Abstract
The C-cyclopropylalkylamide scaffold was previously identified as a new structural framework for antiestrogens. A second generation library provided three compounds that bind estrogen receptor (ER)α. Further screening of this library identified ERβ hits and inspired another round of SAR. A new focused library was tested for binding to the ERs, and for effects on the growth of breast cancer cell lines and protein levels of common cell cycle regulators.
The estrogen receptors (ERs) are ligand activated transcription factors. ERα and ERβ demonstrate ∼56% sequence homology in their ligand binding domains, but their ligand-binding pockets differ by only two amino acids: ERα Leu384 versus ERβ Met336, and ERα Met421 versus ERβ ILe373.1 ERβ's pocket is also about 100 Å3 smaller than that of ERα.2 Additionally, the tissue distributions of these receptors are distinct.3 There is much data demonstrating the benefits of selectively targeting the ERβ. ERβ expression inhibits estradiol (E2)-induced proliferation in T47D and MCF-7 breast cancer cells and leads to a reduction in xenograft volume.4,5,6 ERβ has a growth inhibitory mechanism in medullary thyroid carcinoma, prostate cancer, and ovarian cancer.7,8,9,10 Also, ERβ-selective agonists demonstrate potential as anti-inflammatory agents.11 Several ERβ-selective agents have been identified (Figure 1). The plant-derived ligand genistein (GEN (1)) has also been shown to induce DNA strand breaks and cause chromosomal aberrations.3,12 The Katzenellenbogen group developed a diarylpropionitrile (DPN, 2) that is a 100-fold selective ERβ agonist.13 Recently, biphenyl derivatives have been investigated for ERβ-selectivity. Simple 4-OH-biphenyls demonstrated 20-70 fold selectivity for ERβ over ERα.14 In order to attempt to mimic the C-ring of GEN, an oxime moiety was incorporated, and altering the substitutions on the biphenyl core provided various levels of selectivity: oximes 3 and 4 were 11-fold and 43-fold selective, respectively (Figure 1).15
Figure 1.
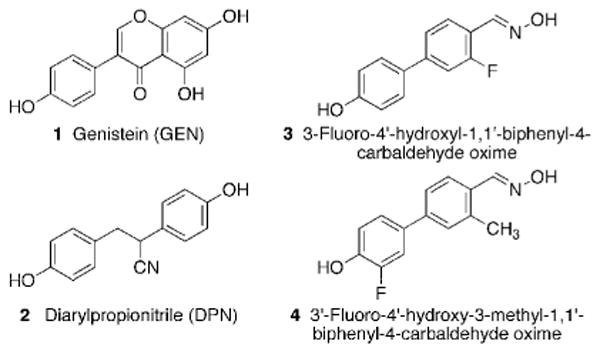
Structures of several known ERβ-selective ligands: GEN (1), DPN (2), and biphenyl carbaldehyde oximes 3 and 4.
Previously, through screening of a library of homoallylic amides, allylic amides and C-cyclopropylalkylamides, we identified a new structural scaffold for antiestrogens.16 The synthesis of a second generation library provided three additional agents that were found to bind tightly to ERα.17 In the present analysis, we screened this second generation library with a fluorescent E2 derivative for competitive binding to ERβ in a fluorescence polarization (FP) assay.18 The percent displacement of the E2 derivative by the test compound was calculated from the difference in measured FP values upon incubation with and without the test sample. The efficiency of displacement demonstrated for ERβ by the initial screening library was significantly lower than that for ERα. In fact, only a single compound (5; Figure 2) demonstrated a concentration-dependent displacement over the concentration range tested and close to 50% displacement at the highest concentration. The displacement values were: 0.2 μM, 26%; 1 μM, 31%; and 5 μM, 46%. Compound 5 was chosen as the lead structure for efforts to improve ERβ-selectivity. Because 5 did not quite reach 50% displacement on ERβ, the selection criteria were modified to include structural features previously known to promote ERβ affinity.15 Screening results on ERα were used as coselection criteria, and 5 was not a hit on ERα.17
Figure 2.
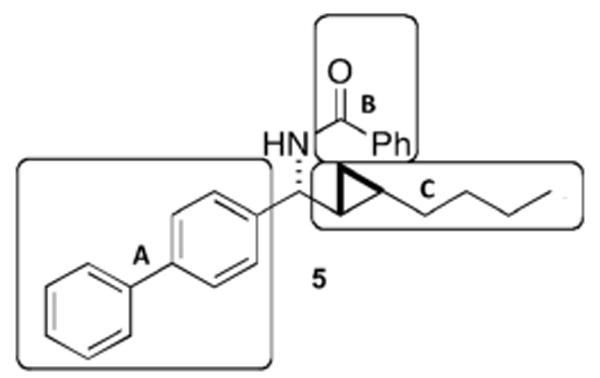
Lead compound 5, and subunits A, B and C used in the SAR.
Figure 2 demonstrates the three regions in 5 that were further evaluated in the next round of our SAR. The biphenyl core A was shown to be beneficial for selective targeting of ERβ, in analogy to Yang et al.15 who used GEN as a lead structure to search for new ERβ-selective ligands. The biphenyl moiety was synthetically readily accessible through diaryl Suzuki cross-couplings, and a large number of substituted phenyl precursors were commercially available. Biphenyl carbaldehyde oxime derivatives were synthesized with various substituents. Upon placement of a fluorine and/or chlorine substituent on the biphenyl core while maintaining the phenol group, agents selective for ERβ were obtained that demonstrated greater than 10-fold (3) and up to 40-fold (4) selectivity.15 Among the second generation library compounds, only one compound containing the biphenyl system satisfied the criteria for hit selection in the screen on ERα; however, this compound failed to inhibit estradiol-stimulated growth of MCF-7 ER positive breast cancer cells.17 Replacement of the biphenyl group with a smaller arene, i.e., p-chlorobenzene, preserved the displacement activity on ERα and recovered the cell-based activity. This substituent effect indicates that the sterically more demanding biphenyl core is less tolerated by ERα if combined with larger substituents on the nitrogen atom in the benzylic position. When the biphenyl group in 5 was replaced with a phenyl ring while keeping the remaining structure constant, the activity at ERβ dropped to ∼10% and activity at ERα was retained. Alterations to region B in 5 also led to detrimental consequences in the ability of the compound to displace ES2 from ERβ. Compound 5 demonstrated that the best-tolerated function in sector B was a phenylamide group, while other rings were better tolerated for ERα. When the phenylamide was replaced with a biphenylamide, activity at ERβ was abolished. Therefore, we concluded that only one biphenyl core was necessary and that increasing the size of the ligand would be detrimental to activity on ERβ. As far as the alkyl chain in region C was concerned, an increase in its size to a cyclohexyl group while keeping the remainder of the molecule constant abolished activity at both receptors. Therefore, region C was not altered in subsequent synthetic efforts. Previous results demonstrated that the cyclopropane ring was important for cellular activity.16 When compounds containing the biphenyl core yet lacking the cyclopropane ring were tested in the FP assay, activity at both receptors was lost. Regions A and B were chosen for further elaboration and the structures were assembled with the biphenyl core, the cyclopropane ring and the amide linkage preserved. The biphenyl group was substituted with a key 4′-OH group expected to promote hydrogen bonding with ERβ Glu305 and ERβ Arg346. A fluorine substituent was chosen based upon published SAR for biphenyl-containing agents.15 The phenylamide was changed into a smaller acetylamide, or completely removed. Six new compounds, analogues of 5, were synthesized, 10a, 11b-d, 12, and 13 (Scheme 1). The palladium catalyst trans-(Cy2NH)2Pd(OAc)2 (DAPCy) was used to perform Suzuki couplings in dioxane because it could be used at room temperature, thereby suggesting functional group tolerance, and its past use in similar couplings.19,20 This catalyst was easily prepared in yellow crystalline form from palladium(II)acetate and dicyclohexylamine.19 The cross-coupling of commercially available benzyl- or methyl-O-protected 4-hydroxybenzeneboronic acid to 4-bromobenzaldehyde using the DAPCy catalyst provided biphenyl aldehydes 6a-c in good to excellent yields (65-88%). Imine formation using titanium(IV) chloride as the Lewis acid initially gave phosphinoylimine 8a in poor to moderate yields (30-47%).17 Changing the workup procedure to a fast filtration over a pad of silica gel and washing with ethyl acetate significantly improved the yields (80%) of the phosphinoylimines 8b-c. One-pot hydrozirconationtransmetallation-aldimine addition in dichloromethane using a microwave (100 °C, 300 W, 10 min) immediately followed by cyclopropanation in the presence of excess diiodomethane at room temperature (16 – 20 h) gave the phosphinoyl-protected biphenyl C-cyclopropylalkylamides 9a-c in moderate overall yields (43-66%). Acidic deprotection followed by acylation gave the final product 10a (61%) or benzyl-protected compounds 10b-d (22-75%). Benzyl ethers were easily removed by hydrogenolysis at 1 atm in the presence of palladium hydroxide on carbon to give the final products 11b-d and 12. Deprotections and acylations were performed in parallel using a Bradley Greenhouse reactor, and final compounds were purified individually by column chromatography. The deacetylated primary amine 13 was purified as its HCl salt.
Scheme 1.
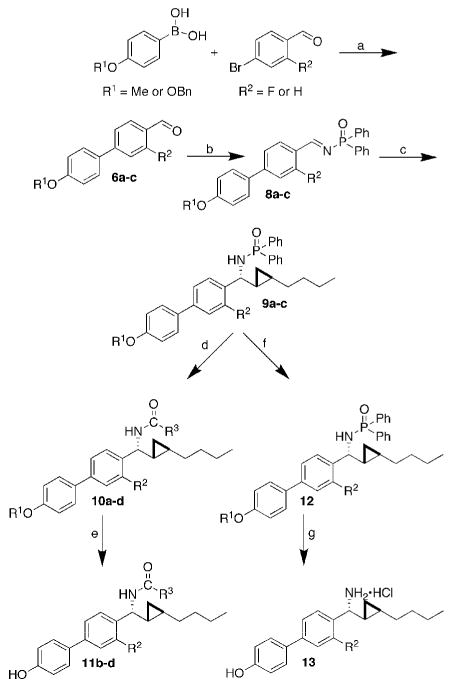
Overview of the synthesis of six new biphenyl substituted C-cyclopropylalkylamides. Reagents and conditions: a, DAPCy (2 mol%), Cs2CO3, dioxane, r.t; b, Ph2P(O)NH2, TiCl4, Et3N, r.t; c, i) Cp2Zr(H)Cl, 1-hexyne, CH2Cl2, ii) Me2Zn, toluene, -78 to 0°C, iii) μW 100 °C, 300 W, 10 min, iv) CH2I2, r.t.; d, (1) HCl, MeOH, (2) R3COCl, DMAP, iPr2EtN, r.t.; e, H2, Pd(OH)2/C, THF, MeOH, r.t.; f, H2, Pd(OH)2/C, THF, MeOH, r.t.; g, HCl, MeOH. Percent yields: 6a, 70; 6b, 88; 6c, 50; 8a, 47; 8b, 67; 8c, 40; 9a, 30; 9b, 68; 9c, 40; 10a, 61; 10b, 57; 10c, 22; 10d, 75; 11b, 69; 11c, 92; 11d, 94; 12, 80; 13, 43. R1 = Me, R2 = F (6a, 8a, 9a); R1 = Bn, R2 = F (6b, 8b, 9b); R1 = Bn, R2 = H (6c, 8c, 9c); R1 = Me, R2 = F, R3 = Ph (10a); R1 = H, R2 = F, R3 = Ph (10b, 11b); R1 = H, R2 = H, R3 = Ph (10c, 11c); R1 = H, R2 = F, R3 = Me (10d, 11d); R2 = F (12, 13).
This synthetic scheme allowed for diversification in two ways. Substituents on the biphenyl core were varied by changing the substitution on the arylboronic acid and aryl bromide coupling components. The amide linkage also allowed for diversification by altering the acylating agents, analogously to the synthesis of the second generation library. After the acidic deprotection step, the amine salt could be coupled without further purification with diverse acyl chlorides, carbamoyl chlorides, and sulfonyl chlorides in the presence of DMAP.17
3-Fluoro-4′-hydroxy-1,1′-biphenyl-4-carbaldehyde oxime 3 served as a positive control. The past synthesis of 3 was modified as shown in Scheme 2.15 Suzuki cross-coupling using DAPCy gave the desired biphenyl aldehyde 6a in 70% yield. The aryl methyl ether was then removed with BBr3 at low temperature, and, after simple basic aqueous work up, the resulting deprotected aldehyde was used without further purification. Biphenyl carbaldehyde oxime 3 was formed in two steps by treating the aldehyde 7 with hydroxyloxime hydrochloride in methanol at room temperature.
Scheme 2.
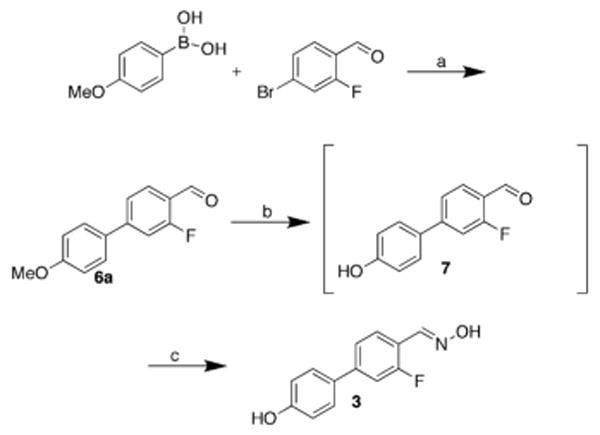
Synthesis of positive control agent, 3-fluoro-4′-hydroxy-1,1′-biphenyl-4-carbaldehyde oxime 3. Reagents and conditions: a, DAPCy (2 mol%), Cs2CO3, dioxane, r.t, 70%; b, BBr3; c, NH2OH•HCl, 42%.
The six new compounds 10a, 11b-d, 12 and 13 and the control compound, oxime 3, were first evaluated for their ability to compete with ES2 for binding to both ERs using the FP assay as described (Figure 3).18
Figure 3.
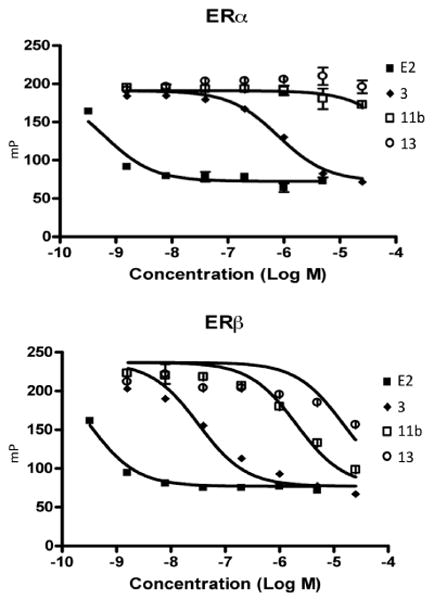
Competition FP assay for 11b and 13 with ES2 for binding to ERα or ERβ. E2 and 3 were used as controls.
Only two compounds, 11b and 13 (Figure 4), significantly competed with ES2 as shown in Figure 3. The curves were constructed from one-site competition best-fit curves of polarization (mP) versus concentration using GraphPad Prism 4 software. The IC50 values determined from this fit are listed in Table 1. Biphenyl carbaldehyde oxime 3 and E2 were used as positive controls, the former giving an IC50 comparable to the previously determined value in a radioligand binding assay.15
Figure 4.
Lead compounds 11b and 13.
Table 1.
IC50 values for competition at both ERs from the FP assay.
Values are means of three experiments, (na = not active).
The compounds were next evaluated for their ability to inhibit E2-induced proliferation of MCF-7 human breast cancer cells and demonstrate their potential as antiestrogens. This was done using two variants of the MCF-7 cell line. The later passage ATCC variant which expresses only ERα, and an earlier passage which expresses both ERα and ERβ. The western blot demonstrating this expression is in the supplementary information. Figure 5 shows the growth inhibition curves generated for compounds determined to be selective for ERβ using the MTS assay.21 Raloxifene (RAL), an antiestrogen known to inhibit the proliferation of MCF-7 cells, was included as a control.22 GraphPad Prism 4 was used to estimate GI50 values using a non-linear best curve fit. In the ERα and ERβ expressing MCF-7 cells, compounds 11b and 13 gave GI50 values of 4.1 and 1.2 μM, respectively.
Figure 5.
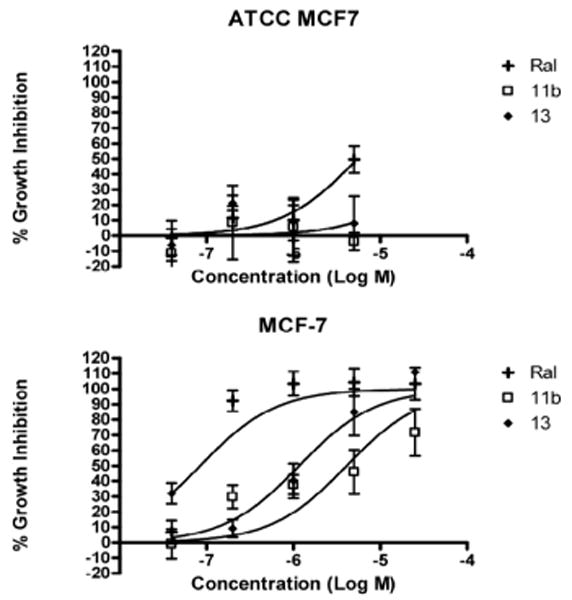
Inhibition of E2-stimulated growth of ATCC MCF7 (Top) or MCF-7 (Bottom) cells by biphenyl C-cyclopropylalkylamides 11b and 13. RAL was used as a positive control. Data represents % growth inhibition (mean ± SD, N = 3).
In the ERα expressing ATCC MCF7 cells, compound 11b did not demonstrate inhibition, while compound 13 gave a GI50 value of 51.7 μM. In these cells, all 3 compounds tested were toxic at a concentration of 25 μM. Additionally, the growth inhibition of ER-negative MDA-MB231 cells was also assayed.23,24 11b, 13, and RAL did not demonstrate significant growth inhibitory activity against this cell line but the compounds again demonstrated toxicity at 25 μM. This can also be seen in the supplementary information. Therefore, this provides support that the compounds are exerting their action through ERβ, because in breast cancer cells expressing both ERβ and ERα a significantly more potent anti-proliferative effect was demonstrated compared to breast cancer cells expressing only ERα or no ERs.
c-Myc is known to play a major role in cell proliferation and malignant transformation in breast cancer.25 E2 has been shown to cause a rapid increase in the level of c-Myc expressed by human breast cancer cells.26 Induction of the expression of ERβ inhibits c-Myc at both the mRNA and protein level, while increasing the levels of its regulators, the cyclin-dependent kinase inhibitors, p21 (Cip1) and p27(Kip1).5 Because of these known links to breast cancer and E2, we chose to investigate how our compounds affected the protein levels of c-Myc, p21, and p27. Western blot analyses (Figure 5) from lysates of MCF-7 cells treated with the indicated compounds and densitometric analyses (Table 2) show compounds 11b and 13 hold promise because of their ability to increase the levels of cyclin-dependent kinase inhibitors and decrease the levels of c-myc, likely inhibiting cell cycle progression and proliferation. PPT, a 410-fold ERα-selective agonist, and DPN, a 70-fold ERβ-selective agonist, were included as controls.27,13
Table 2.
Relative fold changes of protein levels induced by the indicated treatment in MCF-7 cells compared to DMSO vehicle treatment.
| 10 nM E2 | 1 μM PPT | 1 μM DPN | 1 μM 11b | 1 μM 13 | |
|---|---|---|---|---|---|
| cMyca | 3.84 | 2.17 | 2.67 | 0.77 | 0.59 |
| p21a | 1.15 | 4.05 | 3.47 | 3.64 | 1.10 |
| p27a | 1.76 | 2.47 | 3.32 | 3.90 | 1.51 |
ImageJ was used to determine band intensity, and levels were standardized to actin.
In conclusion, compounds 11b and 13 demonstrate selectivity for ERβ as well as promising antiproliferative effects in breast cancer cells. The compounds are likely acting as agonists for ERβ based on what has been shown about ERβ's activity in breast cancer,4,5,6 and this encourages future investigation. Compared to the lead compounds 3 and 5, 11b and 13 show no affinity for ERα and moderate affinity for ERβ. This initial investigation into the biphenyl C-cyclopropylalkylamide pharmacophore demonstrates the potential of this scaffold to deliver ER-selective antiestrogens. Furthermore, the preliminary SAR provides clues for improving selectivity for ERβ. A subtype-selective agent will help to understand the complex biology of the ERs and their interplay. The arrival of new ER ligands with differential subtype selectivity ratios enables the tailoring of antiestrogenic or estrogenic therapy according to the condition and level of receptor isoforms present in patients.
Figure 6.
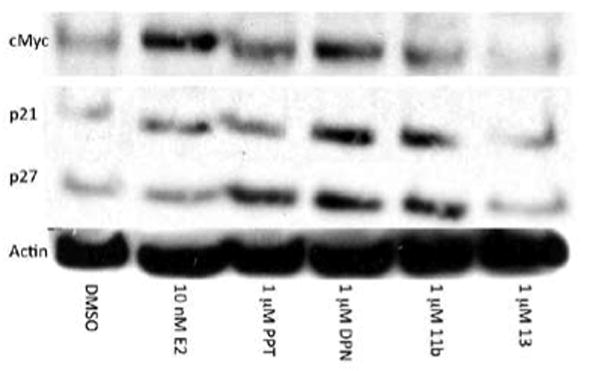
Western blot analysis of the levels of c-Myc, p21, p27, and actin in lysates of MCF-7 cells treated with 10 nM E2, 1 μM PPT, 1 μM 11b and 1 μM 13 for 12 h.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Sun J, Baudry J, Katzenellenbogen JA, Katzenellenbogen BS. Mol Endocrinol. 2003;17:247. doi: 10.1210/me.2002-0341. [DOI] [PubMed] [Google Scholar]
- 2.Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engström O, Ljunggren J, Gustafsson JA, Carlquist M. EMBO J. 1999;18:4608. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuiper GG, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, Gustafsson JA. Endocrinology. 1997;138:863. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 4.Ström A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. Proc Natl Acad Sci USA. 2004;101:1566. doi: 10.1073/pnas.0308319100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. Cancer Res. 2004;64:423. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- 6.Hartman J, Lindberg K, Morani A, Inzunza J, Ström A, Gustafsson JA. Cancer Res. 2006;66:11207. doi: 10.1158/0008-5472.CAN-06-0017. [DOI] [PubMed] [Google Scholar]
- 7.Cho MA, Lee MK, Nam KH, Chung WY, Park CS, Lee JH, Noh T, Yang WI, Rhee Y, Kim SK, Lee HC, Lee EJ. J Endocrinol. 2007;195:255. doi: 10.1677/JOE-06-0193. [DOI] [PubMed] [Google Scholar]
- 8.Ji Q, Liu P, Elshimali Y, Stolz A. Mol Cell Endocrinol. 2005;229:103. doi: 10.1016/j.mce.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Pravettoni A, Mornati O, Martini PG, Marino M, Colciago A, Celotti F, Motta M, Negri-Cesi P. Mol Cell Endocrinol. 2007;263:46. doi: 10.1016/j.mce.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Chan KK, Wei N, Liu SS, Xiao-Yun L, Cheung AN, Ngan HY. Obstet Gynecol. 2008;111:144. doi: 10.1097/01.AOG.0000296715.07705.e9. [DOI] [PubMed] [Google Scholar]
- 11.Harris HA, Albert LM, Leathurby Y, Malamas MS, Mewshaw RE, Miller CP, Kharode YP, Marzolf J, Komm BS, Winnekar RC, Frail DE, Henderson RA, Zhu Y, Keith JC., Jr Endocrinology. 2003;144:4241. doi: 10.1210/en.2003-0550. [DOI] [PubMed] [Google Scholar]
- 12.Boos G, Stopper H. Toxicol Lett. 2000;116:7. doi: 10.1016/s0378-4274(00)00192-2. [DOI] [PubMed] [Google Scholar]
- 13.Meyers MJ, Sun J, Carlson KE, Marriner GA, Katzenellenbogen BS, Katzenellenbogen JA. J Med Chem. 2001;44:4230. doi: 10.1021/jm010254a. [DOI] [PubMed] [Google Scholar]
- 14.Edsall RJ, Jr, Harris HA, Manas ES, Mewshaw RE. Bioorg Med Chem. 2003;11:3457. doi: 10.1016/s0968-0896(03)00303-1. [DOI] [PubMed] [Google Scholar]
- 15.Yang C, Edsall R, Jr, Harris HA, Zhang X, Manas ES, Mewshaw RE. Bioorg Med Chem. 2004;12:2553. doi: 10.1016/j.bmc.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 16.Janjic JM, Mu Y, Kendall C, Stephenson CR, Balachandran R, Raccor BS, Lu Y, Zhu G, Xie W, Wipf P, Day BW. Bioorg Med Chem. 2005;13:157. doi: 10.1016/j.bmc.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 17.Wipf P, Coleman CM, Janjic JM, Iyer PS, Fodor MD, Shafer YA, Stephenson CR, Kendall C, Day BW. J Comb Chem. 2005;7:322. doi: 10.1021/cc049842a. [DOI] [PubMed] [Google Scholar]
- 18.The ER competitor assays were performed according to the manufacturer's (Invitrogen, Carlsbad, CA) recommendations with some modifications. Human recombinant ER was used at the recommended concentration of 15 nM and the Fluormone™ ES2 was used at a concentration of 1 nM. Aliquots (20 μL) of the mixture of ER and Fluormone™ ES2 were distributed in 384-well, black flat-bottom plates and serial dilutions of test compounds in DMSO were added. Each compound was tested in duplicate, and 1 μM E2 was used as a positive control. The DMSO concentration was kept at 1% (v/v) throughout the experiment. After 2 h, the fluorescence polarization (FP) was measured using an Analyst™ AD & HT Assay Detection Systems reader (Molecular Devices, Sunnyvale, CA) equipped with 485 nm excitation and 530 nm emission filters with the appropriate FL505 dichroic mirror. Data was then analyzed by linear regression using GraphPad Prism's one-site competition method. The fit was constrained by the high polarization control, which was the ER/ES2 complex with no competition, and the low polarization control, which was the ER/ES2 complex with 1 μM E2 for 100% competition.
- 19.Tao B, Boykin DW. Tetrahedron Lett. 2003;44:7993. [Google Scholar]
- 20.De Angelis M, Stossi F, Carlson KA, Katzenellenbogen BS, Katzenellenbogen JA. J Med Chem. 2005;11:1132. doi: 10.1021/jm049223g. [DOI] [PubMed] [Google Scholar]
- 21.MCF-7 breast cancer cells were seeded in 96-well plates at a density of 4 × 103 cells per well in phenol redfree RPMI-1640 medium containing 10% charcoal-dextran stripped FBS and allowed to adhere overnight. Test compounds were then added (concentrations ranging from 25 pM to 50 μM) along with 1 nM E2, and cells were incubated for 6 days. Effect of the test compounds on growth was determined using the CellTiter Aqueous One assay system (Promega, Madison, WI), which utilizes the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) dye reduction assay with phenazine methosulfate as the electron acceptor. Absorbance was measured at 490 nm using an M5 plate reader (Perkin Elmer, Boston, MA) and the background absorbance at 630 nm was subtracted after 2 h incubation with the reagents. A plate of control cells (16 wells) was measured at day 0 to establish 0% growth. E2-stimulated growth at day 6 was set to 100% growth. GraphPad Prism 4 (GraphPad Software Inc, La Jolla, CA) software was used for constructing dose-response curves and calculating GI50 values.
- 22.Thompson EW, Reich R, Thomas TB, Albini A, Graf J, Martin GR, Dickson RB, Lippman ME. Cancer Res. 1988;48:6764. [PubMed] [Google Scholar]
- 23.Cailleau R, Olivé M, Cruciger QV. In Vitro. 1978;14:911. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 24.MDA-MB231 cells were plated in 96-well plates at 1 × 103 cells/well and allowed to adhere for 72 h in phenol redfree RPMI-1640 containing 10% FBS. Test agents were added (concentrations ranging from 3.2 nM to 50 μM) and cells were incubated for 72 h. Cell density was determined using the MTS assay as described above.
- 25.Liao DJ, Dickson RB. Endocrine-Related Cancer. 2000;7:143. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 26.Dubik D, Dembiniski TC, Shiu RP. Cancer Res. 1987;47:6517. [PubMed] [Google Scholar]
- 27.Stauffer SR, Coletta CJ, Tedesco R, Nishiguchi G, Carlson K, Sun J, Katzenellenbogen BS, Katzenellenbogen JA. J Med Chem. 2000;43:4934. doi: 10.1021/jm000170m. [DOI] [PubMed] [Google Scholar]


