Abstract
Nedd8 is a small ubiquitin-like protein that can be conjugated to substrate–proteins in a process known as neddylation. Although neddylation plays a critical regulatory role in cell proliferation and development, the spectrum of Nedd8 substrates and its interaction network remain poorly understood. To explore the neddylation pathway at the proteome level, we have affinity purified Nedd8 modified and associated proteins from HEK293 cells stably expressing GST-Nedd8 and employed LC–MS/MS for subsequent protein identification. A total of 496 GST-Nedd8 modified and associated proteins have been identified, including all of the eight cullin family members (i.e., Cul-1, -2, -3, -4A, -4B, -5, -7, and Parc) that are involved in the neddylation and ubiquitin-proteasome degradation pathway. In addition, a group of proteins involved in transcription, DNA repair and replication, cell cycle regulation and chromatin organization, and remodeling have been copurified and identified. Apart from protein identification, the neddylation sites of cullins were determined by MS/MS analysis, which agree well with previous mutagenesis studies. Furthermore, MS analyses revealed that Nedd8 K11, K22, K48, and K60 can form chains in vivo, whereas Nedd8 K22 and K48 can be neddylated in vitro. These results present the first molecular evidence for in vitro and in vivo polyneddylation, suggesting that chain formation of ubiquitin and ubiquitin-like proteins may be a general phenomenon for these modifications. Although much remains to be explored for the biological significance of the observations, this work provides critically important information regarding Nedd8 chain assembly and its interaction network. The vast amount of proteomic information obtained here can provide clues on the biological role of Nedd8 and lay the foundation for an in-depth analysis of the regulation of the Nedd8 pathway.
Keywords: Nedd8, neddylation, ubiquitin-like protein, cullin-containing ubiquitin ligase, ubiquitination, Nedd8 interaction network, affinity purification, polyneddylation, mass spectrometry
Introduction
Protein post-translational modifications play critical roles in regulating protein function. Among the known post-translational modifications, ubiquitination and ubiquitin-like protein (Ubl) modifications have emerged as important regulatory mechanisms in modulating cellular processes. The ubiquitin system is a well-characterized pathway that is involved in regulating nearly every cellular event in eukaryotes including cell proliferation and differentiation.1,2 In the past few years, a number of small ubiquitin-like proteins (e.g., SUMO, Nedd8, and ISG15) have been found to covalently attach to their target proteins similar to ubiquitination.3,4 Ubl-modifications appear to be very important because of their high degree of conservation among diverse eukaryotes. However, the signaling power of Ub and Ubl modifications has yet to be fully appreciated due to the limited understanding of these protein modifier substrates and their interaction networks.
Nedd8, a small ubiquitin-like protein with highest sequence homology to ubiquitin, can be conjugated to substrate–proteins in a process known as neddylation.5 In fission yeast, Nedd8 is essential for cell viability.6 In animals, Nedd8 is required for development as inactivation of the Nedd8 pathway in either mouse7 or Drosophila8 results in embryonic lethality. Inactivation of the SMC1 (APP-BP1 homologue) gene, encoding for a subunit of the Nedd8 activating enzyme, causes cells to undergo multiple S phases without intervening mitosis.9 Attachment of Nedd8 to proteins mimics attachment of ubiquitin, and requires the Nedd8-specific E1 activating enzyme (i.e., APP-P1/Uba3), E2 conjugating enzyme (i.e., Ubc12), and a tentatively identified E3 ligase, Rbx1, to form an isopeptide-bond, which links the ε-amino group of a lysine residue on the substrate to the carboxy-end of Nedd8 Gly-76.5 Interestingly, like ubiquitination, neddylation is a reversible and dynamic process. Although the neddylation pathway shares many mechanistic similarities to the ubiquitination pathway, the biological consequences of such modifications appear to be very different. Unlike ubiquitination, neddylation does not directly affect the stability of substrate–proteins, but instead, modulates their biochemical activity. Compared to the number of identified ubiquitinated substrates, the number of known targets of neddylation has been limited. For a long time, the only identified targets for neddylation have been the cullin family proteins.10 Cullin is the essential component of SCF or SCF-like E3 ubiquitin ligases, which catalyze the ubiquitination of a broad spectrum of proteins involved in a variety of cellular functions such as cell cycle control, transcription, signal transduction, and DNA replication.3,11–13 Neddylation is critical to the functions of cullin-containing ubiquitin ligases,3 and is required to facilitate the processive transfer of ubiquitin from E2 to the target protein,14 thus, increasing the efficiency of polyubiquitin chain assembly through its ligation to cullin molecules. Recently, several noncullin proteins have been identified as Nedd8 substrates such as pVHL, p53, BCA3, and EGF receptor.15–18 Neddylation of pVHL affects the activity of pVHL-containing ubiquitin ligases, whereas neddylation of p53 and BCA3 has an impact on transcriptional activities, and neddylation of EGFR is essential for ligand-induced down-regulation and degradation of EGFR.
Ubiquitin can modify proteins with conjugation of either one ubiquitin molecule (i.e., monoubiquitination) or a polyubiquitin chain (i.e., polyubiquitination). Both monoubiquitination and polyubiquitination serve as distinct recognition signals for modulating protein functions. In general, it is believed that Nedd8 only modifies its substrates, such as cullins, with the attachment of one Nedd8 molecule to a lysine residue. Similar to the cullin family, only one lysine (K159) residue in pVHL15 is the major acceptor site of Nedd8. In contrast, multiple lysines of p53 and EGFR seem to be involved in neddylation.16,18 Interestingly, in vitro neddylation of Cul-1 resulted in hyperneddylation by immunoblotting analysis,19 although it is not known whether the hyperneddylation of Cul-1 is due to the formation of multiple mononeddylation or polyneddylation on Cul-1. Nevertheless, whether and how Nedd8 can form chain assemblies needs to be explored.
Identification of the Nedd8 interaction network and characterization of Nedd8–subtrate linkages are of key importance to understanding the biological significance of Nedd8 modifications. To this end, proteomic analyses of the Nedd8 pathway have been carried out,20,21 but the number of proteins identified in those studies has been limited. Many details of the Nedd8 pathway including neddylation sites remain unsolved. Here, we report a global profiling of Nedd8 modified and associated proteins using affinity purification and tandem mass spectrometry. A 1-D gel/MS-based proteomic approach is used to identify purified proteins and characterize the neddylation sites. The work reported here offers novel insight into neddylation, improves the understanding of protein neddylation pathways, and allows the design of better experiments for further investigation.
Experimental Procedures
1. Chemicals and Reagents
Sequencing grade trypsin was purchased from Promega Corp. (Madison, WI). Endoproteinase Lys-C was from WAKO chemicals (Osaka, Japan). All other general chemicals for buffers and culture media were purchased from Fisher Scientific and/or VWR International.
2. Plasmids
A two-step procedure was used to generate vectors expressing Nedd8 precursor (proNedd8). The proNedd8 sequence was first amplified from pRS-Nedd8p (a kind gift from K. Wilkinson, Emory University) by PCR using the primers: 5′CACC ATG CTA ATT AAA GTG AAG ACG CTG 3′ (start codon in bold letters) and 5′TCA CTT ATC GTC GTC ATC CTT GTA ATC AAC AGA TGC ACG ACG CTG CCT AAG ACC ACC TCC TCC TCT CAG 3′ (stop codon in bold letters, Flag tag sequence underlined, and cAMP-dependent protein kinase recognition sequence in italics). In the second step, the resulting PCR product, containing the proNedd8, Flag tag, and kinase recognition sequences, was inserted into the Gateway system entry vector, pENTR/D-TOPO, per manufacturer’s instructions (Invitrogen) and verified by DNA sequencing.
To generate the pDEST27-proNedd8-PK-Flag expression vector (expressing N-terminal GST-tagged proNedd8-PK-Flag), the proNedd8-PK-Flag sequence was recombined into the pDEST27 Gateway destination vector (Invitrogen) based on the manufacturer’s instructions, generating pDEST27-V5-proNedd8-PK-Flag. For pDEST27-GUS (expressing N-terminal GST-tagged GUS protein), the GUS sequence was recombined into the pDEST27 Gateway destination from the pENTR-GUS (Invitrogen) vector.
3. Cells: Culturing, Transfection, Lysis, and Affinity Purification
HEK293 cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Gibco). For transfection, expression vectors were introduced into HEK293 cells using Lipofectamine 2000 (Invitrogen), according to manufacturer’s protocol.
Cell extracts were prepared as described previously14 and maintained in Lysis Buffer (15 mM Tris-HCl, pH7.4, 0.5 M NaCl, 0.35% NP-40, 1 mM DTT, 1 mM PMSF, 2 μg/mL Antipain, and 2 μg/mL Leupeptin). For affinity purification, lysates were incubated with the appropriate antibody-conjugated beads or glutathione beads overnight at 4 °C. The resulting matrix was washed three times with Lysis Buffer and two times with buffer A50 (25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.01% NP-40, 10% glycerol, 50 mM NaCl, 1 mM DTT, 1 mM PMSF, 2 μg/mL Antipain, and 2 μg/mL Leupeptin), boiled for 3 min at 100 °C in SDS-loading buffer, and subjected to SDS-PAGE.
4. Generation of Stable Cell Lines Expressing GST-Nedd8 and GST-Gus
HEK293 cells, maintained in growth medium (DMEM supplemented with 10% bovine serum and 100 μg/mL of penicillin–streptomycin [GIBCO/BRL]), were plated at an approximate density of 2 × 106 cells per plate (10-cm). After 20–24 h, cells were cotransfected using Lipofectamine 2000 with two vectors: pDEST27 proNedd8-PK-Flag or pD-EST27-Gus, and a plasmid carrying the puromycin resistance gene, pBABE-Puro. After 40 h post-transfection, cells were trypsinized; diluted at a ratio of 1:5, 1:50, or 1:500; and subsequently plated onto 15-cm plates with the addition of selection medium, constituted by the growth medium (20 mL) plus puromycin (2.5 μg/mL). The selection medium was changed daily to remove dead cells in the subsequent 3 days and then every third day for another 2 weeks. Individual colonies were picked by suction with sterile 1 mL pipet tips and placed onto 24-well plates containing the selection medium. The positive clones were identified by screening expression of GST-Nedd8 or GST-Gus using in-cell Westerns on an Odyssey Infrared Imaging System (LI-COR) and confirmed by immunoblot analysis performed on the same device.
5. Affinity Purification and Separation of Nedd8-Modified and Associated Proteins
A stable cell line expressing GST-proNedd8 was grown in 15-cm tissue culture dishes until confluency. Because of low concentration of Nedd8 conjugated proteins, purification was scaled up using 250 plates of the cells that were harvested and lysed as described.14 Lysates (125 mL) were incubated with 4 mL pre-equilibrated Glutathione Sepharose beads overnight at 4 °C, and the beads were then loaded into a chromatography column and washed with 100 mL of lysis buffer followed by 100 mL of buffer A50. The bound proteins were eluted with 100 mL of buffer A50 plus 20 mM glutathione and concentrated 100-fold using 5000 MWCO Amicon Ultra centrifugal concentrators (Millipore). Approximately 2 mg of protein was obtained from 125 mL of lysates. The large-scale purification was duplicated in this study.
6. In Vitro Neddylation
A reaction mixture (300 μL) containing 50 mM Tris-HCl, pH7.4, 0.6 mM DTT, 5 mM MgCl2, 2 mM ATP, 2 mM NaF, 10 nM Okadaic Acid, Nedd8 (7.5 μg), APP-BP1/Uba3 (1.5 pmol), and Ubc12 (250 pmol) was mixed with GST-ROC1-CUL1324–776 (~200 pmol; prepared as described previously14) immobilized on glutathione beads and incubated for 60 min at 37 °C. The beads were then washed, boiled for 3 min in SDS sample buffer, and subjected to SDS-PAGE. The neddylated proteins were visualized by Coomassie blue staining.
7. Protein Digestion
The purified proteins were separated by 1-D SDS-PAGE and stained by Coomassie Blue. Proteins bands were excised, in-gel digested by trypsin, extracted, and concentrated for LC–MS/MS analysis as described.22 In-solution digestion of the purified proteins was performed following a previous protocol.23
8. Mass Spectrometric Analysis by LC–MS/MS
LC–MS/MS was carried out by nanoflow reverse-phase liquid chromatography (RPLC) (Dionex, CA) coupled online with a quadrupole-orthogonal-time-of-flight tandem mass spectrometer (QSTAR XL, Applied Biosystems/MDS Sciex). The experimental conditions were similar to those in the previous work.23,24 Briefly, the LC analysis was performed using a PepMap capillary column (75 μm i.d. × 150 mm long, Dionex, CA), and the peptides were eluted using a linear gradient of 0% B to 35% B in 40 min at a flow of 200 nL/min. Solvent A contained 98% H2O/2% acetonitrile/0.1% formic acid, whereas solvent B was composed of 98% acetonitrile/2% H2O/0.1% formic acid. The QSTAR MS was operated in an information-dependent mode in which each full MS scan was followed by three MS/MS scans where the three most abundant peptide molecular ions were dynamically selected for collision induced dissociation (CID), thus, generating tandem mass spectra.
The tryptic digests were also analyzed by Linear Ion Trap (LTQ) mass spectrometer (Thermo-Electron Corp.), which was equipped with an Eksigent nanoflow LC (Eksigent, CA). The HPLC separation of the peptides is similar to that described earlier. The LTQ was configured for data-dependent CID mode, where the top 5 representative ions from a single MS scan were chosen for CID analysis, and subsequently added to a dynamic exclusion list with a timed expiration of 30 s. To facilitate improved MS/MS spectral acquisition that corresponds to the LC elution peak, data acquisition was further configured to repeat CID on a selected ion within 15 s of the first MS/MS acquisition.
9. Data Extraction
Monoisotopic masses of parent and corresponding fragment ions, parent ion charge states, and ion intensities from the tandem mass spectra (MS/MS) were obtained by using an automated version of the mascot extraction scripts, that is, mascot.dll (v1.6b16) for QSTAR and lcq_dta.exe (Xcalibur 1.3/Bioworks 3.0) for LTQ MS/MS data through the MASCOT Daemon interface (v 2.1.0). For QSTAR data, the parameters for peak extraction were set as default, except (1) mass tolerance for MS/MS spectra group was 0.2 Da; (2) deisotoping of MS/MS data was disabled; (3) only +2, +3, +4, and +5 charged precursor ions were extracted. For LTQ data, extraction parameters are set as follows: first scan, 0; last scan, ’blank’; minimum mass, 300; maximum mass, 5000; grouping tolerance, 1.4; precursor charge, auto; minimum peaks, 10; threshold, 500.
10. Database Searching
Immediately following automated data extraction, resultant peak lists for each LC–MS/MS experiment were submitted to an in-house MASCOT server (v. 2.1.0). UniProt database (06/29/2006, 228 670 sequence entries) was used for database searching, and Homo sapiens was selected as specific species (14 328 sequence entries). Trypsin was set as the enzyme with a maximum of three missed cleavage sites. For QSTAR data, the mass accuracy for parent ions was set as ±100 ppm, and ±0.3 Da was used for the fragment ion mass tolerance. For LTQ data, ±1.2 Da was used for parent ion mass tolerance, whereas ±1 Da tolerance was chosen for the fragment ions. Trypsin was selected as the enzyme, and the maximum number of missed tryptic cleavages was set as 2. Chemical modifications such as protein N-terminal acetylation, methionine oxidation, N-terminal pyro-glutamine, and deamidation of asparagine were selected as variable modifications during database search. These modifications were chosen due to their frequent occurrence during sample preparation. In addition, modification of Lys by GG was selected as a variable modification for identifying neddylated and/or ubiquitinated peptides. To minimize the manual validation regarding the confidence of protein identification, we have examined the relationship of expectation values and false positive rate. Database searching using both normal database and reverse database has been performed. The false positive rate was calculated as hits(reverse)/(hits(reverse) + hits(normal)) assuming all the hits identified in reverse database were false.25 On the basis of the plot of number of peptides versus expectation values obtained in both normal database and reverse database searching, we determined the false positive rate of protein identification. In this work, 1% peptide false positive rate was chosen, which corresponds to an expectation value of ≤0.01 for QSTAR and ≤0.0086 for LTQ results. Only the proteins identified by two or more unique peptides by QSTAR and/or LTQ MS/MS were reported here.
11. Data Mining
Results were gathered via in-house software designed to extract itemized data from each MASCOT results file and then compiled into spreadsheet formatted files. This approach allows for specific and generalized peptide and protein to be reported based on specific criteria, which include single or multiple experiments, variable score or expectation value cutoff, and specific peptide modification. The nature of the experiment described here requires scrutiny of specific peptide assigned spectra with manual validation as well as the description of globally identified proteins with statistical validation.
12. Relative Quantitation of Cullins
Relative abundance of eight cullin proteins in the GST-Nedd8 sample was accomplished through a normalized spectral abundance factor (NSAF) calculated based on the method developed by Wash-burn and co-workers.26 Briefly, we calculated a spectral abundance factor normalized against the total number of proteins identified (NSAF) as follows:
where the total number of tandem mass spectra matching peptides from protein k (SpC) was divided by the protein length (L), then divided by the sum of SpC/L for all N proteins in the GST-Nedd8 sample.
Results
1. Purification of Nedd8 Conjugated and Associated Proteins
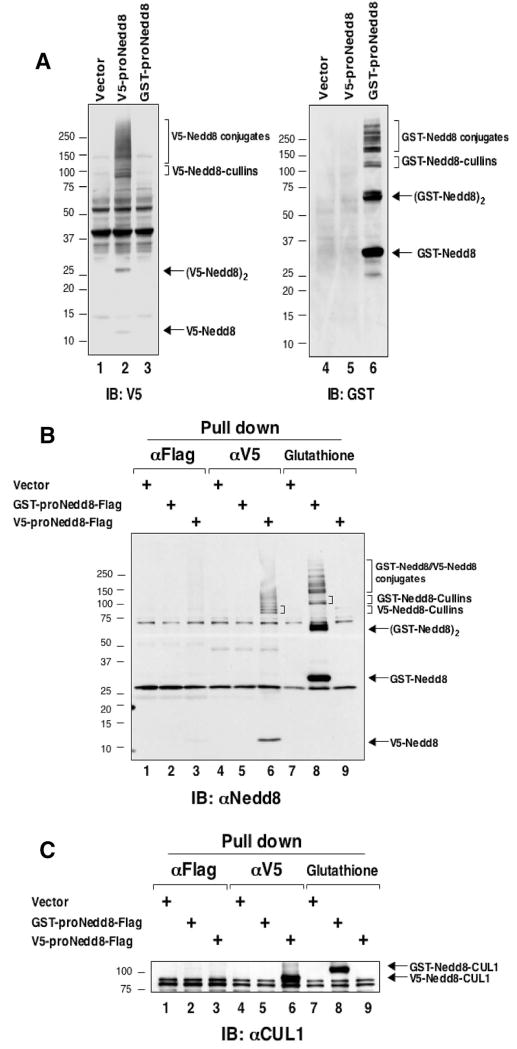
To study neddylation at the proteome scale, precursor forms of Nedd8, N-terminally tagged with GST or V5, were created and introduced into HEK293 cells by transfection. These forms of precursor Nedd8 were used because of their expression at levels significantly higher than those driven by plasmid encoding for mature Nedd8. To evaluate the expression, cell lysates were analyzed by immunoblotting using either anti-V5 or anti-GST antibodies. Figure 1A shows that proteins specifically conjugated to V5-Nedd8 or GST-Nedd8 were detected only in the cells expressing tagged proNedd8. Subsequent affinity pull-down experiments revealed that (1) both V5-Nedd8 and GST-Nedd8 formed conjugates specifically with molecular masses that match cullin-Nedd8 and very slow-migrating species of unknown identity (Figure 1B, lanes 6 and 8) and (2) both V5- and GST-Nedd8 precursors were efficiently processed into mature forms because anti-Flag immunoprecipitation failed to detect the precursor forms (Figure 1B, lanes 1–3). To verify that the neddylated species contain a well-characterized substrate CUL1, precipitates derived from above immunoprecipitation experiments were analyzed by immunoblotting using Cul-1 antibody (Figure 1C). The result showed the formation of either V5-Nedd8-CUL1 or GST-Nedd8-CUL1 of predicted size, indicating that cells expressing either GST-proNedd8 or V5-proNedd8 can be used for the purification of Nedd8 conjugated proteins. To eliminate nonspecifically bound proteins during purification, the isolated proteins are preferred to be eluted from the affinity matrix. Our initial experiments revealed that glutathione-based affinity purification yielded cellular proteins conjugated to, or associated with, Nedd8 with efficiency far greater than that with V5-based approach. Therefore, GST-proNedd8, but not V5-proNedd8, was used for further proteomic analysis. Proteins that were conjugated and/or bound to GST-Nedd8 or GST-Gus (as a negative control) were prepared using large-scale affinity purification from HEK293 cells stably expressing GST-proNedd8 or GST-Gus. Coomassie blue staining analysis of the purified samples revealed the formation of conjugates specifically linked to GST-Nedd8 but not GST-Gus (Figure 2).
Figure 1.
Expression of recombinant Nedd8 in HEK293 cells. (A and B) Expression of V5- or GST-tagged Nedd8 in HEK293 cells. HEK293 cells were transfected with vectors expressing V5-proNedd8 or GST-proNedd8. At 48 h post-transfection, cells were harvested. Extracts (50μg) were used for direct immunoblot analysis with the indicated antibodies (panel A). For pull-down experiments, extracts (500 μg) were used for glutathione matrix purification or immunoprecipitation by anti-V5 or anti-Flag antibodies (panel B). Tagged Nedd8 or its conjugates, separated by 4–20% SDS-PAGE, were detected by anti-GST (panel A), anti-V5 (panel A), or anti-Nedd8 antibodies (panel B). (C)The recombinant Nedd8 is conjugated with Cul-1. The precipitates generated in panel B were subjected to immunoblot analysis with anti-Cul-1 antibodies.
Figure 2.
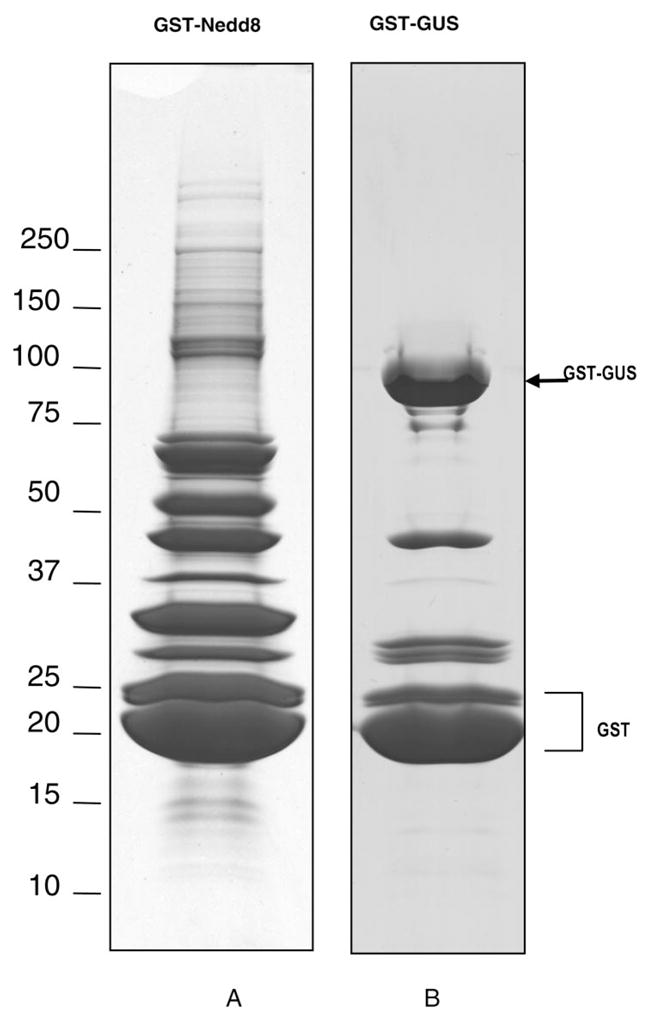
1-D SDS-PAGE gel pictures of the proteins affinity purified from 293 cells stably expressing GST-Nedd8 (A) or GST-Gus (B).
2. Identification of Nedd8 Associated and Conjugated Proteins by LC–MS/MS
Fifty-seven protein bands from GST-Nedd8 purified samples (Figure 2A) and 22 bands from GST-Gus (Figure 2B) were excised for digestion and MS analyses. To improve confidence in protein identification and generate maximum information from a given sample, two different tandem mass spectrometers, QSTAR XL and LTQ MS, were used for LC–MS/MS analyses. The MS/MS spectra from each band were subjected to an in-house MASCOT server for database searching, and the results were complied, validated, and summarized using in-house software. With the use of the criteria of a minimum of two peptides for protein identification with a 1% false positive rate, 392 proteins were identified from a total of 5985 peptides analyzed by QSTAR XL MS; 462 proteins were identified from a total of 11 482 peptides acquired by LTQ MS (Supplemental Table 1 in Supporting Information). More peptides and proteins were determined by LTQ MS due to its high scan speed and sensitivity. As a control, analysis of GST-GUS associated proteins (Figure 2B) yielded a total of 22 proteins by QSTAR XL and 28 proteins by LTQ MS. Presumably, proteins common to both GST-Nedd8 and GST-GUS are considered nonspecific to Nedd8. In comparison, 13 proteins were classified as putative background proteins by QSTAR MS and LTQ MS, whereas an additional one was only identified by LTQ MS (Supplemental Tables 2A and 2B in Supporting Information). It should be noted that there is a possibility that some of the proteins present in GST-GUS purification may still be specific to GST-Nedd8, especially if these proteins are identified with significantly increased number of peptides in the GST-Nedd8 sample, such as DNA damage-binding protein 1 and WD repeat protein 23. Since DNA damage-binding protein 1 is involved in the Nedd8 pathway10 and WD repeat protein 23 contains motif of potentially associating with Cul-4, they are not excluded from the final list. After removing the putative background proteins from the proteins identified in GST-Nedd8 preparation, 335 proteins were identified from both instruments, with 46 proteins unique to QSTAR XL MS and 115 proteins unique to LTQ MS (Supplemental Figure 1 in Supporting Information). In total, 496 proteins identified from the Gel/MS approach represent Nedd8 modified and/or associated proteins (Supplemental Table 1 in Supporting Information). Repetitive purifications confirmed the presence of more than 90% of proteins identified (457 out of 496). The proteins identified in GST-Nedd8 sample can be grouped into six categories based on their biological functions.
(i) Cullins
The first group of proteins identified are the cullin family members, which are known neddylated substrates.10 All of the eight cullin-family members including Cul-1, -2, -3, -4A, -4B, -5, -7, and Parc have been identified in the GST-Nedd8 affinity purified proteins, which has not been accomplished previously.20,21 The detection of Cul-1, -2, -3, -4A/B, and -5 were further confirmed by immunoblotting using specific cullin antibodies, as shown in Figure 3A. The anti-Nedd8 immunoblot revealed the presence of both cullin conjugates and additional high molecular weight species, suggesting additional novel substrates for Nedd8. To verify the presence of multiple GST–Nedd8–cullin conjugates, proteins copurified with GST-Nedd8 were treated with DEN1, which is capable of hydrolyzing the Nedd8–cullin isopeptide bond in vitro.19 The results showed that cleavage by DEN1 resulted in formation of CUL-1, -2, -3, and -4 with sizes of unmodified forms as expected (Figure 3B). Figure 3C displays the relative abundance of all cullin proteins captured during the purification determined by use of normalized spectral abundance factor (NSAF).26 It appears that Cul-2 is slightly more abundant than the others, whereas Cul-7 seems to be the least abundant in HEK293 cells.
Figure 3.
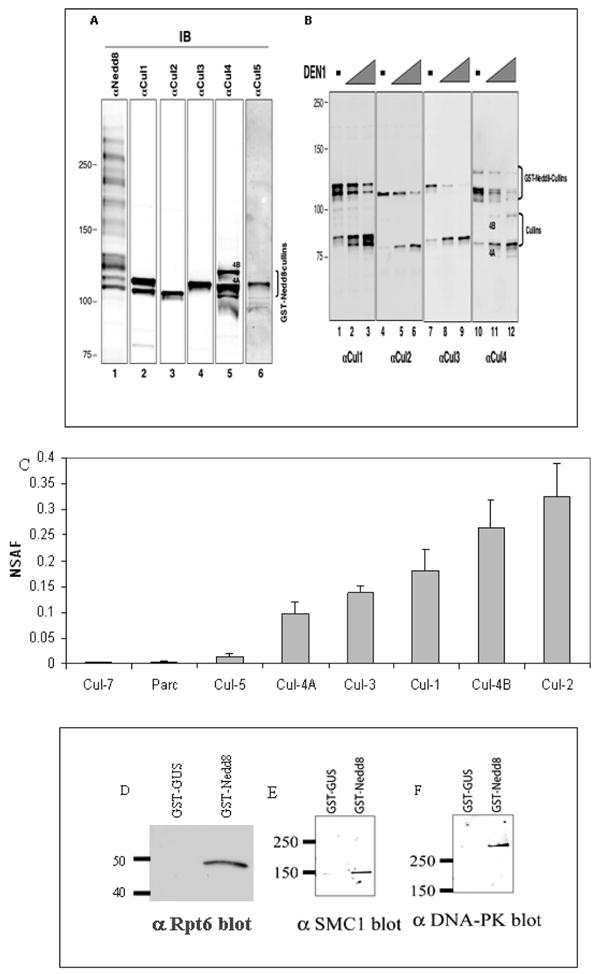
(A) GST-Nedd8 and its conjugates, isolated after glutathione matrix purification of extracts from HEK293 cells constitutively expressing GST-Nedd8, were subjected to immunoblot analysis with indicated antibodies. (B) The precipitates generated in panel A were treated with purified DEN1 to remove GST-Nedd8 from cullin substrates. The treated mixtures were analyzed by immunoblot analysis with antibodies as indicated. (C) Relative abundance plot of the identified cullins using NSAF. The detailed results were shown in Supplemental Table 3 in Supporting Information; D–F) Validation of the selected interactions with GST-Nedd8 in comparison to GST-Gus (negative control) by immunoblotting. (D) Proteasome 19S subunit Rpt6. (E) Structural maintenance of chromosome 1-like 1 protein (SMC1). (F) DNA-dependent protein kinase catalytic subunit (DNA-PK).
(ii) Cullin-Containing Ubiquitin E3 Ligases
In addition to cullin proteins, components of cullin-containing ubiquitin E3 ligases such as ROC1/Rbx1, elongin C/B dimmer, Skp1, F-box proteins, BTB family proteins, DDB1 and DDB2, and additional WD-repeat proteins were identified and summarized in Table 1. Twelve F-box proteins were identified, which are responsible for substrate specificity of SCF E3 ligases. On the basis of spectral counts, it appears that F-box protein 22 and Skp2 are the most abundant F-box proteins detected here. Additionally, four BTB family proteins and seven WD repeat proteins were identified as listed.
Table 1.
Components of Cullin-Containing Ubiquitin Ligase Complexes Identified by LC–MS/MS
| functional category | acc no. | protein name |
|---|---|---|
| F-box proteins | O94952 | F-box only protein 21 |
| Q8NEZ5 | F-box only protein 22 | |
| Q8TB52 | F-box only protein 30 | |
| Q8P3S6 | F-box only protein 42 | |
| Q6PJ61 | F-box only protein 46 | |
| Q9UKT4 | F-box only protein 5 | |
| Q9Y3I1 | F-box only protein 7 | |
| Q96ME1 | F-box/LRR-repeat protein 18 | |
| Q9BSU8 | F-box/WD repeat protein 5 | |
| O60907 | F-box-like/WD repeat protein TBL1X | |
| Q13309 | F-box protein Skp2 | |
| Q9UKC2 | F-box only protein 11 | |
| BTB family proteins | Q86T24 | Zinc finger and BTB domain-containing protein 33 |
| Q9P2N7 | BTB and kelch domain-containing protein 2 | |
| Q86V97 | Kelch repeat and BTB domain-containing protein 6 | |
| Q9H0C5 | BTB/POZ domain-containing protein 1 | |
| WD repeat proteins | Q8N5D0 | WD and tetratricopeptide repeats protein 1 |
| Q8TEB1 | WD repeat protein 23 | |
| Q9NNW5 | WD repeat protein 6 | |
| P61962 | WD repeat protein 68 | |
| Q12788 | WD repeat protein SAZD | |
| Q9BQ67 | Glutamate-rich WD repeat-containing protein 1 | |
| P61964 | WD repeat protein 5 | |
| Other | Q15369 | elongin C |
| Q15370 | elongin B | |
| P62877 | Rbx1/Roc1 | |
| P63208a | Skp1 | |
| Q16531 | DDB1 |
Note: Skp1 was identified by single peptide above the expectation threshold. The peptide report for Skp1 is listed in Supporting Information Table 4.
(iii) Enzymes and Regulators Involved in the Nedd8 Pathway
It is known that the neddylation process requires an E1 activating enzyme (APP-BP1/Uba3 heterodimer) and an E2 conjugating enzyme, Ubc12, for conjugating Nedd8 to a substrate.10 Not surprisingly, these enzymes were identified in the GST-Nedd8 sample. In addition, Nub1 (Supplemental Table 4 in Supporting Information), p120CAND1, DCN-1 like protein, and COP9 signalosome components (Supplemental Table 4 in Supporting Information) were identified as well. Nub1 is a known Nedd8 interacting protein and has been suggested to recruit Nedd8 and its conjugates to the proteasome for degradation through interactions with S5a of the 19S proteasome subunit.27 The identification of p120CAND1 is unexpected because this protein has been shown to selectively bind to cullins free of Nedd8.28,29 We speculate that a fraction of cullin-based E3s probably exist in dimerized forms, in which one cullin molecule is conjugated with Nedd8 and the other cullin molecule remains unmodified and is bound to p120CAND1. Indeed, a recent study has shown that budding yeast SCF acts in dimeric forms.30 It is worth noting that p120CAND2 was also identified here. The association between DCN-1 and Nedd8 is consistent with previous observations, demonstrating that DCN-1 like proteins in Caenorhabditis elegans and Saccharomyces cerevisiae directly bind Nedd8 and physically associate with cullins, which is required for neddylation of cullins in SCF-type E3 ubiquitin ligases.31 With regards to the signalosome components identified, the association between COP9 and Nedd8 is likely indirect as the signalosome binds to both cullin and ROC1/Rbx1.32–34
(iv) Additional Components of the Ubiquitin–Proteasome Pathway
Components of the 19S regulatory proteasome complex are associated with Nedd8, with 11 out of approximately 20 19S subunits identified in the Nedd8 pull down. To validate the identified interactions, immunoblot analysis was carried out to confirm the association between Nedd8 and the 19S complex. As shown in Figure 3D, GST-Nedd8, but not GST-Gus, copurified with 19S subunit Rpt6. No 20S proteasome complex subunits were identified by MS analysis. However, only the 20S, not the 19S, subunits have been reported to copurify with Nedd8 samples previously.20 This discrepancy most likely is due to the use of different affinity purification procedures. In addition, several other components involved in ubiquitin–proteasome degradation pathway were also copurified, which include ubiquitin, ubiquitin-protein ligase BRE1A (Q9NWQ3), ubiquitin-protein ligase EDD1 (O95071), ubiquitin-associated protein 2-like (Q14157) (contains one UBA domain), E3 ubiquitin protein ligase HUWE1 (Q9NSL6), and Baculoviral IAP repeat-containing protein 6 (Q9NR09).
(v) Other Nedd8 Associated Proteins
A broad spectrum of proteins (Supplemental Table 1 in Supporting Information) was identified in the GST-Nedd8 purification and can be classified into biological processes such as signal transduction, metabolism, transcription, cell cycle, and apoptosis. Among them, proteins that are involved in DNA repair, replication, and transcription were also identified and summarized in Table 2. To verify their association with GST-Nedd8, selected proteins were validated by immunoblotting. Figure 3E, F shows GST-Nedd8, but not GST-Gus, copurified with SMC1 and DNA-PK. Since these proteins exhibited gel mobility consistent with forms free of Nedd8, they are most likely associated with GST-Nedd8 noncovalently.
Table 2.
List of the Identified Proteins Known To Be Involved in DNA Repair, Transcription, and Replication
| functional category | acc no. | protein name |
|---|---|---|
| DNA Repair | P43246 | DNA mismatch repair protein Msh2 |
| P52701 | DNA mismatch repair protein MSH6 | |
| Q9UME3 | DNA-dependent protein kinase catalytic subunit | |
| O43502 | DNA repair protein RAD51 homologue 3 | |
| DNA replication | P33993 | DNA replication licensing factor MCM7 |
| Q14566 | DNA replication licensing factor MCM6 | |
| P33992 | DNA replication licensing factor MCM5 | |
| P33991 | DNA replication licensing factor MCM4 | |
| P25205 | DNA replication licensing factor MCM3 | |
| O75419 | CDC45-related protein | |
| P35250 | Replication factor C subunit 2 | |
| P35249 | Replication factor C subunit 4 | |
| P40937 | Replication factor C subunit 5 | |
| P28340 | DNA polymerase delta catalytic subunit | |
| Q07864 | DNA polymerase epsilon catalytic subunit A | |
| O60673 | DNA polymerase zeta catalytic subunit | |
| P11388 | DNA topoisomerase 2-alpha | |
| Q02880 | DNA topoisomerase 2-beta | |
| Histones and Histone modifying enzyme | O75367 | Core histone macro-H2A.1 |
| Q92769 | Histone deacetylase 2 | |
| Q9UBN7 | Histone deacetylase 6 | |
| Q96QV6 | Histone H2A type 1-A | |
| P04908 | Histone H2A type 1-B | |
| P20671 | Histone H2A type 1-D | |
| Q96KK5 | Histone H2A type 1-H | |
| Q8IUE6 | Histone H2A type 2-B | |
| Q16777 | Histone H2A type 2-C (H2A-GL101) (H2A/r) | |
| P0C0S5 | Histone H2A.Z (H2A/z) | |
| Q71UI9 | Histone H2AV (H2A.F/Z) | |
| Q96A08 | Histone H2B type 1-A | |
| P33778 | Histone H2B type 1-B | |
| P62807 | Histone H2B type 1-C/E/F/G/I | |
| Q93079 | Histone H2B type 1-H | |
| P06899 | Histone H2B type 1-J | |
| Q99880 | Histone H2B type 1-L | |
| Q16778 | Histone H2B type 2-E | |
| Q8N257 | Histone H2B type 3-B | |
| Q16695 | Histone H3.4 | |
| P62805 | Histone H4 | |
| Q09028 | Histone-binding protein RBBP4 | |
| O60341 | Lysine-specific histone demethylase 1 | |
| Transcription | O15160 | DNA-directed RNA polymerase I 40 kDa polypeptide |
| P30876 | DNA-directed RNA polymerase II 140 kDa polypeptide | |
| P19387 | DNA-directed RNA polymerase II 33 kDa polypeptide | |
| P24928 | DNA-directed RNA polymerase II largest subunit | |
| P52434 | DNA-directed RNA polymerases I, II, and III 17.1 kDa polypeptide | |
| P18074 | TFIIH basal transcription factor complex helicase subunit | |
| Chromatin | Q14683 | Structural maintenance of chromosome 1-like 1 protein, SMC1 |
| O95347 | Structural maintenance of chromosome 2-like 1 protein, SMC2 | |
| Q9UQE7 | Structural maintenance of chromosome 3, SMC3 | |
| Q9NTJ3 | Structural maintenance of chromosomes 4-like 1 protein, SMC4 | |
| Q14839 | Chromodomain-helicase-DNA-binding protein 4 |
3. Identification of Neddylation Sites by LC–MS/MS
(i) Identification of Neddylation Sites of Cullins
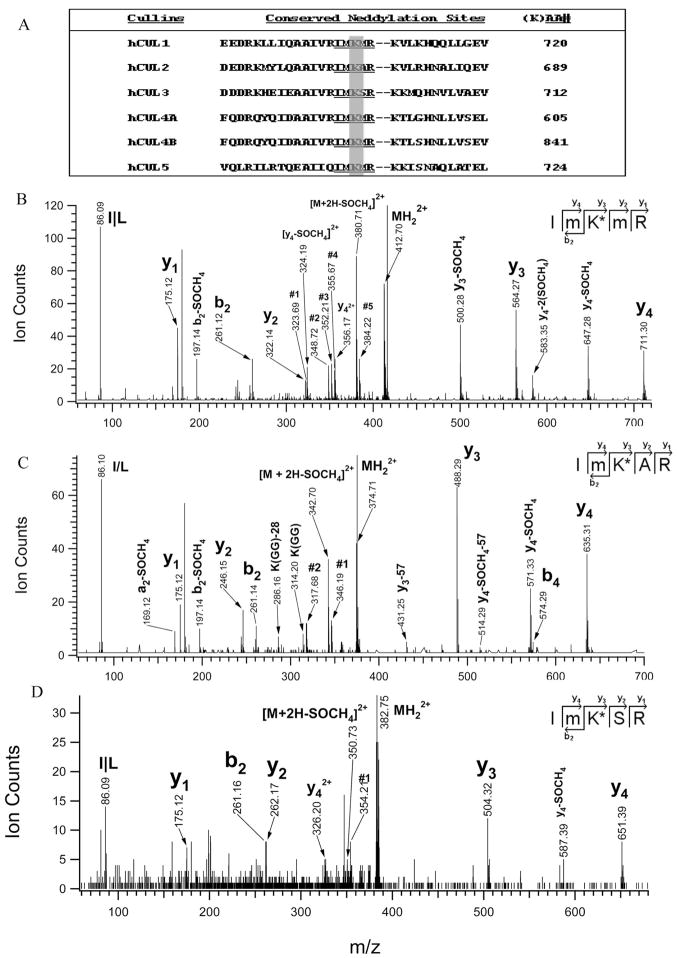
The characteristics of neddylation after trypsin digestion are presented by the remnant of two glycine residues (GG) attached to an internal lysine residue of the substrate. This feature can be determined by MS/MS similarly to that for ubiquitination.35 Previously, it has been shown that cullins have a conserved sequence (IVRIMKMR) for neddylation.10 As shown in Figure 4A, the tryptic peptide having the sequence IMKMR is conserved for Cul-1, -4A/4B, and -5, where the K is known to be neddylated. In our analysis, the neddylated form of IMKMR was identified and its MS/MS spectrum is displayed in Figure 4B. Because of its short sequence, this peptide eluted very early (~9 min) during the LC–MS run. In the MS/MS spectrum (Figure 4B), several unique mass losses of the parent ion (MH22+ 412.70) were observed. Doubly charged ions (380.71, 348.72) were due to the loss of one SOCH4 (−64 Da) and two SOCH4(−128 Da) from the parent ion; whereas doubly charged ions (384.22, 355.67) were the resulting ions after the parent ion lost either one (−57 Da) or two glycine (−114 Da). Doubly charged ions (352.21, 323.69) were attributed to the loss of [SOCH4 + Gly] and [SOCH4 + 2×Gly] from the parent ion. These characteristic losses suggest that this peptide has two oxidized methionines (m) and one GG extension. The detection of b2, b2-SOCH4, y1, y2, y3, y3-SOCH4, y4, y4-SOCH4, and y4-2SOCH4 determined the peptide sequence as ImK*mR, where K* is modified by GG. The identified sequence agrees well with the neddylated sequence of Cul-1, -4A/B, or -5. In addition to this sequence, three related peptides with sequence of ImK*MR (MH22+404.7), IMK*mR (MH22+404.7), and IMK*MR (MH22+ 396.7) were observed and their MS/MS spectra further confirmed the neddylation (data not shown). These peptides were found in all of the bands containing the four different cullins, suggesting that the modification was detected for each cullin with the same neddylation sequences. Similarly, the conserved neddylation sites of Cul-2 and -3 were identified and shown in Figure 4C, D. In Figure 4C, fragment ions (MH22+, 342.70, 346.19, 317.68) due to loss of SOCH4 and/or glycine from parent ion (MH22+ 374.71) were also obtained. The observation of y and b ion series and their satellite ions determined the sequence as ImK*AR for Cul-2. In comparison, the MS/MS spectrum of MH22+ 382.70 (Figure 4D) identified its sequence as ImK*SR for Cul-3.
Figure 4.
Identification of neddylated peptide sequences of cullins. (A) Alignment of the conserved sequences of cullins for neddylation; QSTAR MS/MS spectra of (B) a tryptic peptide MH22+ 412.70. Ion assignments: no. 1 (323.69), [M+2H-SOCH4-2×Gly]2+; no. 2 (348.72), [M+2H-2(SOCH4)]2+; no. 3 (352.21), [M+2H-(SOCH4+ Gly)]2+; no. 4 (355.67), [M+2H-2×Gly]2+; no. 5 (384.22), [M+2H-Gly]2+. The sequence was determined as ImK*mR, corresponding to Cul-1, -4A/B, -5. (C) A tryptic peptide MH22+ 374.71. Ion assignments: no. 1 (346.19), [M+2H-Gly]2+; no. 2 (317.68), [M+2H-2×Gly]2+. The sequence was determined as ImK*AR, corresponding to Cul-2. (D) A tryptic peptide MH22+ 382.70. Ion assignment: no. 1 (354.21), [M+2H-Gly]2+. The sequence was determined as ImK*SR, corresponding to Cul-3. K*-modified with GG; m-oxidized methionine.
(ii) Evidence for Nedd8 Chain Assembly in Vivo
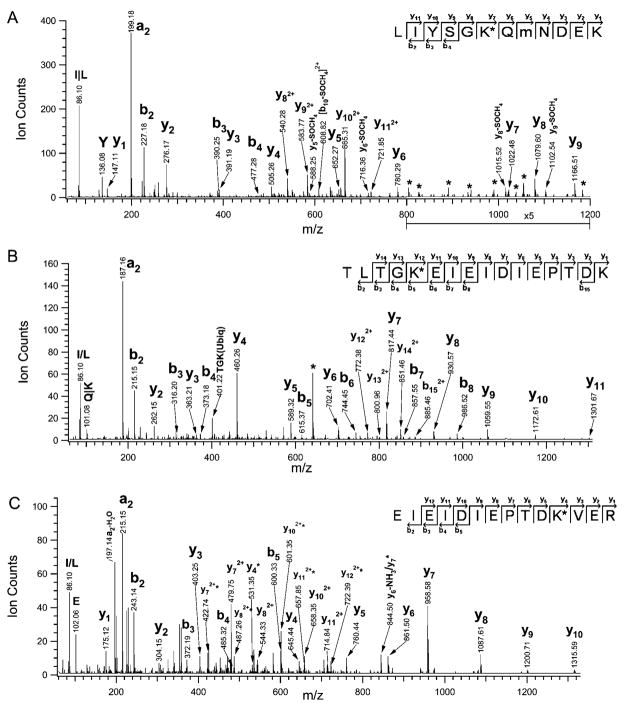
We discovered for the first time that internal lysines of Nedd8 can be modified by the GG motif, indicative of assembly of a Nedd8 chain. Protein neddylation usually involves conjugation of one Nedd8 to a lysine residue of its substrates, and has not been known whether Nedd8 can form chains in vivo. Figure 5A shows a MS/MS spectrum of a triply charged tryptic peptide (MH33+519.24). The observation of y1 ~ y11, and b2 ~ b4 ions determined its sequence as 43LIYSGK*QmNDEK54, corresponding to Nedd8. The unique loss of SOCH4 (−64 Da) from y5, y6, y8, y9, and b10 confirms the presence of oxidized methionine. Because of the characteristic mass addition, that is,114 Da for GG linkage, the K48 of this Nedd8 sequence is determined to be covalently conjugated to another Nedd8 to form the chain assembly, supported by the detection of large internal ions (marked with * in Figure 5A, Supplemental Table 5 in Supporting Information). MS/MS analyses of the same peptide by LTQ MS and a related peptide containing a nonoxidized methionine (43LIYSGK*QMNDEK54, MH33+ 513.93) further confirmed the attachment of GG on K48 (data not shown). Similarly, K11, K22, and K60 of Nedd8 have also been found to be covalently conjugated to GG. MS/MS spectra of their corresponding tryptic peptides are shown in Figure 5B–D. In comparison, the loss of GG (−114 Da) was only detected in the MS/MS spectra of the peptide containing modified K22. This suggests that such a loss is likely dependent on peptide sequence. As a result, the modified Nedd8 peptides have been identified in multiple band regions as shown in Supplemental Figure 3 in Supporting Information, and the frequency of detecting peptides containing the modified K48 appears to be very high. Collectively, these results strongly suggest that Nedd8 can form a chain assembly in vivo.
Figure 5.
Determination of Nedd8 chain assembly in vivo. QSTAR MS/MS spectra of the neddylated Nedd8 peptides. (A) MH33+ 519.24, the sequence was determined as LIYSG(48K)*QmNDEK. Ions labeled with *: internal ions containing K* as displayed in Supplemental Figure 3 in Supporting Information. (B)MH33+ 639.35, the sequence was determined as TLTG(11K)*EIEIDIEPTDK. (C)MH33+ 600.63, the sequence was determined as EIEIDIEPTD(22K)*VER. Ions labeled with *: ions with loss of 114 Da;. (D) LTQ MS/MS spectrum of a neddylated Nedd8 peptide MH33+ 742.16, the sequence was determined as TAADY(60K)*ILGGSVLHLVLALR. K*: modified with GG; m, oxidized methionine.
(iii) Analysis of in Vitro Neddylated Cul-1
(a) Both Cul-1 K720 and K769 Serve as Neddylation Sites in Vitro.
Previously Cul-1 has been shown to be hyperneddylated in vitro.19 However, the nature of the hyperneddylation remains elusive. We sought to analyze in vitro neddylated products of Cul-1. For this purpose, the protein bands corresponding to neddylated products Cul-1 (324–776) (band nos. 1–5, lane 2 in Figure 6A) and the control bands (nos 1′–5′, lane 1 in Figure 6A) were subjected to in-gel digestion and subsequent LC–MS/MS analysis. In bands 1 and 1′, unmodified Cul-1 (323–776) was identified. In band 2–3, both Nedd8 and Cul-1 (324–776) were identified. In addition, MS/MS analyses yielded two peptides (MH22+ 412.71, MH22+ 404.71) of sequences as ImK*mR (Figure 6B) and IMK*mR (Supplemental Figure 2A in Supporting Information), respectively, indicating the neddylation of K720 in Cul-1 (324–776). Their MS/MS spectra are virtually identical to those obtained from the GST-Nedd8 sample (Figure 4B). In addition, K769 of Cul-1 (324–776) was also determined to be neddylated by comparing the LTQ MS/MS spectra of the unmodified (MH22+ 680.8, Figure 6C) and neddylated form (MH22+ 737.8, Figure 6D) of the same peptide, with the sequence determined as VDGEKDTYSYLA, the C-terminal sequence of Cul-1 (324–776). In comparison to the MS/MS spectrum in Figure 6C, the overall fragmentation pattern displayed in Figure 6D is very similar. The b4 ions are the same in both spectra, whereas the other b ions (i.e., b5–b10) in Figure 6D are 114 Da higher than their corresponding b ions in Figure 6C. This suggests that the lysine residue at position 5 (i.e., K769) from the N-terminus was modified by a chemical moiety with a mass of 114 Da, characteristic of GG attachment. Although a keratin peptide (MH22+ 738.4) was coeluted and sequenced with the modified peptide (MH22+ 737.8, Figure 6D) by LTQ MS, the unique fragment ions for each peptide were obtained, which allowed unambiguous identification of their sequences (Supplemetal Figure 2B in Supporting Information). Additionally, the corresponding QSTAR MS/MS spectra of the unmodified and modified peptides shown in Figure 6C, D further supported their sequence assignments (Supplemental Figure 2C, D in Supporting Information), and determined the neddylation on K769 of Cul-1 (324–776). Although K720 of Cul-1 is the prominent site for neddylation, the results suggest that additional lysines in Cul-1 could be targeted as well.
Figure 6.
In vitro neddylation of Cul-1 (324–776). (A)The neddylated proteins were visualized by Coomassie blue staining. Lane 1, negative control-without Nedd8; lane 2, in the presence of Nedd8; a, hyperneddylated Cul-1 (324–776); b, dineddylated Cul-1 (324–776); c, mononeddylated Cul-1(324–776); d, unmodified FLAG-Cul-1 (324–776); e, GST-Roc1. (B) Identification of the known neddylation site (K720) of Cul-1. QSTAR MS/MS spectrum of a tryptic peptide MH22+ 412.70. Ion assignments: no. 1 (323.70), [M+2H-SOCH4-2×Gly]2+; no. 2 (348.73), [M+2H-2(SOCH4)]2+; no. 3 (352.20), [M+2H-(SOCH4+ Gly)]2+; no. 4 (384.20), [M+2H-Gly]2+. The sequence was determined as ImK*mR. (C and D) Identification of a new in vitro neddylation site of Cul-1. (C)LTQ MS/MS spectrum of a tryptic peptide (MH22+ 680.8). The sequence was determined as VDGE(769K)DTYSYLA, corresponding to the C-terminus of Cul-1(324–776), (D) LTQ MS/MS spectrum of a tryptic peptide (MH22+ 737.8). The sequence was determined as VDGE(769K)*DTYSYLA; K*, modified with GG. Ions labeled with “*” represent the sequence ions matched to the coeluting peptide (MH22+ 738.4), matched to Keratin. The details of ion assignment are shown in Supplemental Figure 2B in Supporting Information. K*, modified with GG; m, oxidized methionine. The corresponding QSTAR MS/MS spectra of the peptides shown in panels C and D are illustrated in Supplemental Figure 2C, D in Supporting Information.
(b) Assembly of Nedd8 Chains on Cul-1 in Vitro
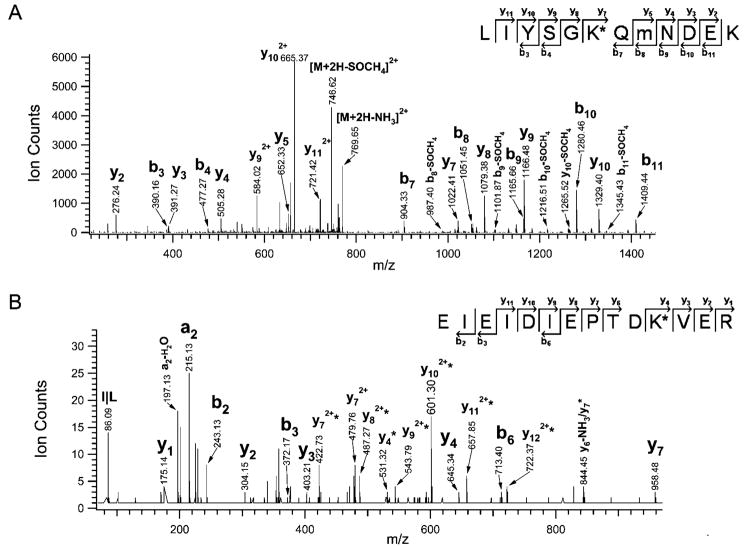
Aside from neddylation of Cul-1 (324–769), Nedd8 chain assembly was observed at a higher molecular weight in lane 2 (Figure 6A). Figure 7A illustrates the MS/MS spectrum of a tryptic peptide (MH22+778.37), in which a series of b ions (i.e., b3, b4, b7~b11) and y ions (i.e., y2 ~ y5, y7 ~ y10) were obtained. The peptide sequence was determined as LIYSGK*QmNDEK, corresponding to Nedd8, where K48 is neddylated. The observation of losses of SOCH4 (−64 Da) from the parent ion as well as fragment ions further confirmed the presence of oxidized methionine in the sequence. Figure 7B displays the MS/MS spectrum of a tryptic Nedd8 peptide (MH33+ 600.63), and the peptide sequence was determined as EIEIDIEPTDK*VER, where K22 was neddylated. Not surprisingly, their fragmentation patterns appear to be very similar to the same peptides detected in the purified GST-Nedd8 sample (Figure 5A, C). It seems that Nedd8 can form a chain assembly in vitro, and both K48 and K22 can be used as the conjugation site for neddylation. Interestingly, K48 appears to be more favorable since the peptide containing the modified K48 has been found in multiple regions of the gel.
Figure 7.
Determination of in vitro Nedd8 chain assembly. (A) LTQ MS/MS spectrum of a tryptic Nedd8 peptide (MH22+778.37). The sequence was determined as LIYSG(48K)*QmNDEK. (B) QSTAR MS/MS spectrum of a tryptic Nedd8 peptide (MH3 3+ 600.63). The sequence was determined as EIEIDIEPTD(22K)*VER. Ions labeled with *, ions with loss of 114 Da; K*, modified with GG; m, oxidized methionine.
Discussion
In this work, we have employed an integrated proteomic approach for the study of Nedd8 associated and conjugated proteins. The combined use of efficient single-step affinity purification, 1-D gel electrophoresis, and two different tandem mass spectrometers, that is, LTQ and QSTAR XL MS, has provided the identification of a broad spectrum of proteins captured during the preparation. The high sensitivity of LTQ MS enables the identification of low-abundance proteins, and the usage of two different mass spectrometers greatly increases the detection confidence, especially for those identified by only one or two peptides. Furthermore, the information generated by both the LTQ and QSTAR XL MS is complimentary, providing the identification of an increased number of proteins. Separation of samples by 1-D SDS-PAGE prior to enzymatic digestion improves the dynamic range because several abundant components in the mixture are separated from the low-abundance protein bands prior to their mass spectrometric analyses. In addition, 1-D gel profiling provides a visual display of copurified proteins and direct observation of the same proteins migrating at different molecular weights due to post-translational modifications and/or processing. Purification of the GST-Gus protein effectively provides the elimination of nonspecific background. Therefore, the methods described here would be a valuable strategy for investigating protein interactions in general, especially for other ubiquitin-like proteins and their modified and associated proteins.
In total, 496 putative Nedd8 modified and associated proteins were identified. Among them are neddylation and deneddylation enzymes, proteasome components, polyubiquitin, transcriptional factors, DNA repair complexes, DNA replication complexes, cell cycle regulatory complexes, and histone complexes. The identification of such a diverse array of proteins is significant since the majority of them have not been reported from previous proteomic analyses of the Nedd8 pathway.20,21 For the first time, all cullin proteins were identified in a single analysis. On the basis of the relative abundance analysis using NSAF, it appears that Cul-2 and Cul-4B are at similar levels, but they are more abundant than Cul-1, Cul-3, and Cul-4A. Cul-5, Parc, and Cul-7 are much less abundant, among which Cul-7 is the least abundant. In addition to cullin proteins, various components of cullin containing E3 ubiquitin ligases were copurified and identified including F-box proteins, BTB family proteins, and WD40 repeat proteins. It is worth noting that several recently assigned and noncullin Nedd8 substrates, that is, pVHL, mdm2, p53, EGFR receptor, and BCA3,15–18 were not identified. Coincidently, they have not been identified by MS in any previous reports.20,21 This may be because (1) the newly assigned substrates have extremely low abundance in their neddylated forms and (2) the highly dynamic nature of neddylation/deneddylation yields very low level of neddylated species at steady state. In this regard, purification of Nedd8 modified proteins under fully denaturing conditions may prove to be beneficial.
The neddylation sites of all of the cullins (i.e., Cul-1, -2, -3, -4A/B, and -5), with the exception of Parc and Cul-7, were determined by MS/MS analyses. The lack of identification of neddylation sites of Parc and Cul-7 is due to their low abundance in the sample. So far, the neddylation sites of cullins have been only determined by mutagenesis.10 Our mass spectrometric data have provided the first evidence to support the mutagenesis results. In addition to cullin neddylation, one of the most exciting findings in this work is the observation of chain assembly on Nedd8 in vivo and in vitro. Four of the nine lysine residues of Nedd8, that is, K11, K22, K48, and K60, have been determined to be modified by GG after the trypsin digestion in GST-Nedd8 sample, suggesting the covalent attachment of another Nedd8 on these lysines for chain assembly in vivo. This view has been further supported by MS analysis of in vitro neddylation of Cul-1(324–776), revealing that both K48 and K22 of Nedd8 are able to covalently link to another Nedd8 and form a chain in vitro. It appears that K48 of Nedd8 is the dominant site for chain formation both in vivo and in vitro due to its occurrence during MS analysis. Previous studies have shown that multiple Nedd8-containing bands were observed when affinity purification of EGFR, p53, or BCA3 was carried out from cells expressing tagged Nedd8.16–18 Although these authors suggested that these bands may be due to multiple mononeddylation events, we speculate polyneddylation is most likely the cause for these observations.
Ubiquitin chain formation is best known for its role in targeting conjugated proteins to the proteasome for degradation, a process that involves chain formation via K48 or K29.1,2 For yeast ubiquitin, it has been reported that chain formation can occur via all seven internal lysines.35 In contrast to the extensive information on ubiquitin chain formation,35–37 very little is known about polymerization of ubiquitin-like proteins. Recent studies have demonstrated that SUMO can form polysumoylation in vitro38,39 as well as in vivo.40 With our results, we speculate that chain formation of ubiquitin-like proteins may potentially be a general phenomenon in cell biology. The biological significance of ubiquitin-like protein chain formation such as SUMO and Nedd8, however, needs to be elucidated.
It is noteworthy that both neddylation and ubiquitination lead to the remnant of GG from either Nedd8 or ubiquitin on the modified internal lysine of their substrates after trypsin digestion. This is due to the high sequence similarity between ubiquitin and Nedd8. Since ubiquitin is present in the GST-Nedd8 sample, we cannot rule out the possibility that Nedd8 could be ubiquitinated, though the low abundance of ubiquitin relative to Nedd8 makes this scenario unlikely. Because of the overexpression of GST-Nedd8 in the 293 stable cell lines, it is possible that Nedd8 may replace ubiquitin to form mixed chains. In addition, it is possible that the Nedd8 chain assembly may be attributed to its overexpression in cells.
In this work, an extensive array of proteins in transcription, replication, DNA repair, and chromatin organization and remodeling have been identified as components that may potentially participate in Nedd8-related regulatory pathways. This is not surprising given the accumulation of evidence that cullin-based E3s are involved in regulating DNA replication and genome stability. However, it should be cautioned that these components were identified in cells that overexpress Nedd8. Further studies to confirm and extend these findings are necessary.
In summary, our proteomic analysis of the Nedd8 pathway has revealed a plethora of proteins involved in this pathway and provided the first hand evidence that Nedd8 can form chains both in vivo and in vitro. The important findings in Nedd8 chain assembly and its interaction network provide a strong basis for future studies to be steered toward understanding the extent of cellular processes influenced by neddylation events and how these events are regulated. Quantitative mass spectrometry such as SILAC41 combined with double affinity purification under fully denaturing conditions36 may be useful for future experiments to examine changes among Nedd8 substrates under different cellular conditions and genetic backgrounds.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants (GM-74830 to L.H., GM61051 and CA095634 to Z.-Q.P.), the Dept. of the Army (PC-041126 to L.H.), a UCI BIT postdoctoral fellowship to J.J., and NCI cancer training grant to K.W. The authors thank Brad Hart and John Nemmers from Thermoelectron Corp. for their generous support on using LTQ MS.
Abbreviations
- LC–MS/MS
liquid chromatography-tandem mass spectrometry
- MS
mass spectrometry
- Ub
Ubiquitin
- Ubl
Ubiquitin-like protein
- m/z
mass/charge
- TOFMS
time-of-flight mass spectrometry
- Gly
glycine
Footnotes
Supporting Information Available: Supplemental Table 1, summary of proteins identified from the GST-Nedd8 sample by LC–MS/MS using QSTAR and LTQ MS; Supplemental Table 2A, B, the putative background proteins identified in both GST-Nedd8 and GST-Gus samples by (A) QSTAR MS, (B) LTQ MS; Supplemental Table 3, NSAF values of cullins for Figure 3C; Supplemental Table 4, detailed peptide report for the proteins known to be involved in Nedd8 pathway but identified with single peptide above the threshold criteria; Supplemental Table 5, internal ion assignments labeled with “*” in Figure 5A; Supplemental Figure 1, comparison of the proteins identified by the two different instruments, LTQ vs QSTAR MS after removing putative background proteins; Supplemental Figure 2 (A) QSTAR MS/MS spectrum of a tryptic peptide MH22+ 404.70: no. 1(376.23), [M+2H-Gly]2+; no. 2(347.70), [M+2H-2×Gly]2+. The sequence was determined as IMK*mR, indicating K720 of Cul 1 is neddylated; (B) the detailed ion assignments in the LTQ MS/MS spectrum of the tryptic peptide (MH22+ 737.8, VDGE(769K)*DTYSYLA) shown in Figure 6D. Ions labeled with “*” matched to the coeluting peptide (MH22+ 738.4) with the sequence of FLEQQNQVLQTK (human keratin type II). The corresponding QSTAR MS/MS spectra of the tryptic peptides shown in Figure 6C, D: (C) MH22+ 680.83, VDGE(769 K)DTYSYLA; (D) MH22+ 737.86, VDGE(769K)*DTYSYLA. K*, modified with GG; m, oxidized methionine.
Supplemental Figure 3, (A) labeling of the protein gel bands analyzed from Figure 2A; (B) the band location of the modified Nedd8 peptides identified by MS/MS. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28(6):321–328. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- 4.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 5.Hochstrasser M. There’s the rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 6.Osaka F, Saeki M, Katayama S, Aida N, Toh-E A, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, Kato S. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19(13):3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155(4):571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou C, Lin Y, Chen Y, Chien C. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16(18):2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handeli S, Weintraub H. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell. 1992;71(4):599–611. doi: 10.1016/0092-8674(92)90594-3. [DOI] [PubMed] [Google Scholar]
- 10.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23(11):1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 11.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 12.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 13.Li B, Yang F, Clapp DW, Chun KT. Enforced expression of CUL-4A interferes with granulocytic differentiation and exit from the cell cycle. Blood. 2003;101(5):1769–1776. doi: 10.1182/blood-2002-05-1517. [DOI] [PubMed] [Google Scholar]
- 14.Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275(41):32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 15.Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, Jr, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24(8):3251–3261. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118(1):83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat Cell Biol. 2006;8(10):1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 18.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, Yarden Y. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem. 2006;281(31):21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 19.Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, Chen A, Lee CG, Wei N, Wilkinson KD, Wang R, Pan ZQ. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278(31):28882–28891. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 20.Norman JA, Shiekhattar R. Analysis of Nedd8-associated polypeptides: a model for deciphering the pathway for ubiquitin-like modifications. Biochemistry. 2006;45(9):3014–3019. doi: 10.1021/bi052435a. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Santockyte R, Shen RF, Tekle E, Wang G, Yang DC, Chock PB. A general approach for investigating enzymatic pathways and substrates for ubiquitin-like modifiers. Arch Biochem Biophys. 2006;453(1):70–47. doi: 10.1016/j.abb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Huang L, Jacob RJ, Pegg SC, Baldwin MA, Wang CC, Burlingame AL, Babbitt PC. Functional assignment of the 20 S proteasome from Trypanosoma brucei using mass spectrometry and new bioinformatics approaches. J Biol Chem. 2001;276(30):28327–28339. doi: 10.1074/jbc.M008342200. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46(11):3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 24.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;5(2):366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2(9):667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 26.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci USA. 2006;103(50):18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276(49):46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10(6):1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 29.Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10(6):1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 30.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129(6):1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 31.Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435(7046):1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 32.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292(5520):1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 33.Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292(5520):1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 34.Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol. 2003;13(11):911–921. doi: 10.1016/s0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- 35.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 36.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5(4):737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8(7):700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 38.Gocke CB, Yu H, Kang J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J Biol Chem. 2005;280(6):5004–5012. doi: 10.1074/jbc.M411718200. [DOI] [PubMed] [Google Scholar]
- 39.Yang M, Hsu CT, Ting CY, Liu LF, Hwang J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J Biol Chem. 2006;281(12):8264–8274. doi: 10.1074/jbc.M510364200. [DOI] [PubMed] [Google Scholar]
- 40.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29(2):124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.