation was performed according to established protocols for estrogen receptor (ER, clone 6F11, dilution 1:200, Novocastra), progesterone receptor (PR, clone PgR636, dilution 1:1,000, Dako), human epidermal growth factor receptor-2 (HER2+, polyclonal, dilution 1:1,000, Dako), epidermal growth factor receptor (EGFR, clone 31G7, dilution 1:500, Zymed), and cytokeratin 5 (CK5, clone XM26, dilution 1:500, Novocastra) [3, 4]. A single pathologist (MES) assessed the stains by examining microscopic TMA slides and scanned images of the TMA slides, using the Aperio TMA Lab system (http://www.aperio.com/), as previously described [4]. Each TMA core was scored for adequacy (satisfactory, suboptimal or unsatisfactory), staining intensity (0 = negative, 1 = weak, 2 = intermediate, and 3 = strong), and percentage of cells stained (0–100%). Results were based on the maximum value of the product of the intensity score (0–3) X percentage of cells stained (0–100%) in adequate cores. Stains for ER and PR were considered positive if the product of the intensity score (0–3) and percentage of cells stained (0–100%) was >10. HER2 was considered positive with 3+ staining in >20% of tumor cells. EGFR and CK5 were positive with 1+ staining in >10% of the tumor cells.
Statistical methods
Incidence rates and incidence rate ratios (IRR) for the 1995 HTR breast cancer cases were obtained with SEER*Stat 6.3.5 (http://seer.cancer.gov/seerstat/). Incidence rates were expressed per 100,000 woman-years (or women per year) and age-adjusted to the 2000 US standard population. Population denominators were not directly available for the TMA cases, but were approximated according to fraction of 1995 HTR cases captured in the TMA (51.5%, 354 of 687). In a sensitivity analysis, a more extensive modeling effort to adjust for missingness of TMA data with other covariates (i.e., missingness at random instead of missingness completely at random [5]) yielded rate estimates similar to the fractional approach (data not shown).
We assessed representativeness of the TMA cases with chi-square tests for heterogeneity, comparing tumor characteristics for the 1995 HTR breast cancer cases in the TMA (TMA cases) to cases not included in the TMA (TMA absent cases). For TMA cases, we used kappa coefficients to measure the concordance between ER and PR expression status recorded in the SEER database with the expression values as measured with IHC stains [6]. Kappa scores ranging from 0.41 to 0.60 demonstrated “moderate agreement”, whereas kappa scores ranging from 0.61 to 80 showed “substantial” concordance.
Two-sample T-Tests were used to compare mean ages at diagnosis and mean tumor sizes. Age-specific incidence rates were calculated by fourteen 5-year age groups (ages 20–24, 25–29 ,…, 80–84, and 85+ years). Kernel density estimation produced “smoothed” age distribution curves at diagnosis (or density plots), as previously described [7]. In brief, the kernel smoother estimated the underlying probability density function for breast cancer incidence by age at diagnosis in single years. Two-sample Kolmogorov-Smirnov (KS) nonparametric tests were used to assess equality of age distributions according to HER2 (±) and/or ER (±) expression [8]. The null hypothesis of equal age distributions was rejected at the 95% confidence level when the P-value of the KS statistic was <0.05.
Results
Demographic and tumor characteristics (Table 1)
Table 1.
Invasive female breast cancer cases from SEER’s Hawaii Tumor Registry (HTR), diagnosed in 1995 and followed through 12/2006
| 1995 HTR | TMA cases | TMA absent | Two-sample T-test | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample size | 687 | 354 | 333 | ||||||
| Percent of total cases | 100.0% | 51.5% | 48.5% | ||||||
| Mean age in years (SE) | 60.3 (0.5) | 60.3 (0.7) | 60.3 (0.8) | P = 0.95 | |||||
| Median age in years | 61 | 62 | 60 | ||||||
| Mean tumor size in cm (SE) | 2.2 (0.9) | 2.1 (0.9) | 2.2 (1.5) | P = 0.48 | |||||
| Median tumor size in cm | 1.5 | 1.5 | 1.5 | ||||||
| Rate | 116.9 | 60.2 | 56.7 | ||||||
| n | % | Rate | IRR | n | % | n | % | P for heterogeneity | |
| Age group | |||||||||
| <50 | 174 | 25 | 41.6 | 1.0 | 82 | 23 | 92 | 28 | |
| 50–59 | 147 | 21 | 261.5 | 6.3 | 80 | 23 | 67 | 20 | |
| 60–69 | 175 | 25 | 356.9 | 8.6 | 100 | 28 | 75 | 23 | |
| 70–79 | 147 | 21 | 393.1 | 9.4 | 76 | 21 | 71 | 21 | |
| 80+ | 44 | 6 | 256.4 | 6.2 | 16 | 5 | 28 | 8 | P = 0.09 |
| Race | |||||||||
| White | 208 | 30 | 144.6 | 1.0 | 102 | 29 | 106 | 32 | |
| Black | 6 | 1 | 194.6 | 1.3 | 5 | 1 | 1 | 0 | |
| API | 469 | 68 | 106.5 | 0.7 | 246 | 70 | 223 | 68 | P = 0.23 |
| Other or unknown | 4 | ~ | ~ | ~ | 1 | ~ | 3 | ~ | |
| Tumor size | |||||||||
| ≤2.0 cm | 403 | 67 | 68.5 | 1.0 | 215 | 66 | 188 | 69 | |
| >2.0 cm | 198 | 33 | 34.2 | 0.5 | 112 | 34 | 86 | 31 | P = 0.51 |
| Other or unknown | 86 | ~ | ~ | ~ | 27 | ~ | 59 | ~ | |
| Lymph nodes (LN) | |||||||||
| LN negative | 439 | 73 | 73.7 | 1.0 | 229 | 71 | 210 | 75 | |
| LN positive | 164 | 27 | 28.8 | 0.4 | 95 | 29 | 69 | 25 | P = 0.24 |
| Other or unknown | 84 | ~ | ~ | ~ | 30 | ~ | 54 | ~ | |
| Historic SEER stage A | |||||||||
| Localized | 457 | 68 | 77.2 | 1.0 | 236 | 67 | 221 | 68 | |
| Regional | 174 | 26 | 30.4 | 0.4 | 97 | 27 | 77 | 24 | |
| Distant | 46 | 7 | 7.6 | 0.1 | 21 | 6 | 25 | 8 | P = 0.42 |
| Other or unknown | 10 | ~ | ~ | 0 | ~ | 10 | ~ | ||
| Tumor grade | |||||||||
| Low | 321 | 59 | 54.5 | 1.0 | 162 | 55 | 159 | 63 | |
| High | 225 | 41 | 38.6 | 0.7 | 130 | 45 | 95 | 37 | P = 0.11 |
| Other or unknown | 141 | ~ | ~ | ~ | 62 | ~ | 79 | ~ | |
| Estrogen receptor (ER) | |||||||||
| ER positive | 438 | 75 | 74.1 | 1.0 | 234 | 73 | 204 | 77 | |
| ER negative | 145 | 25 | 25.1 | 0.3 | 85 | 27 | 60 | 23 | P = 0.32 |
| Other or unknown | 104 | ~ | ~ | ~ | 35 | ~ | 69 | ~ | |
| Progesterone receptor (PR) | |||||||||
| PR positive | 411 | 71 | 69.9 | 1.0 | 218 | 68 | 193 | 73 | |
| PR negative | 171 | 29 | 29.2 | 0.4 | 101 | 32 | 70 | 27 | P = 0.22 |
| Other or unknown | 105 | ~ | ~ | ~ | 35 | ~ | 70 | ~ |
TMA, tissue microarray; n, sample size; %, percent frequency distribution for tumor characteristics with known expression patterns (unknown data were excluded from the calculation), SE, standard error; cm, centimeter; ~, not calculated; Rate, age-adjusted (2000 US standard population) incidence rate expressed per 100,000 woman-years; IRR, incidence rate ratio where a given characteristic is compared to a reference value with an assigned IRR of 1.0 (except for Black compared to White race, all IRRs were statistically significant at the alpha 0.05 level); P values were two-sided and tested statistical significance at the alpha 0.05 level
SEER’s Hawaii Tumor Registry identified 687 invasive female breast cancer cases, newly diagnosed in 1995. The majority (68%) of the women were of Asian or Pacific Islander (API) ethnic origin. Overall incidence rates were 116.9 per 100,000 woman-years. Slightly more than half of the 1995 HTR cases were included in the tissue microarray (TMA cases, 51.5%), leaving n = 333 excluded or absent from the TMA (TMA absent cases, 48.5%).
To test whether the TMA cases were representative of the 1995 HTR, we compared distributions of demographic and tumor characteristics among the TMA cases to the TMA absent cases. We found no evidence to suggest that the TMA study population was not representative of the entire HTR population with respect to measured characteristics, i.e., none of the heterogeneity tests were statistically significant. For example, mean age at diagnosis was 60.3 years in the 1995 HTR, identical to mean ages for the TMA cases and TMA absent cases. Mean tumor sizes ranged from 2.1 to 2.2 cm, P = 0.48 for heterogeneity.
Distribution and reliability of ER, PR, and HER2 measurements (Table 2)
Table 2.
Estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) expression in the 1995 Hawaii Tumor Registry (HTR) tissue microarray
| Sample size | SEER database |
Standard IHC |
Kappa statistic | ||
|---|---|---|---|---|---|
| 354 |
354 |
||||
| n | % | n | % | ||
| Estrogen receptor (ER) | |||||
| ER positive | 234 | 73 | 240 | 72 | |
| ER negative | 85 | 27 | 95 | 28 | 0.67 |
| Other or unknown | 35 | ~ | 19 | ~ | 0.48 |
| Progesterone receptor (PR) | |||||
| PR positive | 218 | 68 | 209 | 62 | |
| PR negative | 101 | 32 | 126 | 38 | 0.72 |
| Other or unknown | 35 | ~ | 19 | ~ | 0.54 |
| HER2 expression | |||||
| HER2 negative | ~ | ~ | 308 | 88 | |
| HER2 positive | ~ | ~ | 41 | 12 | |
| Other or unknown | ~ | ~ | 5 | ~ | |
| Joint HER2 and ER | |||||
| HER2+ER− | ~ | ~ | 28 | 8 | |
| HER2+ER+ | ~ | ~ | 12 | 4 | |
| HER2−ER+ | ~ | ~ | 228 | 68 | |
| HER2−ER− | ~ | ~ | 67 | 20 | |
| Other or unknown | ~ | ~ | 19 | ~ | |
ER and PR expression recorded in the main SEER database was compared to expression patterns as measured with immunohisto-chemical (IHC) stains. SEER does not collect information regarding the human epidermal growth factor receptor 2 (HER2) but was measured with IHC
n, sample size; %, percent; Kappa statistics were calculated with and without ‘other or unknown’ data. Kappa scores ranging from 0.41–0.60 and 0.61–0.88 suggest “moderate” and “substantial” agreement, respectively
We assessed concordance for ER and PR expression as recorded in SEER and as measured with IHC. In the SEER database, “other or unknown” data included missing, borderline, or unknown expression values as recorded in the hospital records. IHC ER or PR expressions were classified as “other or unknown” when all four TMA cores lacked adequate tissue for interpretation. Kappa coefficients for ER and PR expression were calculated with and without “other or unknown” data. Including “other or unknown” expression values, we observed moderate agreement between the SEER database and IHC stains (kappa scores between 0.41 and 0.60). Excluding other or unknown values, there was substantial concordance (0.61–0.80).
HER2 expression was not recorded by SEER, but was determined in the TMA cases with IHC stains. Forty-one (12%) of 349 TMA cases with evaluable tissue cores were HER2+. The joint distributions of HER2 (±) and ER (±) were 28 HER+ER− (HER2 positive), 12 HER+ER+ (double-positive), 228 HER2−ER+ (ER positive), and 67 HER2−ER− (double-negative). It was not possible to classify joint HER2 and ER expression in 19 cases.
Age-specific incidence rates
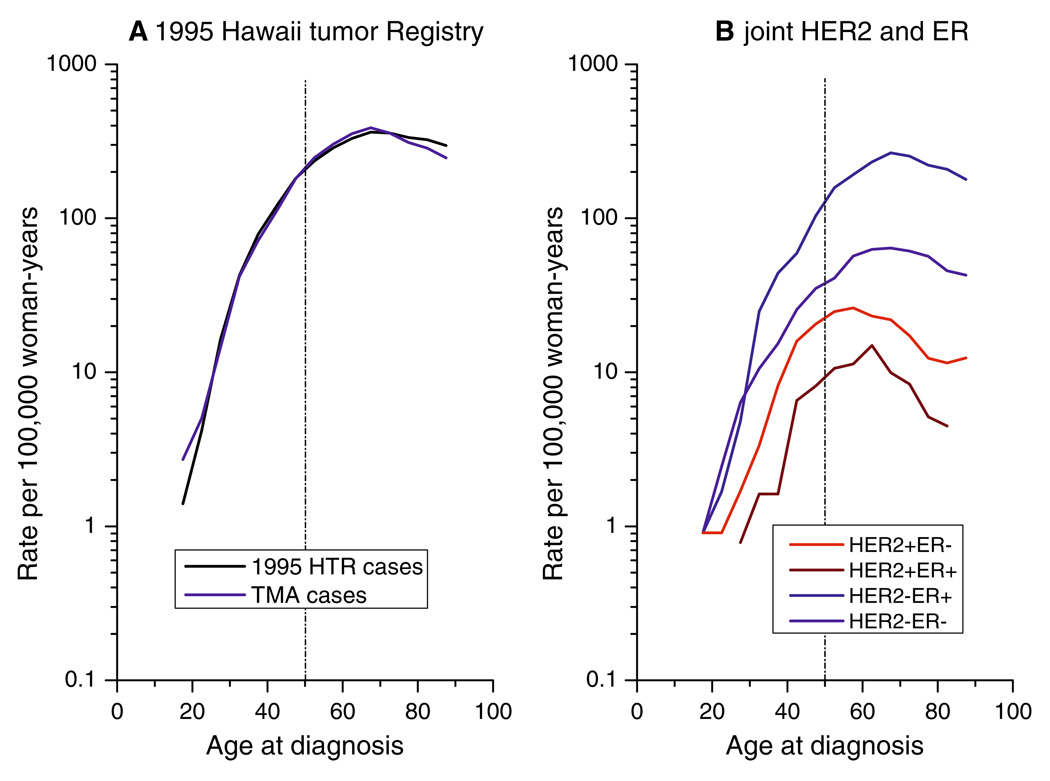
For the 1995 HTR cases (Fig. 1a), age-specific incidence rates increased rapidly until ages 40–50 years, then rose at a slower rate. Rates for the TMA cases were nearly identical to rates for the 1995 HTR cases. Age-specific rates for HER2+ tumors (HER2+ER− and HER2+ER+) tumors increased rapidly until ages 40–50 years, and then fell (Fig. 1b). Age-specific rates for ER+ tumors (HER2−ER + and HER2+ER+) rose rapidly until ages 40–50 years, and then plateaued.
Fig. 1.
Age-specific incidence rates. (a) 1995 HTR cases (n = 687) versus TMA cases (n = 354). (b) Joint HER2 (±) and ER (±) expression
Density plots
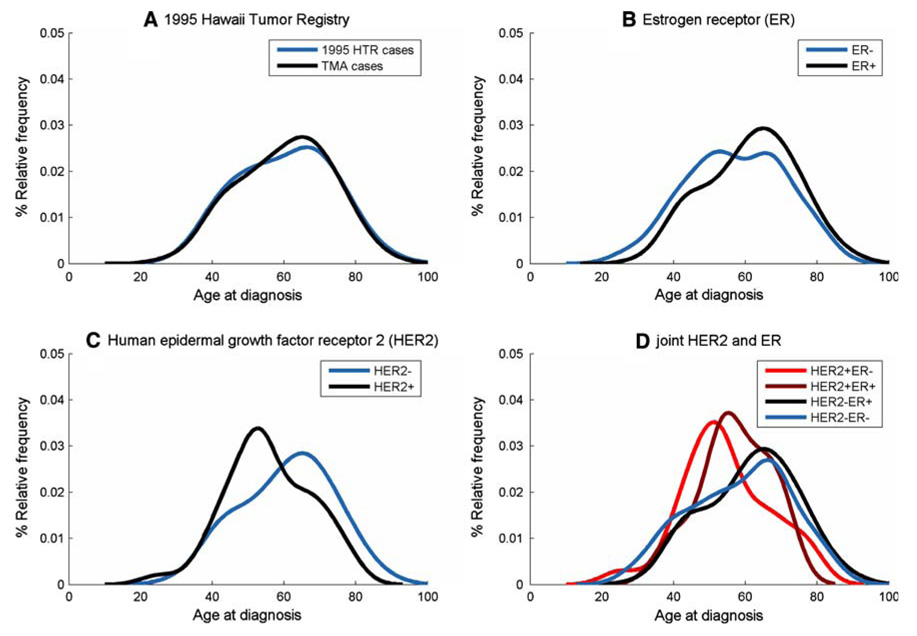
Age distributions at diagnosis demonstrated early-onset and late-onset peak frequencies (or modes) near ages 50 and 70 years, respectively. The density plots for the 1995 HTR and TMA cases had a prominent late-onset mode near age 70 years (Fig. 2a). KS statistics confirmed no difference among the 1995 HTR and TMA cases age distributions, (P = 0.75).
Fig. 2.
Age distributions at diagnosis or density plots. (a) 1995 HTR cases (n = 687) versus TMA cases (n = 354). (b) Estrogen receptor (ER (±)) expression, as measured with standard immunohistochemistry (IHC). (c) Human epidermal growth factor receptor-2 (HER2 (±)). (d) Joint HER2 (±) and ER (±) expression
Age distributions for ER+ breast cancers showed a predominantly late-onset mode, whereas ER- tumors had a bimodal age distribution with slightly dominant early-onset mode (Fig. 2b, P = 0.02). HER2− density plots (Fig. 2c) were similar to ER+ age distributions (Fig. 2b), which might be expected since most HER2− tumors were ER+ (Table 2). In contrast, HER+ tumors had a dominant early-onset mode (KS statistic for HER2− compared to HER2+, P<0.01).
Joint HER2+ER− and HER2−ER+ density plots had dominant early-onset and late-onset modes (Fig. 2d), similar to HER2+ (Fig. 2c) and ER+ (Fig. 2b), respectively. HER2+ER+ was more like HER2+ER− than HER2−ER+, whereas HER2−ER− was more like HER2−ER+. Age distributions were significantly different among HER2+ER−and HER2−ER+ tumors. Comparisons were limited for HER2+ER+ and HER2−ER− due to small sample sizes.
Breast cancer-specific survival and hazard rates for breast cancer death
With a median follow-up of 9.2 years, there were 51 (14.4%) breast cancer deaths. Twenty breast cancer deaths occurred among women with ER- tumors (5.6%), whereas 28 deaths occurred among women with ER+ cancers (7.9%). Forty-two breast cancer deaths were associated with HER2− tumors (11.8%) compared to 9 deaths with HER2+ cancers (2.6%). All HER2+ breast cancer deaths occurred within the first 38 months after breast cancer diagnosis.
Discussion
SEER’s Residual Tissue Repository provided the opportunity to generate initial SEER estimates for the breast cancer molecular subtypes defined by HER (±) and ER (±). A critical component of this effort was to establish that breast cancer cases in the Residual Tissue Repository were representative of the SEER population in Hawaii (Table 1) and reliable (Table 2). Having achieved this goal, we were able to apply tissue microarray technology and descriptive epidemiology to relate prevalence, incidence and prognostic patterns back to the SEER population in Hawaii. Though this study must be viewed as a demonstration project, results provided initial SEER estimates for the distributions of HER2 (±) and/or ER (±) and hypotheses for future analyses.
Frequency distribution for HER2 (±) and/or ER (±) expression
Forty-one (12%) TMA cases were HER2+, which was 2-fold less than initial estimates of 25–30% from high-risk and metastatic breast cancer cohorts [9, 10]. However, these first approximations possibly over stated the true frequency of HER2+ expression in the general breast cancer population [11]. Indeed, Yaziji et al observed 18.6% HER2+ expression in a large volume quality control program [12]. Bilous et al reported 12% HER2+ expression in the HER2000 International Study [13]. Yang et al observed 14% HER2+ in a Poland population-based case control study [3]. Moreover, given the largely favorable prognostic profile for our Hawaii breast cancer population (Table 1), a low prevalence of high-risk HER2+ tumors seems plausible.
Estimates from high risk breast cancer populations also may have exaggerated the proportion of HER2+ER+ (double-positive) status as near 50% [14–16]. Experimental and clinical studies have found that within individual tumors, the strength of HER2+ and ER+ expression is generally inversely related [17–22]. Accordingly, one would anticipate that HER2+ER+ tumors would be relatively uncommon in the general population [23]. The low prevalence of HER2+ER+ tumor in our Hawaii population (4%) also was similar to that found by Yang et al in a Polish case control study [3] as well as by Carey et al. in the Carolina breast cancer study [24].
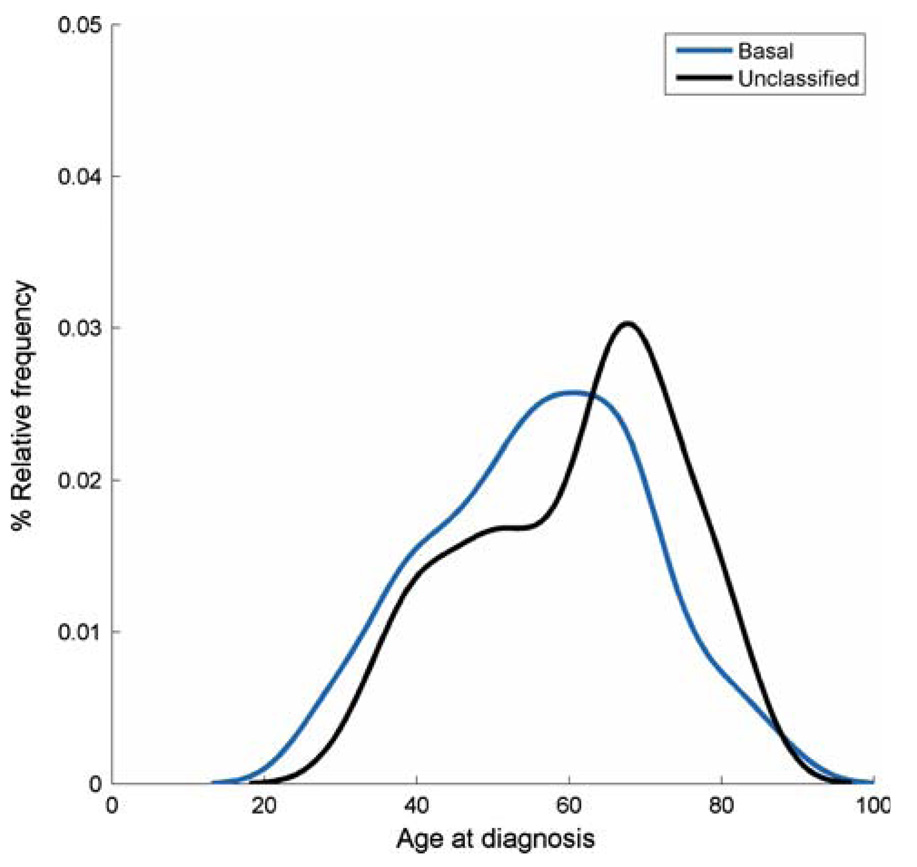
There were 67 (20%) HER2−ER− (double-negative) breast cancers in the Hawaii cohort, but these double-negative tumors appeared to be a mixed phenotype, as recently demonstrated by Kreike et al. [25]. Thirty-four of the 67 HER2−ER− tumors were positive for CK5 and/or EGFR (i.e., basal-like subtype) [26, 27]; yielding an overall distribution of 10%, similar to an API group in California [28]. The remaining 33 of 67 HER2−ER− breast cancers were negative for CK5 and EGFR (i.e., unclassified sub-type). Density plots were shifted to early ages at onset for basal tumors (Fig. 3), whereas unclassified tumors had a dominant late-onset mode. Clearly, future studies are needed to unravel the complexity inherent within the double-negative HER2− and ER− expression pattern.
Fig. 3.
Age distributions at diagnosis or density plots for HER2−ER− (double-negative) breast cancers
Incidence patterns for HER2 (±) and/or ER (±) expression
To our knowledge, this is the first study to provide age-specific incidence rates for HER2 expression. Results confirm the inverse age relationship between HER2+ and ER+ (20). HER2+ (HER2+ER−) breast cancers had early age distributions at diagnosis with a mode near age 50 years (Fig. 2c), and age-specific rates that flattened or fell after age 50 years (Fig. 1b). In contrast, ER+ (HER2− ER+) tumors had late age distributions with a mode near age 70 years (Fig. 2b), and age-specific rates that plateaued after age 50 years. We have previously described similar age interactions for other tumor characteristics as well as for molecular subtypes [29, 30].
Descriptive analyses of registry data are typically limited by non-standardized histopathologic diagnosis and/or ER testing and missing data. However, in this analysis, we avoided some of these concerns by performing IHC on representative archival tissue. Another potential problem is that we used IHC stains rather than the “gold-standard” fluorescence in situ hybridization assay to measure HER2+, but IHC 3+ staining has a reported specificity of 98.8% and a positive predictive value of 91.6% for HER2 overexpression [12]. Our results were based upon relatively small numbers of newly diagnosed breast cancer cases, but the observed age-dependent incidence patterns were consistent with our larger studies [29, 30]. Finally, our analysis reflects characteristics of breast cancer in Hawaii, which differs demographically from the US population overall. However, this analysis provides important population-based data for Asian or Pacific Islanders, a group for whom such information is limited.
Though these limitations should be carefully considered, archival tissues from population-based registries can provide models for future translational studies. The merging of traditional population-based epidemiology with novel molecular techniques can be used to evaluate molecular markers for cancer risk, which were discovered in small-scale molecular studies. The addition of treatment-related data could even complement clinical trial results by assessing “effectiveness” of cancer therapies in the general population. Moreover, methods used for this breast cancer study could be applied to other organ systems in SEER’s RTR.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH/National Cancer Institute, and the Public Health Service contract N01-PC-67001 from DHHS/NIH.
Contributor Information
W. F. Anderson, Email: wanderso@mail.nih.gov, NIH/NCI/DCEG, EPS Room 8036, 6120 Executive Blvd, Bethesda, MD 20852, USA.
S. Luo, NIH/NCI/DCEG, EPS Room 8036, 6120 Executive Blvd, Bethesda, MD 20852, USA
N. Chatterjee, NIH/NCI/DCEG, EPS Room 8036, 6120 Executive Blvd, Bethesda, MD 20852, USA
P. S. Rosenberg, NIH/NCI/DCEG, EPS Room 8036, 6120 Executive Blvd, Bethesda, MD 20852, USA
R. K. Matsuno, NIH/NCI/DCEG, EPS Room 8036, 6120 Executive Blvd, Bethesda, MD 20852, USA
M. T. Goodman, University of Hawaii, Honolulu, HI, USA
B. Y. Hernandez, University of Hawaii, Honolulu, HI, USA
M. Reichman, NIH/NCI/DCCPS, Bethesda, MD, USA
M. P. Dolled-Filhart, HistoRx, Branford, CT, USA
R. M. O’Regan, Emory University, Atlanta, GA, USA
M. Garcia-Closas, NIH/NCI/DCEG, EPS Room 8036, 6120 Executive Blvd, Bethesda, MD 20852, USA
C. M. Perou, University of North Carolina, Chapel Hill, NC, USA
I. Jatoi, National Naval Medical Center and the Uniformed Services University of the Health Sciences, Bethesda, MD, USA
R. W. Cartun, Hartford Hospital, Hartford, CT, USA
M. E. Sherman, NIH/NCI/DCEG, EPS Room 8036, 6120 Executive Blvd, Bethesda, MD 20852, USA
References
- 1.Goodman MT, Hernandez BY, Hewitt S, Lynch CF, Cote TR, Frierson HF, Jr, et al. Tissues from population-based cancer registries: a novel approach to increasing research potential. Hum Pathol. 2005;36(7):812–820. doi: 10.1016/j.humpath.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 2.SEER-13. Surveillance, Epidemiology, and End Results (SEER) SEER*Stat Database: Incidence-SEER 13 Regs Limited-Use, Nov 2006 sub (1992–2004) National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch; 2006. Program http://seer.cancer.gov, released April 2007, based on the November 2006 submission 2007. [Google Scholar]
- 3.Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;18:439–447. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 4.Sherman ME, Rimm DL, Yang XR, Chatterjee N, Brinton LA, Lissowska J, et al. Variation in breast cancer hormone receptor and HER2 levels by etiologic factors: a population-based study. Int J Cancer. 2007;121:1079–1085. doi: 10.1002/ijc.22812. [DOI] [PubMed] [Google Scholar]
- 5.Rubin DB. Multiple imputation for nonresponse in surveys. New York: John Wiley & Sons; 1987. [Google Scholar]
- 6.Fleiss JL. The measurement of interrater agreement. Statistical methods for rates and proportions. 2nd edn. New York: John Wiley & Sons, Inc; 1981. pp. 212–236. [Google Scholar]
- 7.Anderson WF, Chatterjee N, Ershler WB, Brawley OW. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results database. Breast Cancer Res Treat. 2002;76(1):27–36. doi: 10.1023/a:1020299707510. [DOI] [PubMed] [Google Scholar]
- 8.Randles RH, Wolfe DA. Introduction to the theory of nonparametric statistics. New York: John Wiley & Sons; 1979. [Google Scholar]
- 9.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 10.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 11.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 12.Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, Ellis GK, et al. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA. 2004;291(16):1972–1977. doi: 10.1001/jama.291.16.1972. [DOI] [PubMed] [Google Scholar]
- 13.Bilous M, Ades C, Armes J, Bishop J, Brown R, Cooke B, et al. Predicting the HER2 status of breast cancer from basic histopathology data: an analysis of 1500 breast cancers as part of the HER2000 International Study. Breast. 2003;12(2):92–98. doi: 10.1016/s0960-9776(02)00273-4. [DOI] [PubMed] [Google Scholar]
- 14.Burstein HJ. The distinctive nature of HER2-positive breast cancers. N Engl J Med. 2005;353(16):1652–1654. doi: 10.1056/NEJMp058197. [DOI] [PubMed] [Google Scholar]
- 15.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 16.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 17.Zeillinger R, Kury F, Czerwenka K, Kubista E, Sliutz G, Knogler W, et al. HER-2 amplification, steroid receptors and epidermal growth factor receptor in primary breast cancer. Oncogene. 1989;4(1):109–114. [PubMed] [Google Scholar]
- 18.Marsigliante S, Muscella A, Ciardo V, Barker S, Leo G, Baker V, et al. Enzyme-linked immunosorbent assay of HER-2/neu gene product (p185) in breast cancer: its correlation with sex steroid receptors, cathepsin D and histologic grades. Cancer Lett. 1993;75(3):195–206. doi: 10.1016/0304-3835(93)90062-e. [DOI] [PubMed] [Google Scholar]
- 19.Tagliabue E, Menard S, Robertson JF, Harris L. c-erbB-2 expression in primary breast cancer. Int J Biol Markers. 1999;14(1):16–26. doi: 10.1177/172460089901400104. [DOI] [PubMed] [Google Scholar]
- 20.Huang HJ, Neven P, Drijkoningen M, Paridaens R, Wildiers H, Van Limbergen E, et al. Hormone receptors do not predict the HER2/neu status in all age groups of women with an operable breast cancer. Ann Oncol. 2005;16(11):1755–1761. doi: 10.1093/annonc/mdi364. [DOI] [PubMed] [Google Scholar]
- 21.Benz CC, Scott GK, Sarup JC, Johnson RM, Tripathy D, Coronado E, et al. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24(2):85–95. doi: 10.1007/BF01961241. [DOI] [PubMed] [Google Scholar]
- 22.Pietras RJ, Arboleda J, Reese DM, Wongvipat N, Pegram MD, Ramos L, et al. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene. 1995;10(12):2435–2446. [PubMed] [Google Scholar]
- 23.Ciocca DR, Gago FE, Fanelli MA, Calderwood SK. Coexpression of steroid receptors (estrogen receptor alpha and/or progesterone receptors) and Her-2/neu: Clinical implications. J Steroid Biochem Mol Biol. 2006;102(1–5):32–40. doi: 10.1016/j.jsbmb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 25.Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Bartelink H, Van de Vijver MJ. Gene expression profiling and histopathological characterization of triple negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9(5):R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 27.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population- based study from the California cancer Registry. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 29.Anderson WF, Jatoi I, Devesa SS. Distinct breast cancer incidence and prognostic patterns in the NCI’s SEER program: suggesting a possible link between etiology and outcome. Breast Cancer Res Treat. 2005;90(2):127–137. doi: 10.1007/s10549-004-3777-3. [DOI] [PubMed] [Google Scholar]
- 30.Anderson WF, Pfeiffer RM, Dores GM, Sherman ME. Comparison of age frequency distribution patterns for different histopathologic types of breast carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1899–1905. doi: 10.1158/1055-9965.EPI-06-0191. [DOI] [PubMed] [Google Scholar]