Abstract
Macromolecular crowding is expected to have significant effect on protein aggregation. In the present study we analyzed the effect of macromolecular crowding on fibrillation of four proteins, bovine S-carboxymethyl-α-lactalbumin (a disordered form of the protein with reduced three out of four disulfide bridges), human insulin, bovine core histones, and human α-synuclein. These proteins are structurally different, varying from natively unfolded (α- synuclein and core histones) to folded proteins with rigid tertiary and quaternary structures (monomeric and hexameric forms of insulin). All these proteins are known to fibrillate in diluted solutions, however their aggregation mechanisms are very divers and some of them are able to form different aggregates in addition to fibrils. We studied how macromolecular crowding guides protein between different aggregation pathways by analyzing the effect of crowding agents on the aggregation patterns under the variety of conditions favoring different aggregated end products in diluted solutions.
Keywords: Amyloidosis, α-synuclein, insulin, core histone, α-lactabumin, macromolecular crowding, protein aggregation, conformational stability, partially folded intermediate, amyloid fibril, oligomer
INTRODUCTION
Proteins have evolved to fold, associate and function within cells or in other physiological fluid media, where the concentration of macromolecules, including proteins, nucleic acids, and carbohydrates, can be as high as 400 g/L (1). For example, the typical cell contains ∼25 % (v/v) protein, of which about 10 % forms cytoskeletal filaments and 90% is non-aggregating globular proteins. Furthermore, considerable amounts of different RNAs (rRNAs, mRNAs, tRNAs, small RNAs, etc), carbohydrates, and other biopolymers are dissolved in the cell milieu. All these macromolecules collectively occupy between a lower limit of ∼7% and an upper limit of ∼40% of total fluid volume (2, 3), creating a crowded medium, with considerably restricted amounts of free water (1-6). In fact, the cytoplasm of most eukaryotic cells contain numerous compartments and organelles bound by extensively folded membranes, as well as fibrous structures of various types that are thought to occupy between 15–25% of the total cytoplasmic volume (7). This suggests that the fluid phase of the cytoplasm is largely distributed in pores, interstices and channels between fibrous structures (8). Overall, such media are referred as crowded rather than concentrated, as, in general, no individual macromolecular species is present at high concentration, and a special term molecular crowding has been introduced to describe the effect of high solute concentrations on chemical reactions (4, 5). Obviously, the volume occupied by solutes is unavailable to other molecules because two molecules cannot be in the same place at the same time. The thermodynamic consequences of this phenomenon are known as excluded volume effects (3, 9). They manifest themselves by modulating the conformation and stability of biological macromolecules (6, 10-15) and affecting macromolecular equilibria, including protein-protein interactions (3, 5, 16). Furthermore, molecular crowding may lead to the significant alterations of the rates of chemical reactions, protein folding and macromolecular association (1-6, 8, 16-30). Finally, it has been suggested that volume exclusion in physiological media could modulate the rate and extent of amyloid formation in vivo (5, 31). The validity of this hypothesis has been confirmed recently for the in vitro fibrillation of human apolipoprotein C-II (32) and α-synuclein (33-35).
Understanding the effects of macromolecular crowding on protein folding, misfolding and aggregation is of great fundamental and biomedical importance. The exclusion volume effects were predicted to favor the adoption of compact as opposed to expanded macromolecular conformations, resulting in a reduction of total excluded volume (3, 5). This means that macromolecular crowding might stabilize a compact native state relative to a much less compact unfolded state. However, the accumulated to date data on the effect of macromolecular crowding on protein folding are rather controversial. For example, it has been shown that the rate of correct refolding of reduced lysozyme can be increased up to fivefold in the presence of appropriate concentrations of crowding agents compared with the rate in non-crowded environment (29). Similarly, crowding agents were shown to induce folding of the acid-unfolded cytchrome c (15), the intrinsically unstructured reduced and carboxyamidated RNase T1 (24) and of the unfolded RNase A in a 2.4 M urea solution at pH 3.0 (27). On the contrary, it has been shown that high concentrations of crowding agents (200 g/L) had no effect on the refolding of oxidized lysozyme but disrupted the refolding of the reduced protein and caused its aggregation (28, 29). Furthermore, it has been recently established that the ability of dihydrofolate reductase, enolase, and green fluorescent protein to fold spontaneously in dilute solutions was lost in a crowded environment (36). Instead, these proteins were shown to accumulate as protease-sensitive non-native species (36), presumably in a form of soluble oligomers. Furthermore, it has been shown that when the intrinsic folding rate of the protein is relatively slow, crowded environment increases the protein propensity to aggregate (37). Overall, these data directly show that macromolecular crowding might dramatically enhance the competition between protein folding and aggregation, favoring latter when folding is relatively slow. This is rather predictable conclusion as transient association of partially folded intermediates often accompanies the process of protein refolding in diluted solutions. Furthermore, the propensity to aggregate is considered as a general characteristic of the non-native proteins in diluted solutions (38-41). One can expect that crowded environment will further emphasize this tendency.
Protein aggregation represents an essential problem in biomedicine and biotechnology. Particularly, resent studies highlighted increasing recognition of the importance of protein deposition (or conformational) disorders, which include Parkinson’s diseases, Down’s syndrome, Alzheimer’s disease, Hungtington disease, systemic and localized amiloidoses and transmissible encephalopathies (39, 42-46); the formation of inclusion bodies is a major problem in the over-expression of recombinant proteins (39, 47-50); production and in vivo delivery of protein drugs is often complicated by association (50).
The question on the net effect of macromolecular crowding on protein aggregation and fibrillation is not as simple as might appear at first sight due to the complexity of the aggregation process. In fact, the products of protein aggregation are very different morphologically, varying from soluble oligomers of different shapes and oligomerization degree, to insoluble structure-less amorphous aggregates, and to highly-ordered amyloid-like fibrils. As far as the molecular mechanism of protein aggregation is concerned, substantial evidence supports the hypothesis that “pathological” or abnormal aggregation arises from a key partially folded intermediate (39, 44-46, 51). Thus, in protein aggregation the kinetic issue can be broken down into several major steps: (i) structural transformation within monomer leading to the appearance of aggregation-prone partially folded species; (ii) nucleation and/or formation of oligomers; and (iii) formation and growth fibrils or amorphous aggregates. Two latter steps involve association, and the first one involves a conformational change. Obviously, each of these processes is anticipated to be affected by macromolecular crowding in its own way, making the prediction of the net effect of excluded volume on protein aggregation and fibrillation a difficult task. Another complication is the partitioning between different aggregation pathways, which frequently takes place. In other words, the aggregation of a given protein might lead to the appearance or coexistence of different aggregated products. Once again, macromolecular crowding might affect these different aggregation pathways in a different way, favoring some and disfavoring others. All this gives rise to the ambiguity in predictions regarding the effects of macromolecular crowding on protein aggregation.
In the present study we analyzed the effect of macromolecular crowding on fibrillation of four proteins, bovine S-carboxymethyl-α-lactalbumin (a disordered form of the protein with reduced three out of four disulfide bridges), human insulin, bovine core histones, and human α-synuclein. These proteins are very distinctive structurally, with some of them being natively unfolded, whereas others have nicely folded structure. Although all of them were shown to fibrillate, they possess very diverse aggregation mechanisms and, depending on the environmental conditions, are able to form different aggregates in addition to amyloid-like fibril (see below). The main goal of this study was to see how macromolecular crowding guides protein between different aggregation pathways. To this end the aggregation studies were performed under the variety of conditions favoring different aggregated end-products in diluted solutions.
MATERIALS AND METHODS
Materials
Thioflavin T, bovine S-carboxymethyl-α-lactalbumin and bovine core histones were obtained from Sigma, St. Louis, MO. Commercially available sample of histones from calf thymus, representing a mixture of core histones H2A, H2B, H3 and H4, was used without additional fractionation. All other chemicals were of analytical grade from Fisher Chemicals or VWR Scientific. Human insulin (batch HO1713) was obtained from Novo Nordisk A/S, Denmark. The zinc content was 0.4% (w/w of insulin), corresponding to approximately 2 Zn2+ per insulin hexamer.
Expression and purification of human recombinant α-synuclein
Human wild type α-synuclein was expressed using E. coli BL21 (DE3) cell line transfected with pRK172/α-synuclein WT plasmid (generously donated by M. Goedert, MRC Cambridge). Expression and purification of human recombinant α-synuclein and its mutants from E. coli were performed as previously described (52).
Circular dichroism (CD) measurements
CD spectra were obtained on an AVIV 60DS spectrophotometer (Lakewood, NJ, USA). Spectra were recorded in 0.01 cm or 0.1 cm cells from 250 to 190 nm with a step size of 1.0 nm, a bandwidth of 1.5 nm, and an averaging time of 4 s. For all spectra, an average of five or eight scans was obtained. CD spectra of the appropriate buffers were recorded and subtracted from the protein spectra.
FTIR measurements
Data were collected on a Thermo-Nicolet Nexus 670 FTIR spectrometer equipped with a MCT detector and an out-of-compartment 72×10×6 mm, 45° germanium trapezoidal internal reflectance element (IRE). The hydrated thin-films were prepared as described previously (53, 54). Typically 256 interferograms were co-added at 2 cm-1 resolution. Data analysis was performed with GRAMS32 (Galactic Industries). Secondary structure content was determined from curve fitting to spectra deconvoluted using second derivative and Fourier self-deconvolution to identify component band positions.
Thioflavin T fluorescence assays
Assay solutions contained proteins at a protein concentration indicated containing 20 μM ThT with various concentrations of salts and pH as indicated. A volume of 150 μl of the mixture was pipetted into a well of a 96-well plate (white plastic, clear bottom), and a 1/8th in. diameter. Teflon sphere (McMaster-Carr, Los Angeles) was added. Each sample was run in triplicate or quadruplicate. The plates were sealed with Mylar plate sealers (Dynex). The plate was loaded into a fluorescence plate reader (Fluoroskan Ascent) and incubated at 37°C with shaking at 600 rpm with a shaking diameter of 2 mm. The fluorescence was measured at 30 min intervals with excitation at 450 nm and emission at 485 nm, with a sampling time of 100 ms. Data from replicate wells were averaged before plotting fluorescence vs. time. The data were fit to a sigmoidal equation (55) using SigmaPlot software:
| (1) |
where F is the fluorescence intensity and t50 is the time to 50% of maximal fluorescence. The initial baseline during the lag time is described by Fi + mit. The final baseline after the growth phase has ended is described by Ff + mft. The apparent rate constant, kapp, for the growth of fibrils is given by 1/τ, the lag time is calculated as t50 – 2τ and the amplitude, amp, is given by Ff - Fi. Although eq.1 gave very good fits for the ThT kinetic profiles, the expression is strictly a simple empirical means of providing kinetic parameters for comparing rates of fibrillation from different samples and does not directly reflect the underlying complex kinetic scheme.
In a number of cases fibrillation was followed not only by ThT fluorescence, but also by FTIR. Here, 0.5 ml of protein solutions with various concentrations of salts and pH as indicated were stirred at 37°C in glass vials with micro stir-bars. Protein concentrations were kept around 0.5 mg/ml. Fibril formation was monitored with ThT fluorescence: aliquots of 10 μl were removed from the incubated sample and added to 1.0 ml of 20 μM TfT in 20 mM Tris buffer, pH 8.0. At the same time points, sample for FTIR analysis were taken too. The presence of fibrils was confirmed by EM.
Electron Microscopy
Transmission electron micrographs were collected using a JEOL JEM-100B microscope operating with an accelerating voltage of 80 kV. Typical nominal magnification was 75,000x. Samples were deposited on Formvar-coated 300 mesh copper grids and negatively stained with 1% aqueous uranyl acetate.
RESULTS
Figure 1 represents FTIR spectra in the amide I region measured for the four proteins analyzed in this study, S-carboxymethyl-α-lactalbumin, human insulin, bovine core histones and human α-synuclein, in their non-fibrillar and fibril forms. Although these proteins are structurally diverse in their non-fibrillar forms, being fibrillated, all of them are characterized by a very peculiar FTIR spectrum with an intensive peak in the vicinity of 1630 cm-1 attributed to a predominantly β-structural conformation. Formation of extensive β-structure is characteristic for protein aggregation and, particularly, in the formation of β-rich amyloid fibrils.
Figure 1.
FTIR amide I spectra of bovine S-carboxymethyl-α-lactalbumin (A), human insulin (B), bovine core histones (C) and human α-synuclein (D) in non-fibrillar (black circles) and fibrillar forms after the incubation at appropriate conditions at 37°C with stirring (open reversed triangles).
Fibrillation of S-carboxymethyl-α-lactalbumin in crowded milieu
α-Lactalbumin is a small Ca2+-binding protein involved in the regulation of lactose biosynthesis as a component of lactose sythetase (56). Crystallographic analysis revealed that α-lactalbumin is comprised of a large α-helical domain and a small β-sheet domain connected by a Ca2+-binding loop (57, 58). At acidic pH, or at moderate guanidimium-chloride concentrations, or at elevated temperatures this protein is known to adopt the classic molten globule state (59-61). It has been recently shown that similar to several other members of the lysozyme family (62-65), bovine α-lactabumin is able to form amyloid fibrils (66, 67). Importantly, this protein was shown to form amyloid fibrils at low pH, where it adopted the classical molten globule-like conformation. S-carboxymethyl-α-lactalbumin, a disordered form of the protein with reduced three out of four disulfide bridges was even more susceptible to fibrillation. Other partially folded conformations induced in the intact protein at neutral pH, either by the removal of Ca2+ or by the binding of Zn2+ to the Ca2+-protein did not fibrillate, although Zn2+-loaded α-lactalbumin precipitated out of solution as amorphous aggregates. Based on these data it has been concluded that the transformation of a protein into essentially unfolded conformation is required for the successful fibril formation, whereas more rigid species may lead to the formation of amorphous aggregates (66).
As fibrillation of S-carboxymethyl-α-lactalbumin was not complicated by formation of alternative aggregated forms, we have used this protein to analyze the effect of macromolecular crowding on simple fibrillation pathway described by a model:
| (Scheme 1) |
Here U and PF represent unfolded and partially folded conformations, respectively.
Figure 2A compares the fibrillation kinetics of this protein in the absence or presence of high concentrations of model macromolecular crowding agents, such as PEG-3500, Ficoll-70000 and an inert protein, bovine serum albumin. The effect of the PEG-3500 and Ficoll-70000 high concentrations on structural properties of S-carboxymethyl-α-lactalbumin is illustrated by Figures 2B and 2C. Figure 2A shows that the fibrillation of S-carboxymethyl-α-lactalbumin was dramatically accelerated by the addition of all the crowders studied, although the magnitude of the accelerating effect depended on the nature of the polymer [cf. (33)]. In fact, in the absence of crowding agents, it took this protein >200 hrs to show a noticeable increase in ThT fluorescence intensity. This slow fibrillation is rather different from the reported earlier data (66). The discrepancy is likely due to the different settings used in this study. In the presence of PEG, Ficoll and serum albumin, S-carboxymethyl-α-lactalbumin fibrillated essentially faster. We assume that the crowding environment influences the fibrillation of S-carboxymethyl-α-lactalbumin via modulation of its partial folding and promotion of aggregation. Partial folding is expected, as exclusion volume effects are known to favor the adoption of compact as opposed to expanded conformations, resulting in a reduction of total excluded volume (3, 14). Similarly, the rate of aggregation is expected to be accelerated as associated states obviously have smaller volumes in comparison with individual molecules.
Figure 2.
Effect of crowding agents on fibrillation and structural properties of bovine S-carboxymethyl-α-lactalbumin. (A) Fibrillation of bovine S-carboxymethyl-α-lactalbumin (1 mg/ml) monitored by the enhancement of thioflavin T fluorescence intensity at 37°C with stirring in the absence (circles) or presence of high concentrations of model macromolecular crowding agents: PEG-3500 (100 mg/ml, reversed triangles), Ficoll-70000 (150 mg/ml, diamonds) and bovine serum albumin (30 mg/ml, squares). Conditions were 20 mM Tris-HCl, 100 mM NaCl, pH 7.5. (B) Near-UV CD spectra of the native α-lactalbumin (solid black line 1) and its S-carboxymethylated form (solid black line 2) in the absence of crowding agents, as well as in the presence of PEG-3500 (100 mg/ml, solid gray line) and Ficoll-70000 (150 mg/ml, dashed gray line). (C) Far-UV CD spectra of the native (solid black line 1) and its S-carboxymethyl-α-lactalbumin (solid black line 2) in the absence of crowding agents, as well as in the presence of PEG-3500 (100 mg/ml, solid gray line) and Ficoll-70000 (150 mg/ml, dashed gray line).
Importantly, it has been shown that S-carboxymethylated form of α-lactalbumin is significantly unfolded, showing only ∼1/3 of the normal far-UV CD signal (66). In agreement with these earlier observations, Figure 2B shows that S-carboxymethylated form of α-lactalbumin does not possess rigid tertiary structure, whereas Figure 2C reports that the amount of its ordered secondary structure is dramatically depleted in the S-carboxymethylated form in comparison with the non-modified protein. Figures 2B and 2C further illustrates that crowding agents almost do not affect tertiary structure of the protein (it remains completely denatured) and induce some changes in the protein secondary structure. This partial folding, being rather minor, still might represent a crucial aggregation-promoting condition, as reported for many highly unfolded amyloidogenic proteins and peptides (68).
Effect of macromolecular crowding on fibrillation of human insulin
Insulin is a 51-residue hormone that may exist as a mixture of hexamer, tetramer, dimer and monomer in solution, depending on the conditions. The protein is hexameric in the presence of zinc at neutral pH and monomeric in 20% acetic acid (pH 2). Insulin forms amyloid-like fibrils (55) that cause a variety of problems in its biomedical and biotechnological applications (especially in insulin pumps). Amyloid deposits of insulin have been observed both in patients with type II diabetes and in normal ageing, as well as after subcutaneous insulin infusion and after repeated injections (69, 70). Formation of insulin fibrils in diluted solutions in vitro is a physical process in which non-native insulin molecules interact with each other to form linear, biologically inactive aggregates (70). A number of investigations have been undertaken to elucidate the mechanism of insulin fibrillation (55, 69-72). Particularly, it has been established that insulin fibrillation at neutral pH occurs through the dissociation of oligomers into monomers and that the monomer undergoes a structural change to a conformation having strong propensity to fibrillate (55, 69-72). Furthermore, the fibrillation was dramatically accelerated under the conditions favoring monomeric conformation; i.e., in the presence of 20% acetic acid (55, 73), low concentrations of denaturants (55, 73) and for the monomeric insulin analogs (74).
Fibrillation of Insulin at Acidic pH
Based on the previous data, the minimal kinetic model of insulin fibrillation in the presence of 20% acetic acid can be described by a model:
| (Scheme 2) |
Here Nm and PFm represent native and partially folded monomeric conformations, respectively. Figure 3A shows that very fast fibrillation of human insulin in 20% acetic acid is further accelerated in crowded environment. In fact, Figure 3A illustrates that there is a two-fold decrease in the duration of the nucleation stage (i.e., the length of the lag-time) induced by the addition of high concentration of PEG-3500 and almost 4-fold decrease in the duration of lag-time was achieved when high concentrations of Ficoll-70000 were added. Figure 3A’ illustrates that the increase in the ThT fluorescence coincided with a dramatic increase in the intensity of a peak at 1630 cm-1 in the amide I region of the insulin FTIR spectrum. This peak is characteristic of the amyloid-like fibrils with high content of aggregation β-structure. These data taken together show that crowding agents dramatically accelerate the human insulin fibrillation at acidic pH. As insulin is monomeric in the presence of 20% acetic acid, these data further support the hypothesis that crowded environment inevitably accelerates fibrillation uncomplicated by alternative aggregation pathways.
Figure 3.
Fibrillation of human insulin in 20% acetic acid, pH 2.0 (A, A’) or in 25 mM HEPES buffer, pH 7.5 (B. B’) detected by changes in the ThT fluorescence intensity (plots A and B) and by the increase in the b-structure-related peack in the FTIR amide I spectrum (plots A’ and B’). Protein (2 mg/ml) was incubated at 37°C with stirring alone (circles) or in the presence of crowding agents: PEG-3500 (100 mg/ml, reversed triangles), and Ficoll-70000 (150 mg/ml, squares). At appropriate time points, small samples were taken for ThT or FTIR measurements.
Insulin fibrillation at neutral pH
Fibrillation of human Zn2+-loaded insulin at neutral pH represents a more complex case, as it involves dissociation of native hexamers into monomers, which then undergo structural change to a conformation having strong propensity to fibrillate:
| (Scheme 3) |
Here N6, Nm and PFm represent native hexamer, native monomer and partially folded monomer, respectively. Figure 3B shows that at neutral pH macromolecular crowding possessed quite opposite effect on insulin fibrillation, and the addition of both PEG-3500 and Ficoll-70000 significantly slowed down the process of fibril formation. This conclusion is further supported by the FTIR data presented in Figure 3B’. We assume that this retardation of the fibrillation kinetics is due to the molecular crowding-induced stabilization of the native hexameric form of insulin. This suggestion is based on the following observations made in the diluted solutions: (a) Zn2+-loaded insulin is a hexamer at neutral pH; (b) the fibrillation under these conditions requires the dissociation of oligomers into monomers [see Scheme 3, (55, 69-72)].
Effect of crowders on structural properties of human insulin
Figure 4 gives further support to this hypothesis representing near- and far-UV CD spectra of human insulin measured in the native hexameric and native monomeric forms in the absence and presence of Ficoll-70000. Figure 4 shows that the presence of the crowding agent did not affect tertiary and secondary structure of this protein. Earlier it has been established that the near-UV CD spectrum of human insulin is highly sensitive to the oligomerization stage of this protein (75). The fact that the near-UV CD spectra of insulin remain almost unchanged in the presence of Ficoll suggests that the addition of the crowder does affect the native oligomeric state of this protein and insulin remained its monomeric state in 20% acetic acid and mostly hexameric form at neutral pH. This was further supported by the gel-filtration analysis which revealed that at neutral pH insulin is a hexamer both in the absence and in the presence of Ficoll.
Figure 4.
Effect of macromolecular crowding agent Ficoll-70000 (150 mg/ml, dashed lines) on near-UV CD (A) and far-UV CD spectra of human insulin (B) in different conformational states: native hexameric protein in 25 mM HEPES buffer, pH 7.5 (black lines) and in 20% acetic acid, pH 2.0 (gray lines). Solid lines in both plots correspond to spectra measured in the absence of the crowder.
Morphology of insulin fibrils and aggregates
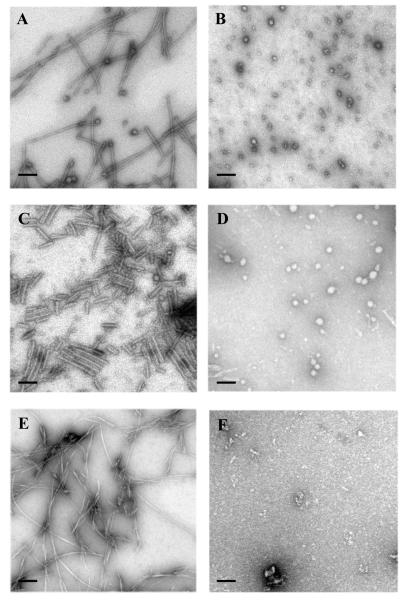
Electron micrographs of the insulin aggregates grown under the different experimental conditions for 20 hrs are present in Figure 5. Figures 5E and 5F clearly show that being incubated in 20% acetic acid (pH 2) in the presence of Ficoll-70000 for 20 hrs insulin formed typical amyloid-like fibrils, whereas oligomers and amorphous aggregates were formed when protein was incubated for same time with Ficoll at neutral pH. Therefore, all these data taken together suggest that the process of fibril formation by a rigid oligomeric protein might be inhibited or slowed down by crowding-induced stabilization of native oligomeric form.
Figure 5.
EM of insulin, α-synuclein, and histone aggregates. A) Fibrils of α-synuclein grown in the presence of Ficoll-70000 and 1 M TMAO (5 hours of incubation); B) oligomers of α-synuclein formed in the presence of Ficoll-70000 and 2.5 M TMAO (5 hours of incubation); C) fibrils of histones grown at pH 2.0 and PEG 10,000 (48 hours of incubation); D) oligomers and amorphous aggregates of histones formed at pH 7.5 and PEG 10,000 (48 hours of incubation); E) insulin fibrils grown at pH 2 and PEG 10,000 (20 hours of incubation); F) oligomers and amorphous aggregates of insulin formed at pH 7.5 and PEG 10,000 (20 hours of incubation). Scale bar is 100 nm.
Fibrillation of bovine core histones in crowded environment
The eukaryotic core nucleosome contains eight histone proteins, with two H2A-H2B dimers that serve as molecular caps for the central (H3-H4)2 tetramer. In the crystal structure, histones are highly helical proteins, with α-helices accounting for 65-70% of the total structure. Only 3% of residues can be assigned to short parallel β-sheets, the remainder is disordered (76, 77). Purified histones dissolved in water with no added salt are in an “extended loose form” (78-87). However, in the presence of salt they adopt a folded conformation (80-87). This salt-induced refolding is a highly cooperative conformational change that is similar to the transitions observed during the renaturation of unfolded globular proteins (88). Furthermore, it has been pointed out that after the addition of salt, histones not only fold but may also aggregate to large structures containing hundreds of protein molecules (88). The conformational and aggregation properties of core histones in diluted solutions in vitro have been recently studied, revealing the existence of different aggregated forms, including soluble oligomers, amorphous aggregates and fibrils, population of which depended on the environmental conditions (89). Particularly, it has been shown that the stabilization of monomeric partially folded conformation under the conditions of low pH and moderate ionic strength led to the accelerated fibrillation, whereas at neutral pH, where histones are in a form of partially folded oligomers, the fibril formation was essentially less effective, requiring the dissociation of oligomers (89). In other words, fibrillation of core histones at acidic pH is described by a model shown in Scheme (1), whereas fibril formation at neutral pH is consistent with the following sequence:
| Scheme (4) |
Here PFo and PFm represent partially folded oligomers and monomers, respectively.
Figure 6 compares the fibrillation kinetics of bovine core histones under conditions favoring partially folded monomeric conformation (low pH) or partially folded oligomers (neutral pH) in the absence or presence of crowding agent. One can see that macromolecular crowding affects the process of fibril formation in a different manner, depending on the histone oligomerization state. In fact, similarly to insulin, the addition of PEG-3500 causes the considerably accelerated fibrillation of monomeric histone, whereas the fibril formation by the oligomeric form was slowed down. This is an important observation because contrarily to insulin, core histones are natively unfolded proteins (89); i.e., they possess little ordered structure due to low overall hydrophobicity and high net charge. Thus, in addition to the described above acceleration of fibril formation in monomeric proteins (folded or partially folded) and inhibition of fibrillation in rigid oligomeric proteins, macromolecular crowding might slow down the fibrillation of partially folded oligomers via their stabilization.
Figure 6.
Fibrillation of bovine core histones at pH 7.0, 200 mM NaCl (reversed triangles) or pH 2.0, 200 mM NaCl (circles) in the absence (black symbols) or presence of 100 mg/ml PEG-3500 (open symbols). Protein at concentration of 0.5 mg/ml was incubated at 37°C with agitation.
Figures 5C and 5D represent electron micrographs of the core histone aggregates formed under the different experimental conditions in the presence of Ficoll-70000. These data provide further support to the hypothesis that fibrillation of an oligomeric partially folded protein might be inhibited or slowed down by crowding-induced stabilization of these partially folded oligomeric forms.
Modulation of α-Synuclein Fibrillation and Aggregation by Crowding Agents
α-Synuclein is a small (14 kDa), highly conserved presynaptic protein that is abundant in various regions of the brain (90). It has been established that the α-synuclein might be involved into the pathogenesis of Parkinson’s disease and several other neurodegenerative disorders (91-95). α-Synuclein belongs to the growing family of natively unfolded proteins (96-98), which have little or no ordered structure under physiological conditions, due to a unique combination of low overall hydrophobicity and large net charge (99-101). The conformational behavior, aggregation and fibrillation of α-synuclein in vitro are well studied [reviewed in (95, 102, 103)]. The protein is known to assemble into the different aggregated forms, including soluble oligomers, amorphous aggregates and fibrils, depending on the environmental conditions (95, 102).
Oligomerization and fibrillation of α-synuclein in TMAO
It has been shown that the natural osmolyte trimethylamine-N-oxide (TMAO) affects the structural properties and fibrillation kinetics of human α-synuclein (104). Importantly, TMAO was shown to cause natively unfolded α-synuclein to fold in a biphasic manner. Moderate TMAO concentrations (between 1 and 2 M) resulted in a partially folded intermediate that possesses structural properties previously described for this protein at high temperatures or low pH (97), in the presence of certain metal cations (105), pesticides/herbicides (106) or in the presence of low concentrations of organic solvents [see below, (107)]. Furthermore, under all the conditions where such intermediate was populated (including moderate TMAO concentrations), α-synuclein was shown to fibrillate significantly faster than in control experiments, consistent with the hypothesis that the formation of a partially folded intermediate represents a critical step in the fibrillation pathway (104). Higher concentrations of TMAO were shown to lead to the formation of the rigid oligomers, probably decamers, with tightly packed globular structure and high helical content. These oligomers do not fibrillate (104). Importantly, the partially folded conformation was not only stabilized in the presence of moderate osmolyte concentrations, but was also transiently populated in the kinetic pathway of oligomer formation at high TMAO concentration (104). Thus, depending on the conditions, the intermediate(s) may self-associate leading to fibrils, amorphous aggregates or soluble globular oligomers. Overall, the fibrillation of α-synuclein in the presence of TMAO can be described by the following model:
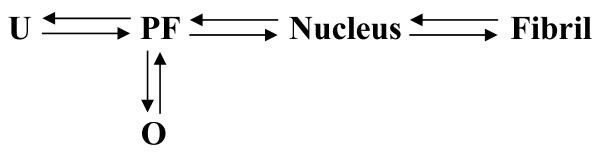 |
(Scheme 5) |
Where U and PF represent unfolded and partially folded monomeric conformations, respectively, whereas O corresponds to rigid well-folded oligomers. We have used these conditions to analyze the effect of macromolecular crowding on fibrillation of α-synuclein in the environment favoring aggregation partitioning between rigid oligomers and fibrils.
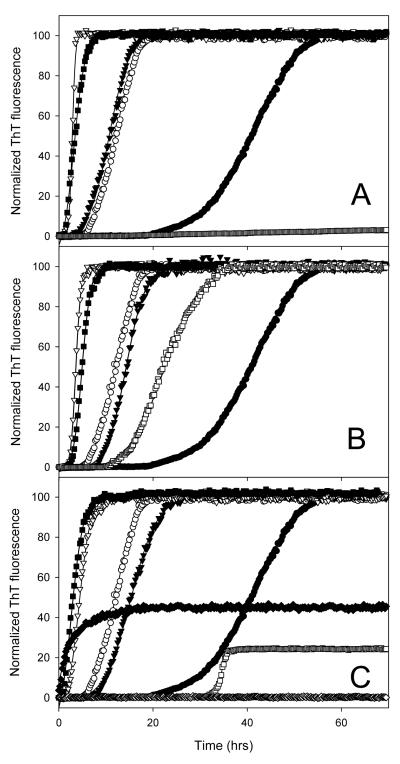
Figures 7 and 8 represents a set of fibrillation curves measured for human α-synuclein in the presence of different TMAO concentrations, which were further modulated by the addition of high concentrations of a crowding agent. In agreement with previous studies, high concentrations of PEG (33-35) and moderate concentrations of TMAO (104) enhances fibrillation of α-synuclein (Figure 7). Similar effect was observed in the presence of high Ficoll-70000 concentrations (Figure 8). Once again Figure 8 illustrates that the increase in the ThT fluorescence occurred simultaneously with the changes in the FTIR spectrum characterizing the formation of the amyloid-like fibrils with high content of aggregation β-structure.
Figure 7.
Fibrillation of human recombinant α-synuclein in the absence (black symbols) or presence of 100 mg/ml PEG-3500 (open symbols). Protein was incubated alone (circles) or in the presence of 1 (reversed triangles), 2 (squares) or 2.5 M TMAO (diamonds). Panel A represents a set of normalized data, whereas Panel B shows the same data in the non-normalized form. In Panel B numbers correspond to: 1, control, no additives; 2, α-synuclein + PEG; 3, 1 M TMAO; 4, 1 M TMAO + PEG; 5, 2 M TMAO; 6, 2 M TMAO + PEG; 7, 2.5 M TMAO; 8, 2.5 M TMAO + PEG. Protein at concentration of 0.5 mg/ml was incubated in 20 mM Tris-HCl, 100 mM NaCl buffer pH 7.5 at 37°C with agitation and fibrillation was monitored by the enhancement of thioflavin T fluorescence intensity.
Figure 8.
Fibrillation of human recombinant α-synuclein in the absence (black circles) and the presence of 150 mg/ml Ficoll-70000 (open reversed triangles). Fibrillation was assessed by the increase in the intensity of the FTIR peak at 1630 cm-1 (plots A, B, and C) or via the characteristic increase in the ThT fluorescence (plots A’, B’, and C’). Protein (0.5 mg/ml) was incubated at 37°C with stirring alone (circles) or in the presence of Ficoll-70000 (150 mg/ml, squares). Panels A and A’ represent data at 0 M TMAO, Panels B and B’ were obtained in the presence of 1 M TMAO, whereas Panels C and C’ show data for 2 M TMAO. At appropriate time points, small samples were taken for ThT or FTIR measurements.
These data show that in the absence of crowding agent different TMAO concentrations are arranged in the following order with respect to their effectiveness in stimulating α-synuclein fibril formation: 1.0 M < 2.0 M > 2.5 M (see Figures 7A and 8). The situation was changed when PEG-3500 or Ficoll-70000 was added, - 1.0 M TMAO was the most effective enhancer of α-synuclein fibrillation in the crowded milieu. In fact, fibril-promoting effect of 1.0 M TMAO in the presence of crowding agents was even more pronounced that the effect of 2.0 M TMAO in the absence of PEG or Ficoll. This means that crowding agents modulates the effect of the different TMAO concentrations in a different manner. Figure 7B shows that not only the rates of fibrillation (i.e., the nucleation and elongation times) but also the yield of fibrils is dramatically affected by PEG and TMAO (which is manifested in large differences in ThT fluorescence intensities measured under the different conditions). In fact, PEG decreases dramatically the amount fibrils formed in the presence of 2.0 M and almost completely eliminated the fibrillation pathway at 2.5 M TMAO.
These data allow some conclusions to be made about the effect of crowding agent on the aggregation and on the partitioning between the rigid oligomers and fibrils. In fact, the analysis of literature data (104) revealed that in the absence of crowders, the population of a partially folded intermediate, being strongly dependent on TMAO concentration, approached maximum at 1.9 M TMAO. Furthermore, at 1 M TMAO α-synuclein represented a 9:1 mixture of unfolded and partially folded conformations, whereas at 2.5 M α-synuclein was a 3:7 mixture of partially folded intermediate and rigid oligomers. This explains why in the absence of PEG or Ficoll the efficiency of TMAO to induce fibrillation follows the trend: 1.0 M (low relative concentration of amyloidogenic intermediate, high relative concentration of natively unfolded protein) < 2.0 M (the population of intermediate is maximal) > 2.5 M TMAO (protein is predominantly in the non-amyloidogenic oligomeric form). Furthermore, our data suggest that PEG and Ficoll change dramatically this picture, enhancing the influence of TMAO and shifting equilibrium toward the lower concentrations of osmolyte.
This hypothesis was further confirmed via the analysis of the Ficoll-70000 (150 mg/ml) on structural properties and TMAO-induced transitions in human recombinant α-synuclein. Figure 9 represents results of this analysis. Figure 9A shows that the addition of 150 mg/ml of Ficoll-7000 does not have a noticeable effect on the natively unfolded conformation of α-synuclein in the absence of TMAO but dramatically enhances the structure-promoting effects of various TMAO concentrations. Importantly, the shape and intensity of far-UV CD spectrum of α-synuclein in the presence of 2.0 TMAO and in the absence of Ficoll were very close to those of the spectrum retrieved at 1.0 M TMAO and 150 mg/ml Ficoll-70000. In their turn, both these spectra resembled far-UV CD spectrum of the amyloidogenic partially folded α-synuclein known to be stabilized at acidic pH (see Figure 9A).
Figure 9.
Effect of TMAO and Ficoll-70000 on structural properties of human α-synuclein. (A) Far-UV CD spectra measured for 0.5/ml α-synuclein in the absence (dark lines) and presence (gray lines) of 150 mg/ml Ficoll-70000. Solid lines correspond to the protein in the absence of TMAO, whereas dotted, short dashed and dash-dotted lines represent data obtained in the presence of 1.0 M, 2.0 M and 2.5 M TMAO, respectively. Black long dashed line corresponds to a-synuclein in the presence of 4.5 M TMAO. Spectrum of the amyloidogenic partially folded conformation induced in α-synucelin at acidic pH is shown as a line with open circles. (B) TMAO-induced changes in the [θ]220 measured in the absence (black circles and black line) or in the presence of 150 mg/ml Ficoll-70000 (gray circles and gray line).
Figure 9B clearly demonstrates that in the presence of Ficoll, the TMAO-induced structural transitions were significantly shifted toward the smaller osmolyte concentrations and shows that the maximal population of the amyloidogenic partially folded intermediate was reached at 1.0 M TMAO when Ficall-70000 was present. Data on Figures 7, 8, and 9 suggest that the addition of crowding agents shifts the maximal population of amyloidogenic conformation from 2.0 to ∼1.0 M TMAO, whereas at 2.0 and especially at 2.5 M TMAO the fibrillation of α-synuclein is dramatically inhibited, most likely due to the preferential crowder-induced stabilization of soluble highly ordered oligomers.
Finally, Figures 5A and 5B demonstrate that aggregates formed by α-synuclein after 5 hours of incubation in the presence of Ficoll-70000 and 1.0 M TMAO (Figure 5A) are typical amyloid-like fibrils, whereas being incubated for same time in the presence of Ficoll-70000 and 2.5 M TMAO (Figure 5B), α-synuclein preferentially assembles into spherical oligomers.
Oligomerization and Fibrillation of α-Synuclein in Alcohols
The structural and fibrillation properties of human α-synuclein in mixtures of water with simple and fluorinated alcohols have been recently analyzed (107). All solvents studied induced folding of α-synuclein, with the common first stage being formation of a partially folded intermediate with an enhanced propensity to fibrillate. Protein fibrillation was completely inhibited due to formation of β-structure-enriched oligomers with high concentrations of methanol, ethanol, propanol and moderate concentrations of trifluoroethanol (TFE), or because of the appearance of a highly α-helical conformation at high TFE and hexafluoroisopropanol (HFiP) concentrations (107). Thus, the overall aggregation behavior of α-synuclein in organic solvents was shown to be consistent with rather complex mechanism described by the following model (107):
 |
(Scheme 6) |
Here U is a natively unfolded conformation of α-synuclein; PF is a partially folded conformation stabilized by low concentrations of any organic solvents. Mα and Oα are α-helical monomer and oligomer, respectively; Mα is semi-stable in high concentrations of TFE and HFiP and with time is transformed into Oα. Mβ and Oβ are β-helical monomer and oligomer, respectively; Mβ is semi-stable in high concentrations of simple alcohols and moderate concentrations of TFE; with time Mβ is transformed into Oβ and then to amorphous aggregates (107).
We took an opportunity to analyze the effect of macromolecular crowding on such a complex aggregation behavior. Figure 10 represents results of these studies for crowded α-synuclein solutions containing simple (ethanol, Figure 10A) or fluorinated alcohols (TFE, Figure 10B; and HFiP, Figure 10C). Figure 10A shows that PEG enhances the propensity of low ethanol concentrations (10 %) to induce partially folded amyloidogenic conformation and to accelerate α-synuclein fibrillation. On the other hand, the addition of crowding agent completely inhibited the fibrillation of α-synuclein at the moderate ethanol concentrations (20%); i.e., under the conditions giving very fast fibrillation of the protein in the absence of PEG. Similarly, PEG was able to super-accelerate fibrillation of α-synuclein at low TFE concentrations (5%), most likely via the preferential stabilization of partially folded conformation, whereas at 10% TFE, the fibrillation was considerably slowed down by the crowding agent (see Figure 10B). It has been previously established that in diluted solutions, high concentrations of simple alcohols (>35%) and moderate concentrations of TFE (15-20%) force α-synuclein to form the non-amyloidogenic β-structure-enriched oligomers (107). We are showing here that macromolecular crowding affects considerably the equilibrium of the system, increasing the propensity of α-synuclein to partially fold and fibrillate at low concentrations of organic solvents and to form β-structural oligomers incapable of fibrillation at their moderate concentrations.
Figure 10.
Effects of different alcohols on the fibrillation of α-synuclein in the absence (black symbols) or presence of 100 mg/ml PEG-3500 (open symbols). Fibrillation was monitored by Thioflavin T. α-Synuclein (0.5 mg/ml) was incubated at 37°C with agitation in the various aqueous-organic solvents. (A) EtOH; (B) TFE; (C) HFiP. EtOH concentrations were 0% ○, 10% (▽), and 20% (□). TFE concentrations were 0% ○, 5% (▽), and 10% (□). HFiP concentrations were 0% ○, 1% (▽), 2.5% (□) and 5% (◇).
Finally, Figure 10C represents fibrillation kinetics determined for α-synuclein in the absence or presence of crowding agent and different concentrations of HFiP. Once again, in the diluted solutions, low HFiP concentrations (1, 2.5 and 5%) led to the very fast fibrillation due to the effective stabilization of partially folded conformation. The addition of PEG further enhances fibrillation of α-synuclein in the presence of 1% HFiP, whereas the process of fibril formation was dramatically slowed down in 2.5% HFiP and completely inhibited when PEG was added to 5% HFiP solutions. Earlier it has been established that in diluted solutions α-synuclein adopts highly helical structure in the presence of the moderate HFiP concentrations. These helical species were unable to fibrillate but had a tendency to oligomerize with time (107). Thus, the analysis of these previous data and our results suggest that macromolecular crowding shifts the conformational and aggregation equilibrium, forcing α-synuclein to fibrillate and adopt folded non-amyloidogenic conformation at lower concentrations of organic solvents.
DISCUSSION
Data presented in this paper suggest that macromolecular crowding has a dramatic effect on conformational and aggregation behavior of fibrillating proteins. We have analyzed four proteins possessing different folding patterns in the diluted solutions: bovine S-carboxymethyl-α-lactalbumin, which is a considerably disordered form of the protein with three reduced disulfide bridges; human insulin, which is a rigid α-helical protein; bovine core histones, which belong to the family of natively unfolded proteins; and human α-synuclein, which is another natively unfolded protein. Furthermore, some of these proteins are known to adopt different oligomeric states in the absence of additives, whereas others are monomeric: S-carboxymethyl-α-lactalbumin and α-synuclein are predominantly monomeric proteins; insulin is a rigid hexamer at neutral pH and a folded monomer at acidic pH; core histones are unfolded monomers at low pH and form partially folded oligomers at neutral pH. Although all these proteins were shown to fibrillate, they are very different in respect of the available aggregation space and the complexity of aggregation patterns, which in some cases possess a number of parallel pathways, giving rise to the obvious rout-partitioning and accumulation of different aggregated species. For example, fibrillation of S-carboxymethyl-α-lactalbumin in diluted solution is described by a simplest kinetic model (see Scheme 1) and is not complicated by alternative aggregation pathways, whereas the aggregation pattern of α-synuclein depends on the environmental conditions and might involve formation of oligomers (varying dramatically in structure, shape and size), amorphous aggregates and fibrils (Scheme 6). Furthermore, under the particular conditions some of these aggregated species may co-exist at least transiently, reflecting a complex equilibrium, which involves a choice between several thermodynamically favorable conformations.
We were studying the effect of macromolecular crowding on these different proteins under the conditions favoring fibrillation in diluted solutions. Overall, our data suggest that crowding agents are able to guide proteins in their choice between different aggregation species. The major conclusions from our studies are:
Irrespectively of the level of structural order in the initial conformation (e.g., rigid insulin hexamer or partially folded oligomers of core histones), fibrillation of oligomeric proteins in crowding environment is slowed down or inhibited, most likely due to the stabilization of the original oligomeric species induced by the excluded volume effects favoring oligomerization;
Fibrillation of monomeric natively unfolded (or substantially disordered) proteins in crowded environment is accelerated. This at least in part due to the excluded volume-driven formation of a partially folded conformation, which is more compact than the original natively unfolded conformation and could be highly amyloidogenic;
Under the conditions favoring multiple folding and aggregation pathways, with the partitioning between these pathways and the potential co-existence of different folded and aggregated species, macromolecular crowding might help system to choose between different routs and drive it toward a single solution. We assume that in these cases macromolecular crowding is working toward the most thermodynamically favorable conformation helping system to eliminate less favorable ones.
Acknowledgements
We express our deepest gratitude to Alexey Uversky for carefully reading and editing the manuscript.
ABBREVIATIONS
- PEG
polyethylene glycol
- TMAO
trimethylamine-N-oxide
- ThT
Thioflavin T
Footnotes
This research was supported in part by grants R01 NS39985 (to L.A.M. and A.L.F.), R01 LM007688-01A1 (V.N.U.) and GM071714-01A2 (V.N.U.) from the National Institutes of Health. We gratefully acknowledge the support of the IUPUI Signature Centers Initiative.
References
- 1.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 2.Fulton AB. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 4.Minton AP. Influence of excluded volume upon macromolecular structure and associations in ‘crowded’ media. Curr Opin Biotechnol. 1997;8:65–69. doi: 10.1016/s0958-1669(97)80159-0. [DOI] [PubMed] [Google Scholar]
- 5.Minton AP. Implications of macromolecular crowding for protein assembly. Curr Opin Struct Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 6.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 7.Gershon ND, Porter KR, Trus BL. The cytoplasmic matrix: its volume and surface area and the diffusion of molecules through it. Proc Natl Acad Sci U S A. 1985;82:5030–5034. doi: 10.1073/pnas.82.15.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Habori M. Macromolecular crowding and its role as intracellular signalling of cell volume regulation. Int J Biochem Cell Biol. 2001;33:844–864. doi: 10.1016/s1357-2725(01)00058-9. [DOI] [PubMed] [Google Scholar]
- 9.Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 10.Eggers DK, Valentine JS. Crowding and hydration effects on protein conformation: a study with sol-gel encapsulated proteins. J Mol Biol. 2001;314:911–922. doi: 10.1006/jmbi.2001.5166. [DOI] [PubMed] [Google Scholar]
- 11.Eggers DK, Valentine JS. Molecular confinement influences protein structure and enhances thermal protein stability. Protein Sci. 2001;10:250–261. doi: 10.1110/ps.36201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison B, Zimmerman SB. T4 polynucleotide kinase: macromolecular crowding increases the efficiency of reaction at DNA termini. Anal Biochem. 1986;158:307–315. doi: 10.1016/0003-2697(86)90555-5. [DOI] [PubMed] [Google Scholar]
- 13.Johansson HO, Brooks DE, Haynes CA. Macromolecular crowding and its consequences. Int Rev Cytol. 2000;192:155–170. doi: 10.1016/s0074-7696(08)60525-2. [DOI] [PubMed] [Google Scholar]
- 14.Minton AP. Effect of a concentrated “inert” macromolecular cosolute on the stability of a globular protein with respect to denaturation by heat and by chaotropes: a statistical-thermodynamic model. Biophys J. 2000;78:101–109. doi: 10.1016/S0006-3495(00)76576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasahara K, McPhie P, Minton AP. Effect of dextran on protein stability and conformation attributed to macromolecular crowding. J Mol Biol. 2003;326:1227–1237. doi: 10.1016/s0022-2836(02)01443-2. [DOI] [PubMed] [Google Scholar]
- 16.Morar AS, Olteanu A, Young GB, Pielak GJ. Solvent-induced collapse of alpha-synuclein and acid-denatured cytochrome c. Protein Sci. 2001;10:2195–2199. doi: 10.1110/ps.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy LD, Zimmerman SB. Macromolecular crowding effects on the interaction of DNA with Escherichia coli DNA-binding proteins: a model for bacterial nucleoid stabilization. Biochim Biophys Acta. 1994;1219:277–284. doi: 10.1016/0167-4781(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerman SB. Macromolecular crowding effects on macromolecular interactions: some implications for genome structure and function. Biochim Biophys Acta. 1993;1216:175–185. doi: 10.1016/0167-4781(93)90142-z. [DOI] [PubMed] [Google Scholar]
- 19.Zimmerman SB, Trach SO. Effects of macromolecular crowding on the association of E. coli ribosomal particles. Nucleic Acids Res. 1988;16:6309–6326. doi: 10.1093/nar/16.14.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman SB, Trach SO. Macromolecular crowding extends the range of conditions under which DNA polymerase is functional. Biochim Biophys Acta. 1988;949:297–304. doi: 10.1016/0167-4781(88)90155-8. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerman SB, Harrison B. Macromolecular crowding increases binding of DNA polymerase to DNA: an adaptive effect. Proc Natl Acad Sci U S A. 1987;84:1871–1875. doi: 10.1073/pnas.84.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galan A, Sot B, Llorca O, Carrascosa JL, Valpuesta JM, Muga A. Excluded volume effects on the refolding and assembly of an oligomeric protein. GroEL, a case study. J Biol Chem. 2001;276:957–964. doi: 10.1074/jbc.M006861200. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Zhang S, Wang C. Effects of macromolecular crowding on the refolding of glucose- 6-phosphate dehydrogenase and protein disulfide isomerase. J Biol Chem. 2001;276:34396–34401. doi: 10.1074/jbc.M103392200. [DOI] [PubMed] [Google Scholar]
- 24.Qu Y, Bolen DW. Efficacy of macromolecular crowding in forcing proteins to fold. Biophys Chem. 2002;101-102:155–165. doi: 10.1016/s0301-4622(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 25.Ren G, Lin Z, Tsou CL, Wang CC. Effects of macromolecular crowding on the unfolding and the refolding of D-glyceraldehyde-3-phosophospate dehydrogenase. J Protein Chem. 2003;22:431–439. doi: 10.1023/b:jopc.0000005458.08802.11. [DOI] [PubMed] [Google Scholar]
- 26.Somalinga BR, Roy RP. Volume exclusion effect as a driving force for reverse proteolysis. Implications for polypeptide assemblage in a macromolecular crowded milieu. J Biol Chem. 2002;277:43253–43261. doi: 10.1074/jbc.M207974200. [DOI] [PubMed] [Google Scholar]
- 27.Tokuriki N, Kinjo M, Negi S, Hoshino M, Goto Y, Urabe I, Yomo T. Protein folding by the effects of macromolecular crowding. Protein Sci. 2004;13:125–133. doi: 10.1110/ps.03288104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Berg B, Ellis RJ, Dobson CM. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 1999;18:6927–6933. doi: 10.1093/emboj/18.24.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Berg B, Wain R, Dobson CM, Ellis RJ. Macromolecular crowding perturbs protein refolding kinetics: implications for folding inside the cell. EMBO J. 2000;19:3870–3875. doi: 10.1093/emboj/19.15.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall D, Minton AP. Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim Biophys Acta. 2003;1649:127–139. doi: 10.1016/s1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]
- 31.Lansbury PT., Jr. Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. Proc Natl Acad Sci U S A. 1999;96:3342–3344. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatters DM, Minton AP, Howlett GJ. Macromolecular crowding accelerates amyloid formation by human apolipoprotein C-II. J Biol Chem. 2002;277:7824–7830. doi: 10.1074/jbc.M110429200. [DOI] [PubMed] [Google Scholar]
- 33.Uversky VN, Cooper EM, Bower KS, Li J, Fink AL. Accelerated alpha-synuclein fibrillation in crowded milieu. FEBS Lett. 2002;515:99–103. doi: 10.1016/s0014-5793(02)02446-8. [DOI] [PubMed] [Google Scholar]
- 34.Shtilerman MD, Ding TT, Lansbury PT., Jr. Molecular crowding accelerates fibrillization of alpha-synuclein: could an increase in the cytoplasmic protein concentration induce Parkinson’s disease? Biochemistry. 2002;41:3855–3860. doi: 10.1021/bi0120906. [DOI] [PubMed] [Google Scholar]
- 35.Munishkina LA, Cooper EM, Uversky VN, Fink AL. The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J Mol Recognit. 2004;17:456–464. doi: 10.1002/jmr.699. [DOI] [PubMed] [Google Scholar]
- 36.Martin J. Requirement for GroEL/GroES-dependent protein folding under nonpermissive conditions of macromolecular crowding. Biochemistry. 2002;41:5050–5055. doi: 10.1021/bi015925l. [DOI] [PubMed] [Google Scholar]
- 37.Kinjo AR, Takada S. Competition between protein folding and aggregation with molecular chaperones in crowded solutions: insight from mesoscopic simulations. Biophys J. 2003;85:3521–3531. doi: 10.1016/S0006-3495(03)74772-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fink AL. Molten globules. Methods Mol Biol. 1995;40:343–360. doi: 10.1385/0-89603-301-5:343. [DOI] [PubMed] [Google Scholar]
- 39.Fink AL. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des. 1998;3:R9–23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 40.Ptitsyn OB. Molten globule and protein folding. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 41.Ptitsyn OB, Bychkova VE, Uversky VN. Kinetic and equilibrium folding intermediates. Philos Trans R Soc Lond B Biol Sci. 1995;348:35–41. doi: 10.1098/rstb.1995.0043. [DOI] [PubMed] [Google Scholar]
- 42.Sipe JD. Amyloidosis. Annu Rev Biochem. 1992;61:947–975. doi: 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- 43.Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- 44.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. I. Systemic amyloidoses. Med Sci Monitor. 1999;5 [Google Scholar]
- 45.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. II. Localized amyloidosis and neurodegenerative disorders. Med Sci Monitor. 1999;5:1238–1254. [Google Scholar]
- 46.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Marston FA. The purification of eukaryotic polypeptides synthesized in Escherichia coli. Biochem J. 1986;240:1–12. doi: 10.1042/bj2400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schein CH. Solubility as a function of protein structure and solvent components. Biotechnology (N Y) 1990;8:308–317. doi: 10.1038/nbt0490-308. [DOI] [PubMed] [Google Scholar]
- 49.Wetzel R, Chrunyk BA. Inclusion body formation by interleukin-1 beta depends on the thermal sensitivity of a folding intermediate. FEBS Lett. 1994;350:245–248. doi: 10.1016/0014-5793(94)00775-6. [DOI] [PubMed] [Google Scholar]
- 50.Lougheed WD, Woulfe-Flanagan H, Clement JR, Albisser AM. Insulin aggregation in artificial delivery systems. Diabetologia. 1980;19:1–9. doi: 10.1007/BF00258302. [DOI] [PubMed] [Google Scholar]
- 51.Wetzel R. For protein misassembly, it’s the “I” decade. Cell. 1996;86:699–702. doi: 10.1016/s0092-8674(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 52.Uversky VN, Yamin G, Souillac PO, Goers J, Glaser CB, Fink AL. Methionine oxidation inhibits fibrillation of human alpha-synuclein in vitro. FEBS Lett. 2002;517:239–244. doi: 10.1016/s0014-5793(02)02638-8. [DOI] [PubMed] [Google Scholar]
- 53.Oberg K, Chrunyk BA, Wetzel R, Fink AL. Nativelike secondary structure in interleukin-1 beta inclusion bodies by attenuated total reflectance FTIR. Biochemistry. 1994;33:2628–2634. doi: 10.1021/bi00175a035. [DOI] [PubMed] [Google Scholar]
- 54.Oberg KA, Fink AL. A new attenuated total reflectance Fourier transform infrared spectroscopy method for the study of proteins in solution. Anal Biochem. 1998;256:92–106. doi: 10.1006/abio.1997.2486. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen L, Khurana R, Coats A, Frokjaer S, Brange J, Vyas S, Uversky VN, Fink AL. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 2001;40:6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 56.Permyakov EA, Berliner LJ. alpha-Lactalbumin: structure and function. FEBS Lett. 2000;473:269–274. doi: 10.1016/s0014-5793(00)01546-5. [DOI] [PubMed] [Google Scholar]
- 57.Acharya KR, Stuart DI, Walker NP, Lewis M, Phillips DC. Refined structure of baboon alpha-lactalbumin at 1.7 A resolution. Comparison with C-type lysozyme. J Mol Biol. 1989;208:99–127. doi: 10.1016/0022-2836(89)90091-0. [DOI] [PubMed] [Google Scholar]
- 58.Acharya KR, Ren JS, Stuart DI, Phillips DC, Fenna RE. Crystal structure of human alpha-lactalbumin at 1.7 A resolution. J Mol Biol. 1991;221:571–581. doi: 10.1016/0022-2836(91)80073-4. [DOI] [PubMed] [Google Scholar]
- 59.Permyakov EA, Yarmolenko VV, Kalinichenko LP, Morozova LA, Burstein EA. Calcium binding to alpha-lactalbumin: structural rearrangement and association constant evaluation by means of intrinsic protein fluorescence changes. Biochem Biophys Res Commun. 1981;100:191–197. doi: 10.1016/s0006-291x(81)80081-2. [DOI] [PubMed] [Google Scholar]
- 60.Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov S, Ptitsyn OB. Alpha-Lactalbumin: compact state with fluctuating tertiary structure? FEBS Lett. 1981;136:311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- 61.Kuwajima K. The molten globule state of alpha-lactalbumin. FASEB J. 1996;10:102–109. doi: 10.1096/fasebj.10.1.8566530. [DOI] [PubMed] [Google Scholar]
- 62.Krebs MR, Wilkins DK, Chung EW, Pitkeathly MC, Chamberlain AK, Zurdo J, Robinson CV, Dobson CM. Formation and seeding of amyloid fibrils from wild-type hen lysozyme and a peptide fragment from the beta-domain. J Mol Biol. 2000;300:541–549. doi: 10.1006/jmbi.2000.3862. [DOI] [PubMed] [Google Scholar]
- 63.Morozova-Roche LA, Zurdo J, Spencer A, Noppe W, Receveur V, Archer DB, Joniau M, Dobson CM. Amyloid fibril formation and seeding by wild-type human lysozyme and its disease-related mutational variants. J Struct Biol. 2000;130:339–351. doi: 10.1006/jsbi.2000.4264. [DOI] [PubMed] [Google Scholar]
- 64.Malisauskas M, Zamotin V, Jass J, Noppe W, Dobson CM, Morozova-Roche LA. Amyloid protofilaments from the calcium-binding protein equine lysozyme: formation of ring and linear structures depends on pH and metal ion concentration. J Mol Biol. 2003;330:879–890. doi: 10.1016/s0022-2836(03)00551-5. [DOI] [PubMed] [Google Scholar]
- 65.Cao A, Hu D, Lai L. Formation of amyloid fibrils from fully reduced hen egg white lysozyme. Protein Sci. 2004;13:319–324. doi: 10.1110/ps.03183404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goers J, Permyakov SE, Permyakov EA, Uversky VN, Fink AL. Conformational prerequisites for alpha-lactalbumin fibrillation. Biochemistry. 2002;41:12546–12551. doi: 10.1021/bi0262698. [DOI] [PubMed] [Google Scholar]
- 67.de Laureto PP, Frare E, Battaglia F, Mossuto MF, Uversky VN, Fontana A. Protein dissection enhances the amyloidogenic properties of alpha-lactalbumin. FEBS J. 2005;272:2176–2188. doi: 10.1111/j.1742-4658.2005.04638.x. [DOI] [PubMed] [Google Scholar]
- 68.Uversky VN. Amyloidogenesis of natively unfolded proteins. Curr Alzheimer Res. 2008;5:260–287. doi: 10.2174/156720508784533312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dische FE, Wernstedt C, Westermark GT, Westermark P, Pepys MB, Rennie JA, Gilbey SG, Watkins PJ. Insulin as an amyloid-fibril protein at sites of repeated insulin injections in a diabetic patient. Diabetologia. 1988;31:158–161. doi: 10.1007/BF00276849. [DOI] [PubMed] [Google Scholar]
- 70.Brange J, Andersen L, Laursen ED, Meyn G, Rasmussen E. Toward understanding insulin fibrillation. J Pharm Sci. 1997;86:517–525. doi: 10.1021/js960297s. [DOI] [PubMed] [Google Scholar]
- 71.Bryant C, Spencer DB, Miller A, Bakaysa DL, McCune KS, Maple SR, Pekar AH, Brems DN. Acid stabilization of insulin. Biochemistry. 1993;32:8075–8082. doi: 10.1021/bi00083a004. [DOI] [PubMed] [Google Scholar]
- 72.Millican RL, Brems DN. Equilibrium intermediates in the denaturation of human insulin and two monomeric insulin analogs. Biochemistry. 1994;33:1116–1124. doi: 10.1021/bi00171a010. [DOI] [PubMed] [Google Scholar]
- 73.Ahmad A, Millett IS, Doniach S, Uversky VN, Fink AL. Partially folded intermediates in insulin fibrillation. Biochemistry. 2003;42:11404–11416. doi: 10.1021/bi034868o. [DOI] [PubMed] [Google Scholar]
- 74.Nielsen L, Frokjaer S, Brange J, Uversky VN, Fink AL. Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry. 2001;40:8397–8409. doi: 10.1021/bi0105983. [DOI] [PubMed] [Google Scholar]
- 75.Uversky VN, Garriques LN, Millett IS, Frokjaer S, Brange J, Doniach S, Fink AL. Prediction of the association state of insulin using spectral parameters. J Pharm Sci. 2003;92:847–858. doi: 10.1002/jps.10355. [DOI] [PubMed] [Google Scholar]
- 76.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 77.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boublik M, Bradbury EM, Crane-Robinson C. An investigation of the conformational changes in histones F1 and F2a1 by proton magnetic resonance spectroscopy. Eur J Biochem. 1970;14:486–497. doi: 10.1111/j.1432-1033.1970.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 79.Boublik M, Bradbury EM, Crane-Robinson C, Johns EW. An investigation of the conformational changes of histone F2b by high resolution nuclear magnetic resonance. Eur J Biochem. 1970;17:151–159. doi: 10.1111/j.1432-1033.1970.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 80.Li HJ, Isenberg I, Johnson WC., Jr. Absorption and circular dichroism studies on nucleohistone IV. Biochemistry. 1971;10:2587–2593. doi: 10.1021/bi00789a027. [DOI] [PubMed] [Google Scholar]
- 81.Li HJ, Isenberg I. The effect of urea on salt-induced changes in histone IV. Biochim Biophys Acta. 1972;285:467–472. doi: 10.1016/0005-2795(72)90334-0. [DOI] [PubMed] [Google Scholar]
- 82.Li HJ, Wickett R, Craig AM, Isenberg I. Conformational changes in histone IV. Biopolymers. 1972;11:375–397. doi: 10.1002/bip.1972.360110206. [DOI] [PubMed] [Google Scholar]
- 83.Wickett RR, Li HJ, Isenberg I. Salt effects on histone IV conformation. Biochemistry. 1972;11:2952–2957. doi: 10.1021/bi00766a005. [DOI] [PubMed] [Google Scholar]
- 84.D’Anna JA, Jr., Isenberg I. A complex of histones IIb2 and IV. Biochemistry. 1973;12:1035–1043. doi: 10.1021/bi00730a003. [DOI] [PubMed] [Google Scholar]
- 85.D’Anna JA, Jr., Isenberg I. Conformational changes of histone ARE(F3, III) Biochemistry. 1974;13:4987–4992. doi: 10.1021/bi00721a018. [DOI] [PubMed] [Google Scholar]
- 86.D’Anna JA, Jr., Isenberg I. Conformational changes of histone LAK (f2a2) Biochemistry. 1974;13:2093–2098. doi: 10.1021/bi00707a015. [DOI] [PubMed] [Google Scholar]
- 87.Smerdon MJ, Isenberg I. Conformational changes in histone GRK(f2a-1) Biochemistry. 1974;13:4046–4049. doi: 10.1021/bi00716a036. [DOI] [PubMed] [Google Scholar]
- 88.Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- 89.Munishkina LA, Fink AL, Uversky VN. Conformational prerequisites for formation of amyloid fibrils from histones. J Mol Biol. 2004;342:1305–1324. doi: 10.1016/j.jmb.2004.06.094. [DOI] [PubMed] [Google Scholar]
- 90.Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 92.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 93.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 94.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 95.Uversky VN. Neuropathology, biochemistry, and biophysics of alpha-synuclein aggregation. J Neurochem. 2007;103:17–37. doi: 10.1111/j.1471-4159.2007.04764.x. [DOI] [PubMed] [Google Scholar]
- 96.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 97.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 98.Eliezer D, Kutluay E, Bussell R, Jr., Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 99.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 101.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 102.Uversky VN. A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 103.Uversky VN, Fink AL. Biophysical properties of human alpha-synuclein and its role in Parkinson’s disease. In: Pandalai SG, editor. Recent Research Developments in Proteins. Transworld Research Network; Kerala, India: 2002. pp. 153–186. [Google Scholar]
- 104.Uversky VN, Li J, Fink AL. Trimethylamine-N-oxide-induced folding of alpha-synuclein. FEBS Lett. 2001;509:31–35. doi: 10.1016/s0014-5793(01)03121-0. [DOI] [PubMed] [Google Scholar]
- 105.Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson’s disease and heavy metal exposure. J Biol Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 106.Uversky VN, Li J, Fink AL. Pesticides directly accelerate the rate of alpha-synuclein fibril formation: a possible factor in Parkinson’s disease. FEBS Lett. 2001;500:105–108. doi: 10.1016/s0014-5793(01)02597-2. [DOI] [PubMed] [Google Scholar]
- 107.Munishkina LA, Phelan C, Uversky VN, Fink AL. Conformational behavior and aggregation of alpha-synuclein in organic solvents: modeling the effects of membranes. Biochemistry. 2003;42:2720–2730. doi: 10.1021/bi027166s. [DOI] [PubMed] [Google Scholar]